Open Access Article
Open Access ArticleIdentification of promising nutraceuticals against filarial immune-modulatory proteins: insights from in silico and ex vivo studies†
Vipin Kumar,
Ayushi Mishra and
Anchal Singh *
*
Dept. of Biochemistry, Institute of Science, Banaras Hindu University, Varanasi, 221005, U.P., India. E-mail: anchalsingh@bhu.ac.in; anchalsinghbhu@yahoo.com
First published on 11th August 2022
Abstract
Lymphatic filariasis is a neglected tropical disease affecting over 863 million people in 47 countries of the world. The anti-filarial drugs, diethylcarbamazine, albendazole, and ivermectin, are effective only at the larval stages and have proven completely ineffective as adulticides. Besides this, a long-term use of these drugs is associated with several side effects including drug toxicity. Nutraceuticals have emerged as better alternatives for long term treatments due to their safety and lesser side effects. In the present work, we have used drug docking analysis and molecular dynamics simulation approaches to explore the effect of anti-inflammatory nutraceuticals against the immune-modulatory proteins of filarial worms. The filarial proteins enolase, ES-62 precursor, serpin, and cystatin, which are highly efficient in host immune modulation were targeted with more than 50 nutraceuticals. In the in silico study nutraceuticals such as naringin, β-carotene, and emodin showed higher binding efficacy and lower dissociation constant as compared to anti-filarial drugs. Molecular dynamics simulation results showed that immune-modulatory proteins formed highly stable complexes with naringin, β-carotene, and emodin over the entire MD simulation run. The nutraceutical emodin formed the most stable system in silico and hence its effect was investigated on adult filarial parasites under ex vivo conditions too. Emodin significantly affected the motility, viability, ROS production, and genomic DNA fragmentation of filarial parasites. Further in vivo and in vitro studies will help in understanding the mechanism of action of emodin at the molecular level and would help in the development of more effective anti-filarial drugs.
Introduction
Lymphatic filariasis (LF) infections are characterized by blockages of lymphatic vessels, culminating in morbidities like lymphedema, elephantiasis, and hydrocele. Chronic patients not only lose their livelihoods but also face social stigma and exclusion. The Global Program to Eliminate Lymphatic Filariasis (GPELF) launched by the WHO targets filarial elimination through mass drug administration (MDA) of drugs diethylcarbamazine citrate (DEC) and albendazole (ALB).1 In 2017 the WHO added ivermectin (IVR) to the ongoing MDA in all endemic countries, except those co-endemic for loiasis and onchocerciasis. Although the drug combination IVR, DEC, and ALB has sufficient effectiveness against larval stages this treatment is completely ineffective against adult filarial worms.2,3 Additionally, IDA administration must be repeated annually for at least 5 years or more for reducing filarial transmission in endemic populations. Side effects including but not limited to nausea, fever, headache, dizziness, and pain of muscle and joints have commonly been reported following IDA administration, which deters people from participating in further rounds of IDA. Hence, research for broad spectrum natural macrofilaricidal agents having minimal side effects is continuously ongoing.Over the last two decades, the use of nutraceuticals (NC) has increased remarkably due to lesser side effects as compared with conventional drugs. NC are being routinely consumed as health supplements as well as for the prevention of diseases/infections. Inclusion of NC in anti-inflammatory therapy not only lowers the anti-inflammatory drug dosage but also minimizes the associated side effects. The NC ginger has strong anti-inflammatory effects and has been shown to reduce the production of TNF-α, IL-1β, and IL-6.4 Naringin can decrease neutrophil infiltration in the kidneys by suppressing the activity of renal myeloperoxidase.5 Ascorbic acid is a known anti-oxidant NC which is also capable of boosting immunity by augmenting phagocytosis and chemotaxis.6 Emodin exhibits anti-proliferative and anti-apoptotic effects on cancer cells and it can regulate PI3K/Akt/NF-κB signaling pathways that are imperative for cell growth and metabolism.7 Since nutraceuticals have potent anti-inflammatory activities8 we tried to harness these benefits for treatment of inflammatory conditions characteristic of chronic lymphatic filarial infections.
Inside the human host, an adult filarial worm can survive nearly 7 years and its microfilariae can remain in circulation for almost a year. The longevity of these parasites is mainly due to re-setting the host's immune system through active immune modulation.9 The filarial worms adapt perfectly to the host environment inundated with antibodies, immune cells, and effector molecules.10 They supplant the host's immune system norms by soothing aggressive immune reactions inducing immunological tolerance and inhibiting inflammatory responses. Several research studies have established the role of immune-modulatory proteins (IMPs) and enzymes in the survival and persistence of filarial parasites. Filarial parasites excrete and secrete immune-modulatory proteins such as ES-62, serpin, cystatin, and enolase which diminish the host's immune response.11,12 IMP ES-62 inhibits CD4+ T-cell and B-cell proliferation as well as IL-4 and IFN-γ production. ES-62 promotes the Th2 response and simultaneously inhibits the Th1 response. Cystatin inhibits key proteases such as cathepsins and endopeptidases, thus affecting the activation of T-cells. Neutrophil serine proteases that mediate extracellular killing of microbes are inhibited by serpins, thus compromising the hosts' immune response. The filarial IMP enolase is important for energy metabolism and is also involved in proteolysis and degradation of the host extracellular matrix. Therefore, in the present work, we chose filarial IMP cystatin, enolase, serpin, and ES-62 precursor as target proteins for docking and MD simulation studies.
The study aims to screen and identify nutraceuticals having potent anti-filarial activity; hence, NC, with known anti-inflammatory activities, were selected for docking against filarial immune-modulatory proteins cystatin, enolase, serpin, and ES-62. More than 50 nutraceuticals having anti-inflammatory activity were screened against filarial IMPs and only those NC which gave better docking scores as compared with the anti-filarial drug DEC were selected for the study. The top scoring NC β-carotene, lycopene, ascorbic acid, zerumbone, naringin, emodin, and citric acid were further docked with filarial IMP using the YASARA tool and PatchDock server. Naringin, β-carotene, and emodin exhibited the highest binding energies in molecular docking and hence were subjected to molecular dynamics simulation. The RMSD, RMSF, and Rg values calculated over the entire MD run confirmed stable and strong interaction of the aforementioned NC with filarial IMPs. The ex vivo work was undertaken to investigate the effect of emodin on bovine filarial parasites S. cervi, which is an established model parasite for LF. We have examined the ex vivo effect of different concentrations of emodin on adult parasites' motility and viability. In addition, the production of reactive oxygen species and genomic DNA fragmentation after treatment of S. cervi parasites with emodin were also examined. To the best of our knowledge, this is the first report of the anti-filarial effects of the nutraceutical emodin on S. cervi worms. The findings of our ex vivo and in silico13–15 studies are promising and the selected NC emodin can be investigated further for its anti-filarial effects.
