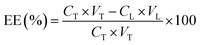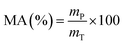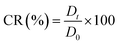Open Access Article
Open Access ArticleAssembly encapsulation of BSA and CCCH-ZAP in the sodium alginate/atractylodis macrocephalae system†
Shuxin Zhanga,
Hai Fan *a,
Chunrong Yia,
Ying Li*a,
Kunmei Yangb,
Shenglong Liub,
Ziqiang Cheng*b and
Jianchao Sun
*a,
Chunrong Yia,
Ying Li*a,
Kunmei Yangb,
Shenglong Liub,
Ziqiang Cheng*b and
Jianchao Sun c
c
aCollege of Chemistry and Material Science, Shandong Agricultural University, Tai'an, 271018, Shandong, PR China. E-mail: fanhai@sdau.edu.cn; ly8049@sdau.edu.cn
bCollege of Veterinary Medicine, Shandong Agricultural University, Tai'an, 271018, Shandong, PR China. E-mail: czqsd@126.com
cSchool of Environment and Materials Engineering, Yantai University, Yantai, 264005, Shandong, PR China
First published on 26th April 2022
Abstract
Zinc finger antiviral proteins (ZAP) can significantly inhibit the replication of avian leukosis virus subgroup J (ALV-J), but the traditional method of ZAP administration is by injection, which can easily cause stress effects in chickens. In this work, we established a sodium alginate/atractylodis macrocephalae system for the encapsulation of CCCH-type zinc finger antiviral protein (CCCH-ZAP). Because of the high cost of ZAP, we first chose bovine serum albumin (BSA) as a model protein to investigate the encapsulation performance. The SEM images clearly confirmed that BSA and the sodium alginate/atractylodis macrocephalae system can assemble easily to form relatively stable nanostructures, and the encapsulation amount of BSA can reach 68%. Subsequently, the encapsulation of ZAP was studied. The SEM and the encapsulation experiments confirmed that ZAP can also be assembly encapsulated in the sodium alginate/atractylodis macrocephalae system with the encapsulation amount of 80%. Release studies showed that the SA/AM-ZAP nanocomposite was able to achieve a release rate of 32% of ZAP. This work successfully confirms the assembly encapsulation of ZAP, which will be beneficial for the usage of ZAP-based animal drugs.
1. Introduction
Zinc finger antiviral proteins (ZAP) are transcription factors that play an important role in gene expression regulation, cell differentiation and many other life processes.1 According to the difference in the binding sequence and structure of the conserved amino acid residues Cys and His of ZAP with Zn2+, Berg et al. divided them into 9 categories, including C2H2, C8, C6, C3HC4, C2HC, C2HC5, C4, C4HC3 and CCCH. Among them, the CCCH-ZAP as an important host antiviral factor, plays a critical role in the natural immune response.2,3CCCH-ZAP can directly or indirectly inhibit the replication of RNA or DNA viruses by preventing RNA synthesis, degrading mRNA or inhibiting mRNA translation, and mediating the ubiquitination and degradation of target proteins, thereby playing a significant role in the host antiviral natural immune response. ALV-J is an RNA virus in the retroviridae family.4,5 Studies by Zhu et al. have shown that ZAP can significantly inhibit the replication of ALV-J. But at present, ZAP is mainly administered by intravenous injection, which is likely to cause irritability in poultry. In order to overcome this problem, the development of other methods, such as oral administration, is necessary. Furthermore, the drug carrier of protein encapsulation with the characteristics of high biocompatibility, low-toxicity, easy degradations urgent.
Sodium alginate (SA), as a linear natural polymer polysaccharide, is a by-product after extracting iodine and mannitol from brown seaweed, kelp, sargassum and other algae plants.6 Alginate contains several active groups, such as carboxyl and alcohol groups, which exhibits polyanionic behavior in aqueous solution and has good film-forming property and stability. SA is composed of β-D-mannuronic acid (M) and α-L-guluronic acid (G) by β-1,4 -glycosidic bond, and it is a copolymer composed of three structural units of GG, MG, and MM in different proportions.7,8 SA shows certain adhesion and obvious pH sensitivity and it can quickly form a gel under extremely mild conditions.9 When divalent cations, such as Sr2+, Ca2+ are added, the Na+ on the G unit undergoes an ion exchange reaction with the divalent cation, and the G unit accumulates to form a cross-linked network structure, thereby forming a hydrogel.10 SA gel can be formed under mild conditions, which can avoid the inactivation of some substances, such as sensitive drugs, proteins, cells and enzymes. Besides, it also exhibits obvious advantages in the encapsulation, preservation and release of drugs.11,12 The application of sodium alginate for drug loading has received extensive attention. Basma et al. used copolymeric nanochitosan grafted with sodium alginate to improve the loading and release properties of streptomycin;13 G Prabha et al. prepared sodium alginate (SA)-polyvinyl alcohol (PVA)-bovine serum albumin (BSA)-coated Fe3O4 nanoparticles (Fe3O4-SA-PVA-BSA) as a drug delivery system for the anticancer drug adriamycin (DOX);14 Meng Xie et al. synthesized graphene oxide nanosheets loaded by magnetic iron oxide nanoparticles (mGO) and successfully generated chitosan/sodium alginate functionalized mGO nanocomposites using layer-by-layer (LbL) self-assembly technique for targeted anticancer drug delivery and photothermal therapy, among others.15
Herbal medicine has attracted widespread attention in the development of healthy foods and medicines since ancient times.16,17 Polysaccharides, a kind of traditional Chinese medicine, are extracted from natural plants. Due to anti-virus, anti-tumor, anti-oxidation, anti-aging and other biological activities,18 polysaccharides have been widely used as immune enhancers, which can enhance or regulate the organism's immune ability.19 Atractylodis macrocephalae (AM) is a perennial herb of the composite family and its rhizome can be used as medicine.20 AM shows various pharmacological activities, including anti-inflammatory, anti-oxidant, anti-aging, anti-tumor, anti-osteoporosis, anti-bacterial, nerve protection and liver protection, et al.21 However, polysaccharides from traditional Chinese medicine cannot achieve specific antiviral activity.
In this context, our main goal is to establish a new type of nanocomposites, which can not only solve the irritability problem caused by ZAP injection, but also enhance immunity of poultry. With CCCH-ZAP as the target protein, sodium alginate/atractylodis macrocephalae (SA/AM) system as carriers, the encapsulation efficiency of ZAP and material availability of SA/AM nanocomposites are evaluated. Since CCCH-ZAP is obtained through the expression of E. coli eukaryotic cells, the acquisition process is time-consuming and ZAP is not easy to be obtained, bovine serum albumin (BSA) was set as a model protein firstly, and the encapsulation of CCCH-ZAP was subsequently studied under the similar conditions. The preliminary results showed that the CCCH-ZAP protein can be encapsulated successfully in the SA/AM carrier with high loading amount. SA/AM system is an easily prepared system in which sodium alginate can form hydrogel by adding Ca2+, and polysaccharide is a general resistance drug with diverse bioactivities, and ZAP can be not only encapsulated in this system, but also administrated by oral, which can solve the problem of irritability caused by injection. Besides, immunity is possibly to be enhanced due to the atractylodis polysaccharide. Preliminary results showed that both BSA and CCCH-ZAP can be encapsulated successfully in the SA/AM system with high loading capacity.
2. Materials and methods
2.1. Materials
Sodium alginate (SA, AR, Shanghai Aladdin Biochemical Technology Co., Ltd), atractylodis macrocephalae polysaccharide (laboratory extraction, 92%),22 bovine serum albumin (BSA, biotechnology grade, 96%), 10 × PBS buffer (0.01 M, pH 7.2–7.4, Beijing Solarbio Science & Technology Co., Ltd), BCA (Tiangen Biotech (Beijing) Co, Ltd), Ca(NO3)2 (AR, Tianjin Kaitong Chemical Reagent Co., Ltd).2.2. Preparation and characterizations of SA/AM carrier
SA/AM carriers were prepared by the solution of sodium alginate (20 mg mL−1) and atractylodis macrocephalae (10 mg mL−1). They were mixed according to a predetermined volume so that the mass ratios of sodium alginate and traditional Chinese medicine polysaccharides were 0.5, 1, and 2 respectively, and the mixtures were stirred for 1 hour under magnetic stirring at a rotational speed of 600 rpm min−1. Then, the solution was added into the Ca(NO3)2 solution (0.1 M) with a syringe with an inner diameter of 0.45 × 15 mm, gelled for 1 hour. Composite carrier was collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 min, and dried under vacuum at 42 °C. Scanning electron microscopy (SEM), EDS energy spectrum analysis and Fourier infrared transform spectroscopy (FT-IR) were tested to characterize the morphology of the carrier and confirmed the assembly.
000 rpm min−1 for 15 min, and dried under vacuum at 42 °C. Scanning electron microscopy (SEM), EDS energy spectrum analysis and Fourier infrared transform spectroscopy (FT-IR) were tested to characterize the morphology of the carrier and confirmed the assembly.
2.3. Encapsulation of BSA
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 minutes to obtain the precipitate, and dried in vacuum at 42 °C.
000 rpm min−1 for 15 minutes to obtain the precipitate, and dried in vacuum at 42 °C.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1 for 15 minutes.
