Open Access Article
Open Access ArticleEngineering globins for efficient biodegradation of malachite green: two case studies of myoglobin and neuroglobin†
Jiao Liu‡
a,
Jia-Kun Xu‡*b,
Hong Yuanc,
Xiao-Juan Wang*ad,
Shu-Qin Gaod,
Ge-Bo Wend,
Xiang-Shi Tan c and
Ying-Wu Lin
c and
Ying-Wu Lin *ad
*ad
aSchool of Chemistry and Chemical Engineering, University of South China, Hengyang 421001, China. E-mail: ywlin@usc.edu.cn
bKey Lab of Sustainable Development of Polar Fisheries, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Lab for Marine Drugs and Byproducts of Pilot National Lab for Marine Science and Technology, Qingdao 266071, China. E-mail: xujk@ysfri.ac.cn
cDepartment of Chemistry, Institute of Biomedical Science, Fudan University, Shanghai 200433, China
dLaboratory of Protein Structure and Function, University of South China, Hengyang 421001, China. E-mail: wxj0207@126.com
First published on 24th June 2022
Abstract
Malachite green (MG)-contaminated wastewater resulting from industrialization causes a global problem because of its toxicity and widespread usage. Compared with traditional physical and chemical approaches, biodegradation provides a new route for the degradation of MG. As promising candidates for native enzymes, artificial enzymes have received tremendous attention for potential applications due to unlimited possibilities based on precise design. In this study, we rationally engineered artificial enzymes based on myoglobin (Mb) and neuroglobin (Ngb). We introduced an aspartic acid (H64D mutation) in the heme pocket of Mb. A distal histidine (F43H mutation) was further introduced into H64D Mb to obtain a double mutant of F43H/H64D Mb. Moreover, we used A15C/H64D Ngb as designed recently for comparison studies. The H64D Mb, F43H/H64D Mb, and A15C/H64D Ngb were found to catalyze MG degradation efficiently, with activities much higher than those of native enzymes, such as dye-decolorizing peroxidase and laccase (83–205-fold). The crystal structure of H64D Mb was solved and the interactions of MG and H64D Mb and A15C/H64D Ngb were investigated by using both experimental and molecular docking studies. The biodegradation products of MG were also revealed by ESI-MS analysis. Therefore, these artificial enzymes have potential applications in the biodegradation of MG in textile industries and fisheries.
Introduction
With the vigorous development of textile industries, more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 commercial dyes have been developed and more than 10% of the dyes are discharged without proper treatment, leading to environmental pollution and causing health risks.1–4 Organic dyes typically have stable aromatic structures, which are difficult to be biodegraded, resulting in widespread environmental contaminations. Malachite green (MG), as a synthetic triphenylmethane dye, is extensively used in the textile industry for its excellent dying capacity. The use of MG has been banned in several countries,5 because MG can irritate the skin and cause permanent damage to the eyes of humans and animals, as well as other health risks.6,7 Meanwhile, MG is still used in aquaculture as a parasiticide and fungicide due to its low cost and good antibacterial effect. Therefore, it is urgent and significant to develop methods for the treatment of MG in the environment.
000 commercial dyes have been developed and more than 10% of the dyes are discharged without proper treatment, leading to environmental pollution and causing health risks.1–4 Organic dyes typically have stable aromatic structures, which are difficult to be biodegraded, resulting in widespread environmental contaminations. Malachite green (MG), as a synthetic triphenylmethane dye, is extensively used in the textile industry for its excellent dying capacity. The use of MG has been banned in several countries,5 because MG can irritate the skin and cause permanent damage to the eyes of humans and animals, as well as other health risks.6,7 Meanwhile, MG is still used in aquaculture as a parasiticide and fungicide due to its low cost and good antibacterial effect. Therefore, it is urgent and significant to develop methods for the treatment of MG in the environment.
To date, several physicochemical methods have been utilized for the removal of MG from the environment, such as acidification, lime flocculation, ozone oxidation, oxidative electrolysis, and adsorbents.8–12 However, each of these methods has limitations, such as high cost,13 more energy consumption, secondary pollution,14 and low efficiency.15 Moreover, tertiary treatments such as photochemistry and ozone oxidation are often involved in this process.16
Remarkably, biological treatment has drawn considerable attention for MG degradation due to the characteristics of being environmental-friendly, efficient, and low-cost. For example, Hao et al. purified a monomeric laccase and showed that it was able to decolorize MG.17 Recently, Guan et al. expressed a mutant of CotA-laccase in E. coli cells and showed that it could decolorize MG under neutral and alkaline conditions.39 Moreover, heme enzymes such as lignin peroxidase, manganese peroxidase, and horseradish peroxidase were applied for the decolorization of MG.18–21,41 Meanwhile, the catalytic efficiency of these native enzymes needs to be improved, and it is still required more efficient enzymes for biodegradation. For example, in a recent study,22 we have realized MG degradation by an artificial heme enzyme engineered in the protein scaffold of myoglobin (Mb), F43H/H64A Mb, with modification of the heme active site.