Materials and methods
3D structure retrieval of filarial immune-modulatory proteins
The 3D structure of the filarial immune-modulatory protein enolase (accession no. AHI18146.1) was retrieved from the PDB (Protein Data Bank) database.16 PDB-BLAST analysis was used for retrieval of 3D structures and maximum score, query cover, and low E-value were selected for the study. As the 3D structures of many immune-modulatory proteins like ES-62 precursor, serpin and cystatin were unavailable in the PDB, the ab initio structure prediction approach was done using the LOMETS (Local Meta-Threading Server)17 server for predicting the homology of ES-62 precursor of Brugia malayi (accession no. CDP95589.1), serpin of Brugia malayi (accession no. XP_001896649.1) and cystatin of Brugia malayi (accession no. XP_001895476.1). The selection of the NCBI reference number by BLAST search was based on maximum score, E-value, and percent identity. The predicted 3D models were refined using ModRefiner18 and the structural qualities of predicted 3D structures were validated using RAMPAGE19 and PROCHECK server.20 Quality assessment and H-bond statistics were confirmed using ERRAT21 and VADAR server.22 The 3D structures of predicted immune-modulatory molecules were visualized using Discovery Studio 3.5. Active sites of predicted 3D models and PDB retrieved structures were predicted using Metapocket 2.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 23 server and Discovery Studio 3.5. The top 3 binding sites were chosen for the identification of active site residues.
23 server and Discovery Studio 3.5. The top 3 binding sites were chosen for the identification of active site residues.
3D structure retrieval of ligands and toxicity prediction23
The 3D structures of ligands β-carotene, lycopene, ascorbic acid, naringin, zerumbone, emodin, and citric acid were retrieved from the PubChem database.24 The structures of anti-filarial drugs DEC and albendazole were also downloaded from the same database and all the retrieved structures were converted into the PDB format using Discovery Studio 3.5. The drug likeness properties were checked using Lipinski rule 5.0.25,26 Further, absorption, distribution, metabolism, excretion, and toxicity (ADMET) were analyzed using admetSAR.27,28 Also, the canonical SMILES of ligands were taken from the PubChem database and copied into the admetSAR server.Docking analysis
Docking analysis of filarial immune-modulatory proteins with various nutraceutical compounds was done using YASARA29 and PatchDock server.30 The default parameter for the RMSD value of the PatchDock server for the protein and ligand complex was 1.5 Å. The docked complexes were visualized using Discovery Studio 3.5. The data interpretation of docking was based on GSC (geometric shape complementary) score and AI (approximate interface) area for the PatchDock server as well as binding energy and dissociation constant for YASARA (Yet Another Scientific Artificial Reality Application).Molecular dynamics simulation analysis
The top docking score as calculated on PatchDock and YASARA sever was given by nutraceutical compounds naringin, β-carotene, and emodin was subjected to MD (molecular dynamics) analysis. MD simulation was carried out using NAMD (nanoscale molecular dynamics version 2.14) software.31 PSF (protein structure file) was generated using VMD32 (visual molecular dynamics) version 1.9.3. The PDB files of nutraceutical compounds were converted into Sybyl Mol2 files using Open Babel Chemical Format Converter (https://www.cheminfo.org/Chemistry/Cheminformatics/FormatConverter/index.html). PSF and force filed parameter files of nutraceutical compounds were generated using CHARMM-GUI input generator and ligand modeler using the Sybyl Mol2 file (https://www.charmm-gui.org/input). Protein and nutraceutical compound complexes were generated by using the VMD dispdev command. The complexes were solvated in X, Y, and Z directions in an orthorhombic water model with a distance of 10 Å in various protein and nutraceutical compound complexes. The solvated box was ionized by adding 0.15 mM NaCl. Before MD run the CHARMM 36 force field file was used with 2 fs time step under 3D boundary condition. The MD run was performed at 310 K temperature, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps for energy minimization to obtain the sterically favored configuration of atoms. After the energy minimization, NVT and NPT ensembles were used for position strained simulation in a different phase. NPT and NVT ensembles were performed for 0.5 ns until a stable RMSD pattern of the complex backbone was observed. The long range electrostatic interaction was calculated by the PME (particle mesh Ewald) method. The MD run was carried out for 50 ns at 310 K temperature. After the MD run, data were visualized using VMD and trajectory was analyzed for root mean square deviation (RMSD) for backbone stability, root mean square fluctuation (RMSF) for amino acid residue variations, and radius of gyration (Rg) for protein localization at the central axis.33,34
000 steps for energy minimization to obtain the sterically favored configuration of atoms. After the energy minimization, NVT and NPT ensembles were used for position strained simulation in a different phase. NPT and NVT ensembles were performed for 0.5 ns until a stable RMSD pattern of the complex backbone was observed. The long range electrostatic interaction was calculated by the PME (particle mesh Ewald) method. The MD run was carried out for 50 ns at 310 K temperature. After the MD run, data were visualized using VMD and trajectory was analyzed for root mean square deviation (RMSD) for backbone stability, root mean square fluctuation (RMSF) for amino acid residue variations, and radius of gyration (Rg) for protein localization at the central axis.33,34
Collection of parasites
Bovine filarial parasites (Setaria cervi) were collected from the peritoneal fold of freshly slaughtered Indian water buffaloes. The parasites were brought to the laboratory in KRB maintenance medium (KRBMM) (KRB supplemented with penicillin, glutamine, streptomycin, and 1% glucose). Further, parasites were washed with PBS, before exposing them to different concentrations of emodin.Exposure of parasites to emodin
An equal number (N = 6) of adult female parasites in 20 mL of KRBMM containing different concentrations of emodin were incubated for 6 h, with 5% CO2 concentration at 37 °C, and 95% humidity. The parasites incubated in KRBMM were assigned as control. After 6 h, the parasites were washed twice in PBS and subjected to further analysis.Parasite's motility and viability
The visual monitoring was done to estimate the movement of the parasite up to 6 h. The motility measurement was based on movement score; the score ‘++++’ means that parasites are highly active, whereas ‘+’ means that parasites are least active and the ‘−’ sign indicates no movement of parasites. Further, after 6 h parasites were transferred to fresh KRBMM for checking the recovery.The viability of the parasites was measured by using an MTT assay according to Sharma et al., 2021, with slight modifications.35 After emodin treatment parasites were incubated in 0.5 mg mL−1 MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] for 2 h at 37 °C. In the next step, parasites were transferred to DMSO (dimethyl sulphoxide) for solubilizing the formazan crystals. The supernatant was gently removed, without disturbing the formazan crystals after 1 h and the absorbance was read at 540 nm wavelength in a Biorad microplate reader. The viability was plotted graphically and the percent reduction in treated parasites compared to control parasites was calculated.
Measurement of ROS
The ROS productions in control and treated parasites were measured by colorimetric assay following Choi et al. (2006) with slight modifications.36 The parasites were incubated in 2% NBT solution for 1 h at 37 °C. After the incubation period the parasites were washed in PBS and in methanol. Furthermore, the parasites were suspended in 2 M KOH for disrupting the cell membrane integrity and incubated in DMSO for solubilizing the formazan crystal for 10 minutes. Finally, the absorbance was recorded at 620 nm.DNA fragmentation analysis
Total genomic DNA of S. cervi was isolated from emodin treated (6 h) and control worms by the standard phenol/chloroform extraction and ethanol precipitation method. In brief, S. cervi were homogenized in 500 μL of lysis buffer (Tris–Cl 40 mM, pH 8.0, EDTA 50 mM, SDS 0.5%, NaCl 100 mM, β-mercaptoethanol 1%, v/v) with proteinase K (0.1 mg mL−1) and incubated in a water bath at 55 °C for 3 h. 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of phenol–chloroform–isoamyl alcohol was used for DNA extraction followed by 3 M sodium acetate and 100% cold ethanol precipitation.37 Precipitated DNA was centrifuged at 10
1 ratio of phenol–chloroform–isoamyl alcohol was used for DNA extraction followed by 3 M sodium acetate and 100% cold ethanol precipitation.37 Precipitated DNA was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and washed twice with 70% ethanol. The final DNA pellet was resuspended in 20 μL of sterile 10 mM Tris–EDTA buffer (pH 8.0), run-on 1% agarose gel, containing 0.1 mg mL−1 of ethidium bromide and visualized under ultraviolet GelDoc system (Bio-Rad, USA).
000 rpm for 10 min and washed twice with 70% ethanol. The final DNA pellet was resuspended in 20 μL of sterile 10 mM Tris–EDTA buffer (pH 8.0), run-on 1% agarose gel, containing 0.1 mg mL−1 of ethidium bromide and visualized under ultraviolet GelDoc system (Bio-Rad, USA).
Statistical analysis
All the experiments were performed in triplicate (n = 3) and the data were expressed as mean ± SEM. The statistical analysis was done by comparing the treated groups with the control using Origin 8.0. The two-tailed Student's t-test was used to calculate the statistical significance between the control and emodin treated filarial worms. P values < 0.05 (*) and < 0.01 (**) were considered as statistically significant.Results and discussion
Molecular structure determination of target proteins
To perform the docking and molecular dynamics (MD) simulation analysis, the 3D structure of enolase was retrieved from the PDB data bank (Fig. 1A), and the structural validation by the Ramachandran plot is given in ESI Fig. 1A.† Enolase is composed of 439 amino acids and ∼99.5% residues are in the allowed region and only 0.5% residues are in the disallowed region. The retrieved 3D structure of enolase was further subjected to VADAR analysis. Enolase had 39% residues as alpha helix, 24% residues in beta-sheet, 36% residues in the random coil, and 20% residue in turn. The H-bond statistics of the enolase structure had almost the same observed and expected values of mean H-bond energy, mean H-bond distance, and residues with the H-bond.Structural determination of filarial immune modulatory proteins
The 3D structures of ES-62 precursor, serpin, and cystatin were not available in any database. Hence, the molecular structure predictions of the aforementioned immune-modulatory proteins were done using the LOMETS server. LOMETS is the next-generation meta-server approach for template-based structure prediction and functional annotation. Deep multiple sequence alignment (MSA) is generated for each target protein sequence by homology search in multiple databases. LOMETS uses nine meta-threading programs to generate the top template model for each protein. The top 10 templates of each protein were subjected to Ramachandran plot analyses using PROCHECK and VADAR software. The PROCHECK server is used for observing and evaluating the stereo-chemical quality of protein models, whereas VADAR (Volume Area Dihedral Angle Reporter) is used for qualitative evaluation of structural parameters of a single residue or entire protein. VADAR parameters include hydrogen bond statistics, steric quality, fold quality, dihedral angle, and Ramachandran plot statistics for predicting the overall quality of proteins. The VADAR server can assess the protein structural quality as determined by homology modeling, NMR spectroscopy, or X-ray crystallography for qualitative and quantitative structural evaluation. The top 3D structures of ES-62 precursor, serpin, and cystatin based on the Ramachandran plot by PROCHECK and VADAR server were subjected to molecular docking and MD simulation (Fig. 1B–D). Structural validation by PROCHECK and VADAR server revealed that 99.3% of residues in ES-62 precursor (a total of 493) were present in the allowed regions (ESI Fig. 1B†). All the 389 amino acids of serpin and 127 amino acids of cystatin were present in the allowed region of the Ramachandran plot (ESI Fig. 1C and D†). The structural quality analysis of the enolase, ES-62 precursor, serpin, and cystatin 3D structures revealed that these models are suitable for docking and MD simulation studies (Tables 1 and 2).| Target protein | Alpha helix | Beta sheet | Coil | Turn |
|---|---|---|---|---|
| Enolase | 172 (39%) | 108 (24%) | 159 (36%) | 92 (20%) |
| ES-62 | 191 (38%) | 147 (29%) | 155 (31%) | 36 (7%) |
| Serpin | 143 (36%) | 134 (34%) | 112 (28%) | 32 (8%) |
| Cystatin | 26 (20%) | 54 (42%) | 47 (37%) | 24 (18%) |
| Target proteins | Mean H-bond distance | Mean H-bond energy | Residue with H-bond | |||
|---|---|---|---|---|---|---|
| Observed | Expected | Observed | Expected | Observed | Expected | |
| Enolase | 2.2 (sd = 0.4) | 2.2 (sd = 0.4) | −1.9 (sd = 1.0) | −2.0 (sd = 0.8) | 351 (79%) | 329 (75%) |
| ES-62 | 2.2 (sd = 0.3) | 2.2 (sd = 0.3) | −1.8 (sd = 1.0) | −2.0 (sd = 0.8) | 365 (74%) | 369 (75%) |
| Serpin | 2.2 (sd = 0.4) | 2.2 (sd = 0.4) | −1.8 (sd = 1.0) | −2.0 (sd = 0.8) | 289 (74%) | 291 (75%) |
| Cystatin | 2.1 (sd = 0.3) | 2.2 (sd = 0.4) | −1.9 (sd = 1) | −2.2 (sd = 0.8) | 94 (74%) | 95 (75%) |
Drug likeness and ADMET analysis of nutraceutical compounds
For this study, nutraceuticals β-carotene, lycopene, ascorbic acid, zerumbone, naringin, emodin, and citric acid (ESI Fig. 2†) were chosen as they are commonly present in numerous fruits and vegetables. Fruits and vegetables can be eaten in their raw form, are a major source of minerals, vitamins, and fibers, and have excellent health benefits. Past research has shown that fruits and vegetables have a substantial activity against many chronic diseases including cancer, diabetes, neurological, cardiac, bacterial, and viral diseases. Nutraceutical compounds of fruits and vegetables can also interfere with the signaling pathways culminating in apoptosis or cancer progression. Nutraceutical compounds are used in their natural/pure form for disease prevention in human beings, although dose standardization may be needed to determine the health benefits. The currently recommended anti-filarial drugs have multiple side effects like headache, fever, chill, nausea and tenderness, but the nutraceutical compounds used in this study are easily available, safe, cheap and have no major side effects. A drug likeness analysis of these nutraceutical compounds was done by Lipinski filter rule 5, which states that orally active compounds must have any two of the following five criteria: molecular mass less than 500 kDa, Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value less than 5, hydrogen bond donors less than 5, hydrogen bond acceptors less than 10, and molar refractivity between 40 and 130. The drug likeness properties of all the nutraceutical compounds used in this study are given in ESI Table 1.†
P value less than 5, hydrogen bond donors less than 5, hydrogen bond acceptors less than 10, and molar refractivity between 40 and 130. The drug likeness properties of all the nutraceutical compounds used in this study are given in ESI Table 1.†
AdmetSAR was used for determining the pharmacokinetic profiles of studied nutraceutical compounds and except for naringin, all other nutraceutical compounds could easily cross the blood–brain barrier. Nutraceuticals can lower neuronal toxicity and provide better mental health because they have potent anti-inflammatory properties. Intestinal absorption of all the chosen nutraceuticals was high, facilitating their easy movement into the systemic circulation. The hERG channel, responsible for conducting the potassium ions from cardiac myocytes, was not affected by the chosen nutraceutical compounds and the compounds were non-AMES toxic as well as non-carcinogenic. The oral acute toxicity was of category III for all NC except ascorbic acid (category IV), indicating that these compounds could be mild irritants at higher dose. Rat acute toxicity defines the standard measurement of drug dosage causing 50% death of experimental animals within a specific period. Rat acute toxicity of anti-filarial drugs was 2.26 mol kg−1 for DEC, 2.075 mol kg−1 for albendazole, 1.598 mol kg−1 for β-carotene, 1.535 mol kg−1 for lycopene, 1.3059 mol kg−1 for ascorbic acid, 2.261 mol kg−1 for naringin, 1.8726 mol kg−1 for zerumbone, 2.526 mol kg−1 for emodin and 1.778 mol kg−1 for citric acid (ESI Table 2†).
Docking analysis of nutraceutical compounds with immune-modulatory proteins
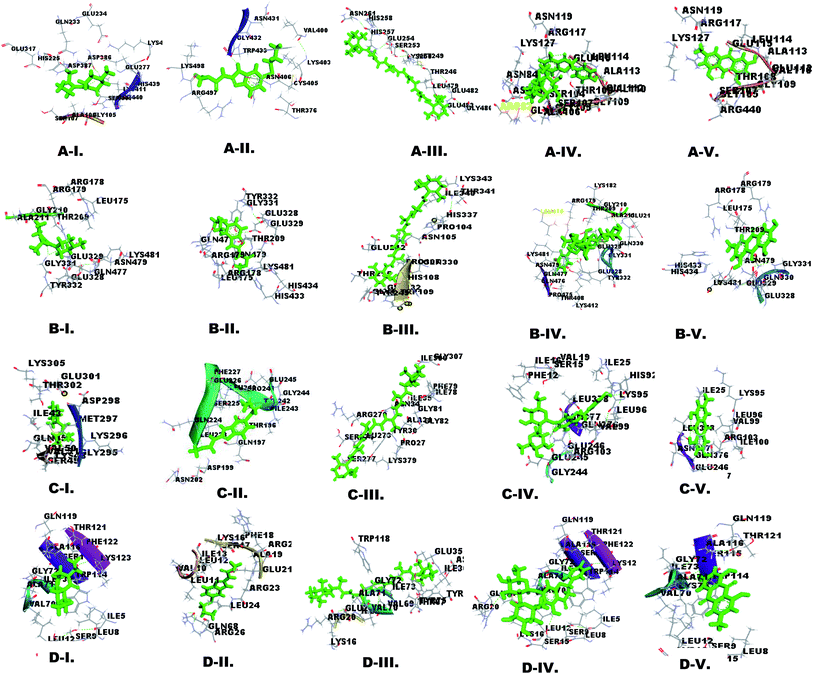
Molecular docking is important for computer-aided designing and in silico testing of drugs.38 The docking properties of nutraceutical compounds with filarial immune-modulatory proteins were characterized using the PatchDock server and YASARA tool. In this work docking analysis was studied with respect to the following parameters: (a) interacting amino acid residue, (b) interacting residue active site number, (c) nutraceutical compounds and immune-modulatory proteins involved in the H-bond, (d) binding energy, (e) dissociation constant, (f) GSC score and (g) AI area. The binding energy (kcal mol−1) and dissociation constant (μM) of docked complexes were analyzed using the YASARA tool, whereas the GSC score and AI area were calculated using the PatchDock server. The interacting residues were identified using the YASARA tool and PatchDock server and additionally prominent binding sites were predicted using Metapocket 2.0 server and Discovery Studio 3.5 (Fig. 2 and ESI Fig. 3†). The retrieved docked complexes were screened for the highest binding energy, lowest dissociation constant, maximum hydrogen bonding, higher GSC score, AI area, and docking within the top 3 binding sites of immune-modulatory proteins, following which only the best-fit complex was chosen for further analysis. Higher binding energy indicates a more favorable interaction of protein and ligand molecules, whereas a smaller value of the dissociation constant (μM) indicates that protein and ligand affinity is higher and chances of separation are low. GSC determines the geometric shape complementary score and generates values and several match features of protein ligand molecules. AI area refers to the approximate interface area covered by the protein ligand complex. The filarial immune modulators enolase, ES-62 precursor, serpin, and cystatin were docked with anti-filarial drugs DEC, albendazole and nutraceuticals β-carotene, lycopene, ascorbic acid, zerumbone, naringin, emodin, and citric acid. The immune modulators help in the survival of filarial parasites, inside the hostile environment of the host, thus allowing the persistence of the parasites for several years.The enolase protein has five active sites as predicted by the Metapocket 2.0 server, but only top three binding sites were docked with nutraceutical compounds. The most prominent interacting residues were GLU, LYS, ARG, and GLY. All the ligands except DEC and lycopene interacted strongly with in the top 3 binding sites. The binding energy of enolase–ligand complexes was in the range of 5.277 kcal mol−1 to 8.508 kcal mol−1 and the highest binding energy was of emodin, whereas albendazole had the lowest binding energy. The dissociation constant calculated for enolase–ligand complexes was lowest for emodin and highest for albendazole. Ascorbic acid, naringin, citric acid, DEC and albendazole could form more stable interactions with enolase by H-bonding (ESI Tables 2 and 3†).
Docking of ES-62 precursor with chosen ligands showed that the most interacting amino acid residues were GLU, ARG, LYS, GLN and GLY. The maximum H-bonds were formed by ascorbic acid (7 H-bonds), citric acid (5 H-bonds), naringin (3 H-bonds), emodin (2 H-bonds), albendazole (2 H-bonds) and DEC (1 H-bond) and all the ligands formed complexes within the top 3 sites (ESI Table 4†). ES-62 had the highest binding energy observed in emodin-protein complexes (7.75 kcal mol−1) and the lowest was observed in DEC–protein complexes (4.69 kcal mol−1) (Table 3).
| Receptor | Name of ligand | Binding energy (kcal mol−1) | Dissociation constant (μm) | GSC score | AI area | Receptor | Name of ligand | Binding energy (kcal mol−1) | Dissociation constant (μm) | GSC score | AI area |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Enolase | DEC | 5.329 | 110.46 | 3884 | 441.20 | Serpin | DEC | 4.320 | 681.393 | 3814 | 442.30 |
| Albendazole | 5.277 | 135.49 | 4114 | 527.40 | Albendazole | 5.481 | 96.0226 | 4288 | 532.00 | ||
| β-Carotene | 6.771 | 10.884 | 7198 | 988.00 | β-Carotene | 6.885 | 8.7979 | 7088 | 960.00 | ||
| Lycopene | 5.42 | 106.435 | 8098 | 1148.00 | Lycopene | 4.374 | 622.035 | 7086 | 992.10 | ||
| Ascorbic acid | 6.00 | 39.989 | 2604 | 284.90 | Ascorbic acid | 5.189 | 157.185 | 2596 | 279.40 | ||
| Zerumbone | 5.784 | 57.800 | 4252 | 460.40 | Zerumbone | 5.625 | 75.304 | 3512 | 443.60 | ||
| Naringin | 8.397 | 0.699 | 6054 | 784.10 | Naringin | 8.034 | 1.219 | 6228 | 806.00 | ||
| Emodin | 8.508 | 0.290 | 3808 | 409.00 | Emodin | 8.217 | 0.917 | 3876 | 446.20 | ||
| Citric acid | 5.612 | 98.651 | 2834 | 297.50 | Cystatin | Citric acid | 5.128 | 105.230 | 2838 | 295.40 | |
| ES-62 | DEC | 4.690 | 364.90 | 3850 | 444.10 | DEC | 5.067 | 193.125 | 3465 | 419.60 | |
| Albendazole | 5.905 | 46.943 | 4364 | 521.60 | Albendazole | 5.307 | 128.800 | 3602 | 441.50 | ||
| β-Carotene | 6.326 | 21.700 | 7364 | 957.30 | β-Carotene | 6.027 | 38.207 | 6066 | 792.60 | ||
| Lycopene | 5.249 | 142.046 | 7784 | 999.90 | Lycopene | 5.655 | 71.586 | 6850 | 817.20 | ||
| Ascorbic acid | 5.568 | 82.909 | 3768 | 313.50 | Ascorbic acid | 5.618 | 76.1993 | 2568 | 277.40 | ||
| Zerumbone | 5.699 | 66.42 | 3746 | 458.60 | Zerumbone | 6.601 | 14.501 | 3612 | 760.60 | ||
| Naringin | 7.5 | 27.731 | 6350 | 847.30 | Naringin | 7.197 | 5.303 | 5534 | 760.60 | ||
| Emodin | 7.75 | 20.853 | 4026 | 475.30 | Emodin | 7.211 | 4.313 | 3450 | 410.10 | ||
| Citric acid | 6.015 | 42.0921 | 3066 | 323.10 | Cirtic acid | 5.752 | 65.124 | 2702 | 293.50 |
Docking analysis of serpin with different ligands showed that most interacting amino acid residues were GLY, GLN, LYS, GLU, and VAL with nutraceuticals and anti-filarial drugs. All the ligands formed complexes within the top 3 active sites except lycopene. Also, ample H-bonding was shown by ascorbic acid (6 H-bonds), naringin (5 H-bonds), citric acid (1 H-bond), albendazole (1 H-bond) and emodin (1 H-bond) (ESI Table 5†). The predicted binding energy for all ligands was in the range of 8.217 kcal mol−1 (emodin) to 4.320 kcal mol−1 (DEC) in serpin complexes, which corroborated well with the AI area, GSC score and dissociation constant (Table 3).
Cystatin docking analysis with ligands showed that most interacting amino acid residues were LEU, ILE, LYS, VAL and ALA. The anti-filarial drug albendazole and NC ascorbic acid failed to bind within top 3 active sites. DEC, albendazole, β-carotene and lycopene did not form any H-bond but naringin (3 H-bonds), ascorbic acid (3 H-bonds), emodin (2 H-bonds) and citric acid (2 H-bonds) formed H-bonds with cystatin (ESI Table 6†). Similar to the results obtained for docking of ES-62 precursor, and serpin, cystatin too showed the highest binding energy with emodin 7.211 kcal mol−1 and least with DEC 5.067 kcal mol−1 (Table 3).
Apart from H-bonds other non-covalent interactions like van der Waals force, hydrophobic and ionic interactions are also important in the stability of the docked complexes. The interactions of nutraceutical compounds at the active site of immune modulatory proteins lead to conformational changes and reduction of functional activities. The most common amino acid residues interacting with ligands were LYS, GLU, LEU, ILE, GLY, and ARG. The interacting amino acid residues determine the strength of interactions and contribute to non-covalent ligand–protein interactions. The active site residues were also involved in the formation of a local interacting environment which is useful in the binding of ligands with filarial immune-modulatory proteins.
Molecular dynamics simulation
Molecular dynamics (MD) simulation can evaluate the stability and flexibility of the docked complexes and provide important dynamic information about the protein–ligand complex at the atomic level.39 For the MD run, we selected β-carotene, naringin, and emodin as these nutraceuticals had stronger interactions and comparatively higher binding energies in molecular docking. The nutraceutical compounds β-carotene, naringin, and emodin are anti-inflammatory in nature and exert their effects by inhibition of TNF-α induced activation of NK-κB. Emodin has therapeutic potential for treatment of inflammatory diseases like pancreatitis, arthritis, myocarditis and Alzheimer's disease.40 Naringin is a major constituent of Painopowder which is used for reduction of inflammation41 in China. β-Carotene can also decrease the transcription of pro-inflammatory cytokine genes interleukin-1β (IL-1β), IL-6 and tumor necrosis factor alpha (TNF-α).42 A reverse correlation between plasma β-carotene and C-reactive protein (CRP) has been reported earlier.43 In LF it has been observed that CRP is over expressed in all clinical stages and the expression in chronic cases is almost double that of normal serum samples.44 The anti-filarial drug albendazole which showed a better docking score as compared to DEC was also included for MD simulation. All the immune-modulatory proteins enolase, ES-62 precursor, serpin, and cystatin were simulated using NAMD software, and visualization was done using VMD software. The study was performed for 50 ns at 310 K and the trajectory was analyzed in terms of RMSD (root mean square deviation), RMSF (root mean square fluctuation), and Rg (radius of gyration).RMSD and Rg analysis
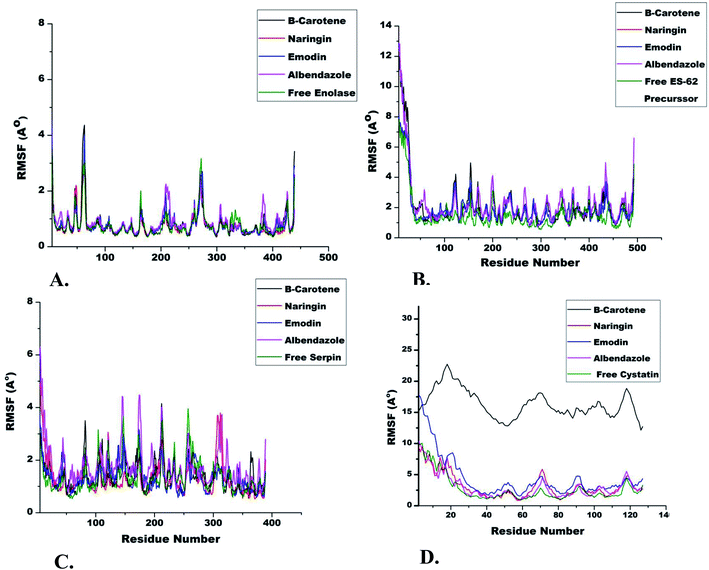
In general terms, the RMSD values represent protein fluctuation and mobility of atoms. Therefore, a higher RMSD value suggests higher atomic mobility and lesser stability of the protein.45 A comparison of RMSD trajectories of all IMPs is shown in Fig. 3. It was observed that initially, all systems fluctuated slightly from 3 ns to 7 ns after which they were stable for the entire run. The IMP enolase complexed with β-carotene, naringin and emodin deviated over a range of 1 to 2.5 Å with an average value of 2 Å. The maximum variability of RMSD was shown in the enolase–albendazole complex with an average RMSD value of 2.2 Å (Fig. 3). Among the ES-62 precursor complexes of nutraceuticals and albendazole, the maximum RMSD value was observed for ES-62 precursor–albendazole complexes ranging from 2 Å to 8 Å. The stable complex of ES-62 precursor, naringin, which had an average RMSD value of 3 Å, was equilibrated during the entire run of 50 ns (Fig. 3B). During MD simulation the serpin complexed with nutraceuticals showed lesser deviation (3 Å to 5 Å), while the serpin–albendazole complex had a maximum RMSD variation of 4 Å with an average RMSD value of 3 Å (Fig. 3C). The MD simulation trajectories of cystatin with all the NC and albendazole fluctuated the most as compared to the complexes of enolase, ES-62 precursor, and serpin. The minimum RMSD value was of cystatin-albendazole (4 Å) and the maximum RMSD was of emodin (12 Å) (Fig. 3D). The radius of gyration (Rg) value shows the compactness of target proteins in the presence or absence of ligands. The time evolution points (Rg) of different immune-modulatory proteins complexed with NC and albendazole are shown in Fig. 4.RMSF analysis
The role of each amino acid residue in the stability of protein–ligand complexes was assessed by RMSF (root mean square fluctuation) calculation.46,47 A stable protein–ligand complex essentially shows lesser fluctuations of amino acid residues during simulation runs. The RMSF trajectories of enolase complexed with different ligands showed fluctuation of amino acid residues from 60 to 70 (3.5 Å), 265 to 285 (2.5 Å) and 200 to 220 (2 Å) respectively. The RMSF of uncomplexed ES-62 precursor (1.5 Å to 2.5 Å) was lower as compared to the complexed ES-62 precursor (1.5 Å to 3.5 Å) for the entire MD run. The nutraceuticals β-carotene, naringin and emodin complexed with serpin showed lesser (3 Å) fluctuations of amino acid residues as compared to free serpin (1.5 Å to 4 Å) and serpin–albendazole (1.5 Å to 4.2 Å). The IMP cystatin complexed with nutraceuticals and albendazole showed the highest RMSF value (10 Å) among all protein targets included in the work (Fig. 5).Ex vivo effect of emodin on adult female S. cervi parasites
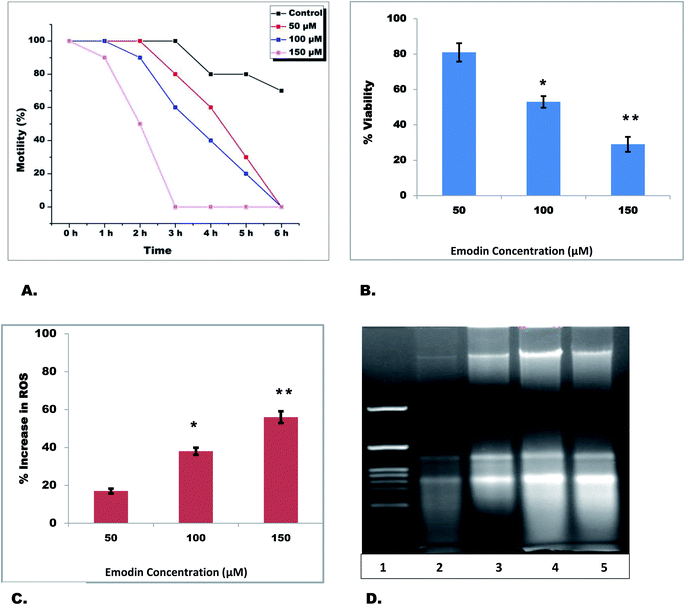
The nutraceutical emodin gave the best docking score and formed the most stable system during the entire MD run with all target proteins. Therefore, the effect of emodin was investigated on adult filarial parasites under ex vivo conditions. Equal numbers of adult S. cervi female worms were incubated with different concentrations of emodin in separate flasks for 6 h. After completion of incubation, recovery of parasites was checked by incubating them in a fresh maintenance medium for an hour. The worms incubated in emodin were sluggish and the activity slowed in comparison to control worms in a time-dependent manner (Table 4). The motility of emodin-treated parasites reduced to zero within 3 h (150 μM), whereas 100 μM and 50 μM emodin treated parasites became completely immotile after 6 h (Fig. 6A) in comparison to control worms (Fig. 7). Likewise the viability of adult female worms reduced by 19%, 47% (p < 0.05) and 71% (p < 0.01) in 50 μM, 100 μM and 150 μM after 6 h of treatment (Fig. 6B). The adult female worms treated with 100 μM and 150 μM emodin did not show any sign of revival after completion of the recovery period; however, 50 μM emodin treated worms did show little movement after an hour of recovery. A significant increase in ROS generation was observed after emodin treatment (Fig. 6C); the total ROS was increased by 56%, 43%, and 17% in 150 μM, 100 μM, and 50 μM emodin treated parasites. Further, the genomic DNA isolation of treated and control parasites was done followed by electrophoretic separation on 1% agarose gel. It was seen that the genomic DNA of control parasites was intact, whereas dose-dependent laddering of DNA was observed in emodin treated filarial worms (Fig. 6D).| Sample | Motility of parasites in the following incubation hours | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 1 h | 2 h | 3 h | 4 h | 5 h | 6 h | Recovery | |
| a The motility of the parasites was visually checked at different time intervals. Adult female worms (n = 6) of equal size were incubated with different concentrations of emodin in 20 mL maintenance medium at 37 °C and 5% CO2 for 6 h. Worms incubated in only maintenance medium served as control. Worm motility was scored as −, no movement; +, least active; ++, less active; +++, moderately active; and ++++, highly active. Worms were transferred into fresh medium (devoid of emodin) after 6 h and motility recovery in the treated group was compared to the control group. The results are from three independent experiments performed in triplicate. | ||||||||
| Control | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++ | ++++ |
| Emodin (50 μM) | ++++ | ++++ | ++++ | ++++ | ++ | − | − | − |
| Emodin (100 μM) | ++++ | ++++ | ++++ | ++ | + | − | − | − |
| Emodin (150 μM) | ++++ | ++++ | +++ | − | − | − | − | − |
 | ||
| Fig. 7 Effect of emodin (50, 100 and 150 μM) on the motility of S. cervi: (A) initial emodin treatment, (B) after 3 h of emodin treatment and (C) after 6 h of emodin treatment. | ||
Conclusion
This study sheds new light on the effect of nutraceuticals on filarial immune-modulatory proteins. To our knowledge, this is the first report of the effect of these nutraceuticals on filarial immune-modulatory proteins. All NC gave better docking scores than the recommended anti-filarial drugs DEC and albendazole. Also, NC β-carotene, naringin, and emodin were stably complexed with IMPs during MD simulation runs. The ex vivo studies with emodin indicate strong anti-filarial potential. The NC emodin is an anthraquinone derivative found in aloe, lichens, etc. Emodin has anti-inflammatory, anti-cancer, anti-viral, and antibacterial effects and can directly inhibit NK-κB, thus controlling the expression and activities of several inflammatory factors. Further in vivo and in vitro studies on S. cervi will help in understanding the anti-filarial effect of emodin at the molecular level. In the next step, the effect of emodin on human filarial worms can be studied to elucidate the molecular mechanism involved in mediating the anti-filarial action. These investigations will certainly prove useful in development of better and more effective anti-filarial drugs in future.Ethical statement
This study does not involve any animals or human samples. The Setaria cervi nematodes used in the study are found in the peritoneal folds of Indian water buffaloes (Bubalus bubalis) which are slaughtered for table purposes. S. cervi worms were collected from a local slaughterhouse and brought to the laboratory in Krebs Ringer bicarbonate (KRB) buffer supplemented with penicillin, glutamine, streptomycin and 1% glucose.Author contributions
Conceptualization, V. K. and A. S.; methodology, V. K. and A. M.; software, V. K. and A. M.; validation, A. M., V. K., and A. S.; formal analysis, A. M. and V. K.; investigation, A. S. and V. K.; resources A. S.; data curation, V. K. and A. S.; writing—original draft preparation, V. K. and A. S.; writing—review and editing, V. K. and A. S.; visualization, A. M. and V. K.; supervision, A. S.; project administration, A. S. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors are declaring that there is no conflict of interest.Acknowledgements
VK is grateful to the Department of Biotechnology (DBT), New Delhi, for providing a Senior Research Fellowship (DBT/JRF/BET-18/I/2018/AL/19). AM is thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, for providing a Senior Research Fellowship (09/013(0832)/2018-EMR-I). The authors are also thankful to DBT-BHU Interdisciplinary School of Life Sciences, Centre for Bioinformatics, School of Biotechnology, Banaras Hindu University, for providing YASARA software, and Supercomputing Center IIT BHU for providing the supercomputing facility.References
- WHO, Ending the neglect to attain the Sustainable Development Goals—A road map for neglected tropical diseases 2021–2030, 2020 Search PubMed.
- K. Y. Won, K. Gass, M. Biamonte, D. A. Dagne, C. Ducker, C. Hanna, A. Hoerauf, P. J. Lammie, S. M. Njenga, R. Noordin, K. D. Ramaiah, R. Ramzy, R. G. C. Scholte, A. W. Solomon, A. A. Souza, J. Tappero, E. Toubali, G. J. Weil, S. A. Williams and J. D. King, PLoS Neglected Trop. Dis., 2021, 15(11), e0009968 CrossRef PubMed.
- WHO, Global programme to eliminate lymphatic filariasis: progress report, 2019, Wkly. Epidemiol. Rec., 2020, 95(43), 509–524 Search PubMed.
- Q. Q. Mao, X. Y. Xu and S. Y. Cao, et al, Foods, 2019, 8(6), 185, DOI:10.3390/foods8060185.
- Y. Chtourou, Z. Kamoun and W. Zarrouk, et al, Food Funct., 2016, 7(1), 183–193, 10.1039/c5fo00871a.
- A. C. Carr and S. Maggini, Nutrients, 2017, 9(11), 1211, DOI:10.3390/nu9111211.
- V. Suvarna, M. Murahari, T. Khan, P. Chaubey and P. Sangave, Front. Pharmacol., 2017, 8, 916, DOI:10.3389/fphar.2017.00916.
- A. Bergamin, E. Mantzioris, G. Cross, P. Deo, S. Garg and A. M. Hill, Pharm. Med., 2019, 33(4), 291–309, DOI:10.1007/s40290-019-00289-w.
- S. Babu and T. B. Nutman, Parasite Immunol., 2014, 36(8), 338–346, DOI:10.1111/pim.12081.
- M. Joshua, EMJ Allergy & Immunology, 2021, 6(1), 71–78 Search PubMed.
- A. Hotterbeekx, J. Perneel, M. K. Vieri, R. Colebunders and S. Kumar-Singh, Front. Cell. Infect. Microbiol., 2021, 11, 662766, DOI:10.3389/fcimb.2021.662766.
- J. P. Hewitson, J. R. Grainger and R. M. Maizels, Mol. Biochem. Parasitol., 2009, 167(1), 1–11, DOI:10.1016/j.molbiopara.2009.04.008.
- M. W. Attwa, A. A. Kadi and A. S. Abdelhameed, RSC Adv., 2020, 10, 5412–5427 RSC.
- A. A. Kadi, S. M. Amer, H. W. Darwish and M. W. Attwa, RSC Adv., 2017, 7(58), 36279–36287 RSC.
- N. S. Al-Shakliah, M. W. Attwa, A. A. Kadi and H. AlRabiah, RSC Adv., 2020, 10, 16231–16244 RSC.
- RCSB Protein Data Bank, 2022, https://www.rcsb.org/pdb/home/sitemap.do.
- W. Zheng, C. Zhang, Q. Wuyun, R. Pearce, Y. Li and Y. Zhang, Nucleic Acids Res., 2019, 47(22), 11746–11754, DOI:10.1093/nar/gkz384.
- D. Xu and Y. Zhang, Biophys. J., 2011, 101(10), 2525–2534, DOI:10.1016/j.bpj.2011.10.024.
- R. A. Laskowski, E. G. Hutchinson, A. D. Michie, A. C. Wallace, M. L. Jones and J. M. Thornton, Trends Biochem. Sci., 1997, 22, 488–490 CrossRef CAS PubMed.
- S. C. Lovell, I. W. Davis, W. B. Arendall, P. I. W. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson and D. C Richardson, Proteins, 2003, 50(3), 437–450, DOI:10.1002/prot.10286.
- C. Colovos and T. O. Yeates, Protein Sci Publ Protein Soc, 1993, 2, 1511–1519 CrossRef CAS.
- L. Willard, A. Ranjan, H. Zhang, H. Monzavi, R. F. Boyko and B. D. Sykes, et al., Nucleic Acids Res., 2003, 31, 3316–3319 CrossRef CAS PubMed . PMID: 12824316.
- B. Huang, OMICS, 2009, 13(4), 325–330, DOI:10.1089/omi.2009.0045 . PMID: 19645590..
- Y. Wang, E. Bolton, S. Dracheva, K. Karapetyan, B. A. Shoemaker, T. O. Suzek, J. Wang, J. Xiao, J. Zhang and S. H. Bryant, Nucleic Acids Res., 2010, D255–D266, DOI:10.1093/nar/gkp965.
- C. A. Lipinski, Drug Discovery Today: Technol., 2004, 1(4), 337–341, DOI:10.1016/j.ddtec.2004.11.007.
- B. Jayaram, T. Singh, G. Mukherjee, A. Mathur, S. Shekhar and V. Shekhar, BMC Bioinf., 2012, 13(Suppl17), S7, DOI:10.1186/1471-2105-13-S17-S7.
- H. Yang, C. Lou, L. Sun, J. Li, Y. Cai, Z. Wang, W. Li, G. Liu and Y. Tang, Bioinformatics, 2019, 35, 1067–1069, DOI:10.1093/bioinformatics/bty707.
- S. Mukherjee, M. Abdalla, M. Yadav, M. Madhavi, A. Bhrdwaj, R. Khandelwal, L. Prajapati, A. Panicker, A. Chaudhary, A. Albrakati, T. Hussain, A. Nayarisseri and S. K. Singh, J. Mol. Model., 2022, 28(4), 100, DOI:10.1007/s00894-022-05081-3.
- H. Land and M. S. Humble, Methods Mol. Biol., 2018, 1685, 43–67, DOI:10.1007/978-1-4939-7366-8_4.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367, DOI:10.1093/nar/gki481.
- J. C. Phillips, R. Braun and W. Wang, et al., J. Comput. Chem., 2005, 26(16), 1781–1802, DOI:10.1002/jcc.20289.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14(1), 33–38, DOI:10.1016/0263-7855(96)00018-5.
- M. Surti, M. Patel and M. Adnan, et al., RSC Adv., 2020, 10(62), 37707–37720, 10.1039/d0ra06379g.
- P. Gopinath and M. K. Kathiravan, RSC Adv., 2021, 11(60), 38079–38093, 10.1039/d1ra07377j.
- S. Sharma, F. Ahmad, A. Singh and S. Rathaur, Vet. Parasitol., 2021, 290, 109357, DOI:10.1016/j.vetpar.2021.109357.
- H. S. Choi, J. W. Kim, Y. N. Cha and C. A. Kim, J. Immunoassay Immunochem., 2006, 27, 31–44 CrossRef CAS PubMed.
- A. Nayak, P. Gayen, P. Saini, N. Mukherjee and S. P. Babu, Parasitol. Res., 2012, 111(3), 1173–1186, DOI:10.1007/s00436-012-2948-0.
- F. Stanzione, I. Giangreco and J. C. Cole, Prog. Med. Chem., 2021, 60, 273–343, DOI:10.1016/bs.pmch.2021.01.004.
- C. B. M. Platania and C. Bucolo, Methods Mol. Biol., 2021, 2253, 245–254 CrossRef CAS PubMed.
- M. Stompor-Gorący, Int. J. Mol. Sci., 2021, 22(17), 9522, DOI:10.3390/ijms22179522.
- A. H. El-Desoky, R. F. Abdel-Rahman, O. K. Ahmed, H. S. El-Beltagi and M. Hattori, Phytomedicine, 2018, 42, 126–134, DOI:10.1016/j.phymed.2018.03.051.
- J. Cheng, E. Balbuena, B. Miller and A. Eroglu, Front. Nutr., 2021, 8, 723480, DOI:10.3389/fnut.2021.723480.
- H. Schultz, G. S. Ying, J. L. Dunaief and D. M. Dunaief, Am. J. Lifestyle Med., 2019, 15(6), 634–643, DOI:10.1177/1559827619894954.
- V. Kumar, A. Mishra, A. K. Yadav, S. Rathaur and A. Singh, PLoS One, 2022, 17(7), e0270635, DOI:10.1371/journal.pone.0270635.
- N. Tembe, K. E. Machaba, U. Ndagi, H. M. Kumalo and N. N. Mhlongo, J. Mol. Model., 2022, 28(2), 35, DOI:10.1007/s00894-021-04993-w.
- L. Shunmugam and M. E. S. Soliman, RSC Adv., 2018, 8(73), 42210–42222, 10.1039/c8ra07346e.
- V. kumar, A. Mishra, V. K. Singh and A. Singh, Lett. Drug Des. Discovery, 2022, 19, 982–995, DOI:10.2174/1570180819666220330122542.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03287b |
| This journal is © The Royal Society of Chemistry 2022 |