Open Access Article
Open Access ArticleEnvironmental protection based on the nanobiosensing of bacterial lipopolysaccharides (LPSs): material and method overview
Ahmad Mobed *abc and
Mohammad Hasanzadeh
*abc and
Mohammad Hasanzadeh *cd
*cd
aAging Research Institute, Faculty of Medicine, Tabriz University of Medical Sciences, Iran. E-mail: Mobeda@tbzmed.ac.ir
bPhysical Medicine and Rehabilitation Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
cPharmaceutical Analysis Research Center, Tabriz University of Medical Sciences, Tabriz 51664, Iran. E-mail: hasanzadehm@tbzmed.ac.ir
dNutrition Research Center, Tabriz University of Medical Sciences, Tabriz, Iran
First published on 28th March 2022
Abstract
Lipopolysaccharide (LPS) or endotoxin control is critical for environmental and healthcare issues. LPSs are responsible for several infections, including septic and shock sepsis, and are found in water samples. Accurate and specific diagnosis of endotoxin is one of the most challenging issues in medical bacteriology. Enzyme-linked immunosorbent assay (ELISA), plating and culture-based methods, and Limulus amebocyte lysate (LAL) assay are the conventional techniques in quantifying LPS in research and medical laboratories. However, these methods have been restricted due to their disadvantages, such as low sensitivity and time-consuming and complicated procedures. Therefore, the development of new and advanced methods is demanding, particularly in the biological and medical fields. Biosensor technology is an innovative method that developed extensively in the past decade. Biosensors are classified based on the type of transducer and bioreceptor. So in this review, various types of biosensors, such as optical (fluorescence, SERS, FRET, and SPR), electrochemical, photoelectrochemical, and electrochemiluminescence, on the biosensing of LPs were investigated. Also, the critical role of advanced nanomaterials on the performance of the above-mentioned biosensors is discussed. In addition, the application of different labels on the efficient usage of biosensors for LPS is surveyed comprehensively. Also, various bio-elements (aptamer, DNA, miRNA, peptide, enzyme, antibody, etc.) on the structure of the LPS biosensor are investigated. Finally, bio-analytical parameters that affect the performance of LPS biosensors are surveyed.
1. Introduction
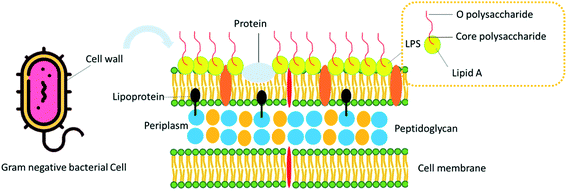
Gram-negative bacteria are described by an envelope that contains two membranes: an outer membrane (OM) that separates the cell from its environment and an inner membrane (IM) that surrounds cytoplasmic components.1,2 Gram-negative bacteria envelope surrounds periplasm that is an aqueous cellular compartment, which encloses the peptidoglycan cell wall. Therefore, OM encompasses a crucial role in the protection of Gram-negative bacteria against environmental treatments. Outstandingly, in distinction to many biological membranes, the OM of most Gram-negative microorganisms is not a lipid bilayer.1 Instead, it is an especially unbalanced bilayer that contains phospholipids within the inner leaflet and lipopolysaccharide (LPS) molecules in the outer leaflet.3 Consistent with previous studies, LPS is the endotoxin portion of the Gram-negative bacterial cell wall. LPS is found in high levels in clinical necrotizing enterocolitis (NEC) that acts as a pro-inflammatory stimulus in the gastrointestinal (GI) tract. LPS performs numerous functions in Gram-negative bacteria. The essential role of LPS is to help as a main structural part of the OM. Also, LPS is a crucial section of the cell wall in some Gram-negative bacteria.4 The results show that the LPS molecules transform OM into a permeable membrane that is effective against small molecules and bacteria that can overcome the phospholipid bilayer, rendering Gram-negative bacteria resistant to natural antibacterial agents of some compounds.5,6 LPS may also play an essential role in bacterial-host interactions by regulating the host immune system.7 LPS is the cause of a wide range of infections in humans, including sepsis, severe fever, and shock sepsis, and is found in different types of water, including groundwater, freshwater, and saline.82. Biological effect of LPS
LPS is a molecule that forms a part of the bacterial cell wall of Gram-negative bacteria and helps to stabilize the bacterial cell wall. The LPS is made up of three parts.9 (A) The O-side (O antigen) is the main site of immunospecificity, (B) a central polysaccharide component close to the lipid A matrix, which is exposed to the extracellular milieu; and (C) lipid A is the real fever component of LPS. Lipid A is surrounded by the cell barrier of Gram-negative bacterium and is the most conserved part of the LPS structure.10–12 The other two components are outside, away from the bacterial cell wall. Lipid A aids as an anchor for the LPS component. LPS acts as an inner and outer portion of the external cell wall and internal membrane of Gram-negative bacterial cells.13 A representative design of the Gram-negative bacteria cell wall structure is presented in Fig. 1.LPS provides a protecting and stabilizing mechanical maintenance to the cell and aids in protecting the integrity of the cell wall, and therefore, survival of the cell mainly in high level stress, such as dehydration. The pathogenicity of the LPS is almost entirely due to the region III component, lipid A (endotoxin), which in several texts is mentioned as catechin.14,15 The main pathogenicity of lipid A may primarily be obvious itself by the increase of alteration of metabolism in small doses and fever.16,17 Therefore, LPS is one of the target antigens for diagnosis and potential candidates for vaccine and drug development. Even though the name “endotoxin” is infrequently used to refer to any cell-associated bacterial toxin, in bacteriology, it is accurately reserved to refer to the LPS complex related to the outer membrane of Gram-negative microorganisms such as Pseudomonas, Bordetella pertussis, Escherichia coli, Salmonella, Haemophilus influenza, Shigella, Neisseria, and Vibrio cholera. Some important endotoxin (LPS) producing bacteria and related diseases are summarized in Table 1.
| Bacterium | Role and diseases | Ref. |
|---|---|---|
| Pseudomonas aeruginosa | Persistent infections, mainly in the genetic ailment cystic fibrosis, facilitating biofilm formation, antibiotic resistance | 18 |
| Escherichia coli and Bacteroides dorei | Neurodegeneration diseases, hemolytic uremic syndrome (HUS), diarrhea that may range from watery to bloody, stomach pains and cramps, loss of appetite or nausea | 19 and 20 |
| Vomiting, cholecystitis, cholangitis, neonatal meningitis | ||
| Haemophilus influenzae | Meningitis, septicemia and pneumonia, Texas Children's Hospital (TCH) disease, meningitis and sepsis, necrotizing myositis and septic shock | 21–23 |
| Salmonella | Bacteremia and meningitis, enterocolitis, tachycardia, fever, hepatomegaly, splenomegaly, diarrhea, vomiting | 24 and 25 |
| Neisseria gonorrhoeae | Ophthalmia neonatorum endocervicitis urethritis, systemic neonatal infection | 25 and 26 |
| Vibrio cholera | Wound infection, watery diarrhea leading to serious dehydration. Gastroenteritis, septicemia | 27 |
| Staphylococcus aureus | Toxic shock syndrome, septic arthritis, pneumonia, endocarditis, pneumonia, boils, folliculitis, impetigo, cellulitis | 28 and 29 |
| Bordetella pertussis | Respiratory tract infection, pertussis, diphtheria | 30 |
Based on Table 1, there are a variety of diseases associated with bacterial endotoxin. Therefore, rapid and specific diagnosis of endotoxin plays a critical role in controlling and treating the disease. In the following, some of the most important and widely used methods of endotoxin diagnosis are discussed. These severe effects instantly demand approaches for sensitive quantification such as endotoxin from pharmaceutical and biological products. Limulus amoebocyte lysate assay (LAL), rabbit pyrogen test, enzyme-linked immunosorbent assay (ELISA), and fluorescence-based methods are the most important and widely used techniques for the determination of LPS.31,32 To strengthen and increase the sensitivity and specificity of routine methods in recent years, various methods have been developed, of which nanotechnology-based methods are one of the most important. The present article aims to investigate the diagnostic methods of lipopolysaccharide, emphasizing biosensors developed in this field. This study also addresses the challenges of routine testing and the importance of developing biosensors. As mentioned in the introduction, endotoxin is one of the most important pathogens associated with Gram-negative bacteria, so that its accurate, specific, and rapid identification can play a crucial role in controlling and treating the disease. In addition, due to the exceptional properties of endotoxin, this biomolecule has been considered by researchers in pharmacy, so the expansion of advanced diagnostic methods will be desirable. In the review, routine diagnostic methods on endotoxin monitoring are introduced, and their strengths/weaknesses are discussed. In addition to the comprehensive introduction of biosensors as a specific and sensitive method for endotoxin detection, the most up-to-date developed biosensors in this field have been reviewed. For the complete nobility of the readers of this article, a table containing general information of the introduced LPS biosensors such as analytical features of used techniques and platforms have been considered appropriately. So in this review, various types of biosensors such as optical (fluorescence, SERS, FRET, and SPR), electrochemical, photoelectrochemical, and electrochemiluminescence were investigated. Also, the critical role of advanced nanomaterials on the performance of the above-mentioned biosensors is discussed. In addition, the application of different labels on the efficient usage of biosensors for LPS is surveyed comprehensively. Also, various bioelements (aptamer, DNA, miRNA, peptide, enzyme, antibody, etc.) on the structure of the LPS biosensor were investigated. Finally, bioanalytical parameters, which affect the performance of LPS biosensors are surveyed.
3. Conventional methods in the detection of endotoxin (LPS)
Culturing and plating techniques are the out-of-date and ancient identified conventional incidental methods that aided as the gold standard method for bacterial endotoxin detection.33–35 The critical limitation is that each microorganism needs its own supplements and also its culture media for optimum growth.36,37 Similarly, new fresh media should be considered for each microbe, and each microbe will develop positive or negative colonies over time.38,39 The limulus amoeba cell lysate (LAL) enzyme assay is usually the preferred gold standard for endotoxin detection.40,41 LAL is obtained from amoebic cells (similar to human leukocytes) of the horseshoe crab family (horseshoe crab family), including the horseshoe crab family (East Asia) and the horseshoe crab family (Atlantic Ocean).37 The LAL test is used both in the industry and in the laboratory to measure bacterial endotoxin in various samples.42 The principle of this technique is based on the coagulation process that occurs in the blood lymph of the horseshoe crab (Limulus polyphemus) in the existence of LPS.43,44 Various types of LAL based on fluorescent and colorimetric methods have been established for the sensitive recognition of LPS. Even though these tests have presented hopeful detection limits and sensitivity, many of them need tedious and complex processes.45,46 LAL assays' sensitivity depends on the processing method, the sample type, and the dilution factor.47 The enzyme-linked immunosorbent assay (ELISA) is an immunological assay frequently used to measure proteins, antibodies, antigens, and glycoproteins in biological samples. Some examples include pregnancy tests, diagnosis of HIV infection, and measurement of cytokines or soluble receptors in serum or cell supernatant. ELISA is another widely used test in bacteriology and endotoxin diagnosis. ELISA method is sensitive, robust, and reliable. Since LPS is a very heterogeneous class of substances, it is still not possible to develop antibodies with sufficient affinity and broad specificity, so this method is not in the market.48Since large-scale commercial production and detection of endotoxin has become an absolute requirement, controlling endotoxin levels is important, primarily because it can cause some severe physiological responses.49 Therefore, it is very important in pharmaceutical manufacturing, life sciences and medical research.43 The commercial excess of various endotoxin detection kits on the market underscores the demand and need for early detection, regulation, and monitoring of endotoxin levels in different sectors. Compared to the official United States Pharmacopeia (USP) rabbit pyrogen test, the LAL test has been shown to be not only more sensitive to endotoxin, but also easier, faster, and cheaper to perform. Commercially available kits are based on various principles. Each strategy has its own advantages over other strategies. Everything is sensitive, reliable, efficient, and usually recommended over the old and slow rabbit pyrogen test. Each method has its own lower bound for endotoxin detection and its own incubation time, depending on the kit used.
Some practical and frequently used methods and also commercial products in the quantification of endotoxin are summarized in Table 2.
| Method | Sample | Technique | LOD/sensitivity | Ref. |
|---|---|---|---|---|
| Cultivation | — | Culture & plating | 1.5 CFU mL−1 | 50 |
| LAL | Milk | Chromogenic assay | ∼103 CFU mL−1 | 45 |
| LAL | Beef | Violet red bile agar overlay | 5067.6 ng g−1 | 51 |
| LAL | Physiological | Violet red bile agar overlay | 7.00–7.49/g−1 | 52 |
| LAL | Milk | — | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 104 to 1 104 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 109 109 |
48 |
| LAL | Plasma | — | 0.005 to 0.001 μg mL−1 | 53 |
| ELISA | Complex mixtures | ELISA using poly-L-lysine | 1 μg mL−1 | 54 |
| ELISA | Murine | Sandwich capture ELISA | 1 ng mL−1 | 55 |
| ELISA | Clinical | ELISA-bacteriophage receptor protein | 0.05 EU mL−1 up to 500 EU mL−1 | 56 |
| ELISA | Human biofluid | ELISA-high coating efficiency | ∼0.2 μg | 57 |
| Kit | Biological | Endpoint chromogenic assay | 0.1 EU mL−1 | 58 |
| Kinetic-QCL | Clinical | Endpoint chromogenic assay | 0.005 EU mL−1 | 59 |
| Pyrosate kit | Clinical | Gel clot assay | 1.0 EU mL−1 | 60 |
| EndoLISA | Biological | Fluorescence | 0.005 EU mL−1 | 56 |
All the above techniques have certain drawbacks, such as attachment of additional agent (BSA), the use of high amounts of the coating antigen, cross-reactive reactions, false-positive results, and the possibility of variation of antigenic sites by chemical treatment. Some critical advantages and limitations of conventional methods are summarized in Table 3.
| Method | Advantages | Limitation | Ref. |
|---|---|---|---|
| Cultivation | It evaluates living (cultureable) microorganisms, recognizes live cells in the sample, easily quantifies the cells in the sample, and achieves high sensitivity in the appropriate medium | Advanced skills are required for optimal results, unique dietary supplements are required, and unique nutritional media are required for optimal growth. Time and resource intensive, relies on phenotypic biochemical characterization | 61 and 62 |
| LAL | Acceptable sensitivity & specificity, cost effective | Time consuming, complicated procedures, serious ecological problems | 63 and 64 |
| ELISA | Rapid, cost saving, acceptable sensitivity, and specificity, flexibility | Difficult antibody pretreatment, cross reactive between the captured antibody and target, need for standard ELISA kit | 65 and 66 |
Due to the high importance of endotoxin (LPS) in various diseases, the development of advanced methods for its accurate and rapid diagnosis is an essential aim of researchers. Due to its exceptional properties, nanotechnology-based biosensing is more attractive to scientists. Biosensors are among the most important types of advanced technologies that have been developed extensively in recent years. The most important and latest diagnostic method (biosensing) for the detection of endotoxin will be discussed in the following subsections.
4. Biosensors; modern methods for the recognition of LPS
A biosensor is an analytical instrument that contains a biological sensor element (bioreceptor) that is closely connected or integrated with a transducer. The first biosensor was developed for the recognition of glucose in 1962.62 Meanwhile, in 1956 Prof. Clark developed the first “real” biosensor (Clark electrode) towards detection of oxygen in biological samples. Professor Clark is known as the “father of biosensors”, but the term “biosensors” was coined by Karl in 1972.67,68 The technology converts biologically induced recognition events into a measurable signal (transduction), and analyses are acquired on screen after a series of processing. Biosensors are classified based on various subjects. As previously mentioned, the transducer is one of the crucial parts of the biosensor. Accordingly, biosensors are classified into three main subclasses, which include electrochemical, optical, and mechanical biosensors. Meanwhile, electrochemical and optical biosensors are very popular in medical diagnoses.Electrochemical (EC) biosensors proposition advantages such as low background noise, high specificity of their biological recognition process, and better signal to noise ratios. Furthermore, measurement with electrochemical method measurement needs small sample volumes. Electrochemical recognition is extensively chosen for the transduction of biosensors due to its simplicity of construction, low cost, portability, and user-friendliness.69 The biosensor has a simple, single overall structure that features direct conversion, high bioselectivity, high sensitivity, miniaturization, electrical/optoelectronic readout, continuous monitoring, ease of use, and cost effectiveness. Electrochemical biosensors are categorized into voltammetric, potentiometric, conductometric, or impedimetric based on transducing methods. Voltammetry sensors apply an electric potential to the working electrode and measure the current by electrochemical reduction or oxidation of the analyte.70 The current response is typically acquired in a peak comparative to the analyte concentration.70 Moreover, voltammetry contributes significantly to the boom in the concept of chemically modified electrodes.71 Findings show numerous kinds of voltammetric biosensors effectively planned over wide surfaces of modifying elements and transducers.71 Potentiometric technique, one of the earliest and most influential methods, occupies a permanent position in sensory analysis. This analytical method is an intelligent instrument for many practical applications because it can detect different ions of different concentrations and can be used in inexpensive measuring instruments.72 A potentiometric biosensor is well-defined as a tool combining a biological sensing component attached to a potential electrochemical transducer. Potentiometric biosensors generally rely on simpler species and biochemical reactions that result in electrochemical detection. The analytical signal produced by a potentiometric biosensor is an electrical potential.73 However, impedance is generated in a complete alternating current (AC) circuit with a voltage–current phase angle gap due to inductive and capacitive effects. In the case of impedimetric biosensors, the impedance consists of capacitive and partial ohm resistance due to complex interactions with small amplitude voltage signals as a function of frequency.74,75 A piezoelectric sensor is one of the most important sensors that can be used in home applications, medical applications, and energy self-sufficiency sensors.76,77 So far, researchers have been able to produce a next-generation piezoelectric sensor that is highly efficient, robust, lightweight, and can be used as an energy self-sufficient sensor.78 Piezoelectric sensors convert mechanical energies such as pressure, force, pressure, and vibration into electrical signals.79 Some of the advantages and disadvantages of different electrochemical transducers are summarized in Table 4.
| Type of detection method | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Potentiometric | Simple assembly technique, mass production, cost-effective, simple monitoring instrument/in field detection, fast operation, high sensitivity towards targeted ions, easy management system for POC measurement | Low selectivity, temperature-dependent system, impact solution changes, enormous mistake caused by logarithmic response, need additional components such as B. For signal amplification as the potential depends on the concentration of the analyte | 80–82 |
| Voltammetric | Real-time detection, simultaneous analysis of several samples | Environmentally sensitive, time consuming, and temperature sensitive | 83 and 84 |
| Impedometric | Small amplitude perturbation from steady state | The sensitivity of measurement depends on the technical accuracy of the equipment and on the operating techniques | 85 and 86 |
| Piezoelectric | High frequency response, high sensitivity, high accuracy, high dynamic range | Poor spatial resolution, dynamic sensing only, charge leakages | 87 |
To date, various types of electrochemical biosensors have been developed for the recognition of LPS. In this section, some EC-based biosensors were surveyed. Also, the role of different nanomaterials on their analytical performance was discussed.
4.1 Optical biosensors for detection of LPS
Another essential type of biosensor is the opto-sensor. Optical biosensors have shown valuable performance in discovering biological systems and promoting primary progress in drug discovery, clinical diagnostics, environmental monitoring, and food process control.88,89 Simplicity in their pretreatment and possible influence on the nature of target molecules and robustness, high sensitivity, reliability, and potential to be incorporated on a single chip are the critical advantages of optical biosensors.88,89 Optical biosensors are categorized into two main groups; labeling and label-free method. Biosensing based on surface plasmon resonance (SPR) and fluorescence is the most popular and extensively used method in research and medical fields, such as pathogen recognition.Examination of optical biosensors shows that despite the many advantages over routine methods, they have limitations and disadvantages that researchers in future studies should consider. Here are some examples of these problems. Optical biosensors have a medium or low selectivity, the main reason for which can be found in the method used. Low reproducibility as well as poor stability are other challenges related to the development of optical biosensors that should be addressed. The development of portable and cellophane-based biosensors can improve their attractiveness and popularity. So, developing such devices should be the goal of future studies.
Interestingly, a smartphone-based biosensor was developed to detect LPS on a grating-coupled surface plasmon resonance. Fixed biodevices provide POC detection for environmental and biomedical targets with attractive simplicity and portability benefits over distance and satisfactory analytical results (linear range; 570–730 ng mL−1 and LOD = 32.5 ng mL−1).90 Streptavidin-Horseradish peroxidase-modified hybridization chain reaction (HCRHRP) nanocomposites have been used as signal amplifiers or signal reporting elements for sensitive quantification of LPS. The planned genosensor quantitatively shows a handheld spectrophotometer with wide linear range and good LOD (1–150 ng mL−1 and 50 pg mL−1, respectively) for future clinical diagnosis, food industry, and environment. It opened up a new perspective on LPS detection in monitoring.91 An interesting immunosensor based on the electrochemiluminescence (ECL) method was fabricated to determine LPS. Planned ECL peptide biosensor offered a novel path in evaluating LPS expression on the bacteria surface and showed huge potential in the clinical analysis of LPS in related diseases. This study (Fig. 2) shows a strong gold–sulfur bond between the gold electrode surface and the thiol group of the peptide. In the presence of LPS, the peptide on the ECL biosensor can effectively capture LPS and produce LPS/peptide/AuE. The proposed system showed a wide linear detection range of 1.0–500 ng mL−1 and a LOD of 0.3 ng mL−1.87
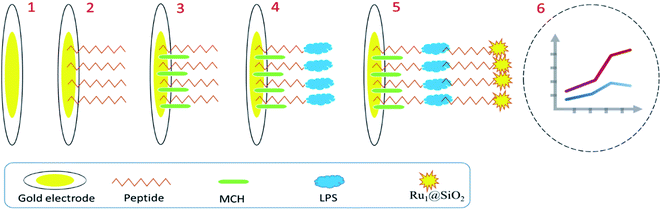 | ||
| Fig. 2 Biosensing of LPS using ECL-based method.87 | ||
Magnetic nanoparticles (MNPs) are useable in optical sensing. The MNPs may improve the refractive index alteration for refractive index sensing, or over with suitable coatings, be turned into fluorescent or plasmonic sensors.92,93 Iron oxide MNPs were employed as prospective therapy platforms, and due to their magnetic property, bacteria detection has drawn excessive consideration. These MNPs have generally been used as drug delivery, magnetic hyperthermia agents, and bioimaging contrast agents to identify and treat bacterial infections.90 Carbon nanotubes (CNTs) have appeared as one of the most intensively explored nanostructured materials.91–93 CNTs are hollow carbon buildings with a nanometer-scale diameter and a relatively more important length, and one or more walls.91,94 The advantage of CNTs over other nanomaterials lies in the extraordinary combination of optical, electrical, magnetic, mechanical, and chemical properties that offers great potential for a wide range of applications, including biosensors.95 Accordingly, several biosensors based on CNTs were developed for identifying pathogenic bacteria. For example, a single-walled carbon nanotube (SWCNT) based biosensor was fabricated to detect Staphylococcus aureus. The fixed tool detected S. aureus with a limit of detection (LOD) of 104 CFU mL−1.96 Also, for rapid and sensitive detection of Yersinia enterocolitica, SWCNT was used appropriately. The concentration range for the SWCNT-based biosensor was 106 to 104 CFU mL−1, and the optimal limit of detection (LOD) of the SWCNT-based biosensor was determined to be 104 CFU mL−1 of Y. enterocolitica in both clinical and phosphate buffer solution (PBS).97 Screen-printed carbon electrodes (SPCE) modified with polymers molecularly imprinted (PMI) were applied to detect P. aeruginosa's LPS. The developed bio-system was able to selectively identify P. aeruginosa's LPS from the similar E. coli endotoxin with good LOD (16.7 μg mL−1).98 The human Toll-Like Receptor-4 (TLR-4) is a protein responsible for sensing the LPS of Gram-negative bacteria. The TLR-4 was immobilized on the micro gold electrode by binding a modified self-assembled monolayer (mSAM). The formation of TLR-4 protein dimers increased resistance to charge transfer from a solution-based redox probe in response to varying concentrations of its target. In this work, linear range and LOD were (104 and 105 cells per mL, 100 cells per mL), respectively.99
A different type of optical biosensor was fabricated to detect Gram-negative LPS based on T4 bacteriophage. This work presented recombinant adhesive phage protein as a receptor molecule in biosensing technology for the first time. Obtained analytical properties were acceptable (linear range: 0–0.07 mg mL−1, LOD: 50 mg L−1).100 Through presenting an LPS specific binding peptide to a usual aggregation-induced emission (AIE) fluorophore, 1-(4-carboxylbenzene)-1,2,2-triphenyl (CTPY), a robust, specific, and sensitive optical biosensor based on the fluorescent method was settled. Such biodevices with rapid clearance and highly specific detection based on LPS specific binding peptides hold significant potential in diverse biological and clinical applications. The developed platform can achieve LPS sensing in a wide linear range of 0.1 to 1 μM and good LOD (6.97 nM).101 Ultra-sensitive detection of LPS based on combined amplification of dual enzymes was designated recently. With the effective signal amplification, the bio-system revealed high sensitivity with wide range linearity (2.5–1000 pg mL−1) and an acceptable LOD1 (pg mL−1). In addition, advanced biosensors can clearly distinguish LPS from interference and have high specificity for LPS detection. This biosensor is also effectively used to measure LPS in actual food samples and has the potential to be applied to detect food safety.102
In early 2001, an original theory termed “aggregation-induced emission (AIE)” was first recommended by Tang et al. AIE luminogens (AIEgens) display negligible or weak emission in dilute solution but are released intensely in the aggregate or solid state.103,104 AIEgens are categorized into two basic biomedical platforms, AIE NP probes and AIE molecular probes, which have been advanced for numerous biomedical applications.105,106 Their exclusive properties in terms of stability, brightness, biocompatibility, and therapeutic functions have increased the constant development of original designs.107,108 Until now, several AIE molecular probes have been established for monitoring bacterial labeling, cancer cell ablation, biological processes, and cellular organelle labelling with low background, high specificity, and tremendous photo stability.109,110 AIE fluorescent platform was planned to detect LPS, photodynamic antibacterial therapy, and bacterial imaging. Created photoelectrochemical biosensor, Fig. 3, presented good analytical output (linear range: 0–2.6 × 108 M and LOD: 2.6 × 108 M), which showed great potential for detecting endotoxin in biological and food samples.107
 | ||
| Fig. 3 AIE fluorescent platform for the detection of lipopolysaccharide, bacterial imaging, and photodynamic antibacterial therapy.107 | ||
Graphene has been used in designing different biosensors of several transduction styles because of its electrical conductivity, capacity to immobilize other molecules, large surface area, and high electron transfer rate.111,112 In addition, the conjugated arrangement of graphene can facilitate electron transfer between the transducer and the bioreceptor, resulting in high signal sensitivity for electrochemical biosensors.113,114 Moreover, graphene-based nanomaterial was employed as a quencher in the transducer to produce florescent biosensors. Findings indicated that reduced graphene oxide (rGO), graphene oxide (GO), and graphene (G) have a very high efficiency of fluorescent quenching.115 Several rGO nanocomposite-based electrochemical biosensors were developed in the past decade for monitoring food-borne pathogenic bacteria.116 A DNA aptamer genosensor was established for Streptococcus pneumonia detection based on the GO combination fluorescent method with exceptional LOD of 15 CFU mL−1.117
Peptide-assembled graphene oxide was employed for endotoxin detection. Engineered fluorescent biosensor, Fig. 4, showed potential in selective detection of LPS from different bacterial strains and LPS on the membrane of living E. coli.115
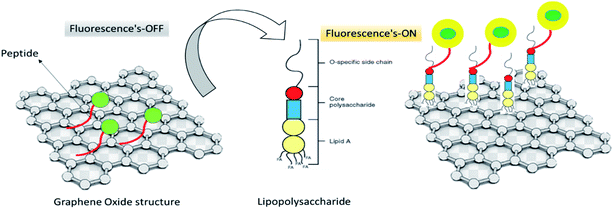 | ||
| Fig. 4 Peptide-based biosensor for detection of LPS.115 | ||
Also, synthesized CdTe quantum dots (QDs) conjugated with concanavalin A (Con A) were proposed as an original bio device for specific and selective quantification of LPS. Wide linearity and acceptable sensitivity were reported in this research.116
Interestingly, a cell-based fluorescent optical biosensor was advanced to detect and recognize LPS, which is carried out in a 96-well microplate with high efficiency, which is user-friendly and nondestructive. This practical study combined the promoter sequence of the critical signaling pathway Enhanced Green Fluorescence Protein (EGFP) and gene ZC3H12A (encoding MCPIP1 protein) with building a recombinant plasmid, which was transferred into 293/hTLR4A-MD2-CD14 cells through lipid-mediated, DNA-transfection technique. LPS could bind to TLR4 and co-receptors-induced signaling pathways could result in green fluorescent protein expression. The developed biosensor showed suitable analytical results (linear range: 5–200 μg mL−1, LOD: 0.075 μg mL−1) applicable for endotoxin screening in food industries and medical approaches.102 Also, an aptamer-based impedance genosensor was established for the detection of negative bacteria LPS. In this report, amine-terminated single-stranded DNA (ssDNA) was used as a probe that displayed high affinity to LPS and was immobilized on a gold electrode via 3-mercaptopropionic acid (MPA) as a linker. The designed system showed acceptable analytical results (linear range: 0.001–1 ng mL−1, LOD: 1 pg mL−1).118 A robust living cell-based optical transcriptional biosensor to detect LPS with a red fluorescent protein reporter system was fabricated properly. In the developed system, 293/hTLR4A-MD2-CD14pGL4.26-mcherry-NF-κB cells were used as a recognition element. The engineered biosensor has great potential for use in bacteria sensitive identification (linear range: 0.01 to 100 ng mL−1 and LOD: 1 ng mL−1), the LPS detection in food industries, biological products, and supporting the control of food-borne diseases.119 Highly sensitive recognition (linearity: 1–105 ng mL−1, LOD: 1.73 ng mL−1) of LPS using a genosensor based on hybridization chain reaction (HCR) was advanced recently. In the presented technique, two complementary stable species of biotinylated DNA hairpins concurred in solution up to the introduction of a recognition probe initiated an HCR cascade. The LPS react specifically with the conjugate DNA, which was captured by the ethanolamine aptamer attached to the reaction fit surface (Fig. 5).120
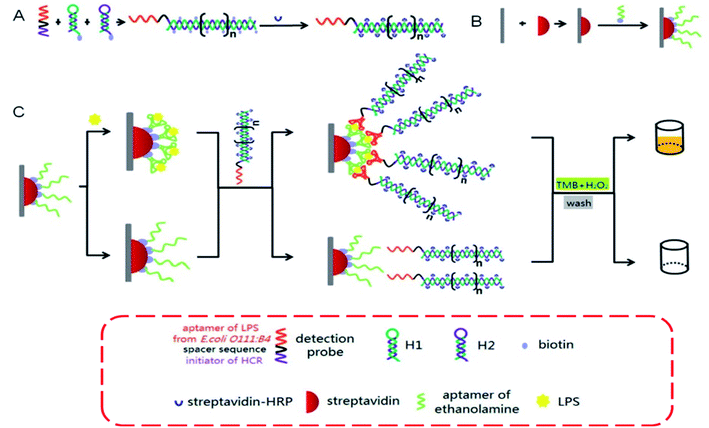 | ||
| Fig. 5 Graphic depiction of HCR-based aptasensor for the sensitive detection of LPS.120 | ||
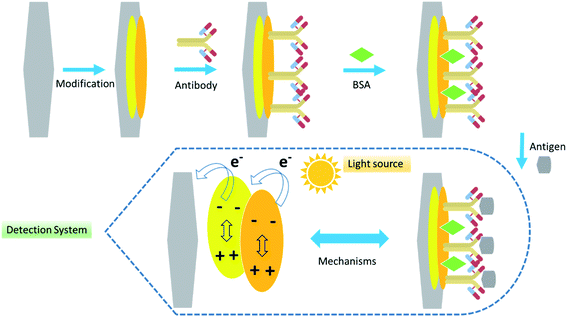
Lipoteichoic acid (LTA) is a surface-binding amphipathic substance derived from Gram-positive bacteria and is a regulator of autolytic wall enzymes. It is generally released from bacterial cells after lysozyme-induced lysis from beta-lactam antibiotics, cationic peptides, and leukocytes.120,121 Original research directly detects bacteremia by using host–pathogen interactions of LTA from Gram-positive bacteria and LPS from Gram-negative bacteria. The developed optical biosensor was sensitive (LOD = 4 ng mL−1), rapid, and achieved an accurate diagnosis of bacteremia at the POC (Fig. 6).122
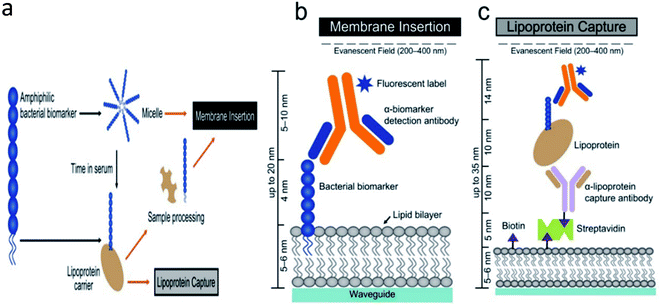 | ||
| Fig. 6 Schematic of the developed optical biosensor.122 | ||
The optical waveguide biosensor offers the facility to recognize the kinetics of the bio-molecular interaction on an online source without the requisite for extrinsic labeling of the biomolecules.123,124 Accordingly, a waveguide-based optical biosensor was settled for the high-sensitive detection of Gram-negative bacteria LPS. Waveguide-based biosystem demonstrated specific and rapid operation in the identification of pathogen biomarkers from small samples of serum or plasma, agreeing for the timely and sensitive analysis (linearity: 3.13 μg mL−1 to 200 μg mL−1, LOD: 25 μg mL−1) of infection at the POC.125 A mechanical QCM biosensor was fabricated to detect the interaction of low molecular weight chitosan and its N-acylated derivatives and chitoliposomes with endotoxin. This work determined the attachment and detachment rate quantities for LPS binding to chitosan. The affinity constants were calculated based on alterations in the oscillation frequency.126
Due to its incredible chemical specificity and capacity to offer a fingerprint-like spectrum for complicated aqueous solutions, Raman spectroscopy (RS) has ended up as one of the maximum promising optical techniques. RS employs easy and reasonably-priced instrumentation, requiring minimum pattern preparation. Raman scattering now has no longer located extensive software with the organic count because of its inherently vulnerable signal. However, to date, techniques to decorate the sensitivity have been employed, namely, resonance Raman effects, which offers a 102–106 enhancement, and surface-enhanced Raman spectroscopy (SERS), which ends up in as much as 108 enhancements.127,128 It is generally accepted that the enhancement is due to the binding of the molecular vibrations of the analyte molecule on the surface of the nanostructure to the locally enhanced electromagnetic field. These fields are created on the surface of metal nanostructures by quasiparticle vibrations called surface polarities and are formed when electrons on the metal surface interact with photons from incident light.129
The endotoxin launched within the side of the antibacterial system was monitored with the aid of the designed SERS detection strategy. The outcomes received with the aid of using SERS evaluation have been steady with the ones of the ELISA kit.130 An innovative work describes the use of SERS for the label-free quantitative nanoscale identification of the LPS adsorbed on the surface of 50 nm AuNPs. This SERS method was an efficient tool to detect LPS on the AuNP surface and the basis for the development of a novel specific and sensitive LPS-detection sensor based on the use of SERS and AuNPs.131 FRET or Förster resonance energy transfer is a flexible and sensitive device for qualitative and quantitative evaluation of organic interactions and processes.132 The access to a wide range of fluorescent materials, in conjunction with improved, easy-to-use, spectrometers and very state-of-the-art microscopes, have made FRET a completely exceptional methodology for biosensing.132 Fluorophores, which can be applied in FRET, now embody natural dyes, semiconductor quantum dots, fluorescent proteins, metallic chelates, several noble metals and different nanoparticles, and also polymers, basically fluorescent amino acids and organic cofactors.132,133 Recently, a fluorescent turn-on sensor for detecting micromolar levels of bacterial LPS has been industrialized. Polydiacetylene liposome surface modified with fabricated pentalysine peptide and histidine have effectively displayed fluorescence signal only in the presence of LPS. A significant increase in the fluorescence signal was perceived at E. coli concentrations as low as 4.91 × 108 CFU mL−1 with liposomes at a concentration of 40.0 μM. The sensor worked on the principle of FRET.134
Highlights of Table 5 include the following: (A) the majority of studies to date have been on ocular bacteria, and other bacteria have received less attention. Therefore, future studies could be aimed at developing biosensors for other bacteria. (B) As shown in the table, the fluorescence technique has been more popular with researchers due to its inherent advantages: simple and reasonably predictive and easily miniaturized. (C) As revealed in Table 5, labeled biosensors have received more attention in most studies. The easy detection method is the main feature of this type of biosensor. In other words, there is no need to interpret the results due to direct diagnosis. (D) The table shows that metal nanoparticles are more prevalent in developing biosensors due to significant features such as low cytotoxicity to biomass cells, ability to bind multiple targeted compounds, and large surface-to-volume ratio. (E) Analytical results of various studies show a linear relationship between μM to nM. Compared to routine methods, these results are acceptable, but their strengthening in future studies seems necessary. According to the findings detailed in Table 5, biosensors based on metal and carbon nanoparticles are more sensitive.138
| Bacterium | Technique | Method | Sample | NPs | Linear range | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| a Abbreviation: (GFLC): gold nanoparticles/ferrocene/liposome cluster, (GE): graphite electrode, (SAW): surface acoustic wave resonator, (EIS): electrochemical impedance spectroscopy (HCR-HRP): streptavidin-horseradish peroxidase modified hybridization chain reaction, (SPCE): screen-printed carbon electrodes, (NTA): nitrilotriacetic acid complex, (AIE) aggregation induced emission, (CTPY): 1-(4-carboxylbenzene)-1,2,2-triphenyl, (SCE): saturated calomel electrode, (QDs): CdTe quantum dots, (Ac-ChLM): N-3-hydroxytetradecanoil chitosan, surface-enhanced Raman spectroscopy (SERS). | |||||||
| E. coli | Grating-coupled SPR | Label-free | Clinical | Metallic | 570–730 nm | 32.5 ng mL−1 | 85 |
| E. coli | UV-Vis spectrophotometry | Colorimetric/immunosensor | Clinical | Nickel | 0.07 mg mL−1 | 50 mg L−1 | 100 |
| S. typhosa, K. pneumonia, E. coli, P. aeruginosa | Fluorescence | Fluorescent/immunosensor | Clinical | CTPY-AIE | 0.1–1 μM | 6.97 nM | 101 |
| Gram-negative bacterium | ChA, SWV, EIS | Labeled genosensor | Food | Gold | 2.5–1000 pg mL−1 | 1 pg mL−1 | 102 |
| E. coli | Fluorescence emission spectra, AIE | Labeled – fluorescence | Culture media | Inorganic nanoparticles | 2.6 × 108 M | 2.6 × 108 M | 107 |
| E. coli | Fluorescence emission spectra, Raman scattering | Labeled – fluorescence | Clinical | GO | 0 to 20 nM | 130 pM | 115 |
| Serratia marcescens | UV-visible spectra | Fluorescence labels-immunosensor | Culture media | QDs | 1 × 10 to 1 × 106 CFU mL−1 | 10–9 ng mL−1 | 116 |
| E. coli | UV-Vis absorption spectra | Fluorescence labels/immunosensor | Clinical | Ru@SiO2 | 1.0–500 ng mL−1 | 0.3 ng mL−1 | 24 |
| E. coli | Cell-based fluorescent | Fluorescence labels/immunosensor | Serum | — | 5–200 μg mL−1 | 0.075 μg mL−1 | 135 |
| E. coli | EIS, CVs | — | — | — | 0.001–1 ng mL | 1 pg mL−1 | 118 |
| E. coli | Fluorescent | Aptamer-based impedance | Serum | — | 0.01 to 100 ng mL−1 | 1 ng mL−1 | 119 |
| E. coli | Fluorescent | Genosensor/labeled | Serum | Streptavidin | 1–105 ng mL−1 | 1.73 ng mL−1 | 136 |
| S. typhimurium and Staphylococcus | Fluorescent | Waveguide-based optical biosensor | Clinical samples | Streptavidin–biotin | — | 4 ng mL−1 | 137 |
| S. typhimurium | Fluorescent, wave guide | Waveguide-based optical biosensor | Serum | — | 3.13 μg mL−1 to 200 μg mL−1 | 6.25 μg mL−1 | 125 |
| P. aeruginosa | SERS | SERS spectra | Biological | AgNPs | 0.10–10.0 μmol L−1 | 6.125 ng mL−1 | 130 |
| E. coli O55:B5 | SERS | Raman and SERS spectra | Biological | AuNPs | 0.1–10 μg mL−1 | 2.6 ± 0.1 ng mL−1 | 131 |
| E. coli | FRET | Fluorescence labels/immunosensor | Clinical | — | 4.91 × 108 CFU mL−1 | 40.0 μM | 134 |
As discussed, optical biosensors are one of the most important and widely used methods of detecting bacterial endotoxin. Due to several advantages, including high specificity, sensitivity, small size, and cost-effectiveness, their development has increased extensively in recent years. In addition, optical biosensors have high environmental compatibility and good migration capability. On the other hand, as shown in the table, optical biosensors have the appropriate analytical properties in detecting real samples. Despite the advantages of optical biosensors to achieve ideal platforms, some of their weaknesses, such as sensitivity to the environment such as pH should be addressed in future studies.
4.2 Developed electrochemical biosensors for detection of LPS
Recently a three-dimensional gold nanoparticles/ferrocene/liposome cluster (GFLC) was developed for electrochemical biosensing of LPS. In this cluster, gold nanoparticles, ferrocene, and liposome clusters act as signal amplification components, signal output components, or molecular recognition components.139 Also, a sensitive DNA-based biosensor (genosensor) was developed for the rapid and selective detection of LPS of E. coli. This system was cost-effective and showed excessive application potential for food safety, environmental monitoring, and real-time diagnosis.140 Magnetic nanoparticles (MNPs) are imperative in sensing as they offer viable solutions to the continuing challenges related to analytical and nonspecific effects. MNPs are applicable to facilitate electrochemical responses via blocking access to an original electrode or after coating to make them conduct.89 Also, an innovative immunosensor based on MNPs was engineered to determine S. typhimurium endotoxin in related samples. The created electrochemical biosensor was selective, specific, and has excellent potential in real-time detection of Gram-negative bacteria LPS.141 For sensing Gram-negative LPS, Cu2+-AuNPs aggregates were appropriately used in the electrochemical platform. The developed biosensor, Fig. 7, showed a high specificity toward LPS in the presence of other common interfering substances and was effortlessly renewed. Moreover, the invented biosensor presented a good practical application for LPS recognition in human serum samples with satisfactory analytical results (linear range: 0.05 pg mL−1 to 10 pg mL−1, LOD: 0.033 pg mL−1).141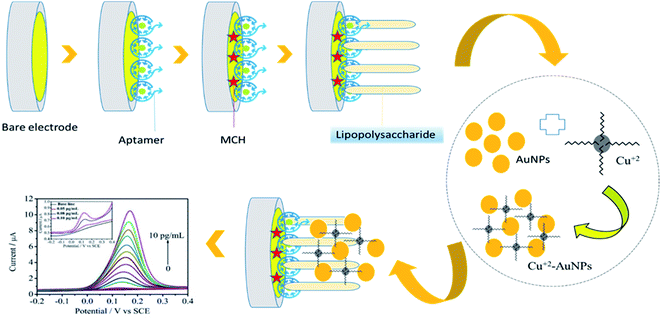 | ||
| Fig. 7 Graphical illustration of the developed aptasensor for the detection of LPS.141 | ||
A sensitive electrochemical platform was established for sensitive determination of LPS based on metal complex immobilized gold electrode. In this research work, electrochemical impedance spectroscopy (EIS) was applied to detect the interaction of nitrilotriacetic acid (NTA)[Cu]2+ complex with endotoxin. In this work, (0.0001–0.1 ng mL−1) and (0.0001 ng mL−1) were reported as linear range and LOD, respectively.142 A three-dimensional cells-in-gels-in-paper miniaturized electrochemical biosensor was fabricated for the rapid and sensitive detection of Gram-negative bacterium LPS. The proposed technique is applicable in food industries with high sensitivity (LOD = 3.5 × 10−3 ng mL−1) and low cost construction.143 Intracellular carbon nanoelectrode was employed as an electrochemical sensor for ultra-sensitive detection (LOD = 1 ng mL−1) of hydrogen peroxide generated intracellularly in response to rough and smooth LPS structures. This work showed different and indirect methods in determining bacterial endotoxin.144 A peptide was used as the recognition molecule in the electrochemical impedance immunosensor to determine LPS. The developed biosystem was used for real-time detection of endotoxin in real serum samples with acceptable linear range (0.01 pg mL−1 to 1.0 ng mL−1) and LOD (2.0 fg mL−1) (Fig. 8).145
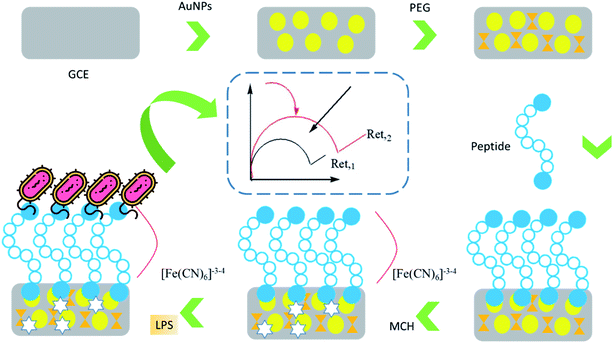 | ||
| Fig. 8 Graphic illustration of the label-free EIS biosensor for LPS detection.145 | ||
A dual electrochemical and mechanical label free lectin platform was developed for highly sensitive detection (LOD = up to 50 cells per mL) of antimicrobial susceptibility and the quantification of bacterial LPS. In this method, a polythiophene interface containing glycosylated fusion quinone units is used to form a carbohydrate platform for immobilization of concanavalin A (Con A), and LPS binding is measured via an orthogonal crystal oscillator microbalance and perform an electrochemical read (EQCM).146 An impedimetric electrochemical biosensor was developed for the quantification of E. coli produced LPS. In the developed system (Fig. 9), concanavalin A (Con A) lectins have been used as a molecular recognition element due to their specific interaction with the carbohydrate groups of LPS. Con A recognizes sigma-bonded mannose as the “core oligosaccharides” of some serum and membrane glycoproteins. The proposed biosensor was constructed based on lectin-functionalized Au–graphene compounds and showed acceptable stability and reproducibility with satisfactory analytical properties (linear range = 7.03 × 10−2 to 1.41 × 10−1 pg L−1 and LOD = 600 pg L−1). Au and graphene nanoparticles have recently been recognized as efficient and most promising electrochemical and luminescence catalysts.147
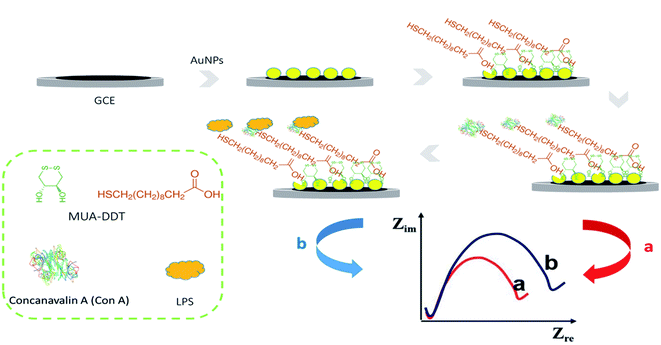 | ||
| Fig. 9 Representation of impedimetric lectin-based biosensors for bacterial LPS.147 | ||
DNAzyme-assisted recycling and Ce-based metal–organic structures dual signal amplifiers for sensitive electrochemical detection of LPS were settled. In the planned biosensor, Ce-MOFs were decorated with AuNPs to achieve AuNPs/Ce-MOFs. In this report, AuNPs/Ce-MOFs acted as catalysts to catalyze the oxidation of ascorbic acid (AA) and nanocarrier to capture –SH terminated hairpin probes 2 (HP2) for acquiring HP2/AuNPs/Ce-MOFs signal probes. Projected aptasensor for LPS showed a wide linear range from 10 fg mL−1 to 100 ng mL−1 with a 3.3 fg mL−1 LOD.148 An innovative electrochemical aptasensing platform based on GCE modified using GO and AuNPs was an assembly for the identification of E. coli endotoxin. The obtained results revealed that the modified GCE has a satisfactory selectivity for LPS over other biomolecules and is positively used to investigate serum of healthy persons and patients. In this work, acceptable linear range (0.1–0.9 pg mL−1) and LOD (30 fg mL−1) were reported.149 An electrochemical impedimetric biosensor based on SAM mixture CramoLL lectin and cysteine-gold nanoparticles (cys-AuNPs) was proposed for bacterial LPS recognition. Findings revealed that LPS from Serratia marcescens, Escherichia coli, Salmonella enterica, and Klebsiella pneumoniae are measurable by the settled system with good LOD (10 mM).150 A gold atomic cluster was applied as an electrochemical aptasensor for the recognition of LPS. The formed gold atomic clusters make Apt/AuAC/Au show an excellent property (LOD = 7.94 × 10−21 M) for the LPS determination in related samples. Also, the designed biosensor demonstrated a simple and easy technique for detecting LPS on the nanoscale amount.151 To complete our discussion, Table 6 summarizes all EC biosensors for the detection of LPS in different samples and compares their analytical parameters.
| Bacterium | Technique | Method | Sample | NPs | Linear range | LOD | Ref. |
|---|---|---|---|---|---|---|---|
| a Abbreviation: (GFLC): gold nanoparticles/ferrocene/liposome cluster, (GE): graphite electrode, (SAW): surface acoustic wave resonator, (EIS): electrochemical impedance spectroscopy (HCR-HRP): streptavidin-horseradish peroxidase modified hybridization chain reaction, (SPCE): screen-printed carbon electrodes, (NTA): nitrilotriacetic acid complex, (AIE) aggregation induced emission, (CTPY): 1-(4-carboxylbenzene)-1,2,2-triphenyl, (SCE): saturated calomel electrode, (QDs): CdTe quantum dots, (Ac-ChLM): N-3-hydroxytetradecanoil chitosan. | |||||||
| P. aeruginosa | DPV, FT-IR spectra | Fluorescence emission spectra | Food | GFLC | 2 × 10−9 μg mL−1 to 8 μg mL−1 | 0.51 × 10−10 μg mL−1 | 139 |
| E. coli | Aptasensor | Labeled-aptasensor | Rat serum | HCR-HRP | 1 to 150 ng mL−1 | 50 pg mL−1 | 152 |
| S. typhimurium | EIS, impedimetric | Immunosensor | Food and water | Magnetic | 0.001–0.1 μg mL−1 | 101 CFU mL−1 | 141 |
| P. aeruginosa | CVs | MIP | Wastewater | MIP | — | 16.7 μg mL−1 | 98 |
| S. typhimurium | EIS | Immunosensor/EDC/NHS | Clinical | — | 104 and 105 cells per mL | 105 cells per mL | 99 |
| E. coli | EIS | Labeled, aptasensor, DPV | Serum | Cu2+-AuNPs | 0.05 pg mL−1 to 10 pg mL−1 | 0.033 pg mL−1 | 145 |
| E. coli | EIS | EIS/CVs | Clinical | Cu2+-NTA | 0.0001–0.1 ng mL−1 | 1 ng mL−1 | 142 |
| S. enterica | CVs, DPV | Electrochemical/EIS | Food | GO | 1 × 101 to 1 × 104 ng mL−1 | 3.5 × 10−3 ng mL−1 | 143 |
| E. coli, S. minnesota | CVs | Labeled electrochemical | Clinical | CNTs | 1 mg mL−1 | 1 ng mL−1 | 144 |
| E. coli | Impedimetric | EIS/immunosensor | Clinical | AuNPs | 0.01 pg mL−1 to 1.0 ng mL−1 | 2.0 fg mL−1 | 145 |
| E. coli | Impedimetric | Impedimetric/labeled | Clinical | AuNPs–graphene | 1.0 × 10−9 to 1.0 × 10−6 g L−1 | 600 pg L−1 | 132 |
| Gram negative bacterium | CVs, EIS | Genosensor/labeled | Serum | AuNPs/Ce-MOFs | 10 fg mL−1 to 100 ng mL−1 | 3.3 fg mL−1 | 148 |
| E. coli | SWV | Aptasensor/label free | Serum | RGO/AuNPs | 0.1–0.9 pg mL−1 | 30 fg mL−1 | 149 |
| Gram negative bacterium | UV-visible absorption spectrum, EIS, DPV | Aptasensor/labeled | Serum | Apt/AuAC/Au | 0.01 attomolar to 1 picomolar | 7.94 × 10−21 M | 151 |
| S. marcescens, E. coli, S. enterica. K. pneumoniae | Impedimetric, CVs | Aptasensor/EIS | Clinical | Cys-Au | — | 10 mM | 150 |
Highlights of Table 6 include the following: (A) as demonstrated in Table 6, similar to optical biosensors, despite the presence of various endotoxin-producing bacteria, most studies have been performed on E. coli. Therefore, future studies should be aimed at detecting endotoxins in other bacteria. (B) In most of the studies summarized in the table, the impedimetric technique has been used mainly because of the simplicity of performing and diagnosis in real-time. (C) Good reducibility, high sensitivity, and specificity are critical properties in aptamer-based biosensors. Therefore, this kind of biosensing method has been considered by researchers to date. (D) Gold nanoparticles have been the most used in the development of LPS biosensors, and the results show that their use has increased the sensitivity and specificity of biosensors. Due to the unique properties of carbon-based nanoparticles, such as low cost and high conductivity, it is suggested that their use be further considered in future studies. (E) Analytical results of various studies indicated a linear relationship between μM to nM. Compared to routine methods, these results are acceptable, but their improvement in future studies seems crucial. According to the findings detailed in Table 6, biosensors based on AuNPs show extra sensitivity and specificity.
Electrochemiluminescence (ECL) based sensors combine electrochemistry and quantity of optical luminescence.153 In this method, when a potential is applied onto an electrode, the electrode surface is excited.153 Therefore, an electron transfer is done between molecules, and the resulting light is measured. ECL biosensors have been established to recognize chemical and bacteriological contaminants in food products.153,154 ECL biosensor was advanced for Escherichia coli O157:H7 quantitative recognition based on nitrogen-doped graphene quantum dots (N-GQDs) and a polydopamine (PDA) surface imprinted polymer (SIP).152
Magnetic resonance elastography (EG) is a valuable noninvasive analytical method of detecting abnormalities, such as liver fibrosis. The method generates tissue deformation by applying external or internal excitation, which causes displacements, and these displacements can be measured using modified MRE pulse sequences. Accordingly, EG as a flexible analytical method was applied for the detection of coagulation. The rapid detection of LPS is achieved by the EG analysis of endotoxin-induced limulus amebocyte lysate coagulation. This method is better than other methods using the same reagents in not only sensitivity but also detection time.155
As mentioned before, various types of biosensors are being developed, among which researchers have given electrochemical and optical biosensors more attention due to their many advantages. In the following, the electrochemical and optical biosensors developed for endotoxin detection are discussed in more detail, respectively. Photoelectrochemical (PEC) biosensors are an imperative branch of electrochemical detection, and PEC sensing has perceived rapid expansion despite the late start.107 Photoelectrochemistry is a dynamic discipline discovering the effect of light on photoactive materials, which involves the transformation of light into electricity and interconversion of electric energy and chemical energy. A schematic illustration of the PEC sensor is presented in Fig. 10.
In recent years, several PEC biosensors have been developed to detect bacteria. For example, an integrated multifunctional PEC platform for rapid detection of pathogenic bacteria was proposed recently. The platform can offer novel approaches for creating advanced green PEC sensors to recognize various pathogenic bacteria. The developed PEC platform showed satisfactory linearity (1 × 102 to 1 × 106 CFU mL−1) and LOD (46 CFU mL−1).156 A layer-by-layer self-assembly technique was employed to create the working electrode of the PEC aptamer sensor. The planned PEC aptamer-based biosensor displayed good stability, detection specificity, and reproducibility for Vibrio parahaemolyticus and provided a simple, cost-saving, and rapid detection approach for screening and controlling food-borne diseases.157 A signal-off PEC aptasensor was projected for pathogenic S. aureus detection based on graphite-like carbon nitride decorated with NiO.83 Despite the unique features of PEC biosensors, according to the studies conducted in this study, to date, this type of platform for LPS detection has not been developed. Therefore, considering the exceptional advantages of PEC biosensors, their development should be considered in future studies.
5. The role of nanoparticles in biosensor development
As reviewed in the present study, various nanoparticles were applied in biosensor development, and some of the presented nanoparticles have been used extensively. The use of NPs, nanotubes, nanowires, etc., in biosensor diagnostic devices is being discovered. With the progression in properties of nanomaterials, with their dimensions at the nanoscale level, smart biosensors that can detect a minute concentration of the desired analyte are developing. Nanomaterials are generally used as transducer materials that are an important part of biosensor development. The advantages and disadvantages of different nanomaterial-based biosensors are presented in Table 7.| Nanomaterial | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| CNTs | Extremely small and lightweight, plentiful resources, resistant to temperature changes, ability to cross cellular membranes and barriers, high chemical and mechanical stability, strong C–H bonds | Expensive nanotube production process, low yield, low hydrogen storage capacity, purification problems | 158 and 159 |
| GO | Water dispersibility, polar functionalization, easy workability, low-cost | Lower electrical and thermal conductibility | 145, 160 and 161 |
| rGO | High electrical and thermal conductibility, good control on functionalization, economical and facile technique | Hydrophobicity, difficult workability, and production methodology are important in their properties | 88 and 89 |
| MNPs | Large surface-to-volume ratio, low cytotoxicity to biomass cell, ease of synthesis, capacity to bind several targeted compounds | High cost of synthesis processes and materials, mobility dependent on environment compatibilities | 162 and 163 |
It has been determined that the usage of nanomaterials can also additionally cause extended biosensor overall performance along with extended sensitivities and occasional limit-of-detection of numerous orders of magnitudes. Nanostructured substances display extended surface-to-extent ratio, mechanical strength, chemical activity, electrocatalytic properties, and more advantageous diffusivity. Nanomaterials were anticipated to play a critical role in the high performance of a biosensor. To probe biomolecules along with bacteria, viruses, DNA, etc., the biocompatibility of nanomaterials has been a serious issue in designing a biosensor.
6. Discoveries of gaps and future outlook
Biosensors establish an interdisciplinary field that is presently one of the critical parts of investigation in analytical chemistry. Expending biosensors naturally reduces the need for sample preparation. The biosensor's performance is typically and experimentally assessed based on its LOD, sensitivity, reproducibility, dynamic and linear ranges, or accuracy of the selectivity response and its response to interferences. Additional frequently compared factors include operational and storage stability, sensor reaction time, easy operation, and portability. Ideally, the sensing surface should be reusable for numerous repeated quantities to be made. For various environmental, clinical, and food applications, the sensor should be capable of constantly monitoring the analyte online. Conversely, single-use biosensors are acceptable for some main applications, such as personal blood glucose monitoring by diabetics. Therefore, future studies should be in line with developing an ideal biosensor with the mentioned features and capabilities. In addition to the above, the methodology is critical in the development of biosensors; for example: as described in the text, LPS are some intercellular components and cell-based biosensors play a critical role in their determination.On the other hand, functionalized CNTs can successfully penetrate individual cells and cross biological barriers like cell membranes.164 This property and the release of CNTs from the cells and mechanism of internalization are of main interest for biological and in particular intracellular biosensing applications. Therefore, the use of CNTs in the development of future biosensors should be given more attention.
7. Conclusion
In this review, the up-to-date scientific literature dealing with biosensors for LPS detection was investigated. Different methods advanced by various studies to detect endotoxin are declared. The biosensors have different sample pretreatment requirements, other detection limits for the target organisms, and different degrees of automation evaluation, complexities, and requisite analysis times. Based on the surveyed literature, biosensors have verified the great potential for LPS analysis, but the following points should be considered in future studies. In conclusion, many of the established biosensing systems target Salmonella, E. coli 0157:H7. At the same time, some of the reported examples are not directed at the main imperative pathogens listed in Table 1. (B) For commercialization, one of the problems still facing the construction of biosensors for direct detection of LPS is the sensitivity of the analyses in the real sample. (C) Specificity is the critical issue in biosensor development and should be improved in future investigations; in other words, a robust biosensor should be able to detect target structures from similar structures in contaminated samples. (D) Simple and low-cost structure, easy to operate and easy interpretation of results, and fast and high sensitivity are the features of LPS biosensors that should be considered in their development. (E) Endotoxins are Gram-negative bacterial wall compounds, so the development of cell-based biosensors is of great importance. (F) As found in reviewing and comparing different studies, various factors play a critical role in constructing an ideal biosensor, the least of which include: the type of the used nanomaterial and techniques. Also, according to the results obtained in this study, the use of appropriate nanomaterials is one of the main factors in achieving improved analytical results. Finally, although biosensors have advantages over routine and older methods, future studies should be developed to address the barriers and weaknesses of biosensors so that researchers should consider the following points: simple and inexpensive structure is one of the strategic goals of biosensor development, so the use of advanced and expensive materials and tools will make the created biosensors not welcome. This may be one of the main reasons for the unsuccessful industrialization and commercialization of biosensors developed to date. Subsequently, according to studies, labeled biosensors, in addition to high cost, have a time-consuming and complex design process, so the development of label-free biosensors should be considered as much as possible. As shown in Table 4, vulnerabilities to temperature changes and the surrounding environment and pH are one of the main problems and limitations in the development of electrochemical biosensors, especially concerning physiological matrices, so eliminating this vulnerability should be highlighted in the future studies.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.Acknowledgements
The authors would like to thank the Tabriz University of Medical Sciences.References
- T. J. Silhavy, D. Kahne and S. Walker, The bacterial cell envelope, Cold Spring Harbor Perspect. Biol., 2010, 2(5), a000414 CrossRef PubMed.
- Y. Kamio and H. Nikaido, Outer membrane of Salmonella typhimurium: accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium, Biochemistry, 1976, 15(12), 2561–2570 CrossRef CAS PubMed.
- A. T. Jan, Outer Membrane Vesicles (OMVs) of Gram-negative Bacteria: A Perspective Update, Front. Microbiol., 2017, 8, 1053 CrossRef PubMed.
- M. P. Bos, V. Robert and J. Tommassen, Biogenesis of the gram-negative bacterial outer membrane, Annu. Rev. Microbiol., 2007, 61, 191–214 CrossRef CAS PubMed.
- G. Zhang, T. C. Meredith and D. Kahne, On the essentiality of lipopolysaccharide to Gram-negative bacteria, Curr. Opin. Microbiol., 2013, 16(6), 779–785 CrossRef CAS PubMed.
- H. Nikaido, Molecular basis of bacterial outer membrane permeability revisited, Microbiol. Mol. Biol. Rev., 2003, 67(4), 593–656 CrossRef CAS PubMed.
- S. M. Galloway and C. R. Raetz, A mutant of Escherichia coli defective in the first step of endotoxin biosynthesis, J. Biol. Chem., 1990, 265(11), 6394–6402 CrossRef CAS.
- E. R. Rojas, G. Billings, P. D. Odermatt, G. K. Auer, L. Zhu, A. Miguel, F. Chang, D. B. Weibel, J. A. Theriot and K. C. Huang, The outer membrane is an essential load-bearing element in Gram-negative bacteria, Nature, 2018, 559(7715), 617–621 CrossRef CAS PubMed.
- X. Su, L. Sutarlie and X. J. Loh, Sensors, Biosensors, and Analytical Technologies for Aquaculture Water Quality, Research, 2020, 2020, 8272705 CrossRef CAS PubMed.
- M. Pither, The structure of Lipid A and composition of the oligosaccharides of the lipopolysaccharides of the bacterial species: Veillonella parvula, Keele University, 2019 Search PubMed.
- G. Datta, B. J. Fuller and B. R. Davidson, Molecular mechanisms of liver ischemia reperfusion injury: insights from transgenic knockout models, World J. Gastroenterol., 2013, 19(11), 1683–1698 CrossRef CAS PubMed.
- F. El-Taboni, E. Caseley, M. Katsikogianni, L. Swanson, T. Swift and M. E. Romero-González, Fluorescence Spectroscopy Analysis of the Bacteria-Mineral Interface: Adsorption of Lipopolysaccharides to Silica and Alumina, Langmuir, 2020, 36(7), 1623–1632 CrossRef CAS PubMed.
- A. Emiola, J. George and S. S. Andrews, A Complete Pathway Model for Lipid A Biosynthesis in Escherichia coli, PLoS One, 2015, 10(4), e0121216 CrossRef PubMed.
- A. P. Moran, M. M. Prendergast and B. J. Appelmelk, Molecular mimicry of host structures by bacterial lipopolysaccharides and its contribution to disease, FEMS Immunol. Med. Microbiol., 1996, 16(2), 105–115 CrossRef CAS PubMed.
- S. M. Huszczynski, J. S. Lam and C. M. Khursigara, The role of Pseudomonas aeruginosa lipopolysaccharide in bacterial pathogenesis and physiology, Pathogens, 2020, 9(1), 6 CrossRef CAS PubMed.
- P. N. A. Harris, P. A. Tambyah, D. C. Lye, Y. Mo, T. H. Lee, M. Yilmaz, T. H. Alenazi, Y. Arabi, M. Falcone, M. Bassetti, E. Righi, B. A. Rogers, S. Kanj, H. Bhally, J. Iredell, M. Mendelson, T. H. Boyles, D. Looke, S. Miyakis, G. Walls, M. Al Khamis, A. Zikri, A. Crowe, P. Ingram, N. Daneman, P. Griffin, E. Athan, P. Lorenc, P. Baker, L. Roberts, S. A. Beatson, A. Y. Peleg, T. Harris-Brown and D. L. Paterson, Effect of Piperacillin-Tazobactam vs. Meropenem on 30-Day Mortality for Patients With E coli or Klebsiella pneumoniae Bloodstream Infection and Ceftriaxone Resistance: A Randomized Clinical Trial, JAMA, 2018, 320(10), 984–994 CrossRef CAS PubMed.
- K. Nesporova, E. R. Wyrsch, A. Valcek, I. Bitar, K. Chaw, P. Harris, J. Hrabak, I. Literak, S. P. Djordjevic and M. Dolejska, Escherichia coli Sequence Type 457 Is an Emerging Extended-Spectrum-β-Lactam-Resistant Lineage with Reservoirs in Wildlife and Food-Producing Animals, Antimicrob. Agents Chemother., 2020, 65(1), e01118–e01120 CrossRef PubMed.
- M. Ibar-Bariain, A. Rodríguez-Gascón, A. Isla, M. Solinís and A. Canut-Blasco, Evaluation of the adequacy of the antimicrobial therapy of invasive Haemophilus influenzae infections: A pharmacokinetic/pharmacodynamic perspective, Enferm. Infecc. Microbiol. Clin., 2021, 39(2), 65–71 CrossRef PubMed.
- J. G. Vallejo, J. C. McNeil, K. G. Hultén, L. M. Sommer, J. J. Dunn and S. L. Kaplan, Invasive Haemophilus influenzae Disease at Texas Children's Hospital, 2011 to 2018, Pediatr. Infect. Dis. J., 2019, 38(9), 900–905 CrossRef PubMed.
- F. Resman, T. Svensjö, C. Ünal, J. Cronqvist, H. Brorson, I. Odenholt and K. Riesbeck, Necrotizing myositis and septic shock caused by Haemophilus influenzae type f in a previously healthy man diagnosed with an IgG3 and a mannose-binding lectin deficiency, Scand. J. Infect. Dis., 2011, 43(11–12), 972–976 CrossRef CAS PubMed.
- J. J. Gilchrist and C. A. MacLennan, Invasive Nontyphoidal Salmonella Disease in Africa, EcoSal Plus, 2019, 8(2), 1–23 CrossRef PubMed.
- R. A. Cheng, C. R. Eade and M. Wiedmann, Embracing Diversity: Differences in Virulence Mechanisms, Disease Severity, and Host Adaptations Contribute to the Success of Nontyphoidal Salmonella as a Foodborne Pathogen, Front. Microbiol., 2019, 10, 1368 CrossRef PubMed.
- A. Lovett and J. A. Duncan, Human Immune Responses and the Natural History of Neisseria gonorrhoeae Infection, Front. Immunol., 2018, 9, 3187 CrossRef CAS PubMed.
- M. A. De Francesco, P. Stefanelli, A. Carannante, S. Corbellini, C. Giagulli, G. Lorenzin, M. Ronconi, E. Arici, M. Cadei, R. Campora and A. Caruso, Management of a Case of Peritonitis Due to Neisseria gonorrhoeae Infection Following Pelvic Inflammatory Disease (PID), Antibiotics, 2020, 9(4), 193 CrossRef PubMed.
- I. Levade, A. I. Khan, F. Chowdhury, S. B. Calderwood, E. T. Ryan, J. B. Harris, R. C. LaRocque, T. R. Bhuiyan, F. Qadri and A. A. Weil, A Combination of Metagenomic and Cultivation Approaches Reveals Hypermutator Phenotypes within Vibrio cholerae-Infected Patients, mSystems, 2021, 6(4), e00889 CrossRef CAS PubMed.
- S. Y. Tong, J. S. Davis, E. Eichenberger, T. L. Holland and V. G. Fowler, Jr, Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management, Clin. Microbiol. Rev., 2015, 28(3), 603–661 CrossRef CAS PubMed.
- M. J. Rybak, J. Le, T. P. Lodise, D. P. Levine, J. S. Bradley, C. Liu, B. A. Mueller, M. P. Pai, A. Wong-Beringer, J. C. Rotschafer, K. A. Rodvold, H. D. Maples and B. M. Lomaestro, Therapeutic monitoring of vancomycin for serious methicillin-resistant Staphylococcus aureus infections: a revised consensus guideline and review by the American Society of Health-System Pharmacists, the Infectious Diseases Society of America, the Pediatric Infectious Diseases Society, and the Society of Infectious Diseases Pharmacists, Am. J. Health-Syst. Pharm., 2020, 77(11), 835–864 CrossRef PubMed.
- J. D. Cherry, The epidemiology of pertussis: a comparison of the epidemiology of the disease pertussis with the epidemiology of Bordetella pertussis infection, Pediatrics, 2005, 115(5), 1422–1427 CrossRef PubMed.
- M. Sakata, Y. Fukuma, M. Todokoro and M. Kunitake, Selective assay for endotoxin using poly(epsilon-lysine)-immobilized Cellufine and Limulus amoebocyte lysate (LAL), Anal. Biochem., 2009, 385(2), 368–370 CrossRef CAS PubMed.
- G. Solano, A. Gómez and G. León, Assessing endotoxins in equine-derived snake antivenoms: Comparison of the USP pyrogen test and the Limulus Amoebocyte Lysate assay (LAL), Toxicon, 2015, 105, 13–18 CrossRef CAS PubMed.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, A review of methods for the detection of pathogenic microorganisms, Analyst, 2019, 144(2), 396–411 RSC.
- L. Váradi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Methods for the detection and identification of pathogenic bacteria: past, present, and future, Chem. Soc. Rev., 2017, 46(16), 4818–4832 RSC.
- A. Mobed, B. Baradaran, M. de la Guardia, M. Agazadeh, M. Hasanzadeh, M. A. Rezaee, J. Mosafer, A. Mokhtarzadeh and M. R. Hamblin, Advances in detection of fastidious bacteria: from microscopic observation to molecular biosensors, TrAC, Trends Anal. Chem., 2019, 113, 157–171 CrossRef CAS.
- A. Saravanan, P. S. Kumar, R. V. Hemavathy, S. Jeevanantham, R. Kamalesh, S. Sneha and P. R. Yaashikaa, Methods of detection of food-borne pathogens: a review, Environ. Chem. Lett., 2020, 19, 189–207 CrossRef.
- C. Y. Park, S. H. Jung, J. P. Bak, S. S. Lee and D. K. Rhee, Comparison of the rabbit pyrogen test and Limulus amoebocyte lysate (LAL) assay for endotoxin in hepatitis B vaccines and the effect of aluminum hydroxide, Biologicals, 2005, 33(3), 145–151 CrossRef CAS PubMed.
- J. Scudder and J. Y. Ye, Limulus amoebocyte lysate test via an open-microcavity optical biosensor, J. Biomed. Opt., 2018, 23(2), 1–6 Search PubMed.
- J. S. Lam, E. M. Anderson and Y. Hao, LPS quantitation procedures, Pseudomonas Methods and Protocols, Springer, 2014, pp. 375–402 Search PubMed.
- R. Blechova and D. Pivodova, Limulus amoebocyte lysate (LAL) test-An alternative method for detection of bacterial endotoxins, Acta Vet. Brno, 2001, 70(3), 291–296 CrossRef CAS.
- R. J. Elin and J. Hosseini, Clinical utility of the Limulus amebocyte lysate (LAL) test, Prog. Clin. Biol. Res., 1985, 189, 307–327 CAS.
- J. C. Hurley, Endotoxemia: methods of detection and clinical correlates, Clin. Microbiol. Rev., 1995, 8(2), 268–292 CrossRef CAS PubMed.
- H. Grallert and S. Leopoldseder, EndoLISA®: a novel and reliable method for endotoxin detection, 2011 Search PubMed.
- R. Pennamareddy, K. Prabakar and J. Pandiyan, Sorting out of interference in detection of endotoxins in biotherapeutic drugs, Indian J. Sci. Technol., 2009, 2(11), 20–22 CrossRef CAS.
- J. L. Ding and B. Ho, Endotoxin detection–from limulus amebocyte lysate to recombinant factor C, Endotoxins: structure, function and recognition, Springer, 2010, pp. 187–208 Search PubMed.
- M. M. M. Masri, M. S. T. Maidin and A. B. Ariff, Impact of single and two-phase dissolved oxygen tension control on bacillus thuringiensis cultivation and δ-endotoxin production, Malays. Appl. Biol., 2021, 50(1), 73–84 CrossRef.
- A. Svensson and B. Hahn-Hägerdal, Comparison of a gelation and a chromogenic Limulus (LAL) assay for the detection of gram-negative bacteria, and the application of the latter assay to milk, J. Dairy Res., 1987, 54(2), 267–273 CrossRef CAS PubMed.
- J. M. Jay, S. Margitic, A. L. Shereda and H. V. Covington, Determining endotoxin content of ground beef by the Limulus amoebocyte lysate test as a rapid indicator of microbial quality, Appl. Environ. Microbiol., 1979, 38(5), 885–890 CrossRef CAS PubMed.
- J. M. Jay, Rapid estimation of microbial numbers in fresh ground beef by use of the Limulus test, J. Food Prot., 1981, 44(4), 275–278 CrossRef PubMed.
- I. Hartman, G. Ziv and A. Saran, Application of the Limulus amoebocyte lysate test to the detection of gram-negative bacterial endotoxins in normal and mastitic milk, Res. Vet. Sci., 1976, 20(3), 342–343 CrossRef CAS PubMed.
- R. B. Reinhold and J. Fine, A technique for quantitative measurement of endotoxin in human plasma, Proc. Soc. Exp. Biol. Med., 1971, 137(1), 334–340 CrossRef CAS PubMed.
- M. M. M. Masri, M. S. T. Maidin and A. B. Ariff, Impact of single and two-phase dissolved oxygen tension control on bacillus thuringiensis cultivation and δ-endotoxin production, Malays. Appl. Biol., 2021, 50(1), 73–84 CrossRef.
- D. Choi, R. Tsang and M. Ng, Sandwich capture ELISA by a murine monoclonal antibody against a genus-specific LPS epitope for the detection of different common serotypes of salmonellas, J. Appl. Bacteriol., 1992, 72(2), 134–138 CrossRef CAS PubMed.
- J. M. Jay, Rapid estimation of microbial numbers in fresh ground beef by use of the Limulus test, J. Food Prot., 1981, 44(4), 275–278 CrossRef PubMed.
- F. C. Martinho, W. M. Chiesa, A. A. Zaia, C. C. Ferraz, J. F. Almeida, F. J. Souza-Filho and B. P. Gomes, Comparison of endotoxin levels in previous studies on primary endodontic infections, J. Endod., 2011, 37(2), 163–167 CrossRef PubMed.
- C. Barro, A. Paul, F. Saleh, T. Chitnis and H. L. Weiner, Validation of Two Kinetic Assays for the Quantification of Endotoxin in Human Serum, Front. Neurol., 2021, 12, 1005 Search PubMed.
- T. N. Soe, Ultrafiltration Membrane Process for Pyrogen Removal in the Preparation of Water for Injection (WFI), Inquiry, 2006, 7(1), 13 Search PubMed.
- R. Wedmann and T. Uhlig, Sensitive and flexible bacterial endotoxin testing with the EndoZyme® II Recombinant Factor C Assay using the Synergy™ HTX Microplate Reader, 2017 Search PubMed.
- F. Mégraud, Advantages and disadvantages of current diagnostic tests for the detection of Helicobacter pylori, Scand. J. Gastroenterol., 1996, 31(suppl 215), 57–62 CrossRef PubMed.
- M. Koskinen, G. Wellenberg, O. Sampimon, J. Holopainen, A. Rothkamp, L. Salmikivi, W. Van Haeringen, T. Lam and S. Pyörälä, Field comparison of real-time polymerase chain reaction and bacterial culture for identification of bovine mastitis bacteria, J. Dairy Sci., 2010, 93(12), 5707–5715 CrossRef CAS PubMed.
- L. R. Stromberg, H. M. Mendez and H. Mukundan, Detection methods for lipopolysaccharides: past and present. Escherichia coli-recent advances on physiology, pathogenesis and biotechnological applications, InTech, 2017, pp. 141–168 Search PubMed.
- S. L. Gaffin, N. Badsha, J. Brock-Utne, B. Vorster and J. Conradie, An ELISA procedure for detecting human anti-endotoxin antibodies in serum, Ann. Clin. Biochem., 1982, 19(3), 191–194 CrossRef CAS PubMed.
- V. Murugan and K. Sankaran, Bacterial lipid modification of ICP11 and a new ELISA system applicable for WSSV infection detection, Mar. Biotechnol., 2018, 20(3), 375–384 CrossRef CAS PubMed.
- G. Banik, Biosensor: Potential and Scope in Agriculture, Chem. Sci. Rev. Lett., 2021, 10(38), 116–122 Search PubMed.
- N. Arora, Recent advances in biosensors technology: a review, Octa J. Biosci., 2013, 1(2), 147–150 Search PubMed.
- K. Srinivas, T. S. Rao and C. B. Kiranmayi, The emergence of biosensors: potentialities and scope in food safety, Everyman’s Science, 1962, 141, 141–144 Search PubMed.
- X.-d. Wang and O. S. Wolfbeis, Fiber-optic chemical sensors and biosensors (2013–2015), Anal. Chem., 2016, 88(1), 203–227 CrossRef CAS PubMed.
- Q. Zhou, W. Zhang, M. Qiu and Y. Yu, Role of oxygen in copper-based catalysts for carbon dioxide electrochemical reduction, Mater. Today Phys., 2021, 20, 100443 CrossRef CAS.
- R. Jain and A. Sinha, Graphene-zinc oxide nanorods nanocomposite based sensor for voltammetric quantification of tizanidine in solubilized system, Appl. Surf. Sci., 2016, 369, 151–158 CrossRef CAS.
- J. Ding and W. Qin, Recent advances in potentiometric biosensors, TrAC, Trends Anal. Chem., 2020, 124, 115803 CrossRef CAS.
- J. Lin, R. Wang, P. Jiao, Y. Li, Y. Li, M. Liao, Y. Yu and M. Wang, An impedance immunosensor based on low-cost microelectrodes and specific monoclonal antibodies for rapid detection of avian influenza virus H5N1 in chicken swabs, Biosens. Bioelectron., 2015, 67, 546–552 CrossRef CAS PubMed.
- E. P. Randviir and C. E. Banks, Electrochemical impedance spectroscopy: an overview of bioanalytical applications, Anal. Methods, 2013, 5(5), 1098–1115 RSC.
- R. Jain and A. Sinha, Graphene-zinc oxide nanorods nanocomposite based sensor for voltammetric quantification of tizanidine in solubilized system, Appl. Surf. Sci., 2016, 369, 151–158 CrossRef CAS.
- S. Akhavan Alavi, M. Mohammadimehr and S. Ejtahed, Vibration analysis and control of micro porous beam integrated with FG-CNT distributed piezoelectric sensor and actuator, Steel Compos. Struct., 2021, 41(4), 595–608 Search PubMed