In addition to Mb, neuroglobin (Ngb) also belongs to the family of globins. Ngb has a similar protein fold to Mb, whereas it has bis-His coordination (His64/His96).23,24 Ngb has been shown to exhibit catalytic functions by alteration of the heme coordination states. For example, Tejero and colleagues rationally site-directed mutagenesis of the heme distal pocket.25 Recently, we have shown that a double mutant of A15C/H64D Ngb exhibits multiple peroxidase activities.26
In this study, we constructed more efficient artificial enzymes based on the globins including both Mb and Ngb by engineering new mutants such as H64D Mb, F43H/H64D Mb and A15C/H64D Ngb. These new artificial enzymes were designed to process a key Asp or His residue in the heme distal site that is responsible for H2O2 activation. We then applied them for the biodegradation of MG. The catalytic efficiencies of these enzymes were further compared to those of other published native and artificial enzymes.
Results and discussion
Structural characterization and substrate binding
To provide structural information for the introduced Asp64 in the heme distal pocket, we crystallized H64D Mb and solved the X-ray crystal structure (PDB entry 7XCQ and Table S1†). As shown in Fig. 1A, the structure revealed that the single mutant provides an environment at the heme distal site, which is similar to that in the native chloroperoxidase, with a distal glutamate responsible for the activation of H2O2 (Fig. 1B).27 Moreover, the introduction of Asp64 in Mb was shown to form a specific binding site for substrates and act as a general acid–base catalytic system to generate an oxoferryl species in reaction with H2O2.28,29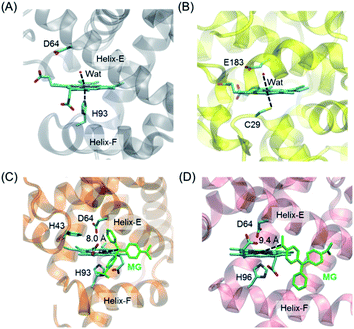 | ||
| Fig. 1 Crystal structures of (A) H64D Mb (PDB entry 7XCQ, this work), and (B) chloroperoxidase (PDB entry 1CPO)27 showing the heme active center; docking structure of the MG binding to (C) H64D Mb and (D) A15C/H64D Ngb. | ||
It is of vital importance for substrate binding to the catalytic site of enzymes. To probe whether MG could bind to the protein scaffold, we first performed molecular docking studies for MG binding to H64D Mb (Fig. 1C) and A15C/H64D Ngb (Fig. 1D), respectively. The overall structure of the MG–H64D Mb complexes revealed that the molecule of MG, like the shape of a fan, binds to the protein cavity between helices E and F. The distance between the heme iron and the central carbon of MG is ∼8.0 Å (Fig. 1C), which may facilitate the oxidative cleavage of MG upon the formation of oxoferryl species. The simulated ten most favorable MG–H64D Mb complexes have similar conformations with the lowest binding energy calculated to −5.92 kcal mol−1 (Table S2†).
The overall structure of the MG–A15C/H64D Ngb complexes was shown in Fig. 1D. It showed that MG bound to similar positions to the protein scaffold whereas it adopted a different conformation concerning that of MG binding to H64D Mb, with a short distance of ∼9.4 Å to the heme iron center. The lowest binding energy of MG–A15C/H64D Ngb complexes was calculated to be −5.42 kcal mol−1 (Table S3†), which is only −0.05 kcal mol−1 higher than that of the MG–H64D Mb complex. These theoretical results suggest that MG may bind to the scaffold of both Mb and Ngb close to the heme active site, albeit with a different conformation.
Experimental studies of MG binding to H64D Mb and A15C/H64D Ngb
The binding of MG to ferric H64D Mb and A15C/H64D Ngb was further suggested by EPR studies as shown in Fig. 2. The EPR spectrum of ferric H64D Mb exhibited the characteristic high-spin signals of Fe3+ (g⊥ ≈ 6.0; g‖ ≈ 2.0, Fig. 2a). The intensity of the g⊥ signal decreased slightly upon the addition of MG to H64D Mb (Fig. 2b), suggesting a slight alteration of the heme coordination. Meanwhile, the ferric A15C/H64D Ngb exhibited typical 6-coordinated high-spin heme signals with g tensor calculated to g⊥ ≈ 6.0 and g‖ ≈ 2.0 (Fig. 2c), which were similar to other globins with H2O/His coordination.30,31 A small fraction of low-spin species were also detected with g ≈ 3.0 and g ≈ 2.2, due to the presence of small amounts of low-heme species. These heme signals almost remained the same in the presence of MG (Fig. 2d), which suggests that the binding of MG does not affect the heme active site.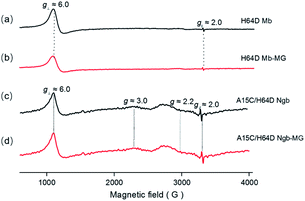 | ||
| Fig. 2 EPR spectra of ferric H64D Mb ((a and b) 0.3 mM, in 100 mM KPi, pH 6.0) and A15C/H64D Ngb ((c and d) 0.3 mM, in 100 mM KPi, pH 7.0) in the absence (a and c) and presence (b and d) of MG. | ||
To further probe the effect of MG binding on the protein secondary structure, we collected the far UV-CD spectra of H64D Mb and A15C/H64D Ngb in the absence and presence of MG. The spectrum of H64D Mb exhibited two major negative absorptions at 209 nm and 222 nm, which indicates that the protein has more α-helix content than β-sheets (Fig. 3A). Upon the addition of MG, only slight increases in ellipticity were observed for the characteristic regions of the secondary structure, suggesting that the binding of Mb almost has little effect on the secondary structure. Moreover, similar results were observed for the spectra of A15C/H64D Ngb (Fig. 3B), suggesting the binding of MG to protein with only slight alteration of the secondary structure.
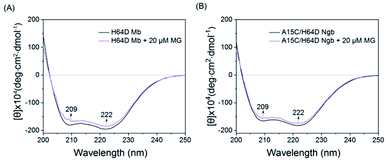 | ||
| Fig. 3 Comparison of the CD spectra of (A) H64D Mb (1 μM) and (B) A15C/H64D Ngb (1 μM) in the absence and presence of MG (20 μM) in 5 mM KPi (pH 6.0 for H64D Mb and pH 7.0 for A15C/H64D Ngb). | ||
MG degradation catalyzed by H64D Mb and A15C/H64D Ngb
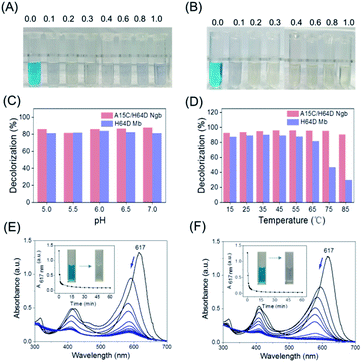
With the information on MG–protein interactions, we then evaluated the dye-decolorizing activity of H64D Mb and A15C/H64D Ngb toward the substrate, MG. The effects of H2O2 concentration, pH values and temperature on the catalytic ability of H64D Mb and A15C/H64D Ngb were studied. As shown in Fig. 4A and B, the blue solution almost faded completely in the presence of 0.2 mM H2O2 for both enzymes. Meanwhile, the reaction was inhibited obviously for H64D Mb compared with A15C/H64D Ngb at higher concentrations of H2O2, which indicates that A15C/H64D Ngb has a high tolerance of H2O2.26As shown in Fig. 4C, at fixed concentrations of MG (10 μM) and H2O2 (0.2 mM), the maximal activity was observed at pH 6.0 and pH 7.0 for H64D Mb and A15C/H64D Ngb, respectively. The temperature had a large influence on the activity of H64D Mb compared to that of A15C/H64D Ngb (Fig. 4D). When the temperature increased from 15 °C to 85 °C, both enzymes exhibited the highest rate of decolorization efficiency at 35–55 °C. Meanwhile, when the temperature was above 55 °C, an inhibition effect was observed for H64D Mb. These observations further indicate the high thermal stability of A15C/H64D Ngb.32
Under the optimal degradation conditions, we monitored the UV-vis spectral changes of MG catalyzed by H64D Mb and A15C/H64D Ngb, respectively (Fig. 4E and F). The absorbance of 617 nm of MG was decreased and blue-shifted gradually, suggesting the generation of degradation products, and the solution color was changed from blue to transparent in 1 h. These observations show that these two heme enzymes are efficient in the degradation of MG.
