Open Access Article
Open Access ArticleSynthesis and antibacterial activity studies in vitro of indirubin-3′-monoximes†
Fen-Fen Yang‡
a,
Ming-Shan Shuai‡a,
Xiang Guana,
Mao Zhanga,
Qing-Qing Zhanga,
Xiao-Zhong Fua,
Zong-Qin Lic,
Da-Peng Wangb,
Meng Zhoua,
Yuan-Yong Yang a,
Ting Liua,
Bin He
a,
Ting Liua,
Bin He a and
Yong-Long Zhao
a and
Yong-Long Zhao *a
*a
aState Key Laboratory of Functions and Applications of Medicinal Plants, School of Pharmacy, and Engineering Research Center for the Development and Application of Ethnic Medicine and TCM (Ministry of Education), Guizhou Medical University, Guiyang 550004, People's Republic of China. E-mail: zhaoyl05@126.com; zhaoyonglong@gmc.edu.cn; 378122847@qq.com
bKey Laboratory of Environmental Pollution Monitoring and Disease Control, Ministry of Education, Guizhou Medical University, Guiyang, 550025, People's Republic of China
cDepartment of Neurology, Sichuan Mianyang 404 Hospital, Mianyang, 621000, People's Republic of China
First published on 2nd September 2022
Abstract
Multi-drug-resistant microbial pathogens are a serious global health problem. New compounds with antibacterial activity serve as good candidates for developing novel antibacterial drugs which is very urgent and important. In this work, based on the unique scaffold of indirubin, an active ingredient of traditional Chinese medicine formulation Danggui Luhui Wan, we synthesized 29 indirubin-3′-monoximes and preliminarily evaluated their antibacterial activities. The antibacterial activity results demonstrated that the synthesized indirubin-3′-monoximes 5a–5z and 5aa–5ad displayed good potency against S. aureus ATCC25923 (MIC = 0.4–25.6 μg mL−1). Among them, we found that the 5-F, 5-Cl and 7-CF3 substituted indirubin-3′-monoximes 5r, 5s and 5aa also showed better antibacterial efficiency for S. aureus (MICs up to 0.4 μg mL−1) than the prototype natural product indirubin (MIC = 32 μg mL−1). More importantly, indirubin-3′-monoxime 5aa has certain synergistic effect with levofloxacin against clinic multidrug-resistant S. aureus (fractional inhibitory concentration index: 0.375). In addition, relevant experiments including electron microscopy observations, PI staining and the leakage of extracellular potassium ions and nucleic acid (260 nm) have been performed after treating S. aureus with indirubin-3′-monoxime 5aa, and the results revealed that indirubin-3′-monoximes could increase the cell membrane permeability of S. aureus. Although indirubin-3′-monoxime 5aa showed some cytotoxicity toward SH-SY5Y cells relative to compounds 5r and 5s, the skin irritation test of male mice after shaving showed that compound 5aa at a concentration of 12.8 μg mL−1 had no toxicity to mouse skin, and it could be used as a leading compound for skin antibacterial drugs.
Introduction
With the indiscriminate use of antibiotics, antibiotic-resistant pathogens (such as ESKAPE pathogens: Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.)1–3 have become an issue of great concern worldwide which seriously threatens global public health safety.1–12 According to statistics, there is an annual death toll worldwide of 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 caused by antimicrobial resistance, and this value is expected to increase to up to 10 million in 2050.4–7 As a consequence, there is an urgent and growing need for the development of novel antibacterial drugs to overcome bacterial resistance.1 For pharmaceutical companies, most of them are not interested in the research and development program of new antibiotics due to their high costs and low profits.1,8 So the development and identification of new compounds with the antibacterial activity that serve as good candidates for developing novel antibacterial drugs are very urgent and important.1,8
000 caused by antimicrobial resistance, and this value is expected to increase to up to 10 million in 2050.4–7 As a consequence, there is an urgent and growing need for the development of novel antibacterial drugs to overcome bacterial resistance.1 For pharmaceutical companies, most of them are not interested in the research and development program of new antibiotics due to their high costs and low profits.1,8 So the development and identification of new compounds with the antibacterial activity that serve as good candidates for developing novel antibacterial drugs are very urgent and important.1,8
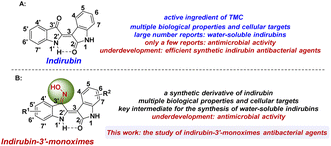
Chinese medicine and pharmacology have a 5000 year-old tradition in China.13,19 Bioactive plant constituents, especially from medicinal plants used in traditional Chinese medicine (TCM), are also playing an increasingly important role in the process of current drug discovery and innovation.6,13–18 A classic success example is the world-famous antimalarial drug named artemisinin, which was also named “Qinghaosu” in China and was isolated and identified by Youyou Tu and coworkers from the TCM plant “Qinghao” via some popular empirical formulas in some regions and the old documentation in the TCM literature. Due to the significant contribution to the discovery of artemisinin and fighting malaria, Youyou Tu became the first Chinese Nobel Laureate in Physiology or Medicine in 2015.14 Similar to bioactive plant constituent artemisinin, indirubin (Fig. 1A) is also the active plant ingredient of indigo naturalis (named Sei Tai in Japanese or Qing Dai in Chinese, a dark blue powder prepared from the leaves and branches of various indigo-producing plants)17,19–27 and Danggui Luhui Wan (a TCM formulation in the treatment of various chronic diseases such as cancer and inflammation, and composed of 11 medicinal herbs including indigo naturalis, etc.).20–23 Indirubin has the interesting 3,2′-bisindole scaffold and shows multiple biological properties,19–23 including hemostatic, anticancer, antipyretic, anti-inflammatory, sedative, antibacterial, and antiviral properties.22,23 However, due to the existence of a hydrogen-bonding framework in the structure of indirubin, the property of extremely poor solubility in most solvents has been observed, which is the main obstacle for the clinical application of indirubin.21,28–36
Up to now, much effort has been focused on indirubin and its derivatives; in particular, various cellular targets have been found including CDKs,19,37,38 GSK-3β,39 VEGFR-2,40 c-Src,41–43 Dyrk1A,44 and other kinases.15,45,46 Despite this, current research on indirubins is still mainly focused on the improvement of their water solubility and cytotoxicity.21,28–36,47 At present, although a few examples of the antimicrobial activity of indirubin and indigo extract from indigo naturalis or Wrightia tinctoria have been reported, post-modified indirubin antibacterial agents have not been reported.48–54 Indirubin-3′-monoxime is a synthetic derivative of indirubin and is usually used as a key intermediate for the synthesis of water-soluble indirubins.22,28,55 In addition, indirubin-3′-monoximes also have various biological properties and can inhibit the activities of CDKs, GSK-3β, 5-lipoxygenase, etc.19,22,29,38,55–59 However, to the best of our knowledge, there are no reports related to antimicrobial research on indirubin-3′-monoximes. With our ongoing interest in the new uses and in-depth development of indirubins,29 we hereby synthesized a series of indirubin-3′-monoximes and investigated their related antibacterial activity (Fig. 1B).
Results and discussion
Chemistry
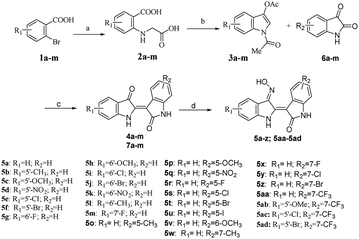
Based on our previous research work on indirubin derivatives29 and related literature reports,19,22,38,55–59 the syntheses of indirubin-3′-monoximes 5a–z and 5aa–5ad have been realized via the related synthesis routes as shown in Scheme 1. Firstly, starting from various substituted 2-bromobenzoic acids 1a–m, corresponding dicarboxylic acids 2a–m were synthesized via copper-catalyzed aromatic amination reaction with glycine (Scheme 1a). Then, various substituted 3-indoxyl acetates 3a–m were synthesized via the intramolecular cyclization of dicarboxylic acids 2a–m in the presence of acetic anhydride (Scheme 1b). Next, the acidic condensation of different substituted 3-indoxyl acetates 3a–m as key intermediates with various isatins 6a–m yielded the corresponding indirubins 4a–m and 7a–m (Scheme 1c). Finally, the target indirubin-3′-monoximes 5a–z and 5aa–5ad could be easily prepared via heating the synthetic indirubins 4a–m and 7a–m with hydroxylamine hydrochloride in pyridine (Scheme 1d).In vitro antibacterial activity of indirubin-3′-monoximes 5a–z and 5aa–5ad
The minimum inhibitory concentrations (MICs) of synthetic indirubin-3′-monoximes were tested against Gram-positive (G+) bacteria (S. aureus ATCC25923 and clinically isolated multidrug-resistant S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077) and Gram-negative (G−) bacteria (E. coli ATCC25922 and Pseudomonas aeruginosa ATCC9027) through broth microdilution procedures described in the Clinical and Laboratory Standards Institute (CLSI) 2018 standard methodology.62,67–69 MIC values for synthetic indirubin-3′-oximes were compared with those for reference drugs levofloxacin and indirubin under the same assay conditions for antimicrobial susceptibility assessment. In general, most of indirubin-3′-monoximes showed stronger antibacterial activities against S. aureus ATCC25923 than indirubin (MIC = 32 μg mL−1 or 12.5 μg mL−1 (ref. 49)). Except that compounds 5c, 5w and 5z showed certain antibacterial activities against P. aeruginosa ATCC9027 (with MIC values of 25.6, 128, and 128 μg mL−1, respectively), most of the indirubin-3′-monoximes have no obvious antibacterial effects on clinically isolated multidrug-resistant S. aureus 20
077) and Gram-negative (G−) bacteria (E. coli ATCC25922 and Pseudomonas aeruginosa ATCC9027) through broth microdilution procedures described in the Clinical and Laboratory Standards Institute (CLSI) 2018 standard methodology.62,67–69 MIC values for synthetic indirubin-3′-oximes were compared with those for reference drugs levofloxacin and indirubin under the same assay conditions for antimicrobial susceptibility assessment. In general, most of indirubin-3′-monoximes showed stronger antibacterial activities against S. aureus ATCC25923 than indirubin (MIC = 32 μg mL−1 or 12.5 μg mL−1 (ref. 49)). Except that compounds 5c, 5w and 5z showed certain antibacterial activities against P. aeruginosa ATCC9027 (with MIC values of 25.6, 128, and 128 μg mL−1, respectively), most of the indirubin-3′-monoximes have no obvious antibacterial effects on clinically isolated multidrug-resistant S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 and G− bacterial strains (E. coli ATCC25922 and P. aeruginosa ATCC9027). For the antibacterial activities against S. aureus ATCC25923, we first investigated the effects of different R1 groups at 5′, 6′ and 7′ positions of indirubin-3′-monoximes (5a–m). The results revealed that indirubin-3′-monoximes with electron-donating (such as OMe and Me) and halide (such as Cl and Br) substitutions at C5′ position showed good antibacterial activities and the MIC values of corresponding 5′-OMe, 5′-Cl and 5′-Br substituted compounds 5c, 5e and 5f up to 0.8 μg mL−1. For R1 substituents at C6′ position, the antibacterial activities of indirubin-3′-monoximes (5g, 5i and 5j) substituted by halogen atoms (such as 5′-F, Cl and Br) were better than those of other indirubin-3′-monoximes (5h, 5k and 5l) substituted with electron-donating (6′-OMe and Me) or electron-withdrawing (6′-NO2) groups. In addition, indirubin-3′-monoximes 5a–z and 5aa with different R2 groups at 5, 6 and 7 positions were also tested. In addition to 5-I substituted compound 5u, whether C5 is an electron-withdrawing (NO2) or electron-donating (Me, OMe) substituent, the antibacterial activities against S. aureus ATCC25923 of indirubin-3′-monoximes substituted by halogen atoms at C5 are better (5r–t vs. 5o–q), and the MIC values of 5-F substituted compound 5r and 5-Cl substituted compound 5s were also up to 0.4 μg mL−1. For R2 substituents at C7 position, indirubin-3′-monoximes substituted with electron-withdrawing substituents showed good antibacterial activities against S. aureus ATCC25923 and the MIC value of 7-CF3 substituted compound 5aa was also up to 0.4 μg mL−1. Based on the size of bacteriostatic ring of synthetic compounds 5r, 5s and 5aa, compound 5aa with an electron-withdrawing CF3 substituent at C7 position was chosen as the model compound for further study (Fig. 2). Considering the effect of R1 and R2 groups in the structure of indirubin-3′-monoximes on antibacterial activities, we further synthesized compounds 5ab–5ad to find indirubin-3′-monoximes with more efficient antibacterial activities. However, it was regrettable that the antibacterial activities against S. aureus ATCC25923 of compounds 5ab–5ad still did not exceed that of compound 5aa.
077 and G− bacterial strains (E. coli ATCC25922 and P. aeruginosa ATCC9027). For the antibacterial activities against S. aureus ATCC25923, we first investigated the effects of different R1 groups at 5′, 6′ and 7′ positions of indirubin-3′-monoximes (5a–m). The results revealed that indirubin-3′-monoximes with electron-donating (such as OMe and Me) and halide (such as Cl and Br) substitutions at C5′ position showed good antibacterial activities and the MIC values of corresponding 5′-OMe, 5′-Cl and 5′-Br substituted compounds 5c, 5e and 5f up to 0.8 μg mL−1. For R1 substituents at C6′ position, the antibacterial activities of indirubin-3′-monoximes (5g, 5i and 5j) substituted by halogen atoms (such as 5′-F, Cl and Br) were better than those of other indirubin-3′-monoximes (5h, 5k and 5l) substituted with electron-donating (6′-OMe and Me) or electron-withdrawing (6′-NO2) groups. In addition, indirubin-3′-monoximes 5a–z and 5aa with different R2 groups at 5, 6 and 7 positions were also tested. In addition to 5-I substituted compound 5u, whether C5 is an electron-withdrawing (NO2) or electron-donating (Me, OMe) substituent, the antibacterial activities against S. aureus ATCC25923 of indirubin-3′-monoximes substituted by halogen atoms at C5 are better (5r–t vs. 5o–q), and the MIC values of 5-F substituted compound 5r and 5-Cl substituted compound 5s were also up to 0.4 μg mL−1. For R2 substituents at C7 position, indirubin-3′-monoximes substituted with electron-withdrawing substituents showed good antibacterial activities against S. aureus ATCC25923 and the MIC value of 7-CF3 substituted compound 5aa was also up to 0.4 μg mL−1. Based on the size of bacteriostatic ring of synthetic compounds 5r, 5s and 5aa, compound 5aa with an electron-withdrawing CF3 substituent at C7 position was chosen as the model compound for further study (Fig. 2). Considering the effect of R1 and R2 groups in the structure of indirubin-3′-monoximes on antibacterial activities, we further synthesized compounds 5ab–5ad to find indirubin-3′-monoximes with more efficient antibacterial activities. However, it was regrettable that the antibacterial activities against S. aureus ATCC25923 of compounds 5ab–5ad still did not exceed that of compound 5aa.
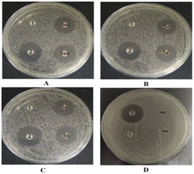 | ||
| Fig. 2 Images of antibacterial zones of DMSO (1), levofloxacin (2), and (D) 4a, (A) 5aa, (B) 5r and (C) 5s against S. aureus ATCC25923. | ||
Determination of antibacterial zone diameter
To further validate the antimicrobial effect of the above-mentioned drug sensitivity test more intuitively, we quantitatively and qualitatively evaluated the antibacterial activities of synthetic indirubin-3′-monoximes 5r, 5s and 5aa on S. aureus ATCC25923 by measuring the diameter of the inhibition zone in vitro. The size of the inhibition ring was positively correlated with the drug concentration, that is, with an increase of the compound concentration, the inhibition zone diameter increased. As shown in Fig. 2, it could be observed that at the same concentration (12.8 μg mL−1) of levofloxacin (2) and compounds 5r, 5s and 5aa, the bacteriostatic ring sizes of levofloxacin (21.7 mm), 5aa (19.0 mm), 5r (18.5 mm) and 5s (18.3 mm) were obtained. However, colonies occurred on the medium containing both negative control DMSO and leading compound indirubin (4a) at the same concentration. Thus, compounds 5r, 5s and 5aa effectively inhibited the emergence of S. aureus ATCC25923.The bactericidal or bacteriostatic assay of indirubin-3′-monoximes
With the better MICs of synthetic indirubin-3′-monoximes 5r, 5s and 5aa against S. aureus ATCC25923 in Table 1, we also tested the MBC (minimal bactericidal concentration) values and obtained the corresponding MBC/MIC ratios of these indirubin-3′-monoximes to further investigate whether their activity was bactericidal or bacteriostatic. According to the CLSI standards (bactericidal behavior: MBC/MIC ratio = 1 to 2; bacteriostatic behavior: MBC/MIC ratio ≥ 8),69,70 the results as shown in Table 2 indicated that indirubin-3′-monoximes 5r, 5s and 5aa were also observed as bacteriostatic agents with MBC/MIC ratios of >8.| Compound | Minimum inhibitory concentration/MIC (μg mL−1) | |||
|---|---|---|---|---|
| S. aureus ATCC25923 | E. coli ATCC25922 | P. aeruginosa ATCC9027 | S. aureusb 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151 151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027 027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 077 |
|
| a Lev. is levofloxacin.b Clinical isolated multidrug-resistant bacterial strain.c MIC value reported in ref. 49. | ||||
| Lev.a | <0.2 | <0.2 | 12.5 | 16 |
| Indirubin (4a) | 32/12.5c | >200 | >200 | >200 |
| 5a | 16 | >200 | >200 | >200 |
| 5b | 1.6 | >200 | >200 | >200 |
| 5c | 0.8 | >200 | 25.6 | >200 |
| 5d | 3.2 | >200 | >200 | >200 |
| 5e | 0.8 | >200 | >200 | >200 |
| 5f | 0.8 | >200 | >200 | >200 |
| 5g | 3.2 | >200 | >200 | >200 |
| 5h | 6.4 | >200 | >200 | >200 |
| 5i | 3.2 | >200 | >200 | >200 |
| 5j | 3.2 | >200 | >200 | >200 |
| 5k | 6.4 | >200 | >200 | >200 |
| 5l | 12.5 | >200 | >200 | >200 |
| 5m | 25.6 | >200 | >200 | >200 |
| 5o | 3.2 | >200 | >200 | >200 |
| 5p | 3.2 | >200 | >200 | >200 |
| 5q | 12.8 | >200 | >200 | >200 |
| 5r | 0.4 | >200 | >200 | >200 |
| 5s | 0.4 | >200 | >200 | >200 |
| 5t | 0.8 | >200 | >200 | >200 |
| 5u | 3.2 | >200 | >200 | >200 |
| 5v | 1.6 | >200 | >200 | >200 |
| 5w | 6.4 | >200 | 128 | >200 |
| 5x | 1.6 | >200 | >200 | >200 |
| 5y | 3.2 | >200 | >200 | >200 |
| 5z | 1.6 | >200 | 128 | >200 |
| 5aa | 0.4 | >200 | >200 | >200 |
| 5ab | 12.8 | >200 | >200 | >200 |
| 5ac | 1.6 | >200 | >200 | >200 |
| 5ad | 3.2 | >200 | >200 | >200 |
| Compound | MBC (μg mL−1) | MIC (μg mL−1) | MBC/MIC |
|---|---|---|---|
| a Bactericidal behavior: MBC/MIC ratio = 1 to 2; bacteriostatic behavior: MBC/MIC ratio ≥ 8. | |||
| Levofloxacin | <0.4 | <0.2 | <2 |
| 5aa | 6.4 | 0.4 | 16 |
| 5r | 12.8 | 0.4 | 32 |
| 5s | 12.8 | 0.4 | 32 |
Indirubin-3′-monoximes in combination with levofloxacin inhibited multidrug-resistant bacterial strain of S. aureus (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) 077)
077)
The in vitro antibacterial activities of compounds indirubin 4a and indirubin-3′-monoximes 5r, 5s and 5aa used alone or in combination with levofloxacin were analyzed by a checkerboard microdilution assay (Table 3). As shown in Table 3, indirubin 4a and indirubin-3′-monoximes 5r, 5s and 5aa generally were inactive in inhibiting the growth of multi-drug-resistant strain S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 (MIC ≥ 200 μg mL−1) when used alone. But, to our delight, indirubin-3′-monoxime 5aa showed a certain synergistic effect with levofloxacin and the MIC value of levofloxacin was reduced from 16 μg mL−1 to 2 μg mL−1, and the corresponding fractional inhibitory concentration index (FICI) value was 0.375.63,71 The results as shown in Table 3 also indicated that 4a, 5r and 5s also displayed additive effects with levofloxacin in inhibiting the growth of multi-drug-resistant strain S. aureus 20
077 (MIC ≥ 200 μg mL−1) when used alone. But, to our delight, indirubin-3′-monoxime 5aa showed a certain synergistic effect with levofloxacin and the MIC value of levofloxacin was reduced from 16 μg mL−1 to 2 μg mL−1, and the corresponding fractional inhibitory concentration index (FICI) value was 0.375.63,71 The results as shown in Table 3 also indicated that 4a, 5r and 5s also displayed additive effects with levofloxacin in inhibiting the growth of multi-drug-resistant strain S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077, and corresponding FICI values of 0.75 and 1 were obtained, respectively. Thus, indirubin-3′-monoxime 5aa showed a certain synergistic effect and represents a lead compound as a potential antibacterial synergist for the combination therapy of multi-drug-resistant S. aureus infections. At the same time, using a similar method, we also investigated the in vitro antibacterial activity of indirubin 4a, and indirubin-3′-monoximes 5r, 5s and 5aa used alone or in combination with levofloxacin against G− bacterium P. aeruginosa ATCC9027 and the results also showed indifference effects (FIC index range: 1.5 to 2, see Table S1† in ESI).
077, and corresponding FICI values of 0.75 and 1 were obtained, respectively. Thus, indirubin-3′-monoxime 5aa showed a certain synergistic effect and represents a lead compound as a potential antibacterial synergist for the combination therapy of multi-drug-resistant S. aureus infections. At the same time, using a similar method, we also investigated the in vitro antibacterial activity of indirubin 4a, and indirubin-3′-monoximes 5r, 5s and 5aa used alone or in combination with levofloxacin against G− bacterium P. aeruginosa ATCC9027 and the results also showed indifference effects (FIC index range: 1.5 to 2, see Table S1† in ESI).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077) by the checkerboard microdilution assay
077) by the checkerboard microdilution assay
| Compound | Levofloxacin | FICIa | Interpretation | |||
|---|---|---|---|---|---|---|
| Alone | Synergetic | Alone | Synergetic | |||
| a The interaction as reflected by FICI values: synergistic (FICI ≤ 0.5), additive (0.5 < FICI ≤ 1), indifferent (1 < FICI ≤ 4) and antagonistic (FICI > 4). | ||||||
| Indirubin (4a) | 256 | 128 | 16 | 8 | 1 | Additive |
| 5r | 256 | 128 | 16 | 4 | 0.75 | Additive |
| 5s | 256 | 128 | 16 | 8 | 1 | Additive |
| 5aa | 256 | 64 | 16 | 2 | 0.375 | Synergistic |
Effects of indirubin-3′-monoxime 5aa on external morphology and cell membrane permeability of S. aureus
Due to its excellent activity against S. aureus (ATCC25923), we further investigated the effect of indirubin-3′-monoxime 5aa on the external morphological features of S. aureus (ATCC25923) cells via transmission electron microscopy (TEM) analysis (Fig. 3). As shown in Fig. 3A, the untreated S. aureus cells show their spherical and plump connatural morphology and appear normal, both the cytoplasmic membrane and the peptidoglycan layer of the cell wall being retained. In contrast, S. aureus cells treated with indirubin-3′-monoxime 5aa appeared to become empty and show rupture and lysis of the membranes (Fig. 3B). For the S. aureus cells treated with 5aa, it could also be clearly observed in Fig. 3B that there was release of cellular contents into the surrounding environment. Therefore, there are various indications that the antibacterial activity of indirubin-3′-monoxime 5aa might be related to changes in the structure and permeability of the S. aureus cell membrane.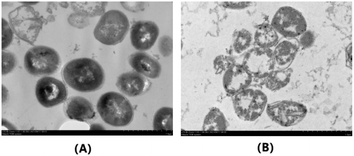 | ||
Fig. 3 TEM images of S. aureus ATCC25923 cells. (B) Treated with 5aa at 12.8 μg/mL for 16 h; (A) blank control (magnification, ×15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000). 000). | ||
