 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Recent advances in fluorescence anisotropy/polarization signal amplification
Xue Xiao
 *a and
Shujun Zhen
*a and
Shujun Zhen
 b
b
aKey Laboratory of Basic Chemistry of the State Ethnic Commission, College of Chemistry and Environment, Southwest Minzu University, 610041, Chengdu, PR China. E-mail: xiaoxue@swun.edu.cn
bKey Laboratory of Luminescence Analysis and Molecular Sensing (Southwest University), Ministry of Education, College of Chemistry and Chemical Engineering, Southwest University, 400715, Chongqing, PR China
First published on 23rd February 2022
Abstract
Fluorescence anisotropy/polarization is an attractive and versatile technique based on molecular rotation in biochemical/biophysical systems. Traditional fluorescence anisotropy/polarization assays showed relatively low sensitivity for molecule detection, because widespread molecular masses are too small to produce detectable changes in fluorescence anisotropy/polarization value. In this review, we discuss in detail how the potential of fluorescence anisotropy/polarization signal approach considerably expanded through the implementation of mass amplification, recycle the target amplification, fluorescence probes structure-switching amplification, resonance energy transfer amplification, and provide perspectives at future directions and applications.
1. Introduction
Fluorescence anisotropy/polarization (FA/FP) was first applied in biochemistry almost 7 decades ago when Gregorio Weber described his studies on bovine serum ovalbumin and albumin conjugated with 1-dimethylaminonaphthalene-5-sulfonyl chloride (dansyl chloride).1–3 The increase in the number and diversity of FA/FP studies are astonishing and the method is extremely widespread in the clinical and biomedical fields following Weber's work.3,4 The utility of FA/FP in clinical and biomedical sciences ultimately rests on the observed polarization on the rotational diffusion rate of molecules.3 The reasons for the popularity of FA/FP assays are manifold. First, FA and FP values are intrinsic parameters, and the ratiometric FA and FP techniques are tolerant to photobleaching of the fluorophore and instrumental parameters.5–8 Second, FA and FP assays are homogeneous; that is, there is no need for separation of substrates.3,9 Third, because of its simplicity and speed, FA/FP assays are especially suitable for an automated high throughput format assay.10–13Earlier FA/FP assays, called fluorescence polarization immunoassays (FPIA), have undoubtedly become predominant methods for the rapid monitoring of small molecules, including abuse of drugs in clinical chemistry and pollutants in environmental and food control.10,14 However, the main limitation of FPIA is that the detection of low mass compounds can be fairly restricted.15–17 Over the last two decades, the investigation using nucleic acid aptamers as recognition units has broadened the applications of FA/FP assays18,19 because of the great stability and high binding affinity of the aptamers and the advantages of aptamers over antibodies in ease of synthesis and labeling.20 Different FA/FP assays have been established based on either direct or indirect aptamer recognition system.20–26 Likewise, the sensitivities of these strategies are relatively low, because masses of widespread molecules are too small to produce detectable changes in FA/FP value.27 Therefore, a general strategy that involves a design for signal amplification will be a highly valued addition to this field. Very recently, the sensitivities of FA/FP assays have been significantly improved by some developed signal amplification approaches such as proteins and nanomaterials mass amplification,28–33 enzyme-catalyzed recycling signal amplification,27,34 competitive displacement and induced-fit binding amplification.35,36
Previous reviews that tackled the FA/FP-based sensing platforms mainly focused on the specific recognition elements and its applicative aspects.3,4,6,16,17,37 Qiu et al. summarized about nanomaterial enhanced FP/FA technology,38 however, there is currently no thorough review dedicated to design for signal amplification about FP/FA. Instead, the present feature review aims at thorough discussing the state of art in signal amplification mechanisms and methods design, with a special emphasis on their analytical performances and implications in terms of scope.
2. The principle of FA/FP assay
The basic principle of measurements relies on the absorption of linearly polarized excitation light by a fluorophore in relation to its orientation.16 The polarization (P) and anisotropy (r) terms, both depending on the measured emission intensities parallel and perpendicular to the plane of the vertically polarized excitation light, can interchangeably be used for determining the polarization degree of a fluorescent rotating species.3,15,39 At the molecular level, P/r is related to the rotational correlation time (θ) and the fluorescence lifetime (τ) of fluorescent molecules, and can be expressed by the modified form of the Perrin formula:
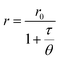 | (1) |
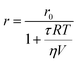 | (2) |
FA or FP, observed when fluorophores are excited by polarized light, is a commonly measured property of fluorescent molecules.6 The FA values (r) can be readily obtained by determining the fluorescence intensities from two polarization planes, normally, calculated using the following eqn (3):
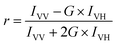 | (3) |
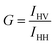 | (4) |
According to eqn (5), the anisotropy (r) and polarization (P) values are interchangeable. As a measure of the ratio of intensities, the anisotropy or polarization is a dimensionless value. In principle, the anisotropy and polarization are independent of the fluorophore concentration and the total intensity of the sample.6
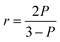 | (5) |
3. Strategies without signal amplification
FA/FP assays can be employed for protein detection under a direct format, because of the relatively large mass or volume of protein. In these approaches, fluorescent dye modified aptamer has been used as FA/FP probe and target recognizing unit. When protein analyte is added, an effective increase in the mass of the aptamer probe occurs upon complex formation, resulting in detectable variations in their FA values. However, most of the methods commonly suffer from poor FA responses, ranging from 0.050 to 0.100.40–43FA is not only influenced by the molecular weight of the formed complex, but also in relation to the nature of the tethered fluorophores.20,44 For example, the electrostatic interaction between the dye and DNA affects the segmental motion of dye, so the choice of the charge characteristics of the fluorophore is crucially important.45 Taking advantage of the electrostatic interaction between fluorescein and lysozyme, Li group developed a simple and sensitive analytical method for lysozyme.46 In this contribution, a coupling of negatively charged fluorescein with positively charged lysozyme upon binding between lysozyme and its aptamer occurs. Such coupling may bring an increased fraction of global rotational movement of the fluorescence species, thus producing a significantly increased FA signal with the maximum changed value of 0.2. However, this method can be only applied to peculiar fluorophore/protein pairs of opposed charge.
In addition, small molecules and metal ions also have been analyzed by a direct FA/FP format.22,24,47–49 Likewise, the FA/FP variation experimentally observed is somewhat negligible. Through changing the structure of aptamer and the coupling mode of aptamer and fluorescent dye, some studies have been developed to overcome this constraint. Peyrin group designed an engineering instability in the secondary structure of an aptameric recognition element that is usually a well-defined stem-loop secondary structure (Fig. 1).26 With this method, the segmental mobility of a fluorophore label, attached to one extremity of the aptamer, can be significantly promoted so that a strong depolarization process will occur. Upon target binding, the structural change of the aptamer will favor the formation of a stable stem-loop structure, inducing an increase in the FA signal. However, this strategy is not widely applicable as it requires a sufficiently large aptamer conformational/structural change or the rational engineering of the aptameric element. Furthermore, taking advantage of competitive reaction between complementary sequences of aptamer and small molecules, their group exploited the small molecules analytical methods with broader applicability.23,50 Zhao et al. utilized intramolecular tetramethylrhodamine (TMR) labeling of ssDNA to achieve a noncompetitive FA assay for the detection of small molecules.20 The intramolecular interaction between labeled TMR and DNA aptamer bases dramatically affects the local rotation and FA of TMR and can be altered by aptamer conformation change upon target binding, leading to a significant change of FA of TMR.
 | ||
| Fig. 1 Proposed representation of the local segmental motion of dye contributed (arrow) to the variation of the FA signal in relation to the aptamer probe structure. rf is the FA in the absence of analyte. Reprinted with permission from ref. 26. Copyright (2010) Elsevier. | ||
Rational design of the linker between fluorophores and target identification unit, appropriate choice of fluorophores, contribute to increase sensitive FA/FP responses. Even so, all these direct size-based assays exhibit modest FA/FP change and sensitivity. Direct assays have the advantage of simple reaction, but its application scope subject to certain limitations, especially for the detection of small molecules and metal ions as they are too small to induce the obvious FA changes.