Catalytic efficiency toward MG
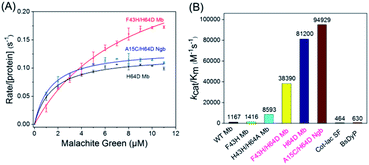
To obtain the kinetic parameters (kcat and Km) for degradation of the substrate by the enzymes, we carried out steady-state kinetic studies with different concentrations of MG and monitored the absorption changes at 617 nm in the UV-vis spectra under the optimal reaction conditions. Moreover, we evaluated the catalytic ability of F43H/H64D Mb by mutation of Phe43 to His43. The values of kobs and the concentrations of MG were then plotted and fitted to the Michaelis–Menten equation (Fig. 5A). The parameters of kcat and Km are listed in Table 1. The results showed that the kcat value of H64D Mb and F43H/H64D Mb are 0.1218 s−1 (87-fold vs. WT Mb22) and 0.3148 s−1 (225-fold vs. WT Mb), respectively. These results indicate that the aspartic acid at the 64 position and the distal His43 could facilitate the catalytic ability, which agree with previous studies.28–30,33,34 It showed that the distal Asp64 or His43 in Mb with a suitable distance to the heme iron efficiently acts as acid–base catalysis in activation of H2O2, generating the reactive compound I (an oxoferryl heme π-cation radical) or compound II (oxoferryl heme). These species may be responsible for the substrate degradation by oxidative cleavage through radical or oxygen transfer.| Proteins | kcat (s−1) | Km (μM) | kcat/Km (M−1 s−1) |
|---|---|---|---|
| WT Mb22 | 0.0014 ± 0.0001 | 1.2 ± 0.2 | 1167 |
| F43H Mb22 | 0.0034 ± 0.0001 | 2.4 ± 0.3 | 1416 |
| F43H/H64A Mb22 | 0.0232 ± 0.0013 | 2.7 ± 0.5 | 8593 |
| H64D Mb | 0.1218 ± 0.0036 | 1.5 ± 0.2 | 81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
| F43H/H64D Mb | 0.3148 ± 0.0185 | 8.2 ± 0.9 | 38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 390 390 |
| A15C/H64D Ngb | 0.1329 ± 0.0037 | 1.4 ± 0.2 | 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 929 929 |
| BsDyP35 | 0.0456 ± 0.0016 | 72.0 ± 7.0 | 630 |
| CotA-lac SF36 | 18.36 | 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 600 |
464 |
Among these enzymes, A15C/H64D Ngb exhibited the highest catalytic efficiency with kcat/Km value up to 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 929 M−1 s−1, which is 151- and 205-fold higher than that reported for dye-decolorizing peroxidase from Bacillus subtilis (BsDyp35) and the mutated CotA-laccase SF enzyme.36
929 M−1 s−1, which is 151- and 205-fold higher than that reported for dye-decolorizing peroxidase from Bacillus subtilis (BsDyp35) and the mutated CotA-laccase SF enzyme.36
Product analysis of MG degradation
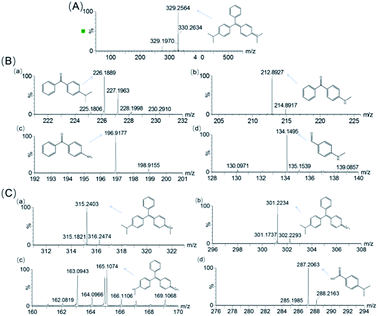
To identify the degradation products of MG catalyzed by, H64D Mb, we conducted ESI-MS studies of the solution after the reaction. As shown in Fig. 5A for the mass of MG (329 m/z), the peak disappeared after the reaction and new peaks were generated. These new peaks included 226,37 212, 197 and 134 m/z, which could be assigned the following species, (Fig. 6B(a)) m/z = 226 is 4-(dimethylamino)benzophenone (DLBP); (Fig. 6B(b)) m/z = 212 is 4-(methylamino)benzophenone (MLBP); (Fig. 6B(c)) m/z = 197 is 4-aminobenzophenone (ABP); (Fig. 6B(d)) m/z = 134 is 4-(methylamino)benzaldehyde (MLBD), respectively. Moreover, the MG degradation products catalyzed by A15C/H64D Ngb produced small molecules such as those detected with molecular weights of 315, 301, 287, and 165 m/z, which are corresponding to (p-dimethylaminophenyl)(p-methylaminopheyl)phenylmethylium (DM-PM), (p-methylaminophenyl)(p-methylaminopheyl)phenylmethylium (MM-PM), (p-methylaminophenyl)phenylmethylium (M-PM) and 4-(dimethylamino)benzoic acid (DM-BA), respectively.Based on the ESI-MS results, we proposed a generation route of the reaction products, which may involve the generation of reactive radical species upon the activation of H2O2 by these enzymes, resulting in different bond cleavages of the substrate.38–40 The cleavage of the C–C bond involving the central carbon by removal of one or two benzenes may generate DLBP and DM-BA. N-Demethylation of DLBP was also observed in a stepwise manner by Du et al.6 Given the identification of MLBP, ABP, MM-PM, DM-PM, and M-PM, the degradation could also be initiated by N-demethylation of MG, followed by oxidation or further N-demethylation.