000 rpm min−1 for 15 minutes.2.4. Extraction of CCCH-ZAP
200 mL of Luria-bertani (LB) liquid medium was added at a ratio of VE. coli solution:VLB = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40, to which ampicillin was added and incubated for 12 hours at 4 °C. The revived E. coli solution was added to 500 mL of LB liquid medium and placed in a constant temperature shaker at 37 °C. It was then centrifuged (5000 rpm min−1, 5 min), the sediment was collected and rinsed 3∼4 times with PBS buffer. The washed precipitate was sonicated in an ice-water bath until the liquid was translucent to obtain inclusion bodies. Lysis Equilibration Buffer (LE Buffer) was added to the inclusion bodies and incubated at 37 °C for 30∼60 min. The lysed inclusion bodies were centrifuged (10
40, to which ampicillin was added and incubated for 12 hours at 4 °C. The revived E. coli solution was added to 500 mL of LB liquid medium and placed in a constant temperature shaker at 37 °C. It was then centrifuged (5000 rpm min−1, 5 min), the sediment was collected and rinsed 3∼4 times with PBS buffer. The washed precipitate was sonicated in an ice-water bath until the liquid was translucent to obtain inclusion bodies. Lysis Equilibration Buffer (LE Buffer) was added to the inclusion bodies and incubated at 37 °C for 30∼60 min. The lysed inclusion bodies were centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm min−1, 15 min) and the supernatant was collected. The lysed inclusion bodies were then purified using High Affinity Ni-NTA resin. The High Affinity Ni-NTA resin was first rinsed with LE Buffer, then the supernatant was added to it and repeated twice, followed by the addition of washing solution to the High Affinity Ni-NTA resin with 3∼4 repeated times, in order to wash out the tramp protein. Finally, the eluent was added to precipitate out the target protein from the High Affinity Ni-NTA resin. The CCCH-ZAP was obtained by using the dialysis method to renature the protein. The concentration of CCCH-ZAP was approximately 400 μg mL−1.
000 rpm min−1, 15 min) and the supernatant was collected. The lysed inclusion bodies were then purified using High Affinity Ni-NTA resin. The High Affinity Ni-NTA resin was first rinsed with LE Buffer, then the supernatant was added to it and repeated twice, followed by the addition of washing solution to the High Affinity Ni-NTA resin with 3∼4 repeated times, in order to wash out the tramp protein. Finally, the eluent was added to precipitate out the target protein from the High Affinity Ni-NTA resin. The CCCH-ZAP was obtained by using the dialysis method to renature the protein. The concentration of CCCH-ZAP was approximately 400 μg mL−1.
2.5. Encapsulation of CCCH-ZAP
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes and then frozen at −80 °C and dried under vacuum.23
000 rpm for 15 minutes and then frozen at −80 °C and dried under vacuum.232.6. Determination of the encapsulation efficiency (EE)
The BSA encapsulation rate (EE) was calculated as follows,24where CL represents concentration of BSA remaining (μg mL−1) in the supernatant, VL represents supernatant volume (mL), CT represents the concentration of BSA loaded (μg mL−1) while VT refers to the volume of BSA loaded (mL).25
Notably, BCA method is used to determine the concentration of BSA and CCCH-ZAP.26 BCA protein quantification kit was used to achieve rapid, stable and sensitive concentration determination of protein. The principle of this kit is that the peptide bond structure in the protein molecule can form a complex with Cu2+ in an alkaline environment and reduce Cu2+ to Cu+, while BCA reagent can sensitively and specifically bind to Cu+ to form a stable and colored complex and has a maximum absorption value at 562 nm.27 The color of the compound is directly proportional to the concentration of protein and the concentration of protein can be determined according to the absorption value.
2.7. Determination of the material availability (MA)
The material availability (MA) of protein encapsulated was calculated as follows,where mT represents the total mass of input raw materials (g), and mP represents the dry composite carrier weight (g).
2.8. Release study
Release study of CCCH-ZAP from SA/AM-ZAP at representative SA to AM w/w of 2 were performed in media with pH 2 to determine its drug release properties. Briefly, 20 mg of the dried SA/AM-ZAP composites was dispersed in 3 mL of media (pH 2) with magnetic stirring (400 rpm) at 42 °C. At predetermined time intervals, 40 μL of the media solution was removed each time and supplemented with an equal volume of fresh media, with the aim of maintaining a constant total volume of solution. Protein concentrations were determined using the BCA protein quantification kit method.The amount of CCCH-ZAP released in the medium have been calculated by the following equation:
3. Results and discussions
3.1. Characterization of sodium SA/AM-BSA composite
Fig. 1a and b present the SEM images of SA/AM with the ratio of 2 (without BSA) in different magnifications, while Fig. 1c and d demonstrate the SEM images of SA/AM with BSA. As shown in Fig. 1c and d, the composite with BSA has a looser sheet-like network structure, which is distributed in crisscross strips while the SEM images of pure SA/AM nanocomposites without BSA (Fig. 1 c and d) show gully-like morphology with a distinctly concave and convex surface.28 The reason may be that the amino acid residues in BSA react with active groups such as carboxyl and hydroxyl groups in sodium alginate, which enhances the cross-linking effect of SA and AM in Ca2+ solution.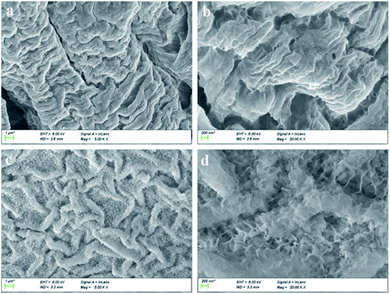 | ||
| Fig. 1 SEM images with mass ratio of SA and AM at 2: (a and b) in the absence and (c and d) in the presence of BSA. | ||
Fig. 2a and b present the SEM image of SA/AM ratio of 0.5 (without BSA) in different magnifications, while Fig. 2c and d are SEM images with BSA. The composite of this ratio has a uniform morphology and presents a network structure intertwined vertically and horizontally in the absence of BSA.29 Compared to composite with BSA (Fig. 2c and d), many small polyhedral structures with regular shapes appear after adding BSA. Fig. 2e and f present the SEM image of sodium alginate/atractylodis macrocephalae (SA/AM) ratio of 1 (without BSA) in different magnifications, while Fig. 2g and h are SEM images with BSA. The surface of the composite is irregular and porous structure can be observed in the absence of BSA while the composite has a dense flake-like stacked structure after adding BSA. Obviously, the morphology of the three composites with BSA is more complex than the morphology without BSA, indicating that BSA has been successfully encapsulated in the sodium alginate-polysaccharide system.
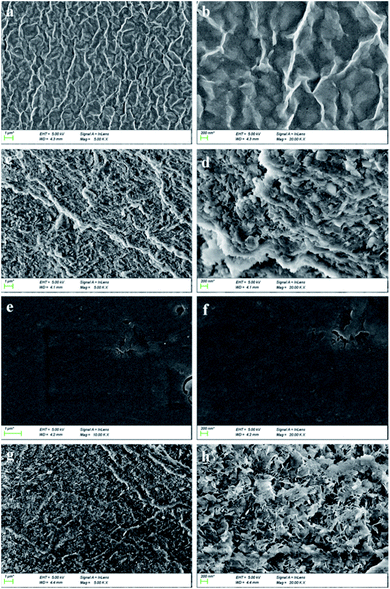 | ||
| Fig. 2 The SEM image of SA/AM composite. SA to AM weight ratio: (a and b) 0.5, (e and f) 1 in the absence of BSA while (c and d) 0.5, (g and h) 1 in the presence of BSA. | ||
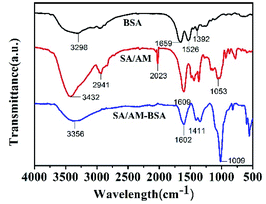
To prove that the BSA was successfully encapsulated in the SA/AM composite, the prepared samples were characterized by FT-IR, SEM, and element mapping. Fig. 3 a presents the FT-IR spectrums of BSA, sodium alginate/atractylodis macrocephalae (SA/AM) composites, and BSA loaded SA/AM (SA/AM-BSA) composites. The typical peaks of BSA are observed at 3298 cm−1 (O–H stretching), 1659 cm−1 (amide I), 1526 cm−1 (amide II), 1392 cm−1 (amide III).30 SA/AM peaks at 3432 cm−1 (O–H stretching), 2941 cm−1 (C–H), 2023 cm−1 (C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C), 1609 cm−1 (C
C), 1609 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1053 cm−1 (C–C).31,32 The FT-IR spectrum of SA/AM-BSA shows peaks at 3356 cm−1 (O–H stretching), 1602 cm−1 (C
O), 1053 cm−1 (C–C).31,32 The FT-IR spectrum of SA/AM-BSA shows peaks at 3356 cm−1 (O–H stretching), 1602 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1411 cm−1 (amide III), 1009 cm−1 (C–C). Comparing the SA/AM with SA/AM-BSA curves, it can be clearly shown that the addition of BSA leads to the blue-shift of the O–H stretching vibration peak from 3432 cm−1 to 3356 cm−1 and the peak is significantly broadened, indicating that intermolecular hydrogen bonding takes place between BSA and the SA/AM composite.32 Besides, the amide III peak at 1392 cm−1 in the spectra of BSA which is absent in SA/AM, is shifted to 1411 cm−1 in SA/AM-BSA, confirming that BSA have been successfully encapsulated into the composite. Element mapping was further used to confirm the existence of BSA in the SA/AM system. As shown in Fig. S1 (in ESI),† the results demonstrated the S element, which only exists in BSA can be clearly observed in the SA/AM-BSA nanocomposite. The results further confirm that the BSA is successfully encapsulated in the SA/AM composite.
O), 1411 cm−1 (amide III), 1009 cm−1 (C–C). Comparing the SA/AM with SA/AM-BSA curves, it can be clearly shown that the addition of BSA leads to the blue-shift of the O–H stretching vibration peak from 3432 cm−1 to 3356 cm−1 and the peak is significantly broadened, indicating that intermolecular hydrogen bonding takes place between BSA and the SA/AM composite.32 Besides, the amide III peak at 1392 cm−1 in the spectra of BSA which is absent in SA/AM, is shifted to 1411 cm−1 in SA/AM-BSA, confirming that BSA have been successfully encapsulated into the composite. Element mapping was further used to confirm the existence of BSA in the SA/AM system. As shown in Fig. S1 (in ESI),† the results demonstrated the S element, which only exists in BSA can be clearly observed in the SA/AM-BSA nanocomposite. The results further confirm that the BSA is successfully encapsulated in the SA/AM composite.
3.2. BSA encapsulation
As shown in Fig. 4a, the encapsulation efficiency (EE) of SA/AM composites for BSA increases, from 53% to 68%, with increasing SA/AM weight ratio from 0.5 to 2. Similarly, the encapsulation efficiency of BSA in sodium alginate increased from 14.45% to 30.35% as the concentration of sodium alginate solution increased in Fig. S2 (see ESI).† As described, the higher ability to encapsulation BSA is not only attributed to the supporting role played by SA under the influence of the cross-linking agent with Ca2+,33 but also adding of AM. As the SA mass increases, the SA/AM nanocomposites become more structurally compact and have stronger interactions among the chains, resulting in a higher trapping and encapsulation capacity for BSA. As shown in Fig. 4b, the material availability (MA) of nanocomposites with different mass ratios of SA/AM vary and increase with the increasing of mass ratio from 0.5 to 2. Apparently, MA of the three mass ratios of nanocomposites increased significantly after the addition of BSA, indicating that BSA has a significant cross-linking effect with SA and AM.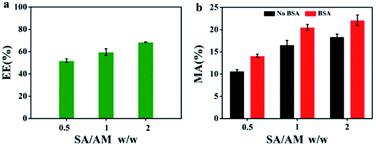 | ||
| Fig. 4 (a) EE of BSA with different SA/AM weight ratio. (b) MA of different mass ratios of SA/AM without BSA and with BSA. | ||
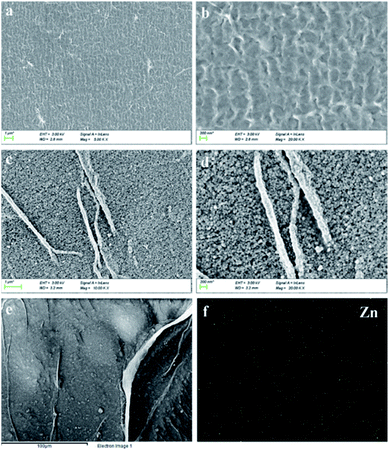
3.3. Characterizations of SA/AM-ZAP composite
As BSA has been successfully encapsulated into the SA/AM system, the similar conditions could be applied in the encapsulation of CCCH-ZAP. Results showed that CCCH-ZAP could be assembly encapsulated. The SA/AM and SA/AM-ZAP composites were first characterized by SEM and element mapping. Compared with SEM image of SA/AM (Fig. 5a and b), the SEM images of SA/AM changed obviously after CCCH-ZAP was added (Fig. 5c and d). The change in morphology from the loose interwoven network structure of SA/AM to the dense and homogeneous spherical nanoparticle structure (with average diameter of 100 nm) of SA/AM-ZAP is related to the strong cross-linking properties between CCCH-ZAP and SA/AM composite.34 TEM images can also confirm that ZAP nanoparticle can be found and linked with SA/AM (Fig. S4, see ESI†). As shown in Fig. 5e and f, Zn element appeared and was observed in the SA/AM-ZAP composite, which further clarifies that CCCH-ZAP has been successfully loaded on the SA/AM composite.353.4. ZAP encapsulation
As shown in Fig. 6a, the encapsulation efficiency (EE) of sodium alginate/atractylodis macrocephalae (SA/AM) composites for BSA has been evaluated, as shown in Fig. 6a. An increase of EE from 43% to 80% can be observed with the increase of the weight ratios of SA/AM from 0.5 to 2. Compared to EE of SA/AM-BSA, EE of SA/AM-ZAP increases obviously which reveals that ZAP can be encapsulated more efficiently in the SA-AM system, which should be attributed to the difference in protein structure between ZAP and BSA. The high encapsulation efficiency of ZAP facilitates the full pharmacological effect of SA/AM-ZAP. And compared with the encapsulation efficiency of ZAP in different concentrations of sodium alginate solution, we can see that ZAP is more efficient in SA/AM (Fig. S3, see ESI†). This result is similar to the encapsulation efficiency determination part of the BSA. As shown in Fig. 6b, similar to MA of SA/AM nanocomposites for BSA, MA of nanocomposites with ZAP also increase significantly with the increase of mass ratios of SA/AM, indicating that ZAP also has high cross-linking properties with SA/AM, which can improve the material utilization of nanocomplexes.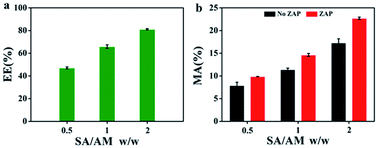 | ||
| Fig. 6 (a) EE of ZAP with different SA/AM weight ratio. (b) MA of different mass ratios of SA/AM without ZAP and with ZAP. | ||
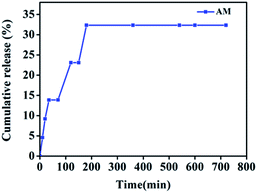
3.5 3.5. Release study
ZAP release performance of SA/AM-ZAP nanocomposite with a sodium alginate to atractylodis macrocephalae mass ratio of 2 was evaluated in a simulated avian gastrointestinal environment, as shown in Fig. 7.The release curve can be divided into two phases: an explosive period and a continuous period immediately afterwards. The first phase is the burst release phase, the intensity of which is related to the intermolecular interactions in SA/AM-ZAP nanocomposites, the complexation of Ca2+ and the hydrogen bonding forces between protein–protein, protein-polysaccharide, and protein-SA. The maximum rate of ZAP release reached at 32% at 180 minutes. After that, the rate of ZAP release tends to a constant value as the polysaccharides in the system dissolve and ZAP continues to dissolve, indicating that the maximum release of ZAP has been reached.
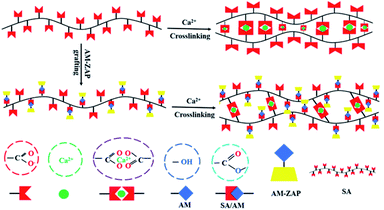
3.6. Encapsulation mechanism of ZAP by SA/AM
As shown in Scheme 1, the mechanism of ZAP encapsulated by SA/AM was proposed according to the assembly of ZAP, SA and AM. SA/AM nanocomposites were prepared by the cross-linking in the presence of Ca2+.36 The assembly of proteins occurs during the formation of SA/AM carriers, and it can be related to the physical encapsulation and spatial conformation of the proteins. The mechanism of ZAP encapsulation by SA/AM was proposed according to the molecular structure of the ZAP, SA and AM. CCCH-ZAP is a single-chain polypeptide containing 776 amino acids (Mw 87 kDa) with four conserved zinc finger motifs at the N-terminal end of the Cys–His repeat in the Cys–Cys–Cys–His (CCCH) conformation, and the 200 amino acids are the main functional domain for antiviral activity.37,38 The molecule of SA contains –COO− groups. When divalent cations Ca2+ were added to the aqueous solution of SA, the Na+ in the G-unit will exchange with these divalent cations, transforming SA solution to a gel. AM contains many active groups such as hydroxyl, carboxyl and amine groups, which can combine with hydroxyl and carboxyl groups in SA through intermolecular hydrogen bonding to form SA/AM composites. ZAP contains a large number of amino acid residues, which offers the possibility to be encapsulated of ZAP in SA/AM nanocomposites. The mechanism of SA/AM-ZAP formation is predicted that ZAP can integrate into SA/AM system in an assembly manner through chemical binding and intermolecular hydrogen bonds, and so that SA/AM-ZAP nanocomposites forms under the cross-linking of Ca2+.39ZAP consists of a single chain polypeptide of 776 amino acids and Mw of 87 kDa while BSA has 607 amino acids and Mw of 66.4 kDa. As shown in Fig. 4 and 6, both EE and MA of SA/AM-ZAP were higher than that of SA/AM-BSA of all the three different weight ratios (SA/AM w/w, 0.5,1 and 2) of nanocomposites. We speculate that the main reasons for the difference due to the disparity in the number of amino acid residues, molecular weight and effect of spatial effects. Furthermore, it should be taken into account that ZAP tends to be able to form more stable nanocomposites with SA.
4. Conclusions
In summary, we successfully designed a sodium alginate/atractylodis macrocephalae system for the encapsulation of BSA and CCCH-ZAP. The SEM images clearly confirmed that BSA and CCCH-ZAP can be assembled easily into SA/AM system to form relatively stable nanostructures, which were confirmed by SEM, element mapping, FTIR. The encapsulation amount can reach 68% for BSA and 80% for ZAP, respectively. In vitro release experiments simulating the avian gastrointestinal environment showed that the SA/AM-ZAP nanocomposite was able to achieve a release rate of 32% of ZAP, indicating that the nanocomposite has the potential to be a potent drug for the treatment of avian ALV-J. The mechanism for the encapsulation of BSA and ZAP by SA/AM has been discussed and the assembly between the molecules of BSA or ZAP and SA/AM is proposed to play a key role for the high encapsulation efficiency. Sodium alginate/atractylodis macrocephalae system is an easily prepared and green system, which will be a potential system for the developing of ZAP based animal drugs by oral administration.Author contributions
Shuxin Zhang: investigation, data curation, writing – original draft preparation. Hai Fan: supervision. Chunrong Yi: data curation. Ying Li: revision. Kunmei Yang: characterizations. Shenglong Liu: sample preparation. Ziqiang Cheng: revision. Jianchao Sun: characterizations.Conflicts of interest
The authors reported no declarations of interest.Acknowledgements
This work was supported by Major Scientific and Technological Innovation Project in Shandong Province (No. 2019JZZY010735), Shandong Provincial Natural Science Foundation of China (No. ZR2019MEM018), Project of Shandong Province Higher Educational Science and Technology Program (No. J18KA008), Key Laboratory of Agricultural Film Application of Ministry of Agriculture and Rural Affairs, Taian 271018, P. R. China.Notes and references
- M. J. Zhu, J. Zhou, X. Q. Ma, G. Li, S. H. He, H. Tang, Y. X. Yao and Z. Q. Cheng, Res. Vet. Sci., 2019, 123, 65–70 CrossRef CAS PubMed.
- X. Luo, X. Wang, Y. Gao, J. Zhu, S. Liu, G. Gao and P. Gao, Cell Rep., 2020, 30, 46–52 CrossRef CAS PubMed.
- M. J. Zhu, X. Q. Ma, X. Y. Cui, J. Zhou, C. G. Li, L. B. Huang, Y. L. Shang and Z. Q. Cheng, Oncotarget, 2017, 8, 58865–58871 CrossRef PubMed.
- Y. Chen, H.-w. Li, F. Cong and Y.-x. Lian, Res. Vet. Sci., 2021, 139, 166–171 CrossRef CAS PubMed.
- G. Mo, B. Hu, G. Wang, T. Xie, H. Fu, Q. Zhang, R. Fu, M. Feng, W. Luo, H. Li, Q. Nie and X. Zhang, Vet. Microbiol., 2021, 261, 109205 CrossRef CAS PubMed.
- Y. Fu, Y. Hou, R. Wang, Y. Wang, X. Yang, Z. Dong, J. Liu, X. Man, W. Yin, B. Yang and H. Tang, Miner. Eng., 2021, 173, 107191 CrossRef CAS.
- H. Pei, F. Chen, X. Niu, Q. Jia, R. Guo, N. Liu and Z. Mo, J. Electroanal. Chem., 2021, 895, 115525 CrossRef CAS.
- N. M. Sanchez-Ballester, B. Bataille and I. Soulairol, Carbohydr. Polym., 2021, 270, 118399 CrossRef CAS PubMed.
- L. Lacerda, A. L. Parize, V. Fávere, M. C. M. Laranjeira and H. K. Stulzer, Mater. Sci. Eng.: C, 2014, 39, 161–167 CrossRef CAS PubMed.
- M. Afshar, G. Dini, S. Vaezifar, M. Mehdikhani and B. Movahedi, J. Drug Delivery Sci. Technol., 2020, 56, 101530 CrossRef CAS.
- R. Ahmad Raus, W. M. F. Wan Nawawi and R. R. Nasaruddin, Asian J. Pharm. Sci., 2021, 16, 280–306 CrossRef PubMed.
- A. Mishra, V. K. Pandey, B. S. Shankar and J. S. Melo, Colloids Surf., B, 2021, 197, 111445 CrossRef CAS PubMed.
- B. E. Jasim, Z. N. Mageed and Z. S. Al-Garawi, J. Phys.: Conf. Ser., 2020, 1660, 012030 CrossRef CAS.
- G. Prabha and V. Raj, Mater. Sci. Eng.: C, 2017, 79, 410–422 CrossRef CAS PubMed.
- M. Xie, F. Zhang, H. Peng, Y. Zhang, Y. Li, Y. Xu and J. Xie, Colloids Surf., B, 2019, 176, 462–470 CrossRef CAS PubMed.
- J. Zhang, K. Hu, L. Di, P. Wang, Z. Liu, J. Zhang, P. Yue, W. Song, J. Zhang, T. Chen, Z. Wang, Y. Zhang, X. Wang, C. Zhan, Y.-C. Cheng, X. Li, Q. Li, J.-Y. Fan, Y. Shen, J.-Y. Han and H. Qiao, Adv. Drug Delivery Rev., 2021, 178, 113964 CrossRef CAS PubMed.
- J. Zhao, Y. Deng and S.-p. Li, TrAC, Trends Anal. Chem., 2017, 96, 138–150 CrossRef CAS.
- Y. Zheng, L. Bai, Y. Zhou, R. Tong, M. Zeng, X. Li and J. Shi, Int. J. Biol. Macromol., 2019, 121, 1240–1253 CrossRef CAS PubMed.
- M. Xie, W. Tao, F. Wu, K. Wu, X. Huang, G. Ling, C. Zhao, Q. Lv, Q. Wang, X. Zhou, Y. Chen, Q. Yuan and Y. Chen, Int. J. Biol. Macromol., 2021, 185, 917–934 CrossRef CAS PubMed.
- W. Xu, S. Fang, X. Cui, R. Guan, Y. Wang, F. Shi and S. Hu, Mol. Immunol., 2019, 111, 19–26 CrossRef CAS PubMed.
- W. Xue, Y. Gao, Q. Li, Q. Lu, Z. Bian, L. Tang, Y. Zeng, C. Chen and W. Guo, Int. J. Biol. Macromol., 2020, 161, 514–524 CrossRef CAS PubMed.
- W. J. Sun, K. Meng, C. H. Qi, X. Y. Yang, Y. G. Wang, W. T. Fan, Z. G. Yan, X. N. Zhao and J. Z. Liu, Carbohydr. Polym., 2015, 126, 91–96 CrossRef CAS PubMed.
- K. Ozaltin, P. S. Postnikov, M. E. Trusova, V. Sedlarik and A. Di Martino, Int. J. Biol. Macromol., 2019, 132, 24–31 CrossRef CAS PubMed.
- O. Sreekanth Reddy, M. C. S. Subha, T. Jithendra, C. Madhavi and K. Chowdoji Rao, J. Pharm. Anal., 2021, 11, 191–199 CrossRef CAS PubMed.
- D. J. McClements, Adv. Colloid Interface Sci., 2018, 253, 1–22 CrossRef CAS PubMed.
- E. Rogatsky, Anal. Biochem., 2021, 631, 114321 CrossRef CAS PubMed.
- J. Cortés-Ríos, A. M. Zárate, J. D. Figueroa, J. Medina, E. Fuentes-Lemus, M. Rodríguez-Fernández, M. Aliaga and C. López-Alarcón, Anal. Biochem., 2020, 608, 113904 CrossRef PubMed.
- G. Navarra, C. Peres, M. Contardi, P. Picone, P. L. San Biagio, M. Di Carlo, D. Giacomazza and V. Militello, Arch. Biochem. Biophys., 2016, 606, 134–142 CrossRef CAS PubMed.
- M. Ghanbari, M. Salavati-Niasari and F. Mohandes, Int. J. Pharm., 2021, 602, 120660 CrossRef CAS PubMed.
- S. F. I. Sofla, M. Abbasian and M. Mirzaei, J. Biomater. Sci., Polym. Ed., 2019, 30, 12–33 CrossRef PubMed.
- L. Fan, Y. Lu, L.-Y. Yang, F. Huang and X.-k. Ouyang, J. Colloid Interface Sci., 2019, 554, 48–58 CrossRef CAS PubMed.
- J.-K. Jiang, Y. Mu and H.-Q. Yu, J. Colloid Interface Sci., 2019, 535, 318–324 CrossRef CAS PubMed.
- W. Feng, C. Yue, Wusigale, Y. Ni and L. Liang, Food Res. Int., 2018, 108, 161–171 CrossRef CAS PubMed.
- B. Khodashenas, M. Ardjmand, A. S. Rad and M. R. Esfahani, Mater. Today Chem., 2021, 20, 100474 CrossRef CAS.
- F. Li, Y. Zhou, S. Wang, H. Yin, Y. Chen, H. Luo and S. Ai, Biosens. Bioelectron., 2020, 151, 111973 CrossRef CAS PubMed.
- L. Xing, J. Sun, H. Tan, G. Yuan, J. Li, Y. Jia, D. Xiong, G. Chen, J. Lai, Z. Ling, Y. Chen and X. Niu, Int. J. Biol. Macromol., 2019, 127, 340–348 CrossRef CAS PubMed.
- S. D. Chen, Y. H. Xu, K. Zhang, X. L. Wang, J. Sun, G. X. Gao and Y. F. Liu, Nat. Struct. Mol. Biol., 2012, 19, 430–435 CrossRef CAS PubMed.
- R. R. Xu, Mol. Genet. Genomics, 2014, 289, 965–979 CrossRef CAS PubMed.
- X. Zhang, Y. Li, Z. Ma, D. He and H. Li, Bioact. Mater., 2021, 6, 3692–3704 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra01767a |
| This journal is © The Royal Society of Chemistry 2022 |