4. Strategies with signal amplification
To enhance FA/FP response, substantial efforts were devoted to the development of signal amplification methodologies. There are mainly creations in two aspects: one is to label the fluorophore with large molecules or nanomaterials, such as proteins, gold nanoparticles (AuNPs), SiO2 nanoparticles (SiO2NPs), or graphene oxide (GO); the other one is to design a specific system to recycle the target molecules, such as enzyme-catalyzed target recycling, target-catalyzed DNA cyclic assembly. Next, we discuss these methods.4.1 Protein amplification strategies
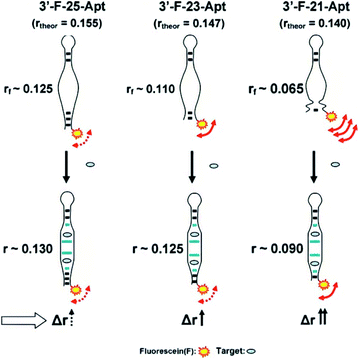
Proteins, which are larger molecular mass or volume than aptamers, can be used to enhance FA/FP signal.51 Famulok et al. used protein-binding aptamers for screening small molecule inhibitors of proteins in an FA/FP approach.52 In addition, Yang group developed a protein mass amplifying strategy to construct FA aptamer probes for small molecule analysis in complex biological samples.28 In this approach (Fig. 2), a mass amplifying probe consists of a targeting aptamer domain against a target molecule and a molecular mass amplifying aptamer domain for the amplifier protein. The probe is initially rendered inactive by a small blocking strand, and will be activated to be bound to the protein only when a target molecule is bound to the probe. Enlarging the molecular mass of the probe/target complex can obtain an obviously increased FA value. In this effect, two probes that constitute a target (ATP and cocaine respectively) aptamer were prepared, and the detection limits for ATP and cocaine were 0.5 μM and 0.8 μM, respectively.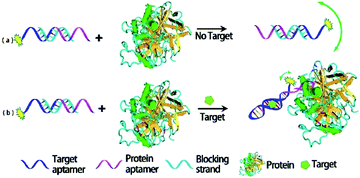 | ||
| Fig. 2 Working principle of protein amplifying FA for sensitive detection of small molecules. Reprinted with permission from ref. 28. Copyright (2012) American Chemical Society. | ||
Significantly, streptavidin as a large-sized protein has been employed as an effective signal amplifier to detect small molecules49,53 and analyze the DNA–protein interactions.54 Unlike other FA/FP signal amplification strategies that use dual binding DNA probes, these methods implement signal amplification through the binding of streptavidin with biotinylated DNA.54 Moreover, to further improve the sensitivity of detection, various signal amplification strategies combining with streptavidin mass amplification have been developed. First, hybridization chain reaction (HCR) and polymerase chain reaction (PCR) was used to construct a target-triggered assembly of DNA–protein hybrid nanowires for detection of small molecules and chloramphenicol respectively.12,55 Second, combining target-triggered enzymatic cleavage protection and the extraordinarily strong interaction between biotin and streptavidin, a novel mass amplification strategy for the sensitive detection of adenosine56 and Pd2+ (ref. 57) in homogeneous solution was reported. Third, Dzantiev et al. developed an FP-based aptamer assay with the use of anchor protein modules, based on the streptavidin and IgG as FP enhancers.51 The change of FP which was dually enhanced by streptavidin and IgG respectively could be used to detect ochratoxin A at the nM level in wine within 15 minutes.
To sum up, FA/FP assay response and sensitivity are improved by protein amplification. However, the limitation for use of protein amplifiers in FA/FP analysis is constituted by their variabilities and the need to operate under precisely defined experimental conditions compatible with the properties and stability of biomolecules.16
4.2 Nanomaterials amplification strategies
Compared with proteins, nanomaterials amplification have high thermal stabilities, easy synthesis and modification, and the FA/FP signal change is more significantly improved.58 Recently, to improve the sensing sensitivity, a number of nanomaterials such as AuNPs, CNTs, GO, metal–organic frameworks (MOFs), SiO2NPs, quantum dots (QDs), 2D transition metal dichalcogenide nanosheets (TMD NSs), were introduced into FA/FP sensor system. Nanomaterials as FA/FP enhancer has brought revolutionary effect on greatly improving detection sensitivity and application range of FA/FP because of its unique structure and extraordinary properties such as simple/controllable synthesis and modification, different sizes, large volumes and masses, excellent optical and chemical properties.38 Furthermore, several nanostructures were specifically bound to ssDNA, such as GO, MOFs and MoS2, giving discriminating effects to the free, unstructured and bound, structured states of a functional nucleic acid. This unique nucleic acid interaction forms the basis for a versatile mass amplifying strategy for the FA/FP technique.31 However, the amplification strategies that utilize nanomaterials are usually compromised by fluorescence quenching and scattering, potentially providing a less accurate measurement.16,54 Moreover, the difficulty to apply to living cells or organisms because of their widespread biotoxicity constitutes another limiting factor. | ||
| Fig. 3 Schematic illustration of the strategy of Hg2+ ion detection using FP enhancement by AuNPs. Reprinted with permission from ref. 65. Copyright (2008) Wiley. | ||
Furthermore, Ye group also employed a novel and practical method relying on metal ion-dependent DNAzyme-based FA via AuNPs enhancement, which was presented for detecting Cu2+ and Pb2+ ions in an aqueous medium at room temperature.61 Ren group used AuNPs as a dual-enhancer of FP for nucleic acid detection.66 In this report, AuNPs increased the volume or mass of the fluorescent dye Alexa fluor 488 dyes (Alexa488) and the AuNPs-mediated nanometal surface energy transfer (NSET) quenching effect decreased the effective concentration of the Alexa488, which caused remarkable dual enhancement of the Alexa488's FP.
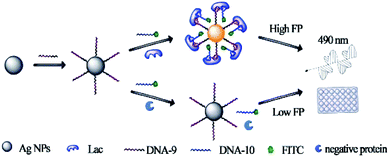 | ||
| Fig. 4 Principle of the dual amplified aptasensor based on bivalent aptamers and Ag10NPs enhancement. Reprinted with permission from ref. 68. Copyright (2017) American Chemical Society. | ||
A multiwalled carbon nanotube as a signal amplifier for FP assay of DNA methyltransferase (MTase) activity has been developed.74 In this work, the introduction of a multiwalled carbon nanotube causes a significant amplification of the detection signal, which substantially improves the detection sensitivity by two orders of magnitude over the reported methods. In addition, by using carbon nanoparticles to enhance FA, an aptamer-based sensor enabling signal-amplification and real-time detection of apyrase is reported.75
Significantly, GO as a two-dimensional (2-D) nanosheet with facile synthesis and high water dispersity, contributes to a slower rotation than that of spherical nanoparticles with the same surface area.76,77 Thus, GO is able to perform as an excellent FA amplifier. Yang et al. reported a FA signal amplification strategy by employing GO as the signal amplifier to detect adenosine triphosphate (ATP).31 Huang and coworkers developed a GO amplifying strategy to construct a DNAzyme-based FA system for Cu2+ analysis.58 Huang group also developed a label-free FA method by using GO as FA amplifier, G-rich single ssDNA as recognition probe, acidine orange as FA reporting fluorophore and potassium ion (K+) as a proof-of-concept target.33 This contribution does not require covalent modification of the recognition probe, thus it is simple and cost-effective. GO-amplified FP assays also were used for the identification of antagonists78 and toxins.79,80 In addition, to further improve the sensitivity of the GO-enhancer FA/FP method, exonuclease and nicking enzymes have been employed to achieve target recycling for signal amplification detection.81,82
Especially, for decreasing the quenching efficiency of GO to the fluorescence of dye-labeled probe DNA to ensure FA detection accuracy, Zhen group reported a novel GO amplified FA assay with improved accuracy and sensitivity.9 In this approach (Fig. 5), a toehold-mediated strand exchange reaction was introduced to detach probe DNA from the GO surface. The probe DNA was indirectly immobilized on GO through the double-stranded region of its hybridization with a capture DNA fragment that contained an A20 tail. Through this design, not only the rotation of fluorescent dye was restricted, but also the quenching efficiency of GO to fluorescent dye was decreased. And in the presence of the target, probe DNA was released from the GO surface more easily through toehold-mediated strand exchange reaction comparing with the method in which probe DNA is directly immobilized on GO, resulting in obviously decreased FA. This strategy has been used for the selective detection of ssDNA, adenosine and thrombin. Besides, based on the above mechanism, to enhance the detection sensitivity, Zhen and coworkers also employed the exonuclease III (Exo III) and target-catalyzed hairpin assembly (CHA) to achieve recycling signal amplification.27,83
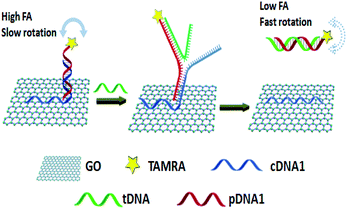 | ||
| Fig. 5 A schematic representation of ssDNA detection using a novel GO amplified FA assay. Reprinted with permission from ref. 9. Copyright (2015) Royal Society of Chemistry. | ||
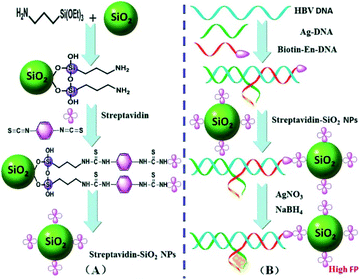 | ||
| Fig. 6 (A) Synthesis procedure for the functionalized streptavidin–SiO2NPs. (B) A working principle of the proposed FP biosensor for HBV DNA detection. Reprinted with permission from ref. 85. Copyright (2015) Royal Society of Chemistry. | ||
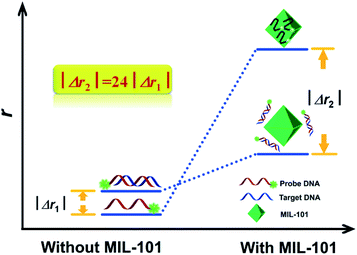 | ||
| Fig. 7 The concept and the principle of MIL-101 amplified fluorescence anisotropy strategy for HIV-DNA detection. Reprinted with permission from ref. 32. Copyright (2014) Royal Society of Chemistry. | ||
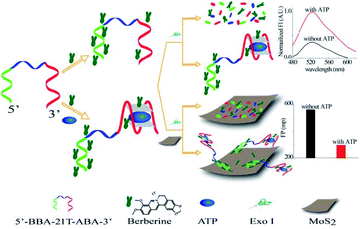 | ||
| Fig. 8 Illustration of a chimeric aptamers-based and MoS2 nanosheet-enhanced label-free FP strategy for ATP detection. Reprinted with permission from ref. 94. Copyright (2018) American Chemical Society. | ||
4.3 Enzyme amplification strategies
The strategies based on the enzyme-catalyzed target amplification have demonstrated great potential. In FA/FP assay, there are two main types of enzyme amplification signal, cleavage enzyme and polymerase. First, Ye group reported a versatile target assisted Exo III-catalyzed amplification FP methodology for the highly sensitive and selective detection of DNA.34 In this contribution, Exo III is employed to recycle the process of target-assisted digestion of probe molecules, thus resulting in significantly improved sensitivity. In the presence of the target DNA, the hybridization of the target and probe strands form a double-stranded structure containing a blunt 3′ terminus and an Exo-III-resistant 3′ protruding terminus (Fig. 9, B). Exo III can catalyze the stepwise removal of mononucleotides from 3′-hydroxyl termini of duplex DNAs with blunt or recessed 3′ termini. The target–probe hybridization triggers the selective enzymatic digestion of the dye-labeled probe, liberating the fluorophore with several nucleotides (low fluorescence polarization value) before ultimately releasing the target (Fig. 9, C). The released intact DNA target then hybridizes with another dye-labeled probe to initiate the cycle digestion of the probe, resulting in the digestion of many probes, generating a substantial decrease of the FP value. In addition, Lee group developed an FP approach for nucleic acid tests that used DNA polymerase to accomplish reverse transcription (RT-) PCR and recycle for the extension reaction on the reporter probes.98 As we mentioned above, different enzymes combined with nanomaterials amplification techniques have been proven to be increasingly valuable for quantitative analysis of various target molecules, such as nicking enzyme assisted GO-based dual signal amplification,82 T7 exonuclease assisted polystyrene nanospheres dual-cycle signal amplification.97 Moreover, enzymes combined with protein amplification FA/FP assays have been exploited to the sense of small molecules.56,99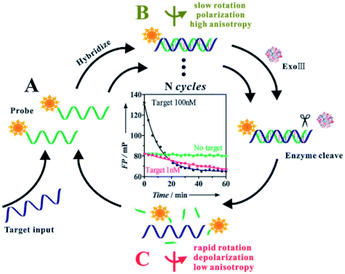 | ||
| Fig. 9 Target assisted Exonuclease III-catalyzed amplification FP for DNA detection. Reprinted with permission from ref. 34. Copyright (2011) Royal Society of Chemistry. | ||
4.4 Target-catalyzed DNA cyclic assembly amplification strategies
The protein enzymes usually can achieve cyclic amplification FA/FP signal. However, as they can be denatured and digested by proteases easily, it is hard to operate them in complex biological samples.100,101 Enzyme-free signal amplification strategies based on toehold-mediated DNA strand displacement have been developed to amplify FA/FP signal, which works under mild conditions without specific demand for either ionic strength or temperature comparing with enzyme amplification. Tan group reported a target-triggered assembly approach based on the formation of DNA–protein hybrid nanowires via HCR, using ATP as a small molecule model.12 This novel dual-amplified, aptamer-based FA assay affords high sensitivity with a detection limit of 100 nM for ATP. Zhen et al. developed the new target-catalyzed hairpin assembly (CHA), enzyme-free DNA circuit, and assisted GO amplified FA strategy for miRNA detection.83,102 Fig. 10 depicts the principles of CHA assay. The fluorescent dye, carboxytetramethylrhodamine (TAMRA), modified probe DNA was first hybridized with capture DNA containing an A20 tail. This design can not only improve the sensitivity of FA detection but also can decrease the fluorescence quenching effect of GO, thus ensuring the accuracy of FA detection.9,27 In the presence of miRNA-21, the CHA was initiated and plenty of H1–H2 duplexes were produced continuously. The obtained H1–H2 duplex could trigger the toehold-mediated strand exchange reaction to form an H1–H2-probe DNA complex that detached the dye-modified probe DNA from the GO surface, leading to a decreased FA of the system. The detection limit of this method was 47 pM, which was 279 times lower than that of the method without CHA. And the selectivity of this method has also been enhanced greatly compared with the method without CHA.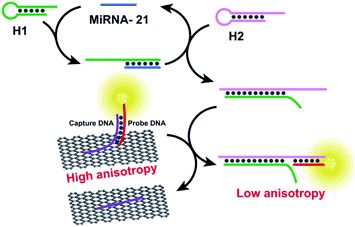 | ||
| Fig. 10 Schematic illustration of enzyme-free FA assay for miRNA-21 detection. Reprinted with permission from ref. 83. Copyright (2017) American Chemical Society. | ||
4.5 Oligonucleotides amplification strategies
The oligonucleotides enhanced FA/FP assay has been generally researched, which is mainly by changing the structure and sequence of the oligonucleotides chain. Zhao group developed a K+-mediated G-quadruplex formation enhancement FP sensor based on CdS–CdTe core–shell quantum dots (QDs) that was constructed for detection of Hg2+ and biothiols.103 In this study (Fig. 11), CdTe/CdS QD functionalized with T-rich DNA with the affinity of biotin–streptavidin as fluorescence polarization probe, and the 50-guanine-rich sequence of G-rich DNA folded into a G-quadruplex in the presence of potassium ions. In the presence of Hg2+, the 3′-thymine-rich sequence of the K+-mediated G-quadruplex (GQ-DNA) and QD-T can form large volume complexes through the strong and specific affinity of T–Hg2+–T. As a consequence, the K+-mediated G-quadruplex leads to dramatic changes in the molecular volume of QDs, thus the FP value of the QDs will significantly increase. Upon the addition of biothiol amino acids, the strong reaction of thiol–Hg2+ prohibits the formation of the T–Hg2+–T complexes and makes the QD-T and the K+-mediated G-quadruplex (GQ-DNA) stay in a free single state. Subsequently, the FP values of the solution greatly decrease. Based on the decrease of FP values, the detection of biothiols can also be realized in this method.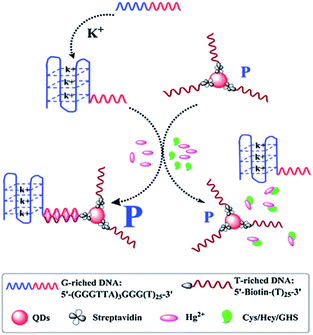 | ||
| Fig. 11 Schematic illustration of the QD FP enhancement homogenous system based on K+-mediated G-quadruplex formation for the determination of Hg2+ and biothiols. Reprinted with permission from ref. 103. Copyright (2014) Royal Society of Chemistry. | ||
In addition, by taking advantage of the binding-induced change of the TMR−G interaction, Wang et al. demonstrated a series of enhanced FA assays for nucleic acid aptamer affinity analysis. These FA strategies allow for analysis of proteins, small molecules, metal ion and DNAzyme activity, showing high sensitivity and selectivity.5,20,36,44,104–106 Peyrin et al. designed a FA aptamer sensing platform dedicated to small molecule detection that relied on enhanced fluctuations of segmental motion dynamics of the aptamer tracer mediated by an unlabelled, partially complementary oligonucleotide.107 Furthermore, by using the accumulation of a lot of DNA in fluorescence indicator-graphene QDs to enhance FA, Hosseini group achieved DNA methyltransferase activity detection.108
4.6 Other amplification
In addition to the signal amplification methods listed above, the enhancing FA/FP assays by resonance energy transfer (RET) and Tween 20 have been used for biomolecules analysis. Taking advantage of the impact of RET on FP, Ren et al. developed a RET-enhanced FA strategy for biomolecules detection.109 This method used streptavidin Alexa Fluors 488 conjugated (SA-488), nanogold and biotinylated substrate peptide (biotin–subpeptide) which connects the SA-488 and nanogold to construct a fluorescence variable probe. When the fluorescence molecule conjugates with the nanogold, its volume and mass will increase. Besides, its fluorescence intensity (FI) will be suppressed by the RET, leading to a decrease in its effective concentration. According to the FP principle, both the decrease in concentration and the increase in volume will lead to an increase in FP. When adding target molecules, the conjugation between nanogold and SA-488 will be blocked, resulting in a decreased FP value. Based on the change in FP, trypsin and biotin were detected in this study. In addition, by using Lissamine Rhodamine B labeled ochratoxin A (OTA) as a fluorescence probe, Zhao et al. developed a competitive aptamer FA assays for OTA.110 In this contribution (Fig. 12), the fluorescent probe can interact with Tween 20. After adding a target, the fluorescent probe shows a higher FA value than that of aptamer–fluorescent probe complex, achieving target sensitive detection through determining the reduced FA value.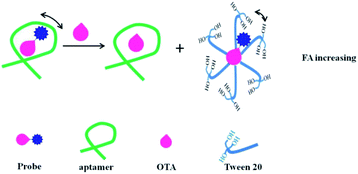 | ||
| Fig. 12 Schematic of competitive aptamer FA assay for the determination of OTA with signal-on responses in a binding buffer that contains a 0.1% Tween 20. Reprinted with permission from ref. 110. Copyright (2020) American Chemical Society. | ||
5. Conclusion and outlook
The measurement ratiometric nature, the homogeneous and high-throughput format, the single dye labeling, and the potential to generate response depending on fluorophore diffusional rotation and/or photochemical changes constitute advantages of the FA/FP technique. Regarding sensitivity, many strategies have been proposed (see Table 1), such as the combination of enzyme-catalyzed target, the application of long-wavelength fluorophores, and signal enhancement approaches using nanomaterials including Ag/Au/SiO2 nanoparticles, GO and others. The development of signal amplification methods of FA/FP effectively expanded the application in clinical chemistry and bioassays.| Targets | Amplification technique | Signal mechanisms | Dynamic ranges | Detection limit | Samples | Ref. |
|---|---|---|---|---|---|---|
| Adenosine | GO amplification | Mass change-based | 60–400 μM | 22 μM | — | 9 |
| Adenosine | Single-stranded DNA binding protein enhancer | Mass change-based | 2–50 μM | 1 μM | Human serum | 11 |
| Adenosine | Phosphodiesterase I and protein amplification | Mass change-based | 0.5–1000 μM | 500 nM | Cell media | 56 |
| Adenosine | GO, nicking enzyme dual amplification | Mass change-based | 4 pM to 10 μM | 2.0 pM | — | 82 |
| Adenosine | Oligonucleotide amplification | Segmental motion change-based | 0–6 μM | ∼1 μM | — | 107 |
| ATP | HCR, protein dual amplification | Mass change-based | 0.2–20 μM | 100 nM | Cell media, human urine, human serum | 12 |
| ATP | Protein amplification | Mass change-based | 1–25 μM | 0.5 μM | Cell media | 28 |
| ATP | GO amplification | Mass change-based | 0.5–250 μM | 100 nM | Human serum | 31 |
| ATP | Digoxin antibody amplification | Mass change-based | 10–350 μM | 3.7 μM | Serum | 111 |
| ATP | Protein, proximity effect amplification | Mass change-based, segmental motion change-based | 0.5–500 μM | 0.5 μM | Human urine | 53 |
| ATP | SiO2NPs enhancement | Mass change-based | 40 pM to 100 μM | 20 pM | — | 84 |
| ATP | MoS2 nanosheet | Mass change-based | 0.3–40 μM | 34.4 nM | Human urine | 94 |
| ATP | DNA nanosheet amplification | Mass change-based | 150–450 μM | 43 μM | — | 95 |
| Thrombin | GO amplification | Mass change-based | 0.5–4 mg L−1 | 0.19 mg L−1 | — | 9 |
| Thrombin | GO, nicking enzyme dual amplification | Mass change-based | 2 fM to 200 nM | 1 fM | Human plasma | 82 |
| Thrombin | TiS2 nanosheet, Exo I amplification | Mass change-based | 0.05 pM to 100 nM | 0.01 pM | Human serum | 93 |
| Thrombin | CSDA and PS NP enhancement | Mass change-based | 50 aM to 100 pM | 28 aM | Human plasma | 96 |
| ssDNA | GO amplification | Mass change-based | 8–40 nM | 4.6 nM | — | 9 |
| ssDNA | AuNPs amplification | Mass change-based | — | 0.95 nM | — | 30 |
| ssDNA | MOF amplification | Mass change-based | 0.3–12 nM | 0.2 nM | — | 32 |
| ssDNA | Exo III amplification | Mass change-based | 0.1 fM to 1 nM | 83 aM | — | 34 |
| ssDNA | AuNPs amplification | Mass change-based | 150 pM to 6 nM | 73 pM | Serum | 60 |
| ssDNA | AuNPs, NSET dual amplification | Fluorescence lifetime-based, mass change-based | — | 372 pM | DMEM | 66 |
| ssDNA | GO, T7 Exo-assisted amplification | Mass change-based | 50–2000 pM | 38.6 pM | Human serum | 81 |
| ssDNA | SiO2NPs amplification | Mass change-based | 1–800 nM | 0.65 nM | Human serum | 85 |
| ssDNA | HCR and SiO2NPs amplification | Mass change-based | 0–2.5 nM | 34 pM | Human serum | 86 |
| ssDNA | MOF amplification | Mass change-based | 1–20 nM | 1 nM | — | 89 |
| ssDNA | DNA nanosheet amplification | Mass change-based | 10–50 nM | 8 nM | — | 95 |
| miRNA | Protein amplification | Mass change-based | 10 pM to 10 nM | 8.5 pM | Cell lysis buffer, cell lysate | 29 |
| miRNA | GO and CHA amplification | Mass change-based | 0–16 nM | 47 pM | Cell extractions | 83 |
| miRNA | T7 Exo, polystyrene nanospheres amplification | Mass change-based | 0.001–10 nM | 0.001 nM | Human serum, cell extractions | 97 |
| Hg2+ | AuNPs amplification | Mass change-based | 1 nM to 1 mM | 1 nM | River water | 65 |
| Hg2+ | AgNPs amplification | Mass change-based | 10–400 nM | 6.6 nM | Tap water | 67 |
| Hg2+ | MOF amplification | Mass change-based | 20–200 nM | 8.66 nM | Tap water | 88 |
| Hg2+ | K+-mediated G-quadruplex enhancement | Mass change-based | 10–800 nM | 8.6 nM | Lake water | 103 |
| Cu2+ | GO and DNAzyme amplification | Mass change-based | 1–32 nM | 1 nM | — | 58 |
| Cu2+ | DNAzyme self-assembled AuNPs amplification | Mass change-based | 0.001–10 μM | ∼1 nM | River water | 61 |
| Cu2+ | DNAzyme self-assembled gold nanorods amplification | Mass change-based | 8–320 pM | 8.40 pM | — | 62 |
| Cu2+ | SiO2 NPs amplification | Mass change-based | 0.050–2.0 μM | 0.0178 μM | River water | 87 |
| Pb2+ | Multiple G bases amplification | Fluorescence lifetime-based | 200 pM to 100 nM | 100 pM | — | 5 |
| Pb2+ | Phosphate-perylene modification G-quadruplex probes amplification | Segmental motion change-based | 25–5000 nM | 24.5 nM | Tap water | 13 |
| Pb2+ | Cleavable DNA–protein hybrid molecular beacon amplification | Mass change-based, segmental motion change-based | 1–20 nM | 0.5 nM | Water samples | 57 |
| Pb2+ | DNAzyme self-assembled AuNPs amplification | Mass change-based | 0.001–10 μM | 1 nM | River water | 61 |
| Ag+ | AuNPs amplification | Mass change-based | 50–750 nM | 9.5 nM | Tap water | 64 |
| Ag+ | MnO2 nanosheet amplification | Mass change-based | 30–240 nM | 9.1 nM | Tap water, lake water | 92 |
| Cocaine | Protein amplification | Mass change-based | 1–100 μM | 0.8 μM | Human urine | 28 |
| Cocaine | Protein, isothermal exponential amplification | Mass change-based | 30 pM to 30 μM | 18 pM | Human serum | 99 |
| Aflatoxin B1 | Protein, proximity effect amplification | Mass change-based, segmental motion change-based | 60 pM to 125 nM | 60 pM | White wine | 53 |
| Aflatoxin B1 | GO amplification | Mass change-based | 0.05–5 nM | 0.05 nM | Rice extract | 78 |
| Aflatoxin B1 | Protein, isothermal exponential amplification | Mass change-based | 0.4 pM to 400 nM | 0.24 pM | — | 99 |
| Ochratoxin A | Protein amplification | Mass change-based | — | 3.6 nM | White wine | 51 |
| Ochratoxin A | Protein, proximity effect amplification | Mass change-based, segmental motion change-based | 1 nM to 5 μM | 1 nM | White wine | 53 |
| Cysteine | AgNPs amplification | Mass change-based | 20–700 nM | 11 nM | — | 67 |
| Cysteine | K+-mediated G-quadruplex enhancement | Mass change-based | 50–2000 nM | 9.9 nM | Human urine | 103 |
| Ricin B-chain | GO, Exo III-assisted amplification | Mass change-based | 1–13.3 μg mL−1 | 400 ng mL−1 | Orange juice | 27 |
| K+ | GO amplification | Mass change-based | 10 μM to 2 mM | 1 μM | — | 33 |
| DNA–protein interactions | Protein amplification | Mass change-based | — | 6.3 nM | — | 54 |
| Chloramphenicol | PCR and protein amplification | Mass change-based | 0.001–200 nM | 0.5 pM | Honey | 55 |
| EcoRI endonuclease | AuNPs amplification | Mass change-based | 5.0 × 10−4 to 10 U mL−1 | 5.0 × 10−4 U mL−1 | — | 59 |
| Lac | MEF, Ag10NPs amplification | Mass change-based, fluorescence lifetime-based | 0.2 ng mL−1 to 25 μg mL−1 | 1.25 pM | Milk powder | 68 |
| DNA MTase activity | Carbon nanotube signal amplification | Mass change-based | — | 1.0 × 10−4 U mL−1 | Human serum | 74 |
| Apyrase | Carbon nanoparticle amplification | Mass change-based | 0.1–0.5 U μL−1 | 0.05 U μL−1 | — | 75 |
| Lipopolysaccharides (Salmonella enterica serotype Typhimurium) | GO amplification | Mass change-based | — | 38.7 ng mL−1 | Sodium chloride injection | 79 |
| I− | MOF amplification | Mass change-based | 0.02–3.0 μM | 17.4 nM | Tap water | 88 |
| PSA | MOG amplification | Mass change-based | 0.5–8 ng mL−1 | 0.33 ng mL−1 | Human serum | 90 |
| Folate receptor | TiS2 nanosheet, exo I amplification | Mass change-based | 0.01–20 ng mL−1 | 0.003 ng mL−1 | Human serum | 93 |
| DNA glycosylase activity | WS2 nanosheet and exo III amplification | Mass change-based | 0.00080–0.40 U mL−1 | ∼0.00030 U mL−1 | Cell extracts | 91 |
At present, most of the developed strategies used aptamer as a target recognition unit. However, only a handful of good nucleic acid aptamers are available. The selection of aptamers binding to small molecules is challenging, partly due to the smaller molecular space for interaction. As a consequence, most of the assays on this topic have repeatedly targeted ATP, adenosine, cocaine, ochratoxin A, and thrombin, because aptamers of high affinity and selectivity are available. To solve this problem, the properties of new technologies should be clear, that is, the approaches for the selection and design of aptamers are needed to generate high-quality aptamers for diverse applications. In addition, almost all the established FA/FP sensors for living cell analysis and reaching the commercial level are still fairly rare because of the complexity of the real biological matrix. Furthermore, the implementation of many amplification mechanisms can further improve both the assay response and analyte detection limit, but at the same time at the expense of the biosensor simplicity in most cases, it will result in increased testing costs. Up to now, most of the developed FA/FP strategies amplify the signal by altering the size of the fluorescent complexes (see Table 1). It is highly demanded to combine with other paths as FA/FP enhancers. Finally, there are few articles focusing on the effect of fluorescence lifetime on FA/FP signal, the research in this direction may further expand the application of FA/FP.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the Fundamental Research Funds for the Central Universities, Southwest Minzu University (No. 2018NQN38) and the National Natural Science Foundation of China (No. 21974109).References
- G. Weber, Biochem. J., 1952, 51, 155–167 CrossRef CAS PubMed.
- G. Weber, Biochem. J., 1952, 51, 145–155 CrossRef CAS PubMed.
- D. M. Jameson and J. A. Ross, Chem. Rev., 2010, 110, 2685–2708 CrossRef CAS PubMed.
- H. Y. Zhang, S. P. Yang, K. De Ruyck, N. V. Beloglazova, S. A. Eremin, S. De Saeger, S. X. Zhang, J. Z. Shen and Z. H. Wang, TrAC, Trends Anal. Chem., 2019, 114, 293–313 CrossRef CAS.
- D. Zhang, R. Fu, Q. Zhao, H. Rong and H. Wang, Anal. Chem., 2015, 87, 4903–4909 CrossRef CAS PubMed.
- Q. Zhao, J. Tao, J. S. Uppal, H. Peng, H. Wang and X. C. Le, TrAC, Trends Anal. Chem., 2019, 110, 401–409 CrossRef CAS.
- T. Jia, C. Y. Fu, C. S. Huang, H. T. Yang and N. Q. Jia, ACS Appl. Mater. Interfaces, 2015, 7, 10013–10021 CrossRef CAS PubMed.
- N. Ehrich, A. L. Christensen and D. Stamou, Anal. Chem., 2011, 83, 8169–8176 CrossRef PubMed.
- X. Xiao, Y. F. Li, C. Z. Huang and S. J. Zhen, Chem. Commun., 2015, 51, 16080–16083 RSC.
- D. S. Smith and S. A. Eremin, Anal. Bioanal. Chem., 2008, 391, 1499–1507 CrossRef CAS PubMed.
- Z. Zhu, C. Ravelet, S. Perrier, V. Guieu, E. Fiore and E. Peyrin, Anal. Chem., 2012, 84, 7203–7211 CrossRef CAS PubMed.
- B. Yang, X.-B. Zhang, L.-P. Kang, G.-L. Shen, R.-Q. Yu and W. Tan, Anal. Chem., 2013, 85, 11518–11523 CrossRef CAS PubMed.
- Z. X. Wang, X. J. Pei, N. Li and X. J. Tang, J. Mater. Chem. B, 2016, 4, 4330–4336 RSC.
- H. R. Zhang, Q. Wu and M. Y. Berezin, Expert Opin. Drug Discovery, 2015, 10, 1145–1161 CrossRef PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, 2006 Search PubMed.
- S. Perrier, V. Guieu, B. Chovelon, C. Ravelet and E. Peyrin, Anal. Chem., 2018, 90, 4236–4248 CrossRef CAS PubMed.
- O. D. Hendrickson, N. A. Taranova, A. V. Zherdev, B. B. Dzantiev and S. A. Eremin, Sensors, 2020, 20, 33 CrossRef PubMed.
- X. A. Ton, V. Acha, K. Haupt and T. S. B. Bernadette, Biosens. Bioelectron., 2012, 36, 22–28 CrossRef CAS PubMed.
- Y.-C. Chen, Z. Wang, M. Yan and S. A. Prahl, Luminescence, 2006, 21, 7–14 CrossRef CAS PubMed.
- Q. Zhao, Q. Lv and H. Wang, Anal. Chem., 2014, 86, 1238–1245 CrossRef CAS PubMed.
- Y. Wang, J. Killian, K. Hamasaki and R. R. Rando, Biochemistry, 1996, 35, 12338–12346 CrossRef CAS PubMed.
- J. A. Cruz-Aguado and G. Penner, Anal. Chem., 2008, 80, 8853–8855 CrossRef CAS PubMed.
- Z. Y. Zhu, T. Schmidt, M. Mahrous, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Chim. Acta, 2011, 707, 191–196 CrossRef CAS PubMed.
- J. Ruta, S. Perrier, C. Ravelet, J. Fize and E. Peyrin, Anal. Chem., 2009, 81, 7468–7473 CrossRef CAS PubMed.
- A. Kidd, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Bioanal. Chem., 2011, 401, 3229–3234 CrossRef CAS PubMed.
- S. Perrier, C. Ravelet, V. Guieu, J. Fize, B. Roy, C. Perigaud and E. Peyrin, Biosens. Bioelectron., 2010, 25, 1652–1657 CrossRef CAS PubMed.
- X. Xiao, J. Tao, H. Z. Zhang, C. Z. Huang and S. J. Zhen, Biosens. Bioelectron., 2016, 85, 822–827 CrossRef CAS PubMed.
- L. Cui, Y. Zou, N. Lin, Z. Zhu, G. Jenkins and C. J. Yang, Anal. Chem., 2012, 84, 5535–5541 CrossRef CAS PubMed.
- Y. C. He, B. C. Yin, L. H. Jiang and B. C. Ye, Chem. Commun., 2014, 50, 6236–6239 RSC.
- X. Wang, M. Zou, H. Huang, Y. Ren, L. Li, X. Yang and N. Li, Biosens. Bioelectron., 2013, 41, 569–575 CrossRef CAS PubMed.
- J. Liu, C. Wang, Y. Jiang, Y. Hu, J. Li, S. Yang, Y. Li, R. Yang, W. Tan and C. Z. Huang, Anal. Chem., 2013, 85, 1424–1430 CrossRef CAS PubMed.
- J. F. Guo, C. M. Li, X. L. Hu, C. Z. Huang and Y. F. Li, RSC Adv., 2014, 4, 9379–9382 RSC.
- S. J. Zhen, Y. Yu, C. M. Li and C. Z. Huang, Analyst, 2015, 140, 353–357 RSC.
- M. Zhang, Y.-M. Guan and B.-C. Ye, Chem. Commun., 2011, 47, 3478–3480 RSC.
- Z. Zhu, T. Schmidt, M. Mahrous, V. Guieu, S. Perrier, C. Ravelet and E. Peyrin, Anal. Chim. Acta, 2011, 707, 191–196 CrossRef CAS PubMed.
- D. Zhang, L. Yin, Z. Meng, A. Yu, L. Guo and H. Wang, Anal. Chim. Acta, 2014, 812, 161–167 CrossRef CAS PubMed.
- Q. Zhao, J. Tao, W. Feng, J. S. Uppal, H. Peng and X. C. Le, Anal. Chim. Acta, 2020, 1125, 267–278 CrossRef CAS PubMed.
- J. Chen, J. Liu, X. Chen and H. Qiu, Chin. Chem. Lett., 2019, 30, 1575–1580 CrossRef CAS.
- G. Mocz, J. Fluoresc., 2006, 16, 511–524 CrossRef CAS PubMed.
- R. A. Potyrailo, R. C. Conrad, A. D. Ellington and G. M. Hieftje, Anal. Chem., 1998, 70, 3419–3425 CrossRef CAS PubMed.
- X. Fang, Z. Cao, T. Beck and W. Tan, Anal. Chem., 2001, 73, 5752–5757 CrossRef CAS PubMed.
- T. G. McCauley, N. Hamaguchi and M. Stanton, Anal. Biochem., 2003, 319, 244–250 CrossRef CAS.
- G. Gokulrangan, J. R. Unruh, D. F. Holub, B. Ingram, C. K. Johnson and G. S. Wilson, Anal. Chem., 2005, 77, 1963–1970 CrossRef CAS PubMed.
- D. Zhang, Q. Zhao, B. Zhao and H. Wang, Anal. Chem., 2012, 84, 3070–3074 CrossRef CAS PubMed.
- H. Huang, H. Wei, M. Zou, X. Xu, B. Xia, F. Liu and N. Li, Anal. Chem., 2015, 87, 2748–2754 CrossRef CAS PubMed.
- M. Zou, Y. Chen, X. Xu, H. Huang, F. Liu and N. Li, Biosens. Bioelectron., 2012, 32, 148–154 CrossRef CAS PubMed.
- X. Y. Hong, H. D. Huang, M. X. Chen, F. Liu and N. Li, Anal. Methods, 2016, 8, 3156–3162 RSC.
- X. Geng, D. P. Zhang, H. L. Wang and Q. Zhao, Anal. Bioanal. Chem., 2013, 405, 2443–2449 CrossRef CAS PubMed.
- Y. P. Li, L. L. Sun and Q. Zhao, Talanta, 2017, 174, 7–13 CrossRef CAS PubMed.
- E. Goux, Q. Lespinasse, V. Guieu, S. Perrier, C. Ravelet, E. Fiore and E. Peyrin, Methods, 2016, 97, 69–74 CrossRef CAS PubMed.
- A. V. Samokhvalov, I. V. Safenkova, S. A. Eremin, A. V. Zherdev and B. B. Dzantiev, Anal. Chim. Acta, 2017, 962, 80–87 CrossRef CAS PubMed.
- M. Hafner, E. Vianini, B. Albertoni, L. Marchetti, I. Grune, C. Gloeckner and M. Famulok, Nat. Protoc., 2008, 3, 579–587 CrossRef CAS PubMed.
- Y. P. Li and Q. Zhao, Anal. Chem., 2019, 91, 7379–7384 CrossRef CAS PubMed.
- J. W. Choi, B. G. Jo, A. J. deMello, J. Choo and H. Y. Kim, Analyst, 2016, 141, 6499–6502 RSC.
- P. F. Ma, H. Ye, J. Y. Deng, I. M. Khan, L. Yue and Z. P. Wang, Talanta, 2019, 205, 7 Search PubMed.
- L. P. Kang, B. Yang, X. B. Zhang, L. Cui, H. M. Meng, L. Mei, C. C. Wu, S. L. Ren and W. H. Tan, Anal. Chim. Acta, 2015, 879, 91–96 CrossRef CAS PubMed.
- P. Hu and B. Yang, Talanta, 2016, 147, 276–281 CrossRef CAS PubMed.
- Y. Yu, Y. Liu, S. J. Zhen and C. Z. Huang, Chem. Commun., 2013, 49, 1942–1944 RSC.
- Y. Huang, S. L. Zhao, Z. F. Chen, Y. C. Liu and H. Liang, Chem. Commun., 2011, 47, 4763–4765 RSC.
- S. P. Liang, G. C. He, J. N. A. Tian, Y. C. Zhao and S. L. Zhao, Microchim. Acta, 2018, 185, 8 CrossRef PubMed.
- B. C. Yin, P. Zuo, H. Huo, X. H. Zhong and B. C. Ye, Anal. Biochem., 2010, 401, 47–52 CrossRef CAS PubMed.
- Y. He, J. Tian, J. Zhang, S. Chen, Y. Jiang, K. Hu, Y. Zhao and S. Zhao, Biosens. Bioelectron., 2014, 55, 285–288 CrossRef CAS PubMed.
- Y. Gao, J. Xu, B. Li and Y. Jin, ACS Appl. Mater. Interfaces, 2016, 8, 13707–13713 CrossRef CAS PubMed.
- G. K. Wang, S. L. Wang, C. L. Yan, G. Y. Bai and Y. F. Liu, Colloids Surf., B, 2018, 167, 150–155 CrossRef CAS PubMed.
- B.-C. Ye and B.-C. Yin, Angew. Chem., Int. Ed., 2008, 47, 8386–8389 CrossRef CAS.
- B. Wang, D. H. Ren, Z. You, Y. Yalikun and Y. Tanaka, Analyst, 2018, 143, 11 Search PubMed.
- Y. X. Jiang, J. N. Tian, K. Hu, Y. C. Zhao and S. L. Zhao, Microchim. Acta, 2014, 181, 1423–1430 CrossRef CAS.
- Z. Chen, H. Li, W. C. Jia, X. H. Liu, Z. M. Li, F. Wen, N. Zheng, J. D. Jiang and D. K. Xu, Anal. Chem., 2017, 89, 5901–5909 Search PubMed.
- N. Li and C. M. Ho, J. Am. Chem. Soc., 2008, 130, 2380–2381 CrossRef CAS PubMed.
- C. H. Lu, H. H. Yang, C. L. Zhu, X. Chen and G. N. Chen, Angew. Chem., Int. Ed., 2009, 48, 4785–4787 CrossRef CAS PubMed.
- Z. Zhu, Z. W. Tang, J. A. Phillips, R. H. Yang, H. Wang and W. H. Tan, J. Am. Chem. Soc., 2008, 130, 10856–10857 CrossRef CAS PubMed.
- X. Y. Ouyang, R. Q. Yu, J. Y. Jin, J. S. Li, R. H. Yang, W. H. Tan and J. L. Yuan, Anal. Chem., 2011, 83, 782–789 CrossRef CAS PubMed.
- Y. Wang, Z. H. Li, D. H. Hu, C. T. Lin, J. H. Li and Y. H. Lin, J. Am. Chem. Soc., 2010, 132, 9274–9276 CrossRef CAS PubMed.
- Y. Huang, M. Shi, L. M. Zhao, S. L. Zhao, K. Hu, Z. F. Chen, J. Chen and H. Liang, Biosens. Bioelectron., 2014, 54, 285–291 CrossRef CAS PubMed.
- J. H. Liu, J. Yu, J. R. Chen, R. H. Yang and K. M. Shih, Mater. Sci. Eng., C, 2014, 38, 206–211 CrossRef CAS PubMed.
- N. Mohanty and V. Berry, Nano Lett., 2008, 8, 4469–4476 CrossRef CAS PubMed.
- F. Kim, L. J. Cote and J. X. Huang, Adv. Mater., 2010, 22, 1954–1958 CrossRef CAS PubMed.
- H. Ye, Q. Q. Lu, N. Duan and Z. P. Wang, Anal. Bioanal. Chem., 2019, 411, 1107–1115 CrossRef CAS PubMed.
- H. Ye, N. Duan, H. J. Gu, H. T. Wang and Z. P. Wang, Microchim. Acta, 2019, 186, 8 CrossRef PubMed.
- L. Qi, Y. Y. Fan, H. Wei, D. Zhang and Z. Q. Zhang, Sens. Actuators, B, 2018, 257, 666–671 CrossRef CAS.
- L. J. Wang, J. N. Tian, W. Yang, Y. C. Zhao and S. L. Zhao, Luminescence, 2016, 31, 573–579 CrossRef CAS PubMed.
- Y. Huang, X. Liu, L. Zhang, K. Hu, S. Zhao, B. Fang, Z.-F. Chen and H. Liang, Biosens. Bioelectron., 2015, 63, 178–184 CrossRef CAS PubMed.
- S. J. Zhen, X. Xiao, C. H. Li and C. Z. Huang, Anal. Chem., 2017, 89, 8766–8771 CrossRef CAS PubMed.
- Y. Huang, S. Zhao, Z.-F. Chen, M. Shi and H. Liang, Chem. Commun., 2012, 48, 7480–7482 RSC.
- J. Chen, Q. Chen, C. J. Gao, M. L. Zhang, B. Qin and H. D. Qiu, J. Mater. Chem. B, 2015, 3, 964–967 RSC.
- J. Zhao, Z. D. Chu, X. Jin and S. L. Zhao, Sens. Actuators, B, 2015, 209, 116–121 CrossRef CAS.
- S. Q. Liao, J. J. Zhao, Y. F. Qin and S. L. Zhao, RSC Adv., 2017, 7, 55668–55672 RSC.
- J. M. Fang, P. F. Gao, X. L. Hu and Y. F. Li, RSC Adv., 2014, 4, 37349–37352 RSC.
- J. F. Guo, R. M. Fang, C. Z. Huang and Y. F. Li, RSC Adv., 2015, 5, 46301–46306 RSC.
- T. T. Zhao, Z. W. Peng, D. Yuan, S. J. Zhen, C. Z. Huang and Y. F. Li, Spectrochim. Acta, Part A, 2018, 192, 328–332 CrossRef CAS PubMed.
- J. Zhao, Y. Ma, R. Kong, L. Zhang, W. Yang and S. Zhao, Anal. Chim. Acta, 2015, 887, 216–223 CrossRef CAS PubMed.
- L. Qi, Z. Yan, Y. Huo, X. M. Hai and Z. Q. Zhang, Biosens. Bioelectron., 2017, 87, 566–571 CrossRef CAS PubMed.
- X. Li, X. L. Ding, Y. F. Li, L. S. Wang and J. Fan, Nanoscale, 2016, 8, 9852–9860 RSC.
- Y. Y. Fan, Z. L. Mou, M. Wang, J. Li, J. Zhang, F. Q. Dang and Z. Q. Zhang, Anal. Chem., 2018, 90, 13708–13713 CrossRef CAS PubMed.
- Y. X. Liu, X. Xiao, C. H. Li, C. Men, Q. C. Ye, W. Y. Lv, Y. F. Li, C. Z. Huang and S. J. Zhen, Talanta, 2020, 211, 120730 CrossRef CAS PubMed.
- Y. Huang, X. Q. Liu, H. K. Huang, J. Qin, L. L. Zhang, S. L. Zhao, Z. F. Chen and H. Liang, Anal. Chem., 2015, 87, 8107–8114 CrossRef CAS PubMed.
- X. T. Li, N. Huang, L. L. Zhang, J. J. Zhao and S. L. Zhao, Talanta, 2019, 202, 297–302 CrossRef CAS PubMed.
- K. S. Park, R. C. Charles, E. T. Ryan, R. Weissleder and H. Lee, Chem.–Eur. J., 2015, 21, 16359–16363 CrossRef CAS PubMed.
- H. K. Huang, J. Qin, K. Hu, X. Q. Liu, S. L. Zhao and Y. Huang, RSC Adv., 2016, 6, 86043–86050 RSC.
- C. Chen, N. Li, J. Lan, X. Ji and Z. He, Anal. Chim. Acta, 2016, 902, 154–159 CrossRef CAS PubMed.
- Y. Kim, J. Gonzales and Y. B. Zheng, Small, 2021, 17, 2004988 CrossRef CAS PubMed.
- J. Tao, X. Xiao, C. Li, C. Men and S. Zhen, Sci. China: Chem., 2018, 48, 85 Search PubMed.
- J. Zhang, J. Tian, Y. He, Y. Zhao and S. Zhao, Chem. Commun., 2014, 50, 2049–2051 RSC.
- D. Zhang, H. Shen, G. Li, B. Zhao, A. Yu, Q. Zhao and H. Wang, Anal. Chem., 2012, 84, 8088–8094 CrossRef CAS PubMed.
- D. P. Zhang and H. L. Wang, Anal. Chem., 2019, 91, 14538–14544 CrossRef CAS PubMed.
- Y. Liu and Q. Zhao, Anal. Bioanal. Chem., 2017, 409, 3993–4000 CrossRef CAS PubMed.
- S. Perrier, P. Bouilloud, G. D. Coelho, M. Henry and E. Peyrin, Biosens. Bioelectron., 2016, 82, 155–161 CrossRef CAS PubMed.
- H. A. Kermani, M. Hosseini, M. Dadmehr, S. Hosseinkhani and M. R. Ganjali, Sens. Actuators, B, 2017, 241, 217–223 CrossRef CAS.
- D. H. Ren, J. Wang, B. Wang and Z. You, Biosens. Bioelectron., 2016, 79, 802–809 CrossRef CAS PubMed.
- Y. P. Li, N. Zhang, H. L. Wang and Q. Zhao, J. Agric. Food Chem., 2020, 68, 4277–4283 CrossRef CAS PubMed.
- J. N. Tian, Y. Wang, S. Chen, Y. X. Jiang, Y. C. Zhao and S. L. Zhao, Microchim. Acta, 2013, 180, 203–209 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |


