Open Access Article
Open Access ArticleAnti-diabetic activities of phenolic compounds of Alternaria sp., an endophyte isolated from the leaves of desert plants growing in Egypt†
Ahmed Elbermawi *a,
Ahmed R. Ali
*a,
Ahmed R. Ali *b,
Yhiya Amen
*b,
Yhiya Amen a,
Ahmed Ashour
a,
Ahmed Ashour a,
Kadria F. Ahmada,
El-Sayed S. Mansoura and
Ahmed F. Halima
a,
Kadria F. Ahmada,
El-Sayed S. Mansoura and
Ahmed F. Halima
aDepartment of Pharmacognosy, Faculty of Pharmacy, Mansoura University, Mansoura, 35516 Egypt. E-mail: asbeder@mans.edu.eg; Tel: +20-10-0481-1533
bDepartment of Medicinal Chemistry, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt. E-mail: ahmed_reda5588@mans.edu.eg; Tel: +20-10-9838-4072
First published on 1st September 2022
Abstract
Six phenolic compounds (talaroflavone (1), alternarienoic acid (2), altenuene (3), altenusin (4), alternariol (5), and alternariol-5-O-methyl ether (6)) were isolated from the solid rice culture media of Alternaria sp., an endophyte isolated from the fresh leaves of three desert plants, Lycium schweinfurthii Dammer (Solanaceae), Pancratium maritimum L. (Amaryllidaceae) and Cynanchum acutum L. (Apocynaceae). Compounds 2, 3, and 4 exhibited potent α-glucosidase and lipase inhibitory activities suggesting that they might act as naturally occurring anti-diabetic candidates. The same compounds showed potent binding in the active site for both enzymes with desirable pharmacokinetic properties. The isolated bioactive compounds were not exclusive to a certain host plant which reveals the dominant ecological standpoints for consequent optimization. This could lead to a cost-effective and reproducible yield applicable to commercial scale-up.
1. Introduction
The increasing prevalence of obesity and physical inactivity, along with aging, are pivotal factors explaining the growing number of diabetic patients. Diabetes mellitus is a chronic metabolic disorder in which the body either does not produce enough insulin or is not able to respond to the blood-circulating insulin, resulting in elevated blood glucose levels. This would ultimately lead to major complications, such as diabetic neuropathy, retinopathy and other cardiovascular diseases.1,2 There are three main types of diabetes: type I which is caused by the inability of the body to produce sufficient insulin due to progressive death of insulin-producing pancreatic β-cells, possibly induced by the accumulated lipids in the pancreas. Therefore, pancreatic lipase inhibitors have attracted much attention as anti-diabetic agents for their inhibitory activity against lipid absorption, thus protecting the pancreas.3 Type II diabetes results from tissue resistance to already existing insulin. A therapeutic approach for type II is via inhibiting the carbohydrate-hydrolyzing enzymes, such as α-glucosidase, which in turn delay the rapid utilization of dietary carbohydrates and suppress postprandial hyperglycemia.4 The third type is gestational diabetes, which occurs in about 2–5% of all pregnancies.5The endophytic fungi are considered a promising source for new and bioactive secondary metabolites in a wide variety of medical and agricultural areas.6 In the last few decades more than 268 metabolites were reported from different species of Alternaria fungi.7 The isolated metabolites belongs to several structural classes, including alkaloids,8 steroids,9 quinones,10 pyranones,11 phenolics and various diverse classes.12 The isolated metabolites showed versatile biological activities including antitumor, antimicrobial, herbicide, antimalarial, and antileishmanial.7,12
In our ongoing research for bioactive metabolites derived from plant endophytes, leaves of ten desert plants were separately collected, subjected to endophytic fungi isolation, purification, fungal identity, and subsequent chemical investigation. Based on Internal Transcribed Spacer (ITS) genomic analysis, Alternaria sp. was isolated from the fresh leaves of three plants: Cynanchum acutum L. (Apocynaceae), Lycium schweinfurthii Dammer (Solanaceae) and Pancratium maritimum L. (Amaryllidaceae). Chemical investigation for Alternaria sp. rice culture medium was carried out and resulted in isolation and identification of six phenolic metabolites, followed by biological assessment for their inhibitory activity against α-glucosidase and pancreatic lipase.
2. Results and discussion
2.1. Phylogenetic identification of the isolated fungal endophytes
The resulting ITS sequences of AS56, AS55 and AS57 isolated from L. schweinfurthii, P. maritimum and C. acutum, respectively, were submitted to the GenBank, for a homology search with Blast. Alignment with published sequences in GenBank showed that the three strains had 99% identity as Alternaria sp. (Genbank accession no. MK680829, MK693741 and MK695948, respectively). The isolated fungal strains were deposited in the Microbiology Department, Faculty of Pharmacy, Mansoura University, Egypt, under voucher specimen numbers AS56-003, AS55-003 and AS57-003, respectively.2.2. Isolation and identification of the compounds
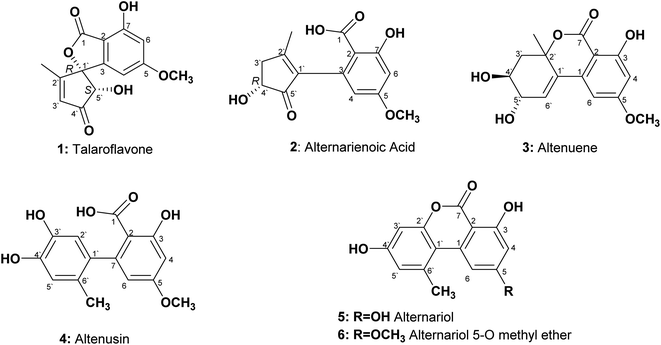
An endophytic fungus was isolated from the fresh leaves of L. schweinfurthii (Solanaceae), P. maritimum L. (Amaryllidaceae) and C. acutum L. (Apocynaceae), and identified as Alternaria sp. according to a molecular biological protocol.13 Investigation of the EtOAc extract of solid rice culture media of Alternaria sp. led to isolation of six previously reported compounds, their structures were elucidated on the basis of 1D, 2D-NMR and HRMS, identified as; talaroflavone (1),14 alternarienoic acid (2),15 altenuene (3),16 altenusin (4),17 alternariol (5)18 and alternariol-5-O-methyl ether (6)19 (Fig. 1).Talaroflavone 1, [α]20D + 42.0 (c 0.05, MeOH), was isolated from the EtOAc extract of rice cultures of Alternaria sp. in the form of white flakes (5.2 mg) with a molecular formula C14H12O6. It displayed UV absorbances at λmax (MeOH) 219.5, 260.2 and 295.1 nm. The HR-ESI-MS+ showed a cationated ion peak at m/z 299.0701 [M + Na]+ (base peak) and HRESI-MS− showed a deprotonated ion peak at m/z 275.0631 [M − H]− indicating a molecular weight of 276 g mol−1. 1H NMR and mass spectra supported a molecular formula of C14H12O6. 1H NMR (600 MHz, CD3OD): δH 1.86 (3H, s, CH3-2′), 3.81 (3H, s, OCH3), 4.74 (1H, s, H-5′), 6.07 (1H, s, H-4), 6.35 (1H, s, H-3′), 6.48 (1H, s, H-6). 13C NMR (150 MHz, CD3OD): δC 13.5 (CH3-2′), 56.5 (OCH3), 79.8 (C-5′), 94.3 (C-1′), 101.1 (C-4), 103.5 (C-6), 106.2 (C-2), 131.5 (C-3′), 150.9 (C-3), 159.9 (C-7), 168.5 (C-5), 170.1 (C-1), 171.5 (C-2′), 202.0 (C-4′).
Alternarienoic acid 2, [α]20D + 26.0 (c 0.03, MeOH), was isolated from the EtOAc extract of rice cultures of Alternaria sp. as viscous yellow oil (1.6 mg). It exhibited UV absorbances at λmax (MeOH) 217.9, 260.8 and 301.7 nm. The HR-FAB-MS+ showed the molecular ion [M]+ at m/z 278.0790 (calculated 278.0790), providing the molecular formula C14H14O6. 1H NMR (600 MHz, CD3OD): δH 2.15 (1H, s, CH3-2′), δH 2.50 (1H, br d, H-3′a), 3.02 (1H, m, H-3′b), 3.81 (3H, s, OCH3), 4.36 (1H, m, H-4′), 6.13 (1H, d, J = 2.4 Hz, H-4), 6.46 (1H, d, J = 2.4 Hz, H-6). 13C NMR (150 MHz, CD3OD 3): δC 17.8 (CH3-2′), 41.8 (C-3′), 56.0 (OCH3), 73.2 (C-4′), 101.6 (C-6), 107.0 (C-2), 111.1 (C-4), 137.5 (C-3), 142.2 (C-1′), 145.9 (C-2′), 165.2 (C-1), 166.7 (C-5), 167.9 (C-7), 208.8 (C-5′).
Altenuene 3, [α]20D − 22.0 (c 0.15, CHCl3), was obtained as viscous yellow oil (18.8 mg). The UV spectrum showed λmax (MeOH) at 243.6, 281.7 and 322.5 nm. The HR-ESI-MS+ showed [M + Na]+ at m/z 315.0972, while HR-ESI-MS− showed [M − H − H2O]− at m/z 273.0488, indicating the molecular formula to be C15H16O6. 1H NMR (600 MHz, CD3OD): δH 1.50 (3H, s, CH3-2′), 1.98 (1H, dd, J = 14.4, 9 Hz, H-3′a), 2.41 (1H, dd, J = 14.4, 3.6 Hz, H-3′b), 3.78 (3H, m, H-4′), 3.88 (3H, s, OCH3), 4.07 (1H, dd, J = 6, 5.4 Hz, H-5′), 6.22 (1H, d, J = 3.0 Hz, H-6′), 6.47 (1H, d, J = 1.8 Hz, H-4), 6.66 (1H, d, J = 1.8, H-6). 13C NMR (150 MHz, CD3OD): δC 28.1 (CH3-2′), 40.9 (C-3′), 56.4 (OCH3), 70.8 (C-4′), 72.3 (C-5′), 82.5 (C-2′), 101.7 (C-2), 101.9 (C-4), 103.8 (C-6), 131.4 (C-6′), 134.8 (C-1′), 140.9 (C-1), 165.3 (C-5), 168.1 (C-3), 170.5 (C-7).
Altenusin 4 was isolated from the EtOAc extracts of rice culture of Alternaria sp. in the form of reddish white prisms (25.4 mg) with a molecular formula C15H14O6. It exhibited UV absorbances at λmax (MeOH) 202.6, 214.6, 256.9 and 293.0 nm. HR-FAB-MS+ showed molecular ion peaks at m/z 290.0971 [M]+ (base peak) (calculated 290.0970). 1H NMR (600 MHz, CD3OD): δH 1.91 (3H, s, CH3-6′), 3.80 (3H, s, OCH3), 6.16 (1H, d, J = 2.4 Hz, H-6), 6.42 (1H, d, J = 2.4 Hz, H-4), 6.48 (1H, s, H-2′), 6.58 (1H, s, H-5′). 13C NMR (150 MHz, CD3OD): δC 19.3 (CH3-6′), 55.9 (OCH3), 100.6 (C-4), 107.7 (C-2), 111.3 (C-6), 116.7 (C-2′), 117.4 (C-5′), 127.4 (C-6′), 135.6 (C-1′), 143.3 (C-3′), 144.9 (C-4′), 148.1 (C-7), 164.9 (C-3), 165.7 (C-5), 174.4 (C-1).
Alternariol 5 was isolated as reddish white needles (7.6 mg) with a molecular formula C14H10O5. It showed UV absorbances at λmax (MeOH) 206.1, 255.8, 299.8 and 339.7 nm. HRFAB+MS showed molecular ion peaks at m/z 258.0528 [M]+ (base peak), calculated 258.0528. 1H NMR (600 MHz, CD3OD): δH 2.87 (3H, s, CH3-6′), 6.39 (1H, d, J = 1.8 Hz, H-4), 6.65 (1H, d, J = 2.4 Hz, H-3′), 6.69 (1H, d, J = 2.4 Hz, H-5′), 7.29 (1H, d, J = 1.8 Hz, H-6). 13C NMR (150 MHz, CD3OD): δC 25.8 (CH3-6′), 98.6 (C-2), 101.9 (C-4), 102.6 (C-3′), 105.5 (C-6), 110.8 (C-1′), 118.2 (C-5′), 139.3 (C-6′), 139.6 (C-1), 153.9 (C-2′), 159.1 (C-4′), 165.5 (C-3), 166.6 (C-5), 166.8 (C-7).
Alternariol-5-O-methyl ether 6 was obtained as a reddish white needles (11.3 mg) with a molecular formula C15H12O5 on the basis of accurate mass measurement (HR-FAB-MS+, m/z 272.0685 [M + H]+), (calculated 272.0685) and the number of signals in both 1H and 13C-NMR, HSQC spectra. It displayed UV absorbances at λmax (MeOH) 214.6, 249.7, 287.6 and 345.1 nm, showing high similarity to UV spectra typical for alternariol derivatives. 1H NMR (600 MHz, CD3OD): δH 2.78 (3H, s, CH3-6′), 3.93 (3H, s, OCH3), 6.55 (1H, d, J = 1.8 Hz, H-4), 6.63 (1H, d, J = 2.4 Hz, H-3′), 6.71 (1H, d, J = 2.4 Hz, H-5′), 7.29 (1H, d, J = 1.8 Hz, H-6). 13C NMR (150 MHz, CD3OD): δC 25.1 (CH3-6′), 56.4 (OCH3), 100.1 (C-2,4), 103.1 (C-3′), 105.0 (C-6), 110.9 (C-1′), 118.9 (C-5′), 139.9 (C-6′), 154.6 (C-2′), 160.2 (C-4′), 166.4 (C-3), 167.0 (C-7), 168.2 (C-5).
2.3. HPLC tracking of isolated compounds in different fungal extracts
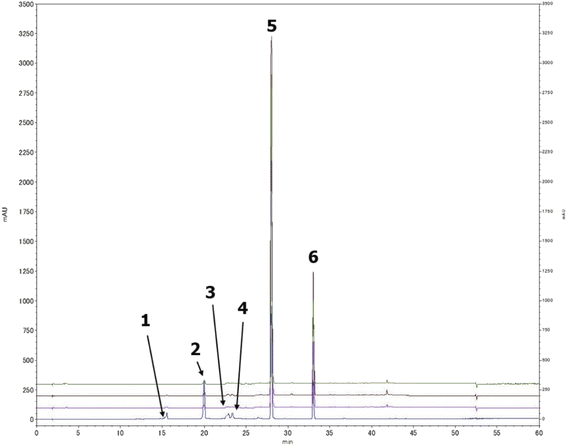
A preliminary chemical study for the three plants fungal secondary metabolites was conducted through a comprehensive HPLC profiling for their different fractions, resulted in their close similarity. As a result, tracking the isolated compounds from L. schweinfurthii isolated Alternaria sp. metabolites was accomplished within the other two similar species isolated from C. acutum and P. maritimum resulted in the existence of the peaks corresponding to each compound guided by their retention time and UV absorbance (Fig. 2) except for talaroflavone (1), which seemed to be minor component in P. maritimum.2.4. Inhibition of α-glucosidase enzyme
An in vitro α-glucosidase inhibitory assay was performed for the isolated compounds as well as the positive control acarbose. The results (Table 1) revealed that compounds 2, 3 and 4 have potent α-glucosidase inhibitory activity with IC50 values 7.95 ± 1.2, 40.38 ± 3.7 and 46.14 ± 0.84 μM, respectively.| Compound | α-Glucosidase IC50 (μM) | Pancreatic lipase IC50 (μM) |
|---|---|---|
| Positive control | Acarbose 283 ± 14.65 | Orlistat 1.35 ± 0.1 |
| Compound 1 | >300 | 73.82 ± 0.82 |
| Compound 2 | 7.95 ± 1.2 | 20.82 ± 0.95 |
| Compound 3 | 40.38 ± 3.7 | 3.18 ± 0.79 |
| Compound 4 | 46.14 ± 0.84 | 21.46 ± 0.97 |
| Compound 5 | 179.88 ± 4.65 | 56.85 ± 0.77 |
| Compound 6 | 236.25 ± 6.22 | 70.60 ± 0.86 |
2.5. Inhibition of pancreatic lipase enzyme
The six isolated compounds together with the positive control orlistat were tested for their inhibitory activity against pancreatic lipase enzyme. Compounds 2, 3 and 4 exhibited a potential for inhibition of pancreatic lipase enzyme with IC50 equal 20.82 ± 0.95, 3.18 ± 0.79 and 21.46 ± 0.97 μM, respectively (Table 1).2.6. Molecular modeling studies
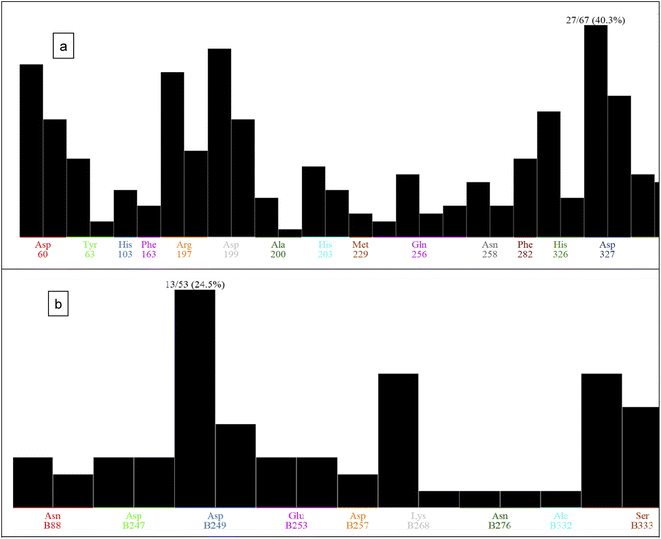
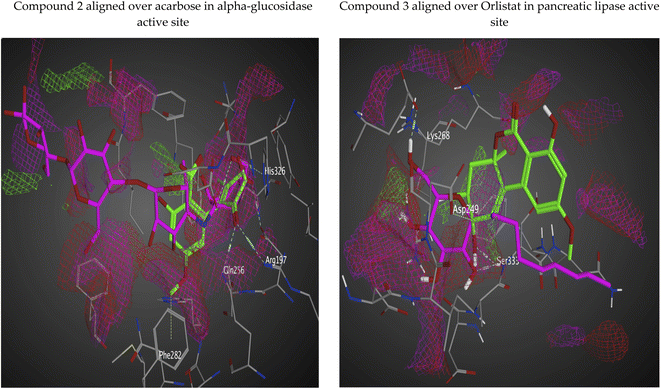
The docking poses of the isolated compounds with α-glucosidase enzyme are shown in (Fig. 3) along with the interacting amino acids residues. Compound 1 showed hydrogen bond interactions with Tyr63, Asp199 and Ala200, while compound 2 interacted with Arg197, Gln256 and His326 via hydrogen bonding and π-H interaction with Phe282. Compound 3 exhibited hydrogen bond interaction with His203 and Asp327, while compound 4 had only hydrogen bonding with Asp327. Compound 5 showed hydrogen bond interactions with Asp60 and π-H interaction with Tyr63, while compound 6 interacted with Asp60, Asp199 via hydrogen bonding. The control compound, acarbose, showed hydrogen bond interactions with Asp199 and Asp327. Compounds 2–4 showed maximum number of interactions with amino acids residues in the active site or was able to maintain interaction with essential key residues as indicated by subsequent PLIF analysis.
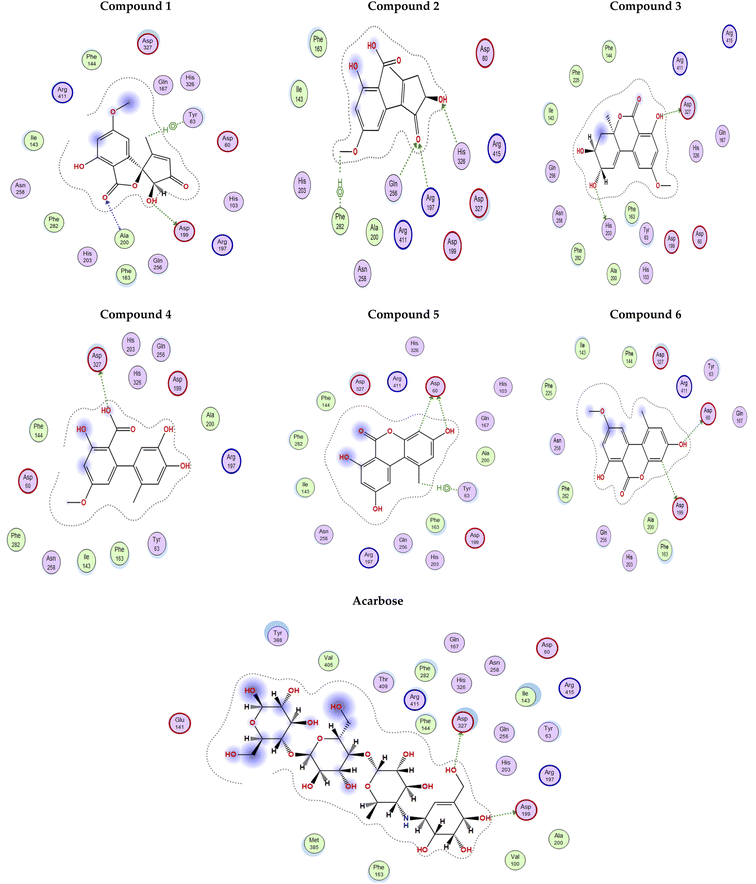 | ||
| Fig. 3 2D Interaction diagram for compounds 1–6 and acarbose with alpha-glucosidase enzyme (PDB id: 5ZCC). | ||
Fig. 4 shows the docking poses for our isolated series of compounds with pancreatic lipase. Compound 1 showed 2 hydrogen bonds via its carbonyl group with Asp249 and its alcoholic hydroxyl group with Lys268. Compound 2 formed hydrogen bond with Ser333 and compound 3 interacted with Asn88, Asp249 and Ser333 through H-bond. We could see that compound 4 interacted with Lys268, while compound 5 formed hydrogen bonds with Glu353 and Ser333. It seems that compound 6 was involved in π-H interaction with Ser333 and the control compound, orlistat, interacted with Asp249 and Lys268 via hydrogen bond. It was found that compound 3 with the most potent activity against pancreatic enzyme showed maximum number of interactions with maintaining of binding with Asp249 (key amino acid residue in the active site).
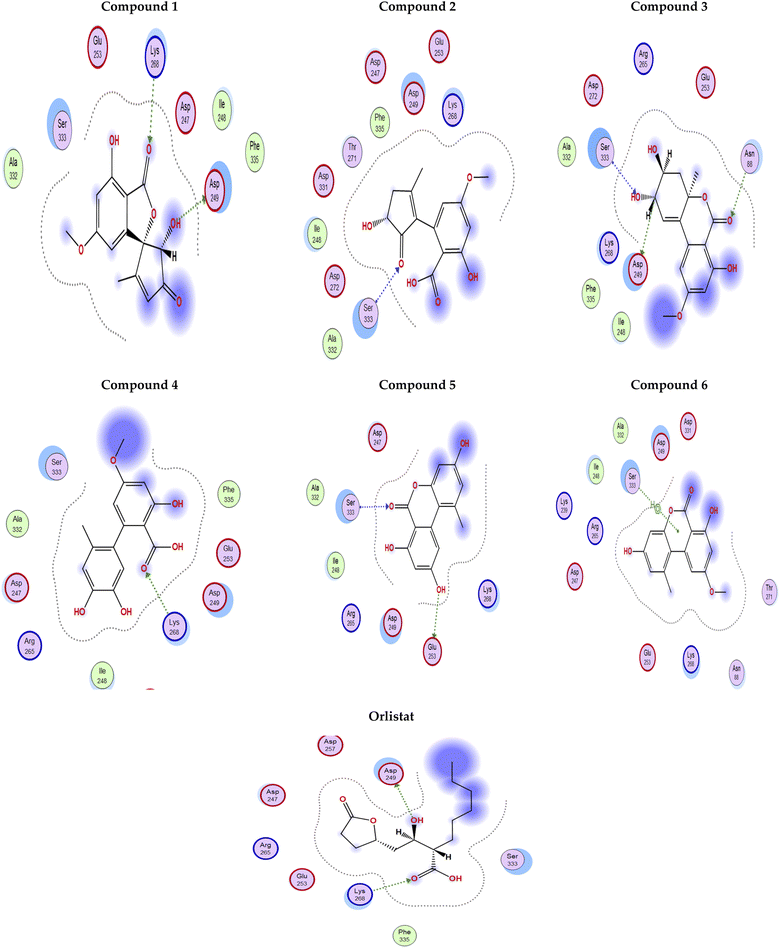 | ||
| Fig. 4 2D Interaction diagram for compounds 1–6 and orlistat with pancreatic lipase enzyme (PDB id: 1LPB). | ||
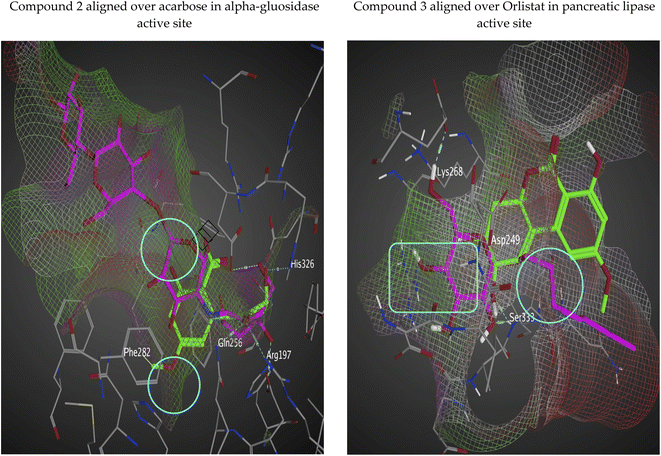 | ||
| Fig. 6 Interaction surface based on van der Waals radius with isolated compounds (green-colored regions are located deep within the active site and red-colored areas are solvent exposed). | ||
The interaction potential surface utilizes three probe types to generate three maps. All three maps are superposed to produce a single map. The probe sliders can be adjusted to produce different energy levels. In our isolated analogs, the maps are made using OH2 probe with default values of −5.5 and −2.5 for the other ones (with some surface transparency for good and efficient visualization). The dry probe is used to generate hydrophobic regions which are shown in green color, hydrogen bond donor is produced by the OH2 probe and shown in purple color, and the N: probe (for hydrogen bond acceptors in red color). We could see form Fig. 7 that our isolated series of compounds fulfills the majority of the patches in the active site even some of un-targeted patches by control compounds (acarbose and orlistat).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value (log(coctanol/cwater)) for a compound is considered a well-validated measure of hydrophilicity for compound series. Poor drug absorption or permeation has been related to low hydrophilicities and high log
P value (log(coctanol/cwater)) for a compound is considered a well-validated measure of hydrophilicity for compound series. Poor drug absorption or permeation has been related to low hydrophilicities and high log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value. In order for compound to have high possibility to be absorbed efficiently, it must maintain log
P value. In order for compound to have high possibility to be absorbed efficiently, it must maintain log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value less than 5.0. We could see from Table 2 that our isolated compound series fulfil this term. Log
P value less than 5.0. We could see from Table 2 that our isolated compound series fulfil this term. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S calculation (aqueous solubility) has significant effect on drug absorption and distribution properties. Poorly water-soluble compounds is associated with bad absorption. Log
S calculation (aqueous solubility) has significant effect on drug absorption and distribution properties. Poorly water-soluble compounds is associated with bad absorption. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values, as calculated by OSIRIS program, is a logarithm of the solubility measured in mol per liter. A histogram representation for the distribution of log
S values, as calculated by OSIRIS program, is a logarithm of the solubility measured in mol per liter. A histogram representation for the distribution of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S values between traded drugs shows that 80% of the traded drugs have an estimated log
S values between traded drugs shows that 80% of the traded drugs have an estimated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S value more than −4. From the results shown in the below table, it is shown that our isolated compounds (1–6) have log
S value more than −4. From the results shown in the below table, it is shown that our isolated compounds (1–6) have log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S greater than −4. To achieve a potent activity with a biological target, structural modifications of lead compounds require installing more and more fragments which comes with increase in molecular weight. On the other hand, increase in the molecular weight of a dug molecule is related with poor absorption and less probability to make to the site of action. Thus, it is a good habit to keep molecular weights as low as possible (less than 500). All the isolated compounds (1–6) maintain low molecular weight.
S greater than −4. To achieve a potent activity with a biological target, structural modifications of lead compounds require installing more and more fragments which comes with increase in molecular weight. On the other hand, increase in the molecular weight of a dug molecule is related with poor absorption and less probability to make to the site of action. Thus, it is a good habit to keep molecular weights as low as possible (less than 500). All the isolated compounds (1–6) maintain low molecular weight.
| Comp. no/ID | c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Log Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
Mol weight | TPSA (Å2) | Druglikeness | Drug-score | Toxicity risks (mutagenicity, tumorigenicity, irritancy, reproductive effects) |
|---|---|---|---|---|---|---|---|
| 1 | 0.52 | −1.98 | 276.0 | 93.06 | −0.15 | 0.69 | None |
| 2 | 0.64 | −1.72 | 278.0 | 104.0 | 1.69 | 0.87 | None |
| 3 | 0.25 | −2.01 | 292.0 | 96.22 | −0.27 | 0.54 | None |
| 4 | 2.04 | −3.19 | 290.0 | 107.2 | −2.52 | 0.23 | Mutagenic |
| 5 | 2.3 | −3.54 | 258.0 | 86.99 | −1.26 | 0.31 | Reproductive effects |
| 6 | 2.57 | −3.86 | 272.0 | 75.99 | −1.06 | 0.19 | Mutagenic, reproductive effects |
| Acarbose | −7.18 | 0.59 | 645.0 | 321.1 | −7.4 | 0.29 | None |
| Orlistat | 1.86 | −2.75 | 272.0 | 83.83 | −20.00 | 0.36 | None |
Fragment-based druglikeness, as defined OSIRIS, lists 5300 diverse substructure fragments with pre-calculated druglikeness scores. The fragment list was generated by shredding 3300 drugs in the market as well as 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 commercial chemicals (Fluka). A histogram representation of the results shows that nearly 80% of drugs molecules have positive druglikeness values, while the majority of Fluka chemicals shows negative values. As a result, it a good idea to maintain your drug candidate in the positive range. We could see that compound 2 shows a drug likeness value of 1.69 and compounds 1 and 3 are near the positive range. We could see those compounds 2 and 3 are the most potent analogs in the enzymatic assay against alpha-glucosidase and pancreatic lipase. Compounds 4–6 are in the negative range. We should keep in mind that a positive value implies that our isolated compound contains mainly fragments which are often show up marketed drugs. We still need to keep balance between these fragments regarding other properties. For example, if a drug candidate could consists of drug-like lipophilic fragments only, this means it will give a high score of druglikeness, but it would fail in subsequent stages due its high lipophilicity. Drug score combines c
000 commercial chemicals (Fluka). A histogram representation of the results shows that nearly 80% of drugs molecules have positive druglikeness values, while the majority of Fluka chemicals shows negative values. As a result, it a good idea to maintain your drug candidate in the positive range. We could see that compound 2 shows a drug likeness value of 1.69 and compounds 1 and 3 are near the positive range. We could see those compounds 2 and 3 are the most potent analogs in the enzymatic assay against alpha-glucosidase and pancreatic lipase. Compounds 4–6 are in the negative range. We should keep in mind that a positive value implies that our isolated compound contains mainly fragments which are often show up marketed drugs. We still need to keep balance between these fragments regarding other properties. For example, if a drug candidate could consists of drug-like lipophilic fragments only, this means it will give a high score of druglikeness, but it would fail in subsequent stages due its high lipophilicity. Drug score combines c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Log
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, log
P, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, molecular weight, druglikeness and toxicity risks in one value. This parameter may be used to tell if the compound would qualify to be a drug molecule. The drug score combines druglikeness, c
S, molecular weight, druglikeness and toxicity risks in one value. This parameter may be used to tell if the compound would qualify to be a drug molecule. The drug score combines druglikeness, c![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Log
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, log
P, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S, molecular weight, and toxicity risks in one handy value that may be used to judge the compound's overall potential to qualify for a drug. We could see that compounds 1–3 exhibit a good drug score. Toxicity risk alerts gives an indication that the supplied structure may show harmful effects concerning specific risk category. The prediction process depends on a precomputed structural fragment sets that yields toxicity alerts in case they are included in the drawn structure. This was based on the assumption that marketed drugs are largely free of toxicity. Any fragment was labeled as a risk factor if it is a substructure of harmful compounds, but never or rarely in marketed drugs. On the other hand, we should not rely completely on the facts that the presence or absence of risk alerts does mean that the compound of interest will or will not show any side effects. We could see that compounds 1–3 showed no toxicity risks of any kind.
S, molecular weight, and toxicity risks in one handy value that may be used to judge the compound's overall potential to qualify for a drug. We could see that compounds 1–3 exhibit a good drug score. Toxicity risk alerts gives an indication that the supplied structure may show harmful effects concerning specific risk category. The prediction process depends on a precomputed structural fragment sets that yields toxicity alerts in case they are included in the drawn structure. This was based on the assumption that marketed drugs are largely free of toxicity. Any fragment was labeled as a risk factor if it is a substructure of harmful compounds, but never or rarely in marketed drugs. On the other hand, we should not rely completely on the facts that the presence or absence of risk alerts does mean that the compound of interest will or will not show any side effects. We could see that compounds 1–3 showed no toxicity risks of any kind.
3. Material and methods
3.1. General
Optical rotations were measured with a Jasco DIP-370 digital polarimeter at 20 °C. 1H and 13C-NMR spectra were obtained on a Bruker DRX 600 NMR spectrometer (Bruker Daltonics Inc., MA, USA) using TMS as internal standard for chemical shifts. Chemical shifts (δ) were expressed in ppm with reference to the TMS resonance. HR-FAB-MS were measured with a JEOL JMS 700 spectrometer (JEOL, Japan) or HR-ESI-MS was determined using LC-MS-IT-TOF (Shimadzu, Tokyo, Japan). The MS instrument was operated using an ESI source in both positive and negative ionization modes with survey scans acquired from m/z 100–2000 for MS and m/z 50–1500 for MS/MS. The ionization parameters were as follows: probe voltage, ±4.5 kV; nebulizer gas flow, 1.5 L min−1; CDL temperature, 200 °C; heat block temperature, 200 °C. MPLC column parameters: GL Sciences Inc. (20 × 250 mm, 5 μm), UV detector at 254 nm, flow rate: 10 mL min−1. Dimethylsulfoxide (DMSO) and other organic solvents were purchased from Wako Pure Chemical Industries (Osaka, Japan). Sephadex LH-20 was purchased from GE Healthcare (Uppsala, Sweden). Diaion HP 20 was purchased from Mitsubishi Chemical Corporation, Japan. Silica gel (75–120 mesh) and RP-C18 silica gel (38–63 μm) were purchased from Wako Pure Chemical Industries (Osaka, Japan). Thin layer chromatography (TLC) silica gel 60 F254 was purchased from Merck (Darmstadt, Germany). The developed chromatograms were visualized under 254 nm. UV light and the spots were made visible by spraying with vanillin/H2SO4 reagent before warming in an oven pre-heated to 110 °C for 5 min.3.2. Endophytic fungal sources
Suaeda vera Forssk. (Amaranthaceae), Heliotropium curassavicum L. (Boraginaceae), Limoniastrum monopetalum L. (Plumbaginaceae), Cynanchum acutum L. (Apocynaceae), Lycium schweinfurthii Dammer (Solanaceae), Pancratium maritimum L. (Amaryllidaceae), Ficus carica L. (Moraceae), Asphodelus microcarpus Salzm. & Viv. (Asphodelaceae), Psidium guajava L. (Myrtaceae) and Thymelaea hirsute L. (Thymelaeaceae) leaves were collected from International Coastal Road, 40 km west from Gamasa City, Egypt on December 2016. The plants' identity was confirmed by Dr Ibrahim Mashaly, Professor of Ecology, Faculty of Sciences, Mansoura University. The fresh leaf fragments, after washing with sterilized water and 70% ethanol, were inoculated in Petri dishes containing malt agar medium18 and incubated for five days. When fungal hyphae almost cover the surface of the plate, cultures were then repeatedly re-inoculated onto fresh malt agar media until achieving a pure homogeneous colony.The isolated strains from L. schweinfurthii, P. maritimum and C. acutum, respectively were submitted to the GenBank, for a homology search with Blast. Alignment with published sequences in GenBank showed that the three strains had 99% identity as Alternaria sp. (Genbank accession no. MK680829, MK693741 and MK695948, respectively).
3.3. Production and isolation of fungal metabolites
The isolated fungus from L. schweinfurthii was grown on solid rice culture media prepared by autoclaving 100 g of rice and 100 mL of water in a 1 L flask, then fermentation was performed in five flasks for 30 days at room temperature under stationary conditions. To each 1 L flask about 400 mL EtOAc was added and left for 3 days with intermittent shaking to speed up the extraction. After filtration, re-extraction with fresh solvent was repeated three times till exhaustion. The combined EtOAc extracts were washed with distilled water and then evaporated under reduced pressure till dryness leaving a sticky extract (3.2 g).3.4. Isolation of compounds
The extract was fractionated on Diaion HP 20 by elution with H2O/MeOH (100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, 50
0, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, 25
50, 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 75 and 0
75 and 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100).
100).
Fractions eluted with 50% MeOH (93.60 mg) were subjected to fine purification using reversed phase open column chromatography, isocratically eluted with H2O/MeOH (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) to afford compounds 1 at Rf value 0.62 & 2 at Rf value 0.74, quenched as dark blue spots under UV light at 254 nm on precoated RP-C18 F254 plates using the solvent system H2O–MeOH (40
60) to afford compounds 1 at Rf value 0.62 & 2 at Rf value 0.74, quenched as dark blue spots under UV light at 254 nm on precoated RP-C18 F254 plates using the solvent system H2O–MeOH (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60).
60).
Fractions eluted with 75% MeOH (74.42 mg) were subjected to chromatographic purification using MPLC gradiently eluted with H2O–MeOH (50–100%, 50 min), compound 3 was purified from the fraction eluted at tR 18.1 min using preparative reversed phase TLC developed with H2O/MeOH (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70) with Rf value 0.6, while compound 4 was isolated in pure form from the fraction eluted at tR 22.2 min.
70) with Rf value 0.6, while compound 4 was isolated in pure form from the fraction eluted at tR 22.2 min.
Fractions eluted with 100% MeOH (400.70 mg) were subjected to chromatographic purification using Sephadex LH-20 (100 g) eluted with MeOH to afford 67 sub-fractions 2 mL each. The combined sub-fraction (33–67, 130 mg) was subjected to fine purification using MPLC, the same condition as mentioned before, to isolate compound 5 at tR 20.4 min & 6 at tR 25.3 min.
3.5. HPLC tracking of isolated compounds in different fungal extracts
Mixture of the isolated compounds together with the MeOH extracts of the three plants-derived fungal metabolites were injected into YMC-Triart C18 column (5 μm, 4.6 × 150 mm) attached to an Agilent 1220 Infinity LC system equipped with a binary solvent delivery system, an autosampler, and a photodiode array detector (Agilent Technologies, California, USA), monitored at 254 and 280 nm., maintained at 40 °C. The mobile phase solvents were solvent A, H2O (0.1% formic acid) and solvent B, MeOH. The gradient program was as follows: 0–40 min (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B 95
B 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 − A
5 − A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B 0
B 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v), 40–50 min (0
100, v/v), 40–50 min (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, v/v), 50–60 min (95
100, v/v), 50–60 min (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v). The flow rate was 1 mL min−1 and the injection volume was 5 μL.
5, v/v). The flow rate was 1 mL min−1 and the injection volume was 5 μL.
3.6. Molecular modeling study methodology
MOE docking protocol was initiated with ligand and receptor preparation for docking. QuickPrep panel was used for protein preparation using the default settings to fix structural problems and add missing hydrogen atoms. The next step involved application of protonate 3D to calculate right protonation state with probability of Asn/Gln/His flip. All water molecules located within 4.5 Å from the ligand or receptor were kept. Only the receptor was held fixed, while the ligand and water molecules were permitted to be fully flexible. Energy minimization process was carried out using Amber10: EHT forcefield considering that atoms farther than 8.0 Å from the ligand have a fixed potential. QuickPrep minimization restraints were kept with refining of final receptor structures to RMS gradient of 0.1 kcal mol−1 Å−1. Ligands conformations to be docked were generated via fragment-based approach. The synthesized ligands were kept in the dominant protonation state at pH = 7. The active site for docking our isolated compounds was defined by 3D coordinates of co-crystallized ligand with the target enzyme. A placement method (Triangle Matcher) is used for generation of a collection of poses (30) generating ligand conformations pool. Poses were created via superposing ligand triplets atoms on triplets of receptor alpha spheres. Individual poses at this stage are given a score relative to its interaction/clashes within the active site. Refinement as final step using GB/WSA dG forcefield-based method was done. This included subjecting the generated poses from the placement stage with keeping the top 10 poses as docking process output.3.7. Enzymatic assays
4. Conclusion
In our course to search for bioactive fungal metabolites derived from plant endophytes, ten desert plant leaves were collected, subjected to endophytic fungi isolation, purification, fungal identity, and subsequent phytochemical investigation. Based on ITS genomic analysis three of them; C. acutum (Apocynaceae), L. schweinfurthii (Solanaceae) and P. maritimum (Amaryllidaceae) were disclosed to inhabit Alternaria sp. in their fresh leaves. Six phenolic compounds; talaroflavone (1), alternarienoic acid (2), altenuene (3), altenusin (4), alternariol (5), alternariol-5-O-methyl ether (6), were isolated from the solid rice culture media of Alternaria sp. The isolated compounds showed an interesting inhibitory activity against both α-glucosidase and pancreatic lipase enzymes designating a promising naturally occurring anti-diabetic candidates.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The first author acknowledges the Ministry of Higher Education, Egypt for scholarship support.References
- A. J. Krentz and C. J. Bailey, Oral Antidiabetic Agents, Drugs, 2005, 65, 385–411 CrossRef CAS PubMed.
- T. Matsui, T. Tanaka, S. Tamura, A. Toshima, K. Tamaya, Y. Miyata, K. Tanaka and K. Matsumoto, α-Glucosidase Inhibitory Profile of Catechins and Theaflavins, J. Agric. Food Chem., 2006, 55, 95–105 Search PubMed.
- M. Nakai, Y. Fukui, S. Asami, Y. Toyoda-Ono, T. Iwashita, H. Shibata, T. Mitsunaga, F. Hashimoto and Y. Kiso, Inhibitory Effects of Oolong Tea Polyphenols on Pancreatic Lipase in Vitro, J. Agric. Food Chem., 2005, 53, 4593–4598 CrossRef CAS PubMed.
- J. Watanabe, J. Kawabata, H. Kurihara and R. Niki, Isolation and Identification of α -Glucosidase Inhibitors from Tochu-cha (Eucommia ulmoides), Biosci., Biotechnol., Biochem., 1997, 61, 177–178 CrossRef CAS PubMed.
- S. Wild, G. Roglic, A. Green, R. Sicree and H. King, Global prevalence of diabetes: estimates for the year 2000 and projections for 2030, Diabetes Care, 2004, 27, 1047–1053 CrossRef PubMed.
- B. Schulz, C. Boyle, S. Draeger, A.-K. Römmert and K. Krohn, Endophytic fungi: a source of novel biologically active secondary metabolites, Mycol. Res., 2002, 106, 996–1004 CrossRef CAS.
- J. Tian, L. Fu, Z. Zhang, X. Dong, D. Xu, Z. Mao, Y. Liu, D. Lai and L. Zhou, Dibenzo-α-pyrones from the endophytic fungus Alternaria sp. Samif01: isolation, structure elucidation, and their antibacterial and antioxidant activities, Nat. Prod. Res., 2017, 31, 387–396 CrossRef CAS PubMed.
- A. T. Bottini and D. G. Gilchrist, Phytotoxins. I. A 1-aminodimethylheptadecapentol from alternaria alternata f. sp. lycopersici, Tetrahedron Lett., 1981, 22, 2719–2722 CrossRef CAS.
- W. Gu, Bioactive metabolites from Alternaria brassicicola ML-P08, an endophytic fungus residing in Malus halliana, World J. Microbiol. Biotechnol., 2009, 25, 1677–1683 CrossRef CAS.
- R. Suemitsu, Y. Yamada, T. Sano and K. Yamashita, Phytotoxic Activities of Altersolanol A, B and Dactylariol, and Activities of Altersolanol A against Some Microorganisms, Agric. Biol. Chem., 1984, 48, 2383–2384 CAS.
- H. Sheridan and A. M. Canning, Novel radicinol derivatives from long-term cultures of Alternaria chrysanthemi, J. Nat. Prod., 1999, 62, 1568–1569 CrossRef CAS PubMed.
- J. Lou, L. Fu, Y. Peng, L. Zhou, J. Lou, L. Fu, Y. Peng and L. Zhou, Metabolites from Alternaria Fungi and Their Bioactivities, Molecules, 2013, 18, 5891–5935 CrossRef PubMed.
- J. Kjer, A. Debbab, A. H. Aly and P. Proksch, Methods for isolation of marine-derived endophytic fungi and their bioactive secondary products, Nat. Protoc., 2010, 5, 479–490 CrossRef CAS PubMed.
- Y. Liu, Y. Wu, R. Zhai, Z. Liu, X. Huang and Z. She, Altenusin derivatives from mangrove endophytic fungus Alternaria sp. SK6YW3L, RSC Adv., 2016, 6, 72127–72132 RSC.
- A. H. Aly, R. Edrada-Ebel, I. D. Indriani, V. Wray, W. E. G. Müller, F. Totzke, U. Zirrgiebel, C. Schächtele, M. H. G. Kubbutat and W. H. Lin, et al., Cytotoxic Metabolites from the Fungal Endophyte Alternaria sp. and Their Subsequent Detection in Its Host Plant Polygonum senegalense, J. Nat. Prod., 2008, 71, 972–980 CrossRef CAS PubMed.
- P. Jiao, J. B. Gloer, J. Campbell and C. A. Shearer, Altenuene Derivatives from an Unidentified Freshwater Fungus in the Family Tubeufiaceae, J. Nat. Prod., 2006, 69, 612–615 CrossRef CAS PubMed.
- S. Nakanishi, S. Toki, Y. Saitoh, E. Tsukuda, K. Kawahara, K. Ando and Y. Matsuda, Isolation of Myosin Light Chain Kinase Inhibitors from Microorganisms: Dehydroaltenusin, Altenusin, Atrovenetinone, and Cyclooctasulfur, Biosci., Biotechnol., Biochem., 1995, 59, 1333–1335 CrossRef CAS PubMed.
- N. Tan, Y. Tao, J. Pan, S. Wang, F. Xu, Z. She, Y. Lin and E. B. Gareth Jones, Isolation, structure elucidation, and mutagenicity of four alternariol derivatives produced by the mangrove endophytic fungus No. 2240, Chem. Nat. Compd., 2008, 44, 296–300 CrossRef CAS.
- J. Sun, T. Awakawa, H. Noguchi and I. Abe, Induced production of mycotoxins in an endophytic fungus from the medicinal plant Datura stramonium L, Bioorg. Med. Chem. Lett., 2012, 22, 6397–6400 CrossRef CAS PubMed.
- S. Fatmawati, K. Shimizu and R. Kondo, Ganoderol B.: A potent α-glucosidase inhibitor isolated from the fruiting body of Ganoderma lucidum, Phytomedicine, 2011, 18, 1053–1055 CrossRef CAS PubMed.
- T. Mizutani, S. Inatomi, A. Inazu and E. Kawahara, Hypolipidemic Effect of Pleurotus eryngii Extract in Fat-Loaded Mice, J. Nutr. Sci. Vitaminol., 2010, 56, 48–53 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: HRMS, 1D and 2D NMR of the isolated metabolites. See https://doi.org/10.1039/d2ra02532a |
| This journal is © The Royal Society of Chemistry 2022 |