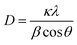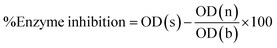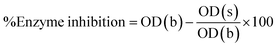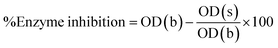Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Potential antimicrobial, antidiabetic, catalytic, antioxidant and ROS/RNS inhibitory activities of Silybum marianum mediated biosynthesized copper oxide nanoparticles
Junaid Iqbal a,
Anisa Andleeba,
Hajra Ashrafa,
Bisma Meera,
Azra Mehmoodb,
Hasnain Janc,
Gouhar Zamana,
Muhammad Nadeemd,
Samantha Drouete,
Hina Fazalf,
Nathalie Giglioli-Guivarc'hg,
Christophe Hano
a,
Anisa Andleeba,
Hajra Ashrafa,
Bisma Meera,
Azra Mehmoodb,
Hasnain Janc,
Gouhar Zamana,
Muhammad Nadeemd,
Samantha Drouete,
Hina Fazalf,
Nathalie Giglioli-Guivarc'hg,
Christophe Hano e and
Bilal Haider Abbasi
e and
Bilal Haider Abbasi *a
*a
aDepartment of Biotechnology, Quaid-i-Azam University, Islamabad 45320, Pakistan. E-mail: juniqbal34@gmail.com; anisaandleeb@bs.qau.edu.pk; hajraashraf67@gmail.com; bismameer786@gmail.com; gzaman@bs.qau.edu.pk; bhabbasi@qau.edu.pk
bStem Cell & Regenerative Medicine Lab, National Centre of Excellence in Molecular Biology, University of Punjab, 87-West Canal Bank Road, Lahore 53700, Pakistan. E-mail: azramehmood@cemb.edu.pk
cInstitute of Biochemical Sciences, National Taiwan University, Taipei City 10617, Taiwan. E-mail: rhasnain849@gmail.com
dInstitute of Integrative Biosciences, CECOS University, Peshawar, 25100, Pakistan. E-mail: m.nadeem@cecos.edu.pk
eLaboratoire de Biologie des Ligneux et des Grandes Cultures (LBLGC), INRAE USC1328, Université d'Orléans, 45067 Orléans Cedex 2, France. E-mail: samantha.drouet@univ-orleans.fr; hano@univ-orleans.fr
fPakistan Council of Scientific and Industrial Research (PCSIR) Laboratories Complex, Peshawar 25120, Pakistan. E-mail: hina_fazalso@yahoo.com
gEA2106 Biomolécules et Biotechnologies Végétales, Université Francois-Rabelais de Tours, Tours, France. E-mail: nathalie.guivarch@univ-tours.fr
First published on 11th May 2022
Abstract
Use of medicinal plants for the biosynthesis of nanoparticles offers several advantages over other synthesis approaches. Plants contain a variety of bioactive compounds that can participate in reduction and capping of nanoparticles. Plant mediated synthesis has the leverage of cost effectiveness, eco-friendly approach and sustained availability. In the current study Silybum marianum, a medicinally valuable plant rich in silymarin content, is used as a reducing and stabilizing agent for the fabrication of nanoparticles. Biosynthesized CuO-NPs were characterized using High Performance Liquid Chromatography (HPLC), Fourier Transform Infrared Spectroscopy (FTIR), X-ray Diffraction (XRD), Scanning Electron Microscopy (SEM), and Dynamic Light Scattering (DLS) techniques. Characterization revealed that CuO-NPs having a crystalline structure showed spherical morphology with an average size of 15 nm. HPLC analysis demonstrated conjugation of various silymarin components, especially the presence of silybin A (705.06 ± 1.59 mg g−1 DW). CuO-NPs exhibited strong bactericidal potency against clinically important pathogenic bacterial strains e.g. Enterobacter aerogenes and Salmonella typhi with an inhibition zone of 18 ± 1.3 mm and 17 ± 1.2 mm, respectively. Synthesized nanoparticles indicated a dose dependent cytotoxic effect against fibroblast cells exhibiting a percentage cell viability of 83.60 ± 1.505% and 55.1 ± 1.80% at 25 μg mL−1 and 100 μg mL−1 concentration, respectively. Moreover, CuO-NPs displayed higher antioxidant potential in terms of (TAC: 96.9 ± 0.26 μg AAE/mg), (TRP: 68.8 ± 0.35 μg AAE/mg), (DPPH: 55.5 ± 0.62%), (ABTS: 332.34 μM) and a significant value for (FRAP: 215.40 μM). Furthermore, enzyme inhibition assays also exhibited excellent enzyme inhibition potential against α-amylase (35.5 ± 1.54%), urease (78.4 ± 1.26%) and lipase (80.50.91%), respectively. Overall findings indicated that biosynthesized CuO-NPs possess immense in vitro biological and biomedical properties and could be used as a broad-spectrum agent for a wider range of biomedical applications.
1. Introduction
Nanotechnology is one of the most dynamic and innovative fields of science and technology, having a multitude of applications ranging from medicines to engineering.1–4 Various methods such as physical, chemical and green routes have been used to fabricate these nanoscale materials, however these methods have a lot of drawbacks. From the last few decades, green approaches are becoming methods of choice for the fabrication of nanoparticles (NPs), because of their biocompatibility, safety, minimal toxicity and cost effectiveness, which makes them more ideal than other counterparts.5,6 Metallic nanoparticles offer a wide variety of applications mainly attributed to their small size and high surface area.7–9 Among other metallic nanoparticles, copper oxide nanoparticles (CuO-NPs) have been used in multiple sectors including the biomedical, textile, catalysis and sensing industries.10–17 Moreover, CuO is comparatively cheaper than silver, can be mixed easily with the polymers and is quite stable regarding physical and chemical properties. Different natural sources are being utilized for the production of CuO-NPs including plants, microbes, and fungi etc.18–21 Plant extract consists of variety of biomolecules and metabolites including vitamins, carbohydrates, phenolics and flavonoids, which can act as a reducing and stabling agent and can covert Cu2+ ions into CuO-NPs.5,22,23Excessive production of free radicals in the body are key contributors of degenerative diseases including cataracts, cardiovascular diseases, cancer, brain dysfunction, and a weakened immune system.24 These free radicals, however, can be deactivated using antioxidants, before they attack body cells and manifest a disease. CuO-NPs are particularly renowned for effectively scavenging free radicals containing oxygen groups.24 CuO and other highly ionic metal oxide nanoparticles may be of significance due to their unusual crystal surface structures and high surface areas.25 CuO-NPs, along with their antioxidant activities, exhibit antibacterial activities against all strains of Gram-positive and -negative bacteria.26–28 Antibacterial activities of CuO-NPs have been reported against Staphylococcus aureus, Enterococcus faecalis, Klebsiella pneumonia, Proteus vulgaris, Salmonella typhimurium, Pseudomonas aeruginosa, Escherichia coli, and Bacillus subtilis. Among these, E. faecalis, and E. coli reported the highest activity upon treatment with CuO-NP.29
Silybum marianum belongs to the family Asteraceae and is enriched with a variety of bio ingredients, in particular silymarin: A major player which have been reported in treating liver30,31, kidney,32 spleen and gallbladder disorders.33 It was a native plant of Asia and Southern Europe however, it is now easily available across the world. Silybum marianum (milk thistle) is 200 cm long plant consist of conical shaped cottony stem, pale green leaves with white veins and purple–red flower. Seed extract of Silybum marianum contains up to 4% of the total silymarin.34 Aqueous extract of Silybum marianum consists of phenolic (silybin A, silybin B, isosilybin A, isosilybin B, silydianin, isosilychristin, silychristin) and flavonoid (taxifolin) which can serve as reducing and capping agent to enhance the biomedical properties of CuO-NPs. Green synthesis of CuO-NPs with number of biomedical importance has been reported previously using different plant parts such as Hull extract of Oak,35 dry black beans,36 peel extract of Punica granatum37 and leaf extract of aloe vera.38
The current study is aimed to use seed extract of Silybum marianum as a reducing/capping agent for biosynthesis of CuO-NPs and to investigate the antibacterial and other biomedical potential of biosynthesized CuO-NPs (Fig. 1). To the best of our knowledge, this is the first report on Silybum marianum mediated biosynthesis of copper oxide nanoparticles. Biosynthesized CuO-NPs were characterized using various spectroscopic and analytical techniques and their biological potential was investigated using various in vitro assays. Moreover, the cytotoxicity of CuO-NPs was analyzed using NIH 3T3 fibroblast cell lines to predict the pharmacological application of CuO-NPs.
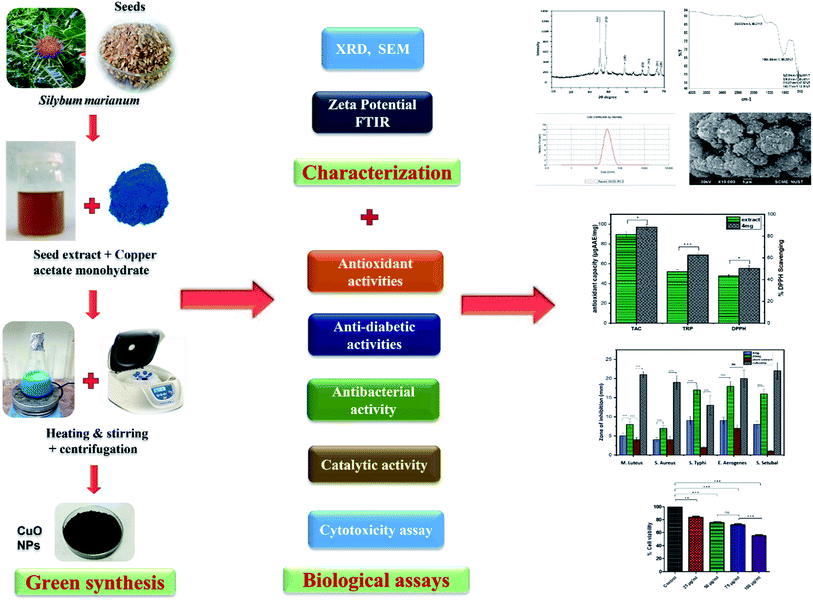 | ||
| Fig. 1 Schematic diagram of green synthesis, characterization and biological applications of CuO-NPs. | ||
2. Materials and methods
2.1 Chemicals
Copper acetate monohydrate (molecular weight: 199.65 g mol−1) was purchased from Sigma Aldrich. Ethanol was purchased from Anal R NORMAPUR (UK). Trypsin-EDTA solution and Dulbecco's Modified Eagle Medium Low Glucose (DMEM-LG) was purchased from Sigma Aldrich USA. Fetal bovine serum (FBS) was acquired from Gibco. NIH 3T3 mouse embryonic fibroblast cell line was a gift from Interdisciplinary Research center in Biomedical materials (IRCBM), COMSATS University Islamabad, Lahore, Pakistan.2.2 Seeds collection and preparation of seed extract
Silybum marianum seeds were obtained from Plant Cell and Tissue Culture Lab (Herbarium code: PSM 04), Department of Biotechnology, Quaid-i-Azam University Islamabad. Seeds were washed with distilled water twice, dried and grinded to form fine powder. 20 gram of seed powder was added to 200 mL of distilled water in a flask and stirred at 6000 rpm for 3 h at 50 °C to prepare aqueous extract. The extract was filtered to remove solid residues by passing through nylon cloth. The filtrate is further purified by passing through Whatman filter paper three times. The prepared extract was stored at 4 °C for further experiments.2.3 CuO-NPs biosynthesis
Biosynthesis of CuO-NPs was conducted by adding copper acetate monohydrate to aqueous seed extract as described by ref. 39. Briefly, 10 gram of salt (copper acetate monohydrate) was dissolved in 100 mL of prepared extract and agitated for 2 h at 50 °C. After the completion of reaction the solution was centrifuged for 10 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to settle down the dissolved precipitate and CuO-NPs in the form of pellet were collected. Pellet was washed thrice with distilled water and twice with ethanol via centrifugation for further purification and allowed to dry at 40 °C for 24 h. The obtained CuO-NPs were calcinated at 500 °C for 3 h to remove further impurities and get crystalline structure.
000 rpm to settle down the dissolved precipitate and CuO-NPs in the form of pellet were collected. Pellet was washed thrice with distilled water and twice with ethanol via centrifugation for further purification and allowed to dry at 40 °C for 24 h. The obtained CuO-NPs were calcinated at 500 °C for 3 h to remove further impurities and get crystalline structure.
2.4 HPLC analysis
Phytochemicals conjugated to CuO-NPs were confirmed though high performance liquid chromatography (HPLC) consists of Varian Prostar 230 pump, Varian Prostar 335 Photodiode Array Detector (PAD), Metachem Degaist degasser, and Varian Prostar 410 autosampler, operated by Galaxie version 1.9.3.2 software (Varian, Le PlessisRobinson, France). During HPLC the standards for compound separation i.e. taxifolin, silychristin, isosilychristin, silydianin, silybin A, silybin B, isosilybin A, isosilybin B. were obtained from sigma Aldrich. For separation, Hypersil PEP 300 C18, 250 × 4.6 mm was used at 35 °C with a particle size of 5 μm prepared by guard column Alltech, 10 × 4.1 mm. Detection was made at wavelength of 320 nm and 520 nm. Mobile phase was composed of solvent A (HCOOH/H2O, pH = 2.1) and solvent B (CH3OH). The composition of mobile phase changes 1 h per run, with nonlinear gradient as follows: 8% B (36 min), 100% B (30–35 min), 33% B (28 min), 30% B (17 min), 12% B (11 min), and 8% B (0 min) at flow rate of 1 mL min−1. Among each run a 10 min re-equilibration time was used. Quantification was made on comparing with reliable reference standards and an assessment of retention time. Analysis was made three times for each sample. The results were measured as μg mg−1 DW of samples.2.5 Physicochemical and morphological characterization of CuO-NPs
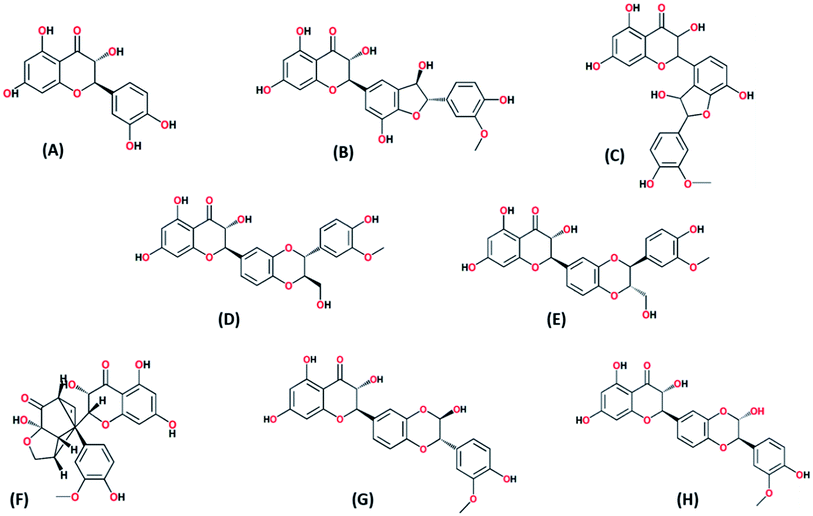
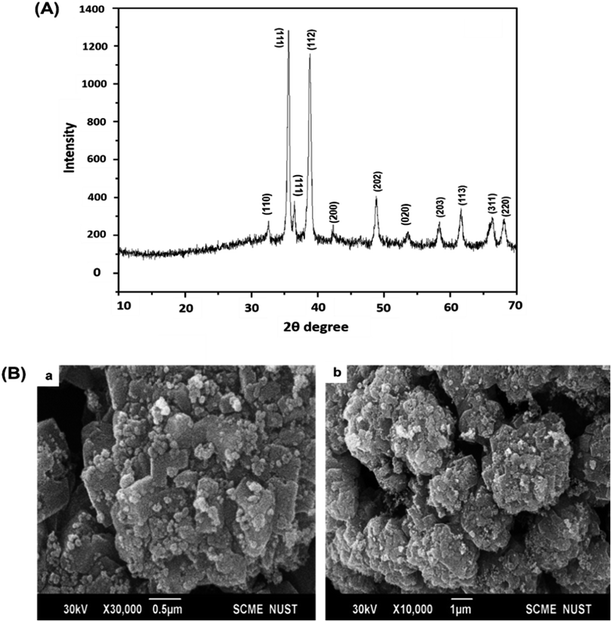
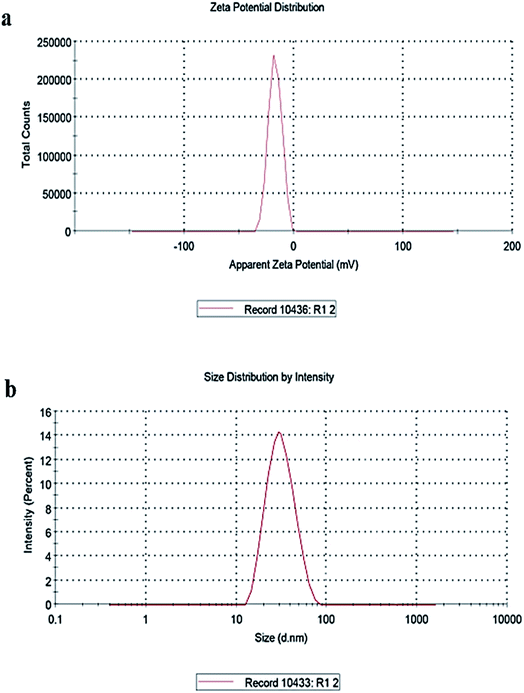
To determine structural, chemical and morphological characteristics of biosynthesized CuO-NPs various characterization techniques were carried out including XRD, FTIR, SEM and DLS. To confirm the synthesis of CuO-NPs, X-ray diffractometer was used. CuO-NPs were subjected to XRD instrument (Model-D8 Advance, Germany) in range of 10–70° with step time of 0.55 seconds. Data was analyzed through Cu Kα radiation with wavelength of 1.5406 Å. Debye–Scherrer equation was used to determine average size of CuO-NPs.where κ = shape factor, λ = X-ray wavelength, θ = Bragg's angle, β = full length at half maximum in radians. Fourier Transform Infrared Radiation (FTIR) was performed with wavelength in range of 515 cm−1 to 4000 cm−1 for determination of attached functional groups that may act as reducing and capping agents. Morphological attributes were examined through scanning electron microscopy (SEM; JEOL, Tokyo, Japan), operated at 20 kV and 0.18 nm point to point resolution. Dynamic light scattering (DLS) of biosynthesized CuO-NPs was performed to determine surface charge and stability as well as to estimate size distribution of nanoparticles using Malvern zeta sizer.
2.6 Anti-diabetic potential of CuO-NPs
Following formula is used to calculate percent enzyme inhibition:
Enzyme inhibition was determined in percentage using following equation.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 minutes and supernatant was collected. The reaction mixture was prepared by adding 150 μL lipase, 350 μL buffer (Tris buffer (100 mM; pH 8.2)) and 50 μL of test sample into Eppendorf. To initiate the reaction 450 μL of substrate (olive oil) was added to each Eppendorf. Orlistat acts as an inhibitor while blank consists of 400 μL buffer, 150 μL lipase, and 450 μL substrate without test sample. The mixture was incubated at 37 °C for 2 h followed by centrifugation at 16
000 rpm for 5 minutes and supernatant was collected. The reaction mixture was prepared by adding 150 μL lipase, 350 μL buffer (Tris buffer (100 mM; pH 8.2)) and 50 μL of test sample into Eppendorf. To initiate the reaction 450 μL of substrate (olive oil) was added to each Eppendorf. Orlistat acts as an inhibitor while blank consists of 400 μL buffer, 150 μL lipase, and 450 μL substrate without test sample. The mixture was incubated at 37 °C for 2 h followed by centrifugation at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. 200 μL supernatant was then poured to each well of microtiter plate and absorbance was recorded at 400 nm wavelength using a microplate reader.
000 rpm. 200 μL supernatant was then poured to each well of microtiter plate and absorbance was recorded at 400 nm wavelength using a microplate reader.Following formula expressed the percent inhibition of enzyme.
2.7 Antioxidant activities of biosynthesized CuO-NPs
In above equation AE indicates absorbance of DPPH solution with sample and AD expresses negative control.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v)]. The reaction mixture was kept at room temperature for 15 min and absorbance was taken at 630 nm. The activity was determined as TEAC and the assay was conducted in triplicates.
1 (v/v/v)]. The reaction mixture was kept at room temperature for 15 min and absorbance was taken at 630 nm. The activity was determined as TEAC and the assay was conducted in triplicates.2.8 Antibacterial activity
Antibacterial potential of CuO-NPs was investigated using five different pathogenic bacterial strains including Salmonella typhi, Enterobacter aerogenes, Salmonella setubal, Micrococcus luteus and Staphylococcus aureus using disc diffusion method.40 Bacterial culture was adjusted to required maximum optical density (OD) of 0.5 by adding sterilized culture broth in a proper ratio. The bacterial strains were then lawn evenly on the plates using autoclaved cotton buds. Sterile discs of filter paper loaded with 5 μL (4 mg mL−1 and 20 mg mL−1 CuO-NPs in DMSO) of samples were applied on the inoculated media and the culture was allowed to incubate for 24 h at 37 °C. Cefexime served as positive control. The appeared zone of inhibitions were measured around samples and control and zone of inhibition ≥12 mm was considered as significant.2.9 Cell viability assay (XTT assay)
Cytotoxicity of biosynthesized CuO-NPs was assessed by XTT assay (2,3-bis[2-methoxy-4-nitro-5-sulfoxyphenyl]-2H-tetrazolium 5-carboxyanilide inner salt), against NIH 3T3 fibroblast cells using XTT assay kit (Roche, Switzerland) as described by ref. 47. Cultured fibroblast cells when reached at 85% confluency were trypsinized (using 1X trypsin-EDTA solution) and were seeded in 96 well plate (3000 cells per well) providing the complete growth medium, Dulbecco's Modified Eagle Medium Low Glucose (DMEM-LG) supplemented with 20% fetal bovine serum (FBS). The plate was incubated at 37 °C in the 5% CO2 incubator for 24 h. After overnight incubation the cells were treated with 20 μg mL−1, 50 μg mL−1, 75 μg mL−1 and 100 μg mL−1 concentration of CuO-NPs suspended in serum free low LG medium for 24 h. The group including cells seeded on simple tissue culture plate without any treatment of CuO-NPs is considered as control. After 24 h treatment, the wells media was aspirated and washed twice with 1XPBS. 50 μL of fresh prepared mixture of XTT (with electron coupling reagents prepared in ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was added to corresponding wells of plate. The plate was then wrapped completely in aluminum foil and kept in 5% CO2 incubator at 37 °C. The absorbance was recorded at wavelength of 450 nm with 630 nm as a reference wavelength. The experiment was repeated twice in a triplicate manner.
1) was added to corresponding wells of plate. The plate was then wrapped completely in aluminum foil and kept in 5% CO2 incubator at 37 °C. The absorbance was recorded at wavelength of 450 nm with 630 nm as a reference wavelength. The experiment was repeated twice in a triplicate manner.
2.10 Measurement of peroxidase-like activity of CuO-NPs
Peroxidase (POD) activity of synthesized CuO-NPs was evaluated by protocol described by ref. 48. Briefly, each well of microtiter plate was filled with 140 μL NaAc–HAc buffer (0.2 M, pH 4.0) and a 20 μL test sample was loaded followed by addition of H2O2 (6 mM, freshly prepared) and 20 μL of TMB (3 mM, freshly prepared). Reaction mixture without sample was considered as a control and absorbance was taken at 652 nm wavelength using spectrophotometer. Enzymatic activity was determined by applying following formula:| A = ELC |
In above equation A represents value of sample absorbance, C represents enzyme concentration (mM min−1 mg−1), E is extinction coefficient and L indicates length of wall, respectively.
2.11 Detection of reactive oxygen/nitrogen species
To determine the level of ROS and RNS generated by application of CuO-NPs, dihydrorhodamine-123 (DHR-123) fluorescent dye was used.46 For this purpose, yeast cells were incubated for overnight in presence of CuO-NPs and DMSO (control cells). Cells were placed in the dark at 30 °C for 10 min after washing with PBS twice and resuspended in PBS with 0.4 μM DHR-123. BioRad VersaFluor Fluorimeter (λex = 505 nm, λem = 535 nm) was used to detect fluorescence after washing with PBS.2.12 Statistical analysis
The experiments were performed in triplicate and the data was evaluated to find out mean values and standard deviation (means ± SD) by using SPSS (Windows version 7.5.1, SPSS Inc., Chicago, IL, USA). One-way ANOVA (analysis of variance) accompanied by unpaired Bonferroni test was used to analyze results of cell viability assay, ROS/RNS activity, peroxidase like activity and lipase, urease and amylase inhibition assays. P value <0.05 indicates that results are statistically significant.3. Results and discussion
3.1 Characterization of biosynthesized CuO-NPs
| Phytochemical compounds | ||||||||
|---|---|---|---|---|---|---|---|---|
| Compounds | Taxifolin | Sily-christin | Iso-silychristin | Silydianin | Silybin A | Silybin B | Iso silybin A | Iso-silybin B |
| Conc. (μg g−1 DW) | 3.70 ± 1.11 | 135.53 ± 1.62 | 266.81 ± 1.33 | 686.31 ± 1.64 | 705.06 ± 1.59 | 451.15 ± 1.76 | 161.83 ± 1.47 | 136.80 ± 1.52 |
| Total silymarin (mg g−1 DW) | 2.55 ± 0.08 | |||||||
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N and N
N and N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S stretch of thiocyanate and isothiocyanate groups. The peak at 1004 cm−1 may corresponds to C–O (C–O–C) stretch indicating ether, pyranose ring and glycosidic linkage.58 The peaks at the fingerprint regions are at 518, 523, 528 and 542 indicating Cu–O stretching. These peaks confirms the formation of CuO nanostructure as reported previously.59,60
S stretch of thiocyanate and isothiocyanate groups. The peak at 1004 cm−1 may corresponds to C–O (C–O–C) stretch indicating ether, pyranose ring and glycosidic linkage.58 The peaks at the fingerprint regions are at 518, 523, 528 and 542 indicating Cu–O stretching. These peaks confirms the formation of CuO nanostructure as reported previously.59,60
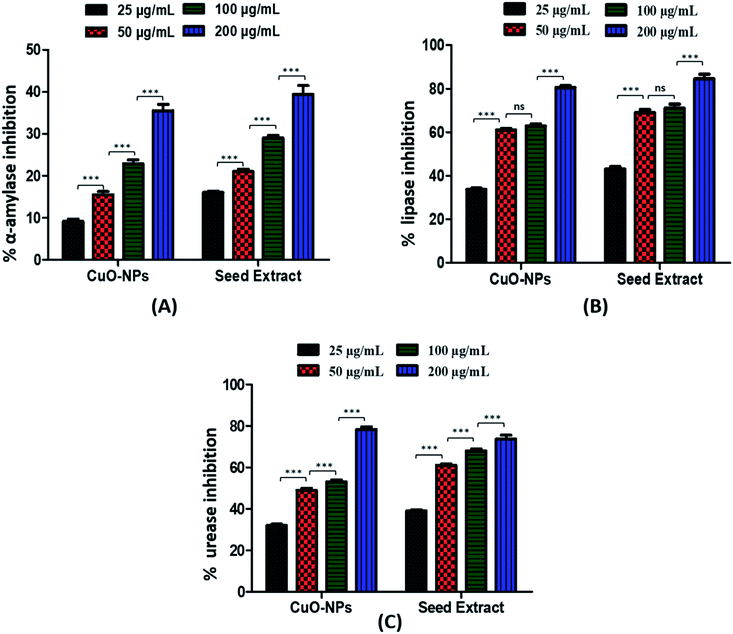
3.2 Anti-diabetic potential of CuO-NPs
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) may be responsible for this inhibitory property.65,67
O) may be responsible for this inhibitory property.65,673.3 Antioxidant potential of CuO-NPs
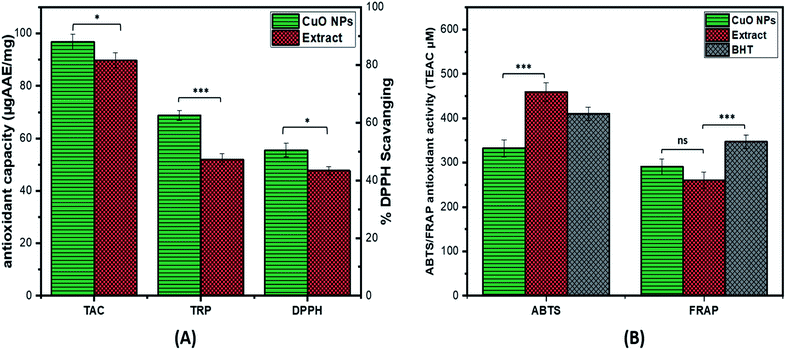
The antioxidant potential of CuO-NPs is investigated by performing TAC, TRP, DPPH, ABTS and FRAP assay (ET based antioxidant activity). The value of TAC was found to be high i.e. 96.9 ± 0.26 μg AAE/mg for CuO-NPs as compared to the extract. The TRP was performed to further assess the antioxidant potential of synthesized CuO-NPs. The reducing potential of CuO-NPs is investigated by assessing the presence of reductones species that are reducing agents and believed to have H-atom donating capacity. The TRP value of CuO-NPs is 68.8 ± 0.35 μg AAE/mg which is higher as compared to plant extract. Furthermore, DPPH assay was carried out in which DPPH moiety accepts electrons from the donor species and get reduced resulting in creation of light-yellow diphenyl picrylhydrazine molecule. At 4 mg mL−1 CuO-NPs concentration high DPPH radical scavenging was observed i.e. 55.5 ± 0.62% as compared to the extract alone (Fig. 7A). The ABTS value for the synthesized CuO-NPs was high i.e. 341.79 μM expressed as TEAC, similarly FRAP results also showed high value for the synthesized CuO-NPs 269.43 μM (Fig. 7B). From the above results it can be assumed that some of the compounds with antioxidant capacity from Silybum marianum seed extract might have involved in reducing and stabilizing the CuO-NPs during the biosynthesis, thus enhancing the antioxidant capacity of synthesized CuO-NPs. These results of antioxidant activities are supported by the previous results.51,683.4 Antibacterial assay
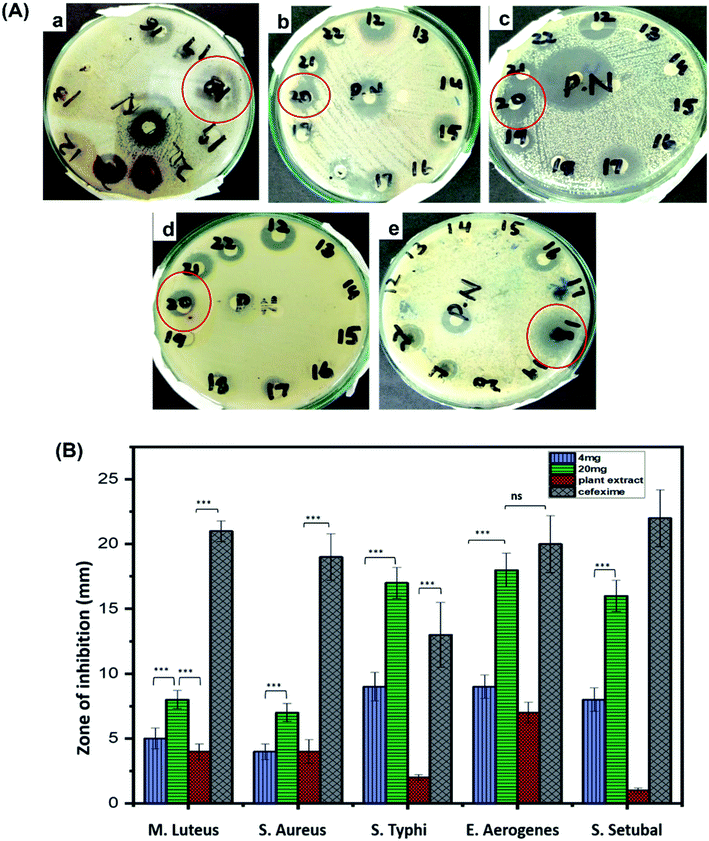
Antibacterial potential of biosynthesized CuO-NPs was investigated against five bacterial strains, using disc diffusion method. Overall, CuO-NPs exhibited efficient antibacterial activity against all tested strains but Enterobacter aerogenes and Salmonella typhi were the most susceptible. Clear zone of inhibitions for these two strains were observed in 24 h incubated cultures supplemented with CuO-NPs loaded discs. The measurement was made in millimeter using vernier caliper calliper. Zone of inhibitations at 4 mg mL−1 were recorded as 5 ± 0.8 mm for Micrococcus luteus, 4 ± 0.6 mm for Staphylococcus aureus, 9 ± 1.1 mm for Salmonella typhi, 9 ± 0.9 mm for Enterobacter aerogenes and 8 ± 0.9 for Salmonella setubal, similarly at 20 mg mL−1 of concentration zone of inhibitations were observed as 8 ± 0.7 mm for Micrococcus luteus, 7 ± 0.7 mm for Staphylococcus aureus, 17 ± 1.2 mm for Salmonella typhi, 18 ± 1.3 mm for Enterobacter aerogenes and 16 ± 1.2 mm for Salmonella setubal, respectively (Fig. 8). At higher concentration of CuO-NPs (20 mg mL−1) applied, the greater bactericidal effect of CuO-NPs was observed compared to 4 mg mL−1 of concentration. The reason for growth inhibition can be the possible interaction between external membrane of bacteria and the CuO-NPs. CuO-NPs may disrupt the integrity of cell membrane causing malfunctioning of enzymes and increasing cell permeability leading to bacterial cell death.25,69 Moreover, CuO-NPs may integrate inside the cell membrane due to smaller size than pores on bacterial cell membrane. CuO-NPs also produce superoxide and hydroxyl free radicals (reactive oxygen species) and damage the cell by oxidizing double bonds in phospholipids and disrupt the cell permeability which results in high osmotic stress and ultimately cause death of bacterial cell.70,713.5 Cytotoxicity of CuO-NPs against NIH3T3 cells
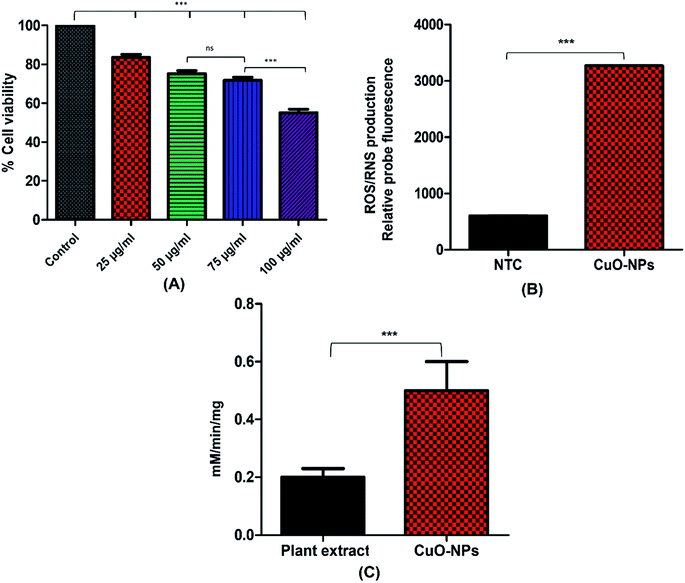
NIH3T3 fibroblast cell lines were used to examine the cytotoxic effect of CuO-NPs. Results showed dose dependent cytotoxicity of CuO-NPs, indicating that with the increase in the concentration of CuO-NPs, the percent cell viability decreases. The XTT assay results indicated %viability (100.0 ± 0.022% in control group vs. 83.60 ± 1.505% in 25 μg mL−1 group, 75.3 ± 1.49% in 50 μg mL−1 group, 71.8 ± 1.59% in 75 μg mL−1 and 55.1 ± 1.80% in 100 μg mL−1) as shown in Fig. 9A. The results showed that 25 μg mL−1 concentration of CuO-NPs offer little toxicity to the cells as compared to control while with the increase in CuO-NPs dose, the cell viability gradually decreases indicating that 50 μg mL−1 and 75 μg mL−1 are slightly more toxic to the cells compared to control and 25 μg mL−1 dose. At 100 μg mL−1 concentration the cell viability becomes reduced to half exhibiting that this dose of CuO-NPs is toxic to fibroblast cells. These results indicated that 100 μg mL−1 and above doses of biosynthesized CuO-NPs can be lethal to the fibroblast cells. With the increase in the use of metallic nanoparticles for biomedical applications, the nanotoxicity associated with nanoparticles is also becoming matter of concern. Over production and extended use of nanoparticles causes serious issues to the human health.72 Similar pattern for cytotoxicity was observed in the previous report,73 however the percent cell viability was higher in previous report, the possible reason for reduced cell viability in the present study might be due to the small size of synthesized CuO-NPs which may result in their more easy penetration inside the cells, causing higher rate of cell death. In case of more nanoparticles penetration inside cells, enzymes leaks out of the cell as the cell membrane integrity damages causing cell death. When cells are treated with high level of CuO-NPs the level of ROS also increase resulting in elevated oxidative stress. The oxidative stress is mainly induced in the mitochondria that leads to initiation of apoptosis inside the cell.733.6 Peroxidase-like activity of CuO-NPs
The ability of green CuO-NPs to break down hydrogen peroxide (H2O2) was determined by performing peroxidase (POD) activity assay. Peroxidases are abundantly present in plants and animals. These enzymes catalyzes the breakdown of H2O2, causing oxidation of many phenolic and non-phenolic compounds. Results showed that the biosynthesized CuO-NPs exhibited lesser catalytic activity as compared to the extract (Fig. 9C). The catalytic activity of CuO-NPs was 0.5 mM min−1 mg−1 while the seed extract showed higher activity of 1.5 mM min−1 mg−1, respectively. Our results are in accordance with the previous study which demonstrated the peroxidase-like catalytic activity of CuO-NPs through formation of blue color product after addition of CuO-NPs to the medium containing TMB as peroxidase substrate.74 This catalytic property of CuO-NPs makes them an ideal candidate as peroxidase mimics for a number of potential applications.3.7 Reactive oxygen and nitrogen species measurement
ROS and RNS are generated as a by-product of metabolism in mitochondria. To evaluate the level of ROS/RNS dihydrorhodamine 123 (DHR 123) probe was used. The results shown in Fig. 9B indicated that CuO-NPs increases the production of ROS/RNS within yeast cells as compared to the NTCs (control). Incubation of CuO-NPs with yeast cells cases the generation of ROS/RNS up to 3265 whereas ROS/RNS production in NTCs was 602 in contrast to the CuO-NPs. Mitochondrial respiration generally generates the free radical species specifically electron transport chain is the site where oxygen leakage and production of ROS occur. Application of metallic nanoparticles enhanced the production of free radicals by Fenton reaction. Also metallic ion of nanoparticles can inhibit electron transport at mitochondria resulting in increased production of ROS. Similar results were reported previously, where application of metallic nanoparticles induced the production of ROS.75 Increase in ROS/RNS causes damage to the cell due to imbalance between free radical species and their biological scavenging activity.4. Conclusion
In current study, CuO-NPs were synthesized via green approach: using seed extract of Silybum marianum. Biosynthesis of CuO-NPs is more environmental friendly, nontoxic, cheap, and easy. The biosynthesized CuO-NPs exhibited well defined physiochemical characteristics, spherical morphology and small size of 15 nm. Moreover, the biosynthesized CuO-NPs revealed broad spectrum antibacterial potency against both Gram positive and Gram-negative bacterial strains used in study, but Enterobacter aerogenes and Salmonella typhi were the most susceptible against CuO-NPs. The effective antimicrobial potential of CuO-NPs suggested their potential use as a precursor novel drug candidate against human pathogenic bacterial strains. CuO-NPs also revealed promising enzymatic inhibitory potential against lipases, ureases and alpha amylase and can be used in anti-diabetic based therapies. In addition, CuO-NPs displayed efficient peroxidase-like catalytic activity and ROS/RNS inhibition potential. Furthermore, CuO-NPs showed significant antioxidant potential and dose dependent cytotoxic effect against fibroblast cells. These results indicated that these biosynthesized CuO-NPs could be a potential candidate for biomedical applications.Funding
No fund was taken from any source.Conflicts of interest
All the authors declared no competing interest.Author contributions
JI and BHA conceptualized the research work. JI, SD, BM and HA performed experiments. GZ assisted in research work and HF and HJ provide reagents. AM and AA performed cell viability assay. JI wrote the manuscript and CH, MN and NG helped in writing and editing manuscript. AA assisted in data analysis and figure preparation. BHA supervised the work.Acknowledgements
We are thankful to Department of Biotechnology, Quaid-i-Azam University, Islamabad, Pakistan for their support.References
- S. E. McNeil, Nanotechnology for the biologist, J. Leukocyte Biol., 2005, 78, 585–594 CrossRef CAS PubMed.
- R. Javed, M. Usman, S. Tabassum and M. Zia, Effect of capping agents: structural, optical and biological properties of ZnO nanoparticles, Appl. Surf. Sci., 2016, 386, 319–326 CrossRef CAS.
- A. Albanese, P. S. Tang and W. C. Chan, The effect of nanoparticle size, shape, and surface chemistry on biological systems, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- S. Laurent, D. Forge, M. Port, A. Roch, C. Robic, L. Vander Elst and R. N. Muller, Magnetic iron oxide nanoparticles: synthesis, stabilization, vectorization, physicochemical characterizations, and biological applications, Chem. Rev., 2008, 108, 2064–2110 CrossRef CAS PubMed.
- M. Naseer, U. Aslam, B. Khalid and B. Chen, Green route to synthesize Zinc Oxide Nanoparticles using leaf extracts of Cassia fistula and Melia azadarach and their antibacterial potential, Sci. Rep., 2020, 10, 1–10 CrossRef PubMed.
- M. I. Din and R. Rehan, Synthesis, characterization, and applications of copper nanoparticles, Anal. Lett., 2017, 50, 50–62 CrossRef CAS.
- C. Wang, X. Gao, Z. Chen, Y. Chen and H. Chen, Preparation, characterization and application of polysaccharide-based metallic nanoparticles: a review, Polymers, 2017, 9, 689 CrossRef PubMed.
- M. Fernandez-Garcia, A. Martinez-Arias, J. Hanson and J. Rodriguez, Nanostructured oxides in chemistry: characterization and properties, Chem. Rev., 2004, 104, 4063–4104 CrossRef CAS PubMed.
- P. Sharma, M. Mehta, D. S. Dhanjal, S. Kaur, G. Gupta, H. Singh, L. Thangavelu, S. Rajeshkumar, M. Tambuwala and H. A. Bakshi, Emerging trends in the novel drug delivery approaches for the treatment of lung cancer, Chem.-Biol. Interact., 2019, 309, 108720 CrossRef CAS PubMed.
- S. Vasantharaj, S. Sathiyavimal, M. Saravanan, P. Senthilkumar, K. Gnanasekaran, M. Shanmugavel, E. Manikandan and A. Pugazhendhi, Synthesis of ecofriendly copper oxide nanoparticles for fabrication over textile fabrics: characterization of antibacterial activity and dye degradation potential, J. Photochem. Photobiol., B, 2019, 191, 143–149 CrossRef CAS PubMed.
- W. M. Rangel, R. A. A. B. Santa and H. G. Riella, A facile method for synthesis of nanostructured copper(II) oxide by coprecipitation, J. Mater. Res. Technol., 2020, 9, 994–1004 CrossRef CAS.
- G. Applerot, J. Lellouche, A. Lipovsky, Y. Nitzan, R. Lubart, A. Gedanken and E. Banin, Understanding the antibacterial mechanism of CuO nanoparticles: revealing the route of induced oxidative stress, Small, 2012, 8, 3326–3337 CrossRef CAS PubMed.
- K. Velsankar, S. Suganya, P. Muthumari, S. Mohandoss and S. Sudhahar, Ecofriendly green synthesis, characterization and biomedical applications of CuO nanoparticles synthesized using leaf extract of Capsicum frutescens, J. Environ. Chem. Eng., 2021, 9, 106299 CrossRef.
- I. M. Chung, A. Abdul Rahuman, S. Marimuthu, A. Vishnu Kirthi, K. Anbarasan, P. Padmini and G. Rajakumar, Green synthesis of copper nanoparticles using Eclipta prostrata leaves extract and their antioxidant and cytotoxic activities, Exp. Ther. Med., 2017, 14, 18–24 CAS.
- A. Waris, M. Din, A. Ali, M. Ali, S. Afridi, A. Baset and A. U. Khan, A comprehensive review of green synthesis of copper oxide nanoparticles and their diverse biomedical applications, Inorg. Chem. Commun., 2021, 123, 1387–7003 CrossRef.
- B. Krishnan and S. Mahalingam, Improved surface morphology of silver/copper oxide/bentonite nanocomposite using aliphatic ammonium based ionic liquid for enhanced biological activities, J. Mol. Liq., 2017, 241, 1044–1058 CrossRef CAS.
- N. Verma and N. Kumar, Synthesis and biomedical applications of copper oxide nanoparticles: an expanding horizon, ACS Biomater. Sci. Eng., 2019, 5, 1170–1188 CrossRef CAS PubMed.
- S. S. Salem and A. Fouda, Green synthesis of metallic nanoparticles and their prospective biotechnological applications: an overview, Biol. Trace Elem. Res., 2021, 199, 344–370 CrossRef CAS PubMed.
- D. Rehana, D. Mahendiran, R. S. Kumar and A. K. Rahiman, Evaluation of antioxidant and anticancer activity of copper oxide nanoparticles synthesized using medicinally important plant extracts, Biomed. Pharmacother., 2017, 89, 1067–1077 CrossRef CAS PubMed.
- A. Singh, N. Singh, I. Hussain and H. Singh, Effect of biologically synthesized copper oxide nanoparticles on metabolism and antioxidant activity to the crop plants Solanum lycopersicum and Brassica oleracea var. botrytis, J. Biotechnol., 2017, 262, 11–27 CrossRef CAS PubMed.
- V. Kalpana and V. Devi Rajeswari, A review on green synthesis, biomedical applications, and toxicity studies of ZnO NPs, Bioinorg. Chem. Appl., 2018, 2018, 1–12 CrossRef PubMed.
- V. Makarov, A. Love, O. Sinitsyna, S. Makarova, I. Yaminsky, M. Taliansky and N. Kalinina, “Green” nanotechnologies: synthesis of metal nanoparticles using plants, Acta Naturae, 2014, 6, 35–44 CrossRef CAS PubMed.
- M. Ovais, A. T. Khalil, A. Raza, N. U. Islam, M. Ayaz, M. Saravanan, M. Ali, I. Ahmad, M. Shahid and Z. K. Shinwari, Multifunctional theranostic applications of biocompatible green-synthesized colloidal nanoparticles, Appl. Microbiol. Biotechnol., 2018, 102, 4393–4408 CrossRef CAS PubMed.
- C. Dipankar and S. Murugan, The green synthesis, characterization and evaluation of the biological activities of silver nanoparticles synthesized from Iresine herbstii leaf aqueous extracts, Colloids Surf., B, 2012, 98, 112–119 CrossRef CAS PubMed.
- G. Ren, D. Hu, E. W. Cheng, M. A. Vargas-Reus, P. Reip and R. P. Allaker, Characterisation of copper oxide nanoparticles for antimicrobial applications, Int. J. Antimicrob. Agents, 2009, 33, 587–590 CrossRef CAS PubMed.
- R. Javed, M. Ahmed, I. ul Haq, S. Nisa and M. Zia, PVP and PEG doped CuO nanoparticles are more biologically active: Antibacterial, antioxidant, antidiabetic and cytotoxic perspective, Mater. Sci. Eng., C, 2017, 79, 108–115 CrossRef CAS PubMed.
- K. G. Germi, F. Shabani, A. Khodayari and Y. Azizian-Kalandaragh, Structural and biological properties of CuO nanoparticles prepared under ultrasonic irradiation, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2014, 44, 1286–1290 CrossRef CAS.
- M. Ahamed, H. A. Alhadlaq, M. Khan, P. Karuppiah and N. A. Al-Dhabi, Synthesis, characterization, and antimicrobial activity of copper oxide nanoparticles, J. Nanomater., 2014, 2014, 4 Search PubMed.
- R. Katwal, H. Kaur, G. Sharma, M. Naushad and D. Pathania, Electrochemical synthesized copper oxide nanoparticles for enhanced photocatalytic and antimicrobial activity, J. Ind. Eng. Chem., 2015, 31, 173–184 CrossRef CAS.
- R. Jayaraj, U. Deb, A. Bhaskar, G. Prasad and P. L. Rao, Hepatoprotective efficacy of certain flavonoids against microcystin induced toxicity in mice, Environ. Toxicol., 2007, 22, 472–479 CrossRef CAS PubMed.
- X. Wang, Z. Zhang and S.-C. Wu, Health benefits of Silybum marianum: Phytochemistry, pharmacology, and applications, J. Agric. Food Chem., 2020, 68, 11644–11664 CrossRef CAS PubMed.
- C. Soto, J. Pérez, V. García, E. Uría, M. Vadillo and L. Raya, Effect of silymarin on kidneys of rats suffering from alloxan-induced diabetes mellitus, Phytomedicine, 2010, 17, 1090–1094 CrossRef CAS PubMed.
- E. Shaker, H. Mahmoud and S. Mnaa, Silymarin, the antioxidant component and Silybum marianum extracts prevent liver damage, Food Chem. Toxicol., 2010, 48, 803–806 CrossRef CAS PubMed.
- M. Bahmani, H. Shirzad, S. Rafieian and M. Rafieian-Kopaei, Silybum marianum: beyond hepatoprotection, J. Evidence-Based Complementary Altern. Med., 2015, 20, 292–301 CrossRef CAS PubMed.
- M. Sorbiun, E. S. Mehr, A. Ramazani and S. T. Fardood, Green synthesis of zinc oxide and copper oxide nanoparticles using aqueous extract of oak fruit hull (jaft) and comparing their photocatalytic degradation of basic violet 3, Int. J. Environ. Res., 2018, 12, 29–37 CrossRef CAS.
- P. Nagajyothi, P. Muthuraman, T. Sreekanth, D. H. Kim and J. Shim, Green synthesis: In-vitro anticancer activity of copper oxide nanoparticles against human cervical carcinoma cells, Arabian J. Chem., 2017, 10, 215–225 CrossRef CAS.
- A. Y. Ghidan, T. M. Al-Antary and A. M. Awwad, Green synthesis of copper oxide nanoparticles using Punica granatum peels extract: Effect on green peach Aphid, Environ. Nanotechnol., Monit. Manage., 2016, 6, 95–98 Search PubMed.
- P. V. Kumar, U. Shameem, P. Kollu, R. Kalyani and S. Pammi, Green synthesis of copper oxide nanoparticles using Aloe vera leaf extract and its antibacterial activity against fish bacterial pathogens, BioNanoScience, 2015, 5, 135–139 CrossRef.
- S. Naz, S. Tabassum, N. Freitas Fernandes, M. Mujahid, M. Zia and E. J. Carcache de Blanco, Anticancer and antibacterial potential of Rhus punjabensis and CuO nanoparticles, Nat. Prod. Res., 2020, 34, 720–725 CrossRef CAS PubMed.
- G. Bibi, N. Ullah, A. Mannan and B. Mirza, Antitumor, cytotoxic and antioxidant potential of Aster thomsonii extracts, Afr. J. Pharm. Pharmacol., 2011, 5, 252–258 Search PubMed.
- M. Biglar, K. Soltani, F. Nabati, R. Bazl, F. Mojab and M. Amanlou, A preliminary investigation of the jack-bean urease inhibition by randomly selected traditionally used herbal medicine, Iran. J. Pharm. Res., 2012, 11, 831 Search PubMed.
- G. J. McDougall, N. N. Kulkarni and D. Stewart, Berry polyphenols inhibit pancreatic lipase activity in vitro, Food Chem., 2009, 115, 193–199 CrossRef CAS.
- H. Fatima, K. Khan, M. Zia, T. Ur-Rehman, B. Mirza and I.-u. Haq, Extraction optimization of medicinally important metabolites from Datura innoxia Mill.: an in vitro biological and phytochemical investigation, BMC Complementary Altern. Med., 2015, 15, 1–18 CrossRef PubMed.
- H. Zafar, A. Ali, J. S. Ali, I. U. Haq and M. Zia, Effect of ZnO nanoparticles on Brassica nigra seedlings and stem explants: growth dynamics and antioxidative response, Front. Plant Sci., 2016, 7, 535 Search PubMed.
- M. Shah, M. A. Ullah, S. Drouet, M. Younas, D. Tungmunnithum, N. Giglioli-Guivarc'h, C. Hano and B. H. Abbasi, Interactive Effects of Light and Melatonin on Biosynthesis of Silymarin and Anti-Inflammatory Potential in Callus Cultures of Silybum marianum (L.) Gaertn, Molecules, 2019, 24, 1207 CrossRef PubMed.
- M. Nazir, D. Tungmunnithum, S. Bose, S. Drouet, L. Garros, N. Giglioli-Guivarc'h, B. H. Abbasi and C. Hano, Differential production of phenylpropanoid metabolites in callus cultures of Ocimum basilicum L. with distinct in vitro antioxidant activities and in vivo protective effects against UV stress, J. Agric. Food Chem., 2019, 67, 1847–1859 CrossRef CAS PubMed.
- M. S. Choudhery, M. Khan, R. Mahmood, A. Mehmood, S. N. Khan and S. Riazuddin, Bone marrow derived mesenchymal stem cells from aged mice have reduced wound healing, angiogenesis, proliferation and anti-apoptosis capabilities, Cell Biol. Int., 2012, 36, 747–753 CrossRef CAS PubMed.
- L. Lagrimini, Plant peroxidases: under-and over-expression in transgenic plants and physiological consequences, Plant Peroxidases., 1980, 1990, 59–69 Search PubMed.
- S. Drouet, B. H. Abbasi, A. Falguieres, W. Ahmad, C. Ferroud, J. Doussot, J. R. Vanier, E. Lainé and C. Hano, Single laboratory validation of a quantitative core shell-based LC separation for the evaluation of silymarin variability and associated antioxidant activity of pakistani ecotypes of milk thistle (Silybum marianum L.), Molecules, 2018, 23, 904 CrossRef.
- J. Fehér and G. Lengyel, Silymarin in the prevention and treatment of liver diseases and primary liver cancer, Curr. Pharm. Biotechnol., 2012, 13, 210–217 Search PubMed.
- M. Shah, S. Nawaz, H. Jan, N. Uddin, A. Ali, S. Anjum, N. Giglioli-Guivarc'h, C. Hano and B. H. Abbasi, Synthesis of bio-mediated silver nanoparticles from Silybum marianum and their biological and clinical activities, Mater. Sci. Eng., C, 2020, 112, 110889 CrossRef CAS PubMed.
- D. O. B. Apriandanu and Y. Yulizar, Tinospora crispa leaves extract for the simple preparation method of CuO nanoparticles and its characterization, Nano-Struct. Nano-Objects, 2019, 20, 100401 CrossRef CAS.
- E. S. Mehr, M. Sorbiun, A. Ramazani and S. T. Fardood, Plant-mediated synthesis of zinc oxide and copper oxide nanoparticles by using ferulago angulata (schlecht) boiss extract and comparison of their photocatalytic degradation of Rhodamine B (RhB) under visible light irradiation, J. Mater. Sci.: Mater. Electron., 2018, 29, 1333–1340 CrossRef.
- Y. Yulizar, I. Latifah, R. Bakri and D. Apriandanu, Plants extract mediated synthesis of copper (II) oxide nanoparticles using Oldenlandia corymbosa L. leaf, AIP Conf. Proc., 2018, 2023, 020097 CrossRef.
- K. Cheirmadurai, S. Biswas, R. Murali and P. Thanikaivelan, Green synthesis of copper nanoparticles and conducting nanobiocomposites using plant and animal sources, RSC Adv., 2014, 4, 19507–19511 RSC.
- K. Rajesh, B. Ajitha, Y. A. K. Reddy, Y. Suneetha and P. S. Reddy, Assisted green synthesis of copper nanoparticles using Syzygium aromaticum bud extract: Physical, optical and antimicrobial properties, Optik, 2018, 154, 593–600 CrossRef CAS.
- S. Honary, H. Barabadi, E. Gharaei-Fathabad and F. Naghibi, Green synthesis of copper oxide nanoparticles using Penicillium aurantiogriseum, Penicillium citrinum and Penicillium waksmanii, Dig. J. Nanomater. Bios., 2012, 7, 999–1005 Search PubMed.
- R. Cuevas, N. Durán, M. Diez, G. Tortella and O. Rubilar, Extracellular biosynthesis of copper and copper oxide nanoparticles by Stereum hirsutum, a native white-rot fungus from chilean forests, J. Nanomater., 2015, 2015, 7 Search PubMed.
- V. V. T. Padil and M. Černík, Green synthesis of copper oxide nanoparticles using gum karaya as a biotemplate and their antibacterial application, Int. J. Nanomed., 2013, 8, 889 Search PubMed.
- S. T. Fardood and A. Ramazani, Black tea extract mediated green synthesis of copper oxide nanoparticles, Journal of Applied Chemical Research, 2018, 12, 8–15 Search PubMed.
- N. Silva, S. Ramírez, I. Díaz, A. Garcia and N. Hassan, Easy, quick, and reproducible sonochemical synthesis of CuO nanoparticles, Materials, 2019, 12, 804 CrossRef CAS PubMed.
- A. Nair, C. Jayakumari, P. Jabbar, R. Jayakumar, N. Raizada, A. Gopi, G. S. George and T. Seena, Prevalence and associations of hypothyroidism in Indian patients with type 2 diabetes mellitus, J. Thyroid Res., 2018, 2018, 7 Search PubMed.
- U. Shwetha, M. Latha, C. R. Kumar, M. Kiran, H. Onkarappa and V. S. Betageri, Potential antidiabetic and anticancer activity of copper oxide nanoparticles synthesised using Areca catechu leaf extract, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2021, 12, 025008 CAS.
- S. Ghosh, P. More, R. Nitnavare, S. Jagtap, R. Chippalkatti, A. Derle, R. Kitture, A. Asok, S. Kale and S. Singh, Antidiabetic and antioxidant properties of copper nanoparticles synthesized by medicinal plant Dioscorea bulbifera, J. Nanomed. Nanotechnol., 2015, S6, 1–9 Search PubMed.
- N. U. Islam, R. Amin, M. Shahid, M. Amin, S. Zaib and J. Iqbal, A multi-target therapeutic potential of Prunus domestica gum stabilized nanoparticles exhibited prospective anticancer, antibacterial, urease-inhibition, anti-inflammatory and analgesic properties, BMC Complementary Altern. Med., 2017, 17, 1–17 CrossRef PubMed.
- S. Sachan and A. Singh, Lipase enzyme and its diverse role in food processing industry, Everyman's Sci., 2015, 4, 214–218 Search PubMed.
- M. I. Choudhary, M. Ali, A.-t. Wahab, A. Khan, S. Rasheed, S. L. Shyaula and A.-u. Rahman, New antiglycation and enzyme inhibitors from Parmotrema cooperi, Sci. China: Chem., 2011, 54, 1926–1931 CrossRef CAS.
- A. Muthuvel, M. Jothibas and C. Manoharan, Synthesis of copper oxide nanoparticles by chemical and biogenic methods: photocatalytic degradation and in vitro antioxidant activity, Nanotechnol. Environ. Eng., 2020, 5, 1–19 CrossRef.
- I. Sondi and B. Salopek-Sondi, Silver nanoparticles as antimicrobial agent: a case study on E. coli as a model for Gram-negative bacteria, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS PubMed.
- S. Jadhav, S. Gaikwad, M. Nimse and A. Rajbhoj, Copper oxide nanoparticles: synthesis, characterization and their antibacterial activity, J. Cluster Sci., 2011, 22, 121–129 CrossRef CAS.
- Y.-N. Chang, M. Zhang, L. Xia, J. Zhang and G. Xing, The toxic effects and mechanisms of CuO and ZnO nanoparticles, Materials, 2012, 5, 2850–2871 CrossRef CAS.
- S. Galdiero, A. Falanga, M. Vitiello, M. Cantisani, V. Marra and M. Galdiero, Silver nanoparticles as potential antiviral agents, Molecules, 2011, 16, 8894–8918 CrossRef CAS PubMed.
- R. Mukhopadhyay, J. Kazi and M. C. Debnath, Synthesis and characterization of copper nanoparticles stabilized with Quisqualis indica extract: evaluation of its cytotoxicity and apoptosis in B16F10 melanoma cells, Biomed. Pharmacother., 2018, 97, 1373–1385 CrossRef CAS PubMed.
- M. N. Karim, M. Singh, P. Weerathunge, P. Bian, R. Zheng, C. Dekiwadia, T. Ahmed, S. Walia, E. Della Gaspera and S. Singh, Visible-light-triggered reactive-oxygen-species-mediated antibacterial activity of peroxidase-mimic CuO nanorods, ACS Appl. Nano Mater., 2018, 1, 1694–1704 CrossRef CAS.
- C. A. Sousa, H. M. Soares and E. V. Soares, Nickel oxide (NiO) nanoparticles induce loss of cell viability in yeast mediated by oxidative stress, Chem. Res. Toxicol., 2018, 31, 658–665 Search PubMed.
| This journal is © The Royal Society of Chemistry 2022 |