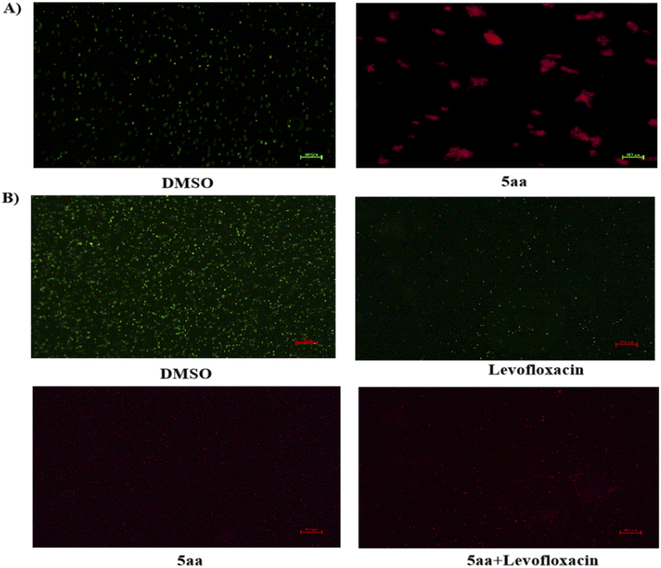
The antibacterial activity of indirubin-3′-monoxime 5aa indicates that the target of 5aa may be the cell membrane of bacteria. Thus, the possibility of 5aa interacting with the bacterial membrane was further investigated by fluorescence microscopy using a double staining approach. S. aureus ATCC25923 cells were treated with 5aa and then costained with Syto9 and PI. PI enters the cell and subsequently intercalates DNA upon loss of membrane integrity.74 As shown in Fig. 4A, S. aureus cells were incubated with a solvent negative control: bacteria were stained using Syto9 showing green fluorescence, while few of them had PI staining (red fluorescence). However, S. aureus cells showed a strong red fluorescence as a result of PI staining after incubation with 5aa for 2 h (Fig. 4A), pointing to the fact that compound 5aa causes pronounced membrane permeabilization and the bacteriostatic activity of the compound was not related to DNA damage. Membrane permeability by 5aa combined with levofloxacin against multi-drug-resistant S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 was further evaluated via PI internalization assay. The effects of solvent, levofloxacin, and 5aa and 5aa combined with levofloxacin on cells of multi-drug-resistant S. aureus 20
077 was further evaluated via PI internalization assay. The effects of solvent, levofloxacin, and 5aa and 5aa combined with levofloxacin on cells of multi-drug-resistant S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 incubated with Syto9/PI are depicted in Fig. 4B. 5aa and 5aa combined with levofloxacin instigated a gradual, yet conspicuous increase in fluorescence intensity, showing an ability to permeabilize the bacterial membrane. In contrast, cells incubated with control group solvent and levofloxacin (antibiotic impeding bacterial growth by inhibiting DNA gyrase synthesis) showed very minor to no fluorescence as expected. This result is consistent with the fluorescence microscopy observations.
077 incubated with Syto9/PI are depicted in Fig. 4B. 5aa and 5aa combined with levofloxacin instigated a gradual, yet conspicuous increase in fluorescence intensity, showing an ability to permeabilize the bacterial membrane. In contrast, cells incubated with control group solvent and levofloxacin (antibiotic impeding bacterial growth by inhibiting DNA gyrase synthesis) showed very minor to no fluorescence as expected. This result is consistent with the fluorescence microscopy observations.
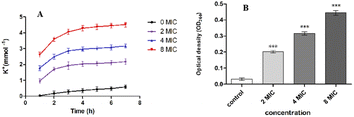
To further confirm the effect of indirubin-3′-monoxime 5aa on S. aureus cell membrane, we also investigated the leakage of cytoplasmic constituents (such as potassium and nucleic acids) of S. aureus (ATCC25923) treated with 5aa. As shown in Fig. 5, the extracellular K+ (Fig. 5A) and nucleic acid concentrations (Fig. 5B) were also increased significantly after S. aureus (ATCC25923) was treated with different concentrations (2MIC, 4MIC and 8MIC) of 5aa. These results were consistent with several previous reports, which all revealed that the membrane structure of microorganisms could be damaged by natural components, and these natural components can also promote the loss of cellular components, such as ions, proteins, and nucleic acids.60,61,64–66,72,73 Therefore, indirubin-3′-monoxime 5aa might show its antibacterial activity by damaging the cell membrane structure of S. aureus (ATCC25923), changing the cell membrane permeability, and eventually leading to the leakage of cytoplasmic constituents such as K+, nucleic acid, etc.
Cytotoxicity results of indirubin-3′-monoximes
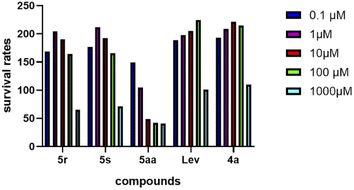
To evaluate the safety of these indirubin-3′-monoximes, we investigated the cytotoxicity of compounds 5aa, 5r and 5s on hepatocytes and SH-SY5Y cells. Unfortunately, these indirubin-3′-monoximes showed greater hepatotoxicity. The effect of indirubin-3′-monoximes 5r, 5s and 5aa on SH-SY5Y cells' survival rate at 0.1, 1, 10, 100, 1000 μM is shown in Fig. 6. It can be seen that indirubin-3′-monoximes 5r and 5s at 0.1–100 μM have also no obvious cytotoxicity to SH-SY5Y cells. However, indirubin-3′-monoxime 5aa has a concentration-dependent effect on the survival rate of SH-SY5Y cells. With the increase of 5aa concentration, the survival rate of SH-SY5Y cells decreased, and the cell survival rate was below 50% at a concentration of 10 μM 5aa.Skin irritation test
To further investigate the safety of these indirubin-3′-monoximes, we conducted a skin irritation test of indirubin-3′-monoxime 5aa on male mice after shaving. Table 4 shows skin tolerance test findings. Mice showed no irritation signs or skin edema after treatment with 12.8 μg mL−1 5aa. The treated skin was intact, with no inflammation and erythema compared to the untreated site (Fig. 7). No erythema or edema was observed with PII equal to “0” in each mouse at any time of the observation after removing the test material, which indicates that indirubin-3′-monoxime 5aa is safe for topical use.76,77 In addition, for the cytotoxicity in hepatocyte cell line, we also analyzed the histopathological section of mice liver tissue after skin irritation test via the treatment of 5aa (12.8 μg mL−1) for 1–72 h and the results also revealed that indirubin-3′-monoxime 5aa is safe for topical use and has no obvious cytotoxic effects on liver tissue (see ESI†). Nevertheless, due to the multiple biological activities of indirubin-3′-monoximes that we mentioned in the introduction, and the above-mentioned cytotoxicity in hepatocyte cell line and SH-SY5Y cells of 5aa, the above results only suggest that indirubin-3′-monoxime 5aa could be used as a leading compound of skin antibacterial drugs, and further in-depth and systematic research (such as in-depth structure–activity relationship research) on antibacterial activity, and thorough and further toxicological studies (such as the mutagenic (genotoxic) potential, carcinogenic, reproductive and developmental toxicity, and neurotoxicity) are still needed for other types of antibacterial uses of indirubin-3′-monoximes.| Reaction | 1 h | 24 h | 48 h | 72 h | ||||
|---|---|---|---|---|---|---|---|---|
| Con | Trt | Con | Trt | Con | Trt | Con | Trt | |
| a Primary Irritation Index (PII) = 0/3; PII = 0, based on PII irritation category for indirubin-3′-monoxime 5aa being negligible. | ||||||||
| Erythema | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Edema | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
 | ||
| Fig. 7 Skin irritation in mice. (a) Untreated mouse. (b) Mouse treated with 12.8 μg mL−1 5aa for 1 h. (c) Mouse treated with 12.8 μg mL−1 5aa for 24 h. | ||
Conclusions
In this work, we synthesized a series of indirubin-3′-monoximes and preliminarily evaluated their antibacterial activities. Among them, we found that the synthesized indirubin-3′-monoximes 5r, 5s and 5aa displayed better anti-S. aureus ATCC25923 (MIC up to 0.4 μg mL−1) activity than indirubin (MIC = 32 μg mL−1). More importantly, indirubin-3′-monoxime 5aa showed a certain synergistic effect with levofloxacin against clinic multidrug-resistant S. aureus 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 (FICI: 0.375), which could be used as a lead compound as a synergistic antibacterial agent of levofloxacin for the combination therapy of multi-drug-resistant S. aureus infections. In addition, relevant experiments of S. aureus treated with indirubin-3′-monoxime 5aa, including TEM analysis, Syto9/PI staining, and the leakage of extracellular potassium ions and nucleic acid (260 nm), have been performed after treatment of S. aureus ATCC25923 with indirubin-3′-monoxime 5aa. The results revealed that indirubin-3′-monoxime 5aa might show its antibacterial activity by targeting the bacterial membrane and changing the cell membrane permeability of S. aureus ATCC25923. The results of cytotoxicity tests of indirubin-3′-monoximes 5aa, 5r and 5s on hepatocytes and SH-SY5Y cells and the skin irritation test of 5aa on male mice after shaving indicate that indirubin-3′-monoxime 5aa should be suitable for topical use and could be used as a lead compound of skin antibacterial drugs. This research also represents a typical example of post-modified indirubin derivatives in the field of antibacterial agent research, and further optimization of molecular structure and antibacterial effect, and more thorough and further toxicological studies (such as the mutagenic (genotoxic) potential, carcinogenic, reproductive and developmental toxicity, and neurotoxicity) of indirubin-3′-monoximes are ongoing in our lab.
077 (FICI: 0.375), which could be used as a lead compound as a synergistic antibacterial agent of levofloxacin for the combination therapy of multi-drug-resistant S. aureus infections. In addition, relevant experiments of S. aureus treated with indirubin-3′-monoxime 5aa, including TEM analysis, Syto9/PI staining, and the leakage of extracellular potassium ions and nucleic acid (260 nm), have been performed after treatment of S. aureus ATCC25923 with indirubin-3′-monoxime 5aa. The results revealed that indirubin-3′-monoxime 5aa might show its antibacterial activity by targeting the bacterial membrane and changing the cell membrane permeability of S. aureus ATCC25923. The results of cytotoxicity tests of indirubin-3′-monoximes 5aa, 5r and 5s on hepatocytes and SH-SY5Y cells and the skin irritation test of 5aa on male mice after shaving indicate that indirubin-3′-monoxime 5aa should be suitable for topical use and could be used as a lead compound of skin antibacterial drugs. This research also represents a typical example of post-modified indirubin derivatives in the field of antibacterial agent research, and further optimization of molecular structure and antibacterial effect, and more thorough and further toxicological studies (such as the mutagenic (genotoxic) potential, carcinogenic, reproductive and developmental toxicity, and neurotoxicity) of indirubin-3′-monoximes are ongoing in our lab.
Experimental section
Chemistry
The chemical solvents and reagents used in synthesis and separation and purification processes were purchased from commercial suppliers (such as Energy-chemical, Alfa, Aladdin). Solvents were of reagent grade, and, when necessary, were purified by standard procedures as specified in Purification of Laboratory Chemicals, 4th edn (Armarego, W. L. F.; Perrin, D. D., Butterworth Heinemann: 1997). All reactions were monitored by analytical thin-layer chromatography (TLC). TLC was performed on silica gel plates with F-254 indicator and compounds were visualized by irradiation with UV light and/or by treatment with a solution of phosphomolybdic acid in ethanol followed by heating. Flash column chromatography was carried out over silica gel (200–300 mesh).Nuclear magnetic resonance (1H NMR, 13C NMR) spectra were obtained with a JNM-ECS400 (400 M) or 600 MHz Bruker Avance III HD spectrometer (400 or 600 MHz 1H, 101 or 151 MHz 13C). The spectra were recorded in CDCl3 or DMSO-d6 as the solvents at room temperature. 1H and 13C NMR chemical shifts are reported in ppm relative to the residual solvent peak or TMS as an internal standard: CDCl3 (1H NMR: δ 7.26, singlet; 13C NMR: δ 77.0, triplet), DMSO-d6 (1H NMR: δ 2.50, singlet, 13C NMR: δ 39.5, multiplet). Data for 1H NMR are reported as follows: chemical shift (δ ppm), multiplicity (s = singlet, d = doublet, t = triplet, m = multiplet, dd = doublet of doublet), integration, coupling constant (Hz) and assignment. Data for 13C NMR are reported as chemical shift.
High-resolution mass spectra (HRMS) were recorded with a WaterXevo G2S QTOF mass spectrometer (ESI) or Thermofisher (Vanquish (UPLC)-Q-Exactive Plus) mass instrument (ESI) and methanol was used to dissolve the samples. HPLC was employed to confirm the purity of compounds to be >95%.
Indirubin-3′-monoximes 5a–z and 5aa–5ad were prepared by the following published procedures.19,22,29,38,55–59
General procedure for the synthesis of indirubin-3′-monoximes 5a–z and 5aa–5ad
To an aqueous solution of KOH (3.83 g, 68 mmol, 2.3 equiv.) and K2CO3 (4.14 g, 30 mmol, 1.0 equiv.) were added the corresponding 2-bromobenzoic acid (6.03 g, 30 mmol, 1.0 equiv.), glycine (3.38 g, 45 mmol, 1.5 equiv.) and copper powder (10.8 mg, 0.15 mmol) under stirring. After the mixture was refluxed for 3–12 h (as judged by TLC analysis), the precipitate was filtered off, and then the filtrate was diluted with 1 N HCl. The precipitate obtained was collected and washed with cold water. The corresponding product 2 was afforded with 48–96% yields through collecting and drying the obtained precipitate.To a solution of 2 (20 mmol, 1.0 equiv.) and Ac2O (30 mL, 60 mmol, 3.0 equiv.) was added anhydrous NaOAc (2.46 g, 30 mmol, 1.5 equiv.) under stirring. Then, the above mixture was refluxed for 2–6 h (as judged by TLC analysis) and then cooled to 60 °C to remove Ac2O under reduced pressure. The corresponding crude product was extracted with ethyl acetate. The collected organic layer was washed with saturated NaHCO3 and brine solution, dried over anhydrous Na2SO4 and concentrated in vacuo after filtration to give crude product. The crude product was directly purified by flash column chromatography to yield the corresponding 1-acetyl-1H-indol-3-yl acetates 3 with 48–80% yields.
Under nitrogen atmosphere, corresponding 3 (3.66 mmol, 1.0 equiv.) and isatin 6 (4.1 mmol, 1.1 equiv.) were dissolved in AcOH (20 mL) and HCl (3.1 mL, 1 mol l−1) under stirring at room temperature. Then, the above reaction mixture was stirred for 12 h at room temperature (as judged by TLC analysis). After the reaction was completed, the mixture was quenched with H2O. The reddish precipitate was filtered off and recrystallized from 1 N HCl/EtOH solution to afford the corresponding indirubins 4 or 7 with 40–90% yields.
The corresponding indirubin 4 or 7 (5 mmol, 1.0 equiv.) and hydroxylamine hydrochloride (110 mmol, 22.0 equiv.) were dissolved in pyridine (7 mL), and the reaction mixture was refluxed for several hours until no starting material was detected by TLC. Then, the above mixture was cooled to room temperature and was diluted with 2 N HCl (50 mL) to afford the corresponding reddish solid 5a–z and 5aa–5ad with 59–97% yields.
Antibacterial susceptibility assays
The MICs of the test compounds were determined using the broth microdilution procedure described in the Clinical and Laboratory Standards Institute (CLSI) guidelines.62 Strains of E. coli ATCC 25922, S. aureus ATCC 25923, P. aeruginosa ATCC9027, and clinically isolated multi-drug-resistant strain of S. aureus (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 151
151![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 027
027![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077) were incubated in MHB for 16–24 h at 37 °C and were diluted to 1 × 106 CFU mL−1. The starting concentration of indirubin-3′-oxime derivatives was set as 204.8 μg mL−1, then two-fold dilution of samples was made in 96-well plates (the ending concentration was 0.2 μg mL−1, 100 μL in each well), then 100 μL test microorganism was added to the wells and was incubated for 16 h at 37 °C. A final concentration of 0.25% Resaxurin (7-sodiooxy-3H-phenoxazin-3-one 10-oxide) in wells as microorganism growth indictor was added and continuously incubated for 3 h before MIC reading. Equivalent amounts of DMSO (5%) and medium were successively used as negative controls and blank control. Levofloxacin was used as a positive drug. All experiments were performed in three replicates.
077) were incubated in MHB for 16–24 h at 37 °C and were diluted to 1 × 106 CFU mL−1. The starting concentration of indirubin-3′-oxime derivatives was set as 204.8 μg mL−1, then two-fold dilution of samples was made in 96-well plates (the ending concentration was 0.2 μg mL−1, 100 μL in each well), then 100 μL test microorganism was added to the wells and was incubated for 16 h at 37 °C. A final concentration of 0.25% Resaxurin (7-sodiooxy-3H-phenoxazin-3-one 10-oxide) in wells as microorganism growth indictor was added and continuously incubated for 3 h before MIC reading. Equivalent amounts of DMSO (5%) and medium were successively used as negative controls and blank control. Levofloxacin was used as a positive drug. All experiments were performed in three replicates.
MBC assay
MBC assay was conducted using the broth microdilution assay described in the preceding section.69,70 After the 24 h incubation period, aliquots from microtiter wells were plated onto MHA. The colonies that grew after 24 h of incubation were counted, with MBC being defined as the lowest compound concentration resulting in a ≥3-log reduction in the number of colony forming units (CFU).Drug combination of indirubin-3′-oxime derivatives and levofloxacin on multi-drug-resistant strain of S. aureus
The combination effects between indirubin-3′-oxime derivatives and antibiotic (levofloxacin) were determined with a checkerboard assay.63,71 Two-fold serial dilutions of indirubin-3′-oxime derivatives (50 μL) were prepared in vertical rows of a 96-well plate, and two-fold serial dilutions of the antibiotic (50 μL) were added in the horizontal orientation rows of the 96-well plate. The concentrations of each compound and antibiotic ranged from 2 to 1/32 of their MICs. Subsequently, 100 μL of 1 × 106 CFU mL−1 bacterial inoculum was added in each well. Then, 100 μL of each MHB and 1 × 106 CFU mL−1 bacterial inoculum were added in wells as reference. The combinatory effects were then determined based on the FICI values calculated according to the formula: FICI = FIC(A) + FIC(B), where FIC is defined as the MIC of samples/drugs in combination divided by the MIC of the samples/drugs used alone. The results were considered to be synergistic when FICI ≤ 0.5, additive when 0.5 ≤ FICI ≤ 1, indifferent when 1 ≤ FICI ≤ 4, and antagonistic when FICI > 4.TEM analysis of S. aureus treated with 5aa
TEM assays were performed to elucidate the impact of indirubin-3′-oxime derivative 5aa on S. aureus microbial cells.64 After the antibacterial assay, S. aureus treated with 12.8 μg mL−1 5aa was centrifuged at 5000×g and 27 °C for 10 min, while the suspension untreated with 5aa was used as a control. The cells were washed with phosphate buffer (0.1 M PBS, pH = 7.4) for 3 times and then resuspended. Then, the samples were fixed in 2.5% buffered glutaraldehyde solution at 4 °C for 4 h. Finally, the samples were cut with an ultramicrotome (Ultracut UCT, Leica, Wetzlar, Germany). Each sample was evaluated by TEM (HT7800, HITACHI, Japan).Fluorescence microscope observations of S. aureus in the presence and absence of 5aa
50 mL cultures of S. aureus bacteria were grown to late log phase in MHB broth with or without 5aa. Then, 30 mL of the bacterial culture was concentrated by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 5–10 minutes. The supernatant was removed and the pellet resuspended in 3 mL of 0.85% NaCl. Then, 1.5 mL of this suspension was added to each of two 30–40 mL centrifuge tubes containing 30 mL of 0.85% NaCl. Both samples were incubated at room temperature for 1 hour, mixing every 15 minutes. Pellets of both samples were obtained by centrifugation at 10
000×g for 5–10 minutes. The supernatant was removed and the pellet resuspended in 3 mL of 0.85% NaCl. Then, 1.5 mL of this suspension was added to each of two 30–40 mL centrifuge tubes containing 30 mL of 0.85% NaCl. Both samples were incubated at room temperature for 1 hour, mixing every 15 minutes. Pellets of both samples were obtained by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10–15 minutes. The pellets were resuspended in 30 mL of 0.85% NaCl and centrifuged again at 10
000×g for 10–15 minutes. The pellets were resuspended in 30 mL of 0.85% NaCl and centrifuged again at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 10–15 minutes. Two pellets were resuspended in separate tubes with 10 mL of 0.85% NaCl each. The optical density was determined at 670 nm (OD670) of a 3 mL aliquot of the bacterial suspensions in glass or acrylic absorption cuvettes (1 cm pathlength). A sample of the 2× stock Syto9/PI staining solution was combined with an equal volume of the bacterial suspension. The final concentration of each dye was 6 μM SYTO 9 stain and 30 μM propidium iodide. Samples were mixed thoroughly and incubated at room temperature in the dark for 15 minutes. Then, 5 μL of the stained bacterial suspension was trapped between a slide and an 18 mm square coverslip. Observation was made with a fluorescence microscope and the excitation/emission maxima for these dyes are about 480/500 nm for SYTO 9 stain and 490/635 nm for propidium iodide. The background remains virtually nonfluorescent.74
000×g for 10–15 minutes. Two pellets were resuspended in separate tubes with 10 mL of 0.85% NaCl each. The optical density was determined at 670 nm (OD670) of a 3 mL aliquot of the bacterial suspensions in glass or acrylic absorption cuvettes (1 cm pathlength). A sample of the 2× stock Syto9/PI staining solution was combined with an equal volume of the bacterial suspension. The final concentration of each dye was 6 μM SYTO 9 stain and 30 μM propidium iodide. Samples were mixed thoroughly and incubated at room temperature in the dark for 15 minutes. Then, 5 μL of the stained bacterial suspension was trapped between a slide and an 18 mm square coverslip. Observation was made with a fluorescence microscope and the excitation/emission maxima for these dyes are about 480/500 nm for SYTO 9 stain and 490/635 nm for propidium iodide. The background remains virtually nonfluorescent.74
Assay of extracellular potassium ions
The amount of potassium ion (K+) outflow was determined according to the method reported in the literature.65 S. aureus cell suspensions were prepared as previously described. Then, different amounts of compound 5aa were added to the suspension so that the final compound concentrations were 0, 2 MIC, 4 MIC and 8 MIC, respectively. The bacterial suspension was centrifuged. The extracellular potassium concentrations were measured in the supernatant by using a microplate reader and a potassium assay kit following the manufacturer's protocols.Determination of nucleic acid concentration
Leakages of nucleic acid from S. aureus cells were monitored by measuring ultraviolet absorbance at 260 nm using a Shimadzu UV-1800 spectrometer.66 In brief, after incubating for 8 h treated with 0, 2 MIC, 4 MIC and 8 MIC of 5aa, the samples were centrifuged and the supernatant was taken. The supernatant was filtered through a microporous membrane and measured for OD260 nm using a UV spectrophotometer. The difference between four recordings of data at 260 nm is the final result of nucleic acid leakage from S. aureus.Determination of antibacterial zone diameter
The antibacterial zone diameter test was performed according to the CLSI method.62Cell safety evaluation in SH-SY5Y cells
Using indirubin and levofloxacin as positive drugs, CCK-8 assay was used to test the in vitro inhibitory activity of the target molecule against SH-SY5Y cells. The SH-SY5Y cells were seeded in 96-well plates and incubated for 24 h. Medium containing test drugs was added according to groups, and solvent group and positive control group were set up. After 72 h of treatment, 10 μL CCK-8 solution was added to each well and incubated for 3 h. The absorbance value (OD) of each well was measured at 450 nm with a microplate reader, and the cell survival rate was calculated.75Skin irritation test
Skin tolerance tests were done using the Organization for Economic Cooperation and Development guidelines with slight modification. Twenty-four hours before the experiment, fur from the backs of all mice was clipped at different sites (Fig. 7). 12.8 μg mL−1 5aa was evenly and gently applied in a test site while untreated skin areas served as the control. The test sites were then examined critically at 1 h after removing the test material and at 24 h, 48 h and 72 h for dermal reaction using Draize scoring criteria.76 All experiments were performed according to the national and institutional guidelines and approved by the Animal Care Welfare Committee of Guizhou Medical University (no. 2100168). The experimental mice were Kunming mice purchased from Guizhou Laboratory Animal Engineering Technology Center (purchase order no. 20002).Conflicts of interest
All the authors declare that they have no conflict of interest.Acknowledgements
We thank the following for the financial support of this work: National Natural Science Foundation of China (no. 22061011, U1812403-05 and 81960628), Guizhou Science and Technology Department of China (no. QKHJC[2020]1Y031 and QKHRCPT[2017]5101), Guizhou Health Committee (gzwjkj2020-1-209), the State Key Laboratory of Functions and Applications of Medicinal Plants (no. FAMP201904K), and the Cultivation Project of National Natural Science Foundation of Guizhou Medical University (no. 20NSP054).Notes and references
- G. Dhanda, P. Sarkar, S. Samaddar and J. Haldar, J. Med. Chem., 2019, 62, 3184 CrossRef CAS PubMed.
- J. N. Pendleton, S. P. Gorman and B. F. Gilmore, Expert Rev. Anti-Infect. Ther., 2013, 11, 297 CrossRef CAS PubMed.
- E. Montassier, R. Valdés-Mas, E. Batard, N. Zmora, M. Dori-Bachash, J. Suez and E. Elinav, Nat. Microbiol., 2021, 6, 1043 CrossRef CAS PubMed.
- Interagency Coordination Group on Antimicrobial Resistance, Report to the Secretary-General of the United Nations, 2019 Search PubMed.
- E. Montassier, R. Valdés-Mas, E. Batard, N. Zmora, M. Dori-Bachash, J. Suez and E. Elinav, Nat. Microbiol., 2021, 6, 1043 CrossRef CAS PubMed.
- B. Kongkham, D. Prabakaran and H. Puttaswamy, Fitoterapia, 2020, 147, 104762 CrossRef CAS PubMed.
- B. Casciaro, L. Mangiardi, F. Cappiello, I. Romeo, M. R. Loffredo, A. Iazzetti, A. Calcaterra, A. Goggiamani, F. Ghirga, M. L. Mangoni, B. Botta and D. Quaglio, Molecules, 2020, 25, 3619 CrossRef CAS PubMed.
- S. N. Khadake, S. Karamathulla, T. K. Jena, M. Monisha, N. K. Tuti, F. A. Khan and R. Anindya, Bioorg. Med. Chem. Lett., 2021, 39, 127883 CrossRef CAS PubMed.
- D. Hughes and A. Karlen, Upsala J. Med. Sci., 2014, 119, 162 CrossRef PubMed.
- J. O'Neill, Review on Antimicrobial Resistance, 2016 Search PubMed.
- F. J. Álvarez-Martínez, E. Barrajón-Catalán and V. Micol, Biomedicines, 2020, 8, 405 CrossRef PubMed.
- R. Tommasi, D. G. Brown, G. K. Walkup, J. I. Manchester and A. A. Miller, Nat. Rev. Drug Discovery, 2015, 14, 529 CrossRef CAS PubMed.
- T. Efferth, P. C. H. Li, V. S. B. Konkimalla and B. Kaina, Trends Mol. Med., 2007, 13, 353 CrossRef CAS PubMed.
- Y.-Y. Tu, Angew. Chem., Int. Ed., 2016, 55, 10210 CrossRef CAS PubMed.
- D. J. Newman, Adv. Pharmacol., 2020, 87, 113 CAS.
- F. J. Álvarez-Martínez, E. Barrajón-Catalán and V. Micol, Biomedicines, 2020, 8, 405 CrossRef PubMed.
- Q.-Y. Yang, T. Zhang, Y.-N. He, S.-J. Huang, X. Deng, L. Han and C.-G. Xie, Chin Med., 2020, 15(1), 127 CrossRef PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2012, 75, 311 CrossRef CAS PubMed.
- R. Hoessel, S. Leclerc, J. A. Endicott, M. E. Nobel, A. Lawrie, P. Tunnah, M. Leost, E. Damiens, D. Marie, D. Marko, E. Niederberger, W. Tang, G. Eisenbrand and L. Meijer, Nat. Cell Biol., 1999, 1, 60 CrossRef CAS PubMed.
- N. Stasiak, W. Kukula-koch and K. Glowniak, Acta Pol. Pharm., 2014, 71, 215 Search PubMed.
- X. Cheng and K. H. Merz, Adv. Exp. Med. Biol., 2016, 929, 269 CrossRef CAS PubMed.
- H.-Z. Wang, Z.-Y. Wang, C.-Y. Wei, J. Wang J, Y.-S. Xu, G.-H. Bai, Q.-Z. Yao, L. Zhang and Y.-Z. Chen, Eur. J. Med. Chem., 2021, 223, 113652 CrossRef CAS PubMed.
- B. Sun, J.-H. Wang, L.-H. Liu, L.-F. Mao, L.-Z. Peng and Y.-W. Wang, Chem. Biol. Drug Des., 2021, 97, 565 CrossRef CAS PubMed.
- W. L. Hsieh, Y.-K. Lin, C. N. Tsai, T.-M. Wang, T. Y. Chen and J. H. S. Pang, J. Dermatol. Sci., 2012, 67, 140 CrossRef CAS PubMed.
- J.-L. Lai, Y.-H. Liu, C. Liu, M.-P. Qi, R.-N. Liu, X.-F. Zhu, Q.-G. Zhou, Y.-Y. Chen, A.-Z. Guo and C.-M. Hu, Inflammation, 2017, 40, 1 CrossRef CAS PubMed.
- T.-J. Qi, H.-T. Li and S. Li, Oncotarget, 2017, 8, 6658 Search PubMed.
- M. H. Kim, Y. Y. Choi, G. Yang, I. H. Cho, D. Nam and W. M. Yang, J. Ethnopharmacol., 2013, 145, 214 CrossRef CAS PubMed.
- X. Cheng, K. H. Merz, S. Vatter, J. Zeller, S. Muehlbeyer, A. Thommet, J. Christ, S. Wölfl and G. Eisenbrand, J. Med. Chem., 2017, 60, 4949 CrossRef CAS PubMed.
- Z.-X. Cao, F.-F. Yang, J. Wang, Z.-C. Gu, S.-X. Lin, P. Wang, J.-X. An, T. Liu, Y. Li, Y.-J. Li, H.-N. Lin, Y.-L. Zhao and B. He, J. Med. Chem., 2021, 60, 15280 CrossRef PubMed.
- H. Pandraud, Bull. Soc. Chim. Fr., 1961, 7, 1257 Search PubMed.
- S. J. Choi, J. E. Lee, S. Y. Jeong, I. Im, S. D. Lee, E. J. Lee, S. K. Lee, S. M. Kwon, S. G. Ahn, J. H. Yoon, S. Y. Han, J. I. Kim and Y. C. Kim, J. Med. Chem., 2010, 53, 3696 CrossRef CAS PubMed.
- K. Vougogiannopoulou, Y. Ferandin, K. Bettayeb, V. Myrianthopoulos, O. Lozach, Y. Fan, C. H. Johnson, P. Magiatis P, A. L. Skaltsounis, E. Mikros and L. Meijer, J. Med. Chem., 2008, 51, 6421 CrossRef CAS PubMed.
- Y. Ferandin, K. Bettayeb, M. Kritsanida, O. Lozach, P. Polychronopoulos, P. Magiatis, A. L. Skaltsounis and L. Meijer, J. Med. Chem., 2006, 49, 4638 CrossRef CAS PubMed.
- F. P. Guengerich, J. L. Sorrells, S. Schmitt, J. A. Krauser, P. Aryal and L. Meijer, J. Med. Chem., 2004, 47, 3236 CrossRef CAS PubMed.
- P. Polychronopoulos, P. Magiatis, A. L. Skaltsounis, V. Myrianthopoulos, E. Mikros, A. Tarricone, A. Musacchio, S. M. Roe, L. Pearl, M. Leost, P. Greengard and L. Meijer, J. Med. Chem., 2004, 47, 935 CrossRef CAS PubMed.
- C. L. Woodard, Z. Li, A. K. Kathcart, J. Terrell, L. Gerena, M. Lopez-Sanchez, D. E. Kyle, A. K. Bhattacharjee, D. A. Nichols, W. Ellis, S. T. Prigge, J. A. Geyer and N. C. Waters, J. Med. Chem., 2003, 46, 3877 CrossRef CAS PubMed.
- E. Gerometta, I. Grondin, J. Smadja, M. Frederich and A. Gauvin-Bialecki, J. Ethnopharmacol., 2020, 253, 112608 CrossRef CAS PubMed.
- X. Cheng, P. Rasqué, S. Vatter, K. H. Merz and G. Eisenbrand, Bioorg. Med. Chem., 2010, 18, 4509 CrossRef CAS PubMed.
- L. Meijer, A. L. Skaltsounis, P. Magiatis, P. Polychronopoulos, M. Knockaert, M. Leost, X. P. Ryan, C. A. Vonica, A. Brivanlou, R. Dajani, C. Crovace, C. Tarricone, A. Musacchio, S. M. Roe, L. Pearl and P. Greengard, Chem. Biol., 2003, 10, 1255 CrossRef CAS PubMed.
- Y. K. Chan, H. H. Kwok, L. S. Chan, K. S. Leung, J. Shi, N. K. Mak, R. N. Wong and P. Y. Yue, Biochem. Pharmacol., 2012, 83, 598 CrossRef CAS PubMed.
- S. Nam, R. Buettner, J. Turkson, D. Kim, J. Q. Cheng, S. Muehlbeyer, F. Hippe, S. Vatter, K. H. Merz, G. Eisenbrand and R. Jove, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 5998 CrossRef CAS PubMed.
- S. Nam, A. Scuto, F. Yang, W. Chen, S. Park, H. S. Yoo, H. Konig, R. Bhatia, X. Cheng, K. H. Merz, G. Eisenbrand and R. Jove, Mol. Oncol., 2012, 6, 276 CrossRef CAS PubMed.
- S. Nam, W. Wen, A. Schroeder, A. Herrmann, H. Yu, X. Cheng, K. H. Merz, G. Eisenbrand, H. Li, Y. C. Yuan and R. Jove, Mol. Oncol., 2013, 7, 369 CrossRef CAS PubMed.
- A. Walter, A. Chaikuad, R. Helmer, N. Loaec, L. Preu, I. Ott, S. Knapp, L. Meijer and C. Kunick, PLoS One, 2018, 13(5), e0196761 CrossRef PubMed.
- W. L. Hsieh, Y.-K. Lin, C. N. Tsai, T.-M. Wang, T. Y. Chen and J. H. S. Pang, J. Dermatol. Sci., 2012, 67, 140 CrossRef CAS PubMed.
- T. Kunikata, T. Tatefuji, H. Aga, K. Iwaki, M. Ikeda and M. Kurimoto, Eur. J. Pharmacol., 2000, 410, 93 CrossRef CAS PubMed.
- R. Jautelat, T. Brumby, M. Schafer, H. Briem, G. Eisenbrand, S. Schwahn, M. Kruger, U. Lucking, O. Prien and G. Siemeister, ChemBioChem, 2005, 6, 531 CrossRef CAS PubMed.
- P. Kannan, B. Shanmugavadivu, C. Petchiammal and W. Hopper, Acta Bot. Hung., 2006, 48, 323 CrossRef.
- P. Kannan, R. Mohankumar, S. Ignacimuthu and M. G. Paulraj, Scand. J. Infect. Dis., 2010, 42, 500 CrossRef PubMed.
- K. Ponnusamy, C. Petchiammal, R. Mohankumar and W. Hopper, J. Ethnopharmacol., 2010, 132, 349 CrossRef CAS PubMed.
- Y. R. Chiang, A. Li, Y. L. Leu, J. Y. Fang and Y. K. Lin, Molecules, 2013, 18, 14381 CrossRef PubMed.
- S. K. Mahendra and P. V. Nityanand, J. Coastal Life Med., 2014, 2(10), 826 Search PubMed.
- K. V. Devika, T. Sabarinathan and S. Shamala, J. Orofacial Sci., 2021, 13, 67 CrossRef CAS.
- B. Sun, J.-H. Wang, L.-H. Liu, L.-F. Mao, L.-Z. Peng and Y.-W. Wang, Chem. Biol. Drug Des., 2021, 97, 565 CrossRef CAS PubMed.
- C. Pergola, N. Gaboriaud-Kolar, N. Jestadt, S. Konig, M. Kritsanida, A. M. Schaible, H. Li, U. Garscha, C. Weinigel, D. Barz, K. F. Albring and O. Huber, J. Med. Chem., 2014, 57, 3715 CrossRef CAS PubMed.
- V. Myrianthopoulos, P. Magiatis, Y. Ferandin, A. L. Skaltsounis, L. Meijer and E. Mikros, J. Med. Chem., 2007, 50, 4027 CrossRef CAS PubMed.
- T. Blažević, E. H. Heiss, A. G. Atanasov, J. M. Breuss, V. M. Dirsch and P. Uhrin, Evid. Based Complementary Altern. Med., 2015, 2015, 654098 Search PubMed.
- T. R. Kameswaran and R. Ramanibai, Biomed. Pharmacother., 2009, 63, 146 CrossRef CAS PubMed.
- X.-M. Liao and K.-N. Leung, Oncol Rep., 2013, 29(1), 371–379 CrossRef CAS PubMed.
- B. S. Jenie, B. P. Priosoeryanto, R. Syarief and G. T. Rekso, Hayati, 2008, 15, 56 Search PubMed.
- V. K. Bajpai, K. H. Baek and S. C. Kang, Food Res. Int., 2012, 45, 722 CrossRef CAS.
- M. A. Wikler, Approved Standard, 2009, M7-A8 Search PubMed.
- S. Mulyaningsih, F. Sporer, S. Zimmermann, J. Reichling and M. Wink, Phytomedicine, 2010, 17, 1061 CrossRef CAS PubMed.
- W.-C. Zeng WC, Q. He, Q. Sun, K. Zhong and H. Gao, Int. J. Food Microbiol., 2012, 153, 78 CrossRef PubMed.
- A. Sharma, V. K. Bajpai and K. Baek, J. Food Saf., 2013, 33, 197 CrossRef.
- H.-Y. Cui, C.-H. Zhang, C.-Z. Li and L. Lin, Ind. Crops Prod., 2019, 139, 111498 CrossRef CAS.
- Clinical and Laboratory Standards Institute, CLSI, 2018, M07Ed11 Search PubMed.
- M. M. F. Ismail, H. G. Abdulwahab, E. S. Nossier, N. G. E. Menofy and B. A. Abdelkhalek, Bioorg. Chem., 2020, 94, 103437 CrossRef CAS PubMed.
- F.-C. Bi, D. Song, N. Zhang, Z.-Y. Liu, X.-J. Gu, C.-Y. Hu, X.-K. Cai, H. Venter and S.-T. Ma, Eur. J. Med. Chem., 2018, 159, 90 CrossRef CAS PubMed.
- D. Song, F.-C. Bi, N. Zhang, Y.-H. Qin, X.-B. Liu, Y.-T. Teng and S.-T. Ma, Bioorg. Med. Chem., 2020, 28(21), 115729 CrossRef CAS PubMed.
- Z. Li, J. Tu, G.-Y. Han, N. Liu and C.-Q. Sheng, J. Med. Chem., 2021, 64(2), 1116 CrossRef CAS PubMed.
- G.-H. Li, Y.-F. Xu, X. Wang, B.-G. Zhang, C. Shi, W.-S. Zhang and X.-D. Xia, Foodborne Pathog. Dis., 2014, 11, 314 Search PubMed.
- A. Sharma, V. K. Bajpai and K. Baek, J. Food Saf., 2013, 33, 197 CrossRef.
- D. Meier, M. V. Hernández, L. Geelen, R. Muharini, P. Proksch and J. E. Bandow, Bioorg. Med. Chem., 2019, 27, 115151 CrossRef CAS PubMed.
- J. Zhang, Z. Zhang, W. Song and J. Liu, Mol. Cell. Probes, 2020, 52, 101566 CrossRef CAS PubMed.
- V. A. Nyigo, R. Mdegela, F. Mabiki and H. M. Malebo, Eur. J. Med. Plants, 2015, 10, 1 CrossRef.
- A. Mekonnen, S. Tesfaye, S. G. Christos, K. Dires, T. Zenebe, N. Zegeye, Y. Shiferaw and E. Lulekal, J. Toxicol., 2019, 2019, 1 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra01035f |
| ‡ These authors made equal contributions to this work. |
| This journal is © The Royal Society of Chemistry 2022 |