As shown in Scheme 1, we proposed a plausible biodegradation mechanism of MG catalyzed by H64D Mb and A15C/H64D Ngb. The biodegradation mechanism for MG catalyzed by H64D Mb probably underwent pathway I, where the oxidation reaction occurred firstly, followed by N-demethylation. Conversely, the biodegradation mechanism for MG catalyzed by A15C/H64D Ngb (pathway II) may include three steps of N-demethylation,41 with a last step of the oxidation reaction. We noticed that the degradation pathways of MG catalyzed by H64D Mb and A15C/H64D Ngb are different, which may due to their different binding models of MG binding to the heme active site. Interestingly, both oxidative cleavage and demethylation were involved in MG degradation catalyzed by H64D Mb and A15C/H64D Ngb.
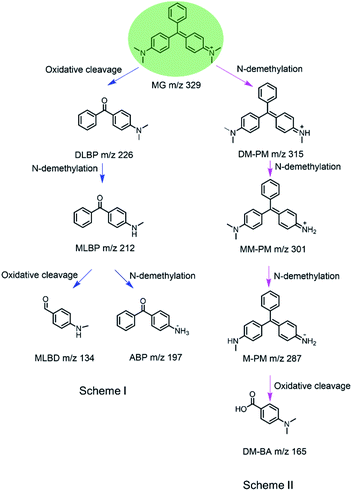 | ||
| Scheme 1 Proposed mechanism for biodegradation of MG catalyzed by H64D Mb (Scheme I) and A15C/H64D Ngb (Scheme II) based on the MS analysis. | ||
Conclusions
In this study, we introduced an aspartate residue, by H64D mutation, into the heme pocket of Mb and obtained the crystal structure of the single mutant. We also engineered a double mutant of F43H/H64D Mb by introducing a distal histidine to the single mutant. Moreover, we used the recently designed A15C/H64D Ngb as a control. EPR studies showed that the binding of the enzyme to MG does not affect the heme active site. At the same time, CD studies showed that the binding of the substrate to the enzyme has little effect on the secondary structure of the enzyme. In addition, we found that A15C/H64D Ngb is highly tolerant and thermally stable to H2O2 compared to that of H64D Mb. Kinetic UV-vis studies further showed that H64D Mb, F43H/H64D Mb and A15C/H64D Ngb effectively catalyze the degradation of MG, with much higher activity than natural enzymes such as decolorization peroxidase and laccase (83–205-fold). Finally, by ESI-MS analysis, we proposed a plausible pathway for the generation of MG biodegradation. This study suggests the potential application of artificial enzymes for the biodegradation of MG in the textile industry and fisheries.Materials and methods
Materials
Sperm whale wild-type (WT) Mb, H64D Mb, F43H/H64D Mb and A15C/H64D Ngb were expressed in BL21 (DE3) cells and purified as previously reported.42 The genes of F43H/H64D Mb were constructed by using the QuikChange Site-Directed Mutagenesis Kit (Stratagene) using the WT Mb gene as a template, and the mutations were confirmed by a DNA sequencing assay.43 All other chemical reagents are commercially available and of analytical grade.Crystal X-ray diffraction of H64D Mb
The protein crystal was shaped by ferric H64D Mb with high purity (A409 nm/A280 nm > 3.0) and was exchanged into 20 mM potassium phosphate buffer. Single-crystal X-ray diffraction data of H64D Mb were collected at the BL16B1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF) using a MAR mosaic 225 CCD detector with a wavelength of 0.9791 Å at 100 K. The diffraction data were processed and scaled with HKL-2000.44 The structure was determined by the molecular replacement method, and the 1.6 Å structure of WT Mb [PDB entry 1JP6 (ref. 45)] was used as the starting model. Manual adjustment of the model was carried out using COOT46 and the models were refined by PHENIX47 and Refmac5.48 The stereochemical quality of the structures was checked using PROCHECK.49 All the residues were in the favored and allowed regions and none in the disallowed region.Molecular modeling
The X-ray structure of H64D Mb was used as the initial structure for docking with MG using the Autodock 4.2.3. The heme iron was set to be the center, with a box size of 60 Å × 60 Å × 60 Å, which covered most of the protein surface. MG as a substrate for docking was generated using the Dundee Prodrg2 server. Docked conformations were ranked automatically by Autodock 4.2 using a binding-energy scoring function. The docking results with the ten most favorable conformations after 2000 steps were then visualized and analyzed using VMD 1.9.2.EPR studies
Electron paramagnetic resonance (EPR) spectra of H64D Mb (0.3 mM, in 100 mM KPi buffer, pH 6.0) and A15C/H64D Ngb (0.3 mM, in 100 mM KPi buffer, pH 7.0) in the absence or presence of MG (0.3 mM) were recorded on a Bruker A300 spectrometer (X-band) equipped with Bruker ER4141VTM liquid nitrogen system. The sample was transferred into an EPR tube with a volume of 300 μL. The spectrum was measured at 100 K, with a frequency of 9.27 GHz, centerfield 2750 G and sweep width 4500 G, microwave power 0.595 mW, and modulation amplitude 3.0 G.CD spectroscopy
The circular dichroism (CD) studies were performed using a J-Model-1500 spectrophot-ometer (JASCO) equipped with a Peltier temperature control system. The far-UV CD spectra were recorded in the range of 190–250 nm for H64D Mb and A15C/H64D Ngb (1 μM in 5 mM KPi buffer) in the concentration of MG (20 μM). The conformational stability of protein was assessed by recording the spectra at different concentrations (0, 20 μM). The averaged protein spectrum was corrected by reducing the buffer blank baseline spectrum.UV-vis spectroscopy
UV-vis spectra were recorded on a Hewlett-Packard 8453 diode array spectrometer. The final concentration of proteins was decided by extinction coefficient, like H64D Mb with ε409 nm of 165 mM−1 cm−1, F43H/H64D Mb with ε407 nm of 159 mM−1 cm−1, A15C/H64D Mb with ε406 nm of 160 mM−1 cm−1, MG with ε617 nm of 148.9 mM−1 cm−1.50Kinetic UV-vis studies
The UV-vis spectra of the reaction were recorded using a Hewlett-Packard 8453 diode array spectrometer UV-vis spectra. The reaction was carried out in a 2 mL cuvette, in which, H64D Mb, F43H/H64D Mb (2 mM in 100 mM KPi buffer, pH 6.0) or A15C/H64D Ngb (2 mM in 100 mM KPi buffer, pH 7.0) was added by varying the concentration of MG (0–16 mM). The reaction was initiated by the addition of H2O2 (0.2 mM), and the decrease of absorption at 617 nm was monitored to determine the initial rate.ESI-MS
The mass of H64D Mb, A15C/H64D Ngb and the products after the reaction H64D, A15C/H64D Ngb and MG were determined on a G2-XS QToF mass spectrometer (Waters), which was then transferred into the mass spectrometer chamber under positive mode. The mass of H64D Mb and A15C/H64D Ngb multiple m/z peaks were transformed to molecular weight by using MaxEnt1 software. And after this reaction, several peaks of transformation products were observed in the MS spectrum of the reaction solution.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully thank Prof. S. G. Sligar and Prof. Y. Lu of the University of Illinois at Urbana-Champaign, USA, for the gift of the Mb gene. X-ray diffraction data were collected at Shanghai Synchrotron Radiation Facility, China. This work was supported by the National Natural Science Foundation of China (32171270, 21977042, 21977017 and 21807058), Special Scientific Research Funds for Central Nonprofit Institutes, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences (20603022016011), the Financial Fund of the Ministry of Agriculture and Rural Affairs, China (NFZX2018) and Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2020TD67).References
- M. P. Shah, J. Biorem. Biodegrad., 2018, 9(1), 1000427 CrossRef.
- B. B. Wang, J. Lu, J. F. Zheng and Z. S. Yu, J. Microbiol., 2021, 59, 142–150 CrossRef CAS PubMed.
- R. Yadav, T. S. Chundawat, P. Rawat, G. K. Rao and D. Vaya, Bull. Mater. Sci., 2021, 44, 250 CrossRef CAS.
- S. Clemmensen, J. C. Jensen, N. J. Jensen, O. Meyer, P. Olsen and G. Würtzen, Arch. Toxicol., 1984, 56, 43–45 CrossRef CAS PubMed.
- W. C. Andersen, C. R. Casey, T. J. Nickel, S. L. Young and S. B. Turnipseed, J. AOAC Int., 2018, 101, 1927–1939 CrossRef CAS PubMed.
- L.-N. Du, M. Zhao, G. Li, F.-C. Xu, W.-H. Chen and Y.-H. Zhao, Int. Biodeterior. Biodegrad., 2013, 78, 108–116 CrossRef CAS.
- A. Stammati, C. Nebbia, I. D. Angelis, A. G. Albo, M. Carletti, C. Rebecchi, F. Zampaglioni and M. Dacasto, Toxicol. in Vitro, 2005, 19, 853–858 CrossRef CAS PubMed.
- C. Muthukumaran, V. M. Sivakumar and M. Thirumarimurugan, J. Taiwan Inst. Chem. Eng., 2016, 63, 354–362 CrossRef CAS.
- A. D. Watharkar, R. V. Khandare, A. A. Kamble, A. Y. Mulla, S. P. Govindwar and J. P. Jadhav, Environ. Sci. Pollut. Res., 2013, 20, 939–949 CrossRef CAS PubMed.
- M. A. M. Salleh, D. K. Mahmoud, W. A. W. A. Karim and A. Idris, Desalination, 2011, 280, 1–13 CrossRef CAS.
- V. K. Gupta, S. Khamparia, I. Tyagi, D. Jaspal and A. Malviya, Global J. Environ. Sci. Manage., 2015, 1, 71–94 CAS.
- Y. Y. Ling and F. B. Mohd Suah, J. Environ. Chem. Eng., 2017, 5, 785–794 CrossRef CAS.
- R. G. Saratale, G. D. Saratale, J. S. Chang and S. P. Govindwar, J. Taiwan Inst. Chem. Eng., 2011, 42, 138–157 CrossRef CAS.
- R. N. Bharagava, S. Mani, S. I. Mulla and G. D. Saratale, Ecotoxicol. Environ. Saf., 2018, 156, 166–175 CrossRef CAS PubMed.
- A. Anastasi, F. Spina, A. Romagnolo, V. Tigini, V. Prigione and G. C. Varese, Bioresour. Technol., 2012, 123, 106–111 CrossRef CAS PubMed.
- F. P. van der Zee and S. Villaverde, Water Res., 2005, 39, 1425–1440 CrossRef CAS PubMed.
- J. Z. Hao, W. W. Zhang, H. X. Wang, N. Ziya, Y. Luo, P. S. Jia, G. Q. Zhang and T. Ng, Biotechnol. Appl. Biochem., 2021, 68, 297–306 CrossRef CAS PubMed.
- A. K. Chauhan and B. Choudhury, Chemosphere, 2021, 273, 129671 CrossRef CAS PubMed.
- M. Bilal, H. M. N. Iqbal, H. Hu, W. Wang and X. Zhang, J. Environ. Manage., 2017, 188, 137–143 CrossRef CAS PubMed.
- J. Yang, J. Zhao, J. Jiang, H. Xu, N. Zhang, J. Xie and M. Wei, Front. Energy Res., 2021, 9, 807286 CrossRef.
- P. Chowdhary, G. Shukla, G. Raj, L. F. R. Ferreira and R. N. Bharagava, SN Appl. Sci., 2019, 1, 45 CrossRef.
- H. F. Xiang, J. K. Xu, J. Liu, X. Z. Yang, S. Q. Gao, G. B. Wen and Y. W. Lin, RSC Adv., 2021, 11, 16090–16095 RSC.
- P. B. Tiwari, L. Astudillo, K. Pham, X. Wang, J. He, S. Bernad, V. Derrien, P. Sebban, J. Miksovska and Y. Darici, Inorg. Chem. Commun., 2015, 62, 37–41 CrossRef CAS.
- B. Vallone, K. Nienhaus, A. Matthes, M. Brunori and G. U. Nienhaus, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 17351–17356 CrossRef CAS PubMed.
- J. Tejero, C. E. Sparacino-Watkins, V. Ragireddy, S. Frizzell and M. T. Gladwin, Biochemistry, 2015, 54, 722–733 CrossRef CAS PubMed.
- S.-F. Chen, X.-C. Liu, J.-K. Xu, L. Li, J.-J. Lang, G.-B. Wen and Y.-W. Lin, Inorg. Chem., 2021, 60, 2839–2845 CrossRef CAS PubMed.
- M. Sundaramoorthy, J. Terner and T. L. Poulos, Structure, 1995, 3, 1367–1378 CrossRef CAS PubMed.
- T. Matsuo, K. Fukumoto, T. Watanabe and T. Hayashi, Chem.–Asian J., 2011, 6, 2491–2499 CrossRef CAS PubMed.
- T. Matsui, S.-i. Ozaki and Y. Watanabe, J. Am. Chem. Soc., 1999, 121, 9952–9957 CrossRef CAS.
- L.-L. Yin, H. Yuan, C. Liu, B. He, S.-Q. Gao, G.-B. Wen, X. Tan and Y.-W. Lin, ACS Catal., 2018, 8, 9619–9624 CrossRef CAS.
- L.-L. Li, H. Yuan, F. Liao, B. He, S.-Q. Gao, G.-B. Wen, X. Tan and Y.-W. Lin, Dalton Trans., 2017, 46, 11230–11238 RSC.
- H.-X. Liu, L. Li, X.-Z. Yang, C.-W. Wei, H.-M. Cheng, S.-Q. Gao, G.-B. Wen and Y.-W. Lin, RSC Adv., 2019, 9, 4172–4179 RSC.
- T. Murakami, I. Morishima, T. Matsui, S.-i. Ozaki, I. Hara, H.-J. Yang and Y. Watanabe, J. Am. Chem. Soc., 1999, 121, 2007–2011 CrossRef CAS.
- T. Matsui, S. Ozaki, E. Liong, G. N. Phillips Jr and Y. Watanabe, J. Biol. Chem., 1999, 274, 2838–2844 CrossRef CAS PubMed.
- L. Xu, K. Sun, F. Wang, L. Zhao, J. Hu, H. Ma and Z. Ding, J. Environ. Manage., 2020, 270, 110904 CrossRef CAS PubMed.
- K. K. Navada and A. Kulal, J. Environ. Chem. Eng., 2020, 8, 103550 CrossRef CAS.
- E. Kusvuran, O. Gulnaz, A. Samil and O. Yildirim, J. Hazard. Mater., 2011, 186, 133–143 CrossRef CAS PubMed.
- L. Xu, J. Sun, M. A. Qaria, L. Gao and D. Zhu, Catalysts, 2021, 11(8), 955 CrossRef.
- K.-Z. Xu, H. Ma, Y.-J. Wang, Y.-J. Cai, X.-R. Liao and Z.-B. Guan, Ecotoxicol. Environ. Saf., 2020, 193, 110335 CrossRef CAS PubMed.
- X. Yang, J. Zheng, Y. Lu and R. Jia, Environ. Sci. Pollut. Res., 2016, 23, 9585–9597 CrossRef CAS PubMed.
- J. Yang, X. Yang, Y. Lin, T. B. Ng, J. Lin and X. Ye, PLoS One, 2015, 10, e0127714 CrossRef PubMed.
- B. A. Springer and S. G. Sligar, Proc. Natl. Acad. Sci. U. S. A., 1987, 84, 8961–8965 CrossRef CAS PubMed.
- P. Zhang, H. Yuan, J. Xu, X.-J. Wang, S.-Q. Gao, X. Tan and Y.-W. Lin, ACS Catal., 2019, 10, 891–896 CrossRef.
- Z. Otwinowski and W. Minor, in Methods in Enzymology, Academic Press, 1997, vol. 276, pp. 307–326 Search PubMed.
- P. Urayama, G. N. Phillips and S. M. Gruner, Structure, 2002, 10, 51–60 CrossRef CAS PubMed.
- P. Emsley and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 2126–2132 CrossRef PubMed.
- P. D. Adams, R. W. Grosse-Kunstleve, L. W. Hung, T. R. Ioerger, A. J. McCoy, N. W. Moriarty, R. J. Read, J. C. Sacchettini, N. K. Sauter and T. C. Terwilliger, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2002, 58, 1948–1954 CrossRef PubMed.
- G. N. Murshudov, A. A. Vagin and E. J. Dodson, Acta Crystallogr., Sect. D: Biol. Crystallogr., 1997, 53, 240–255 CrossRef CAS PubMed.
- R. A. Laskowski, M. W. MacArthur, D. S. Moss and J. M. Thornton, J. Appl. Crystallogr., 1993, 26, 283–291 CrossRef CAS.
- G. A. Kraus, I. Jeon, M. Nilsen-Hamilton, A. M. Awad, J. Banerjee and B. Parvin, Molecules, 2008, 13, 986–994 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Data collection and refinement statistics and docking results. See https://doi.org/10.1039/d2ra02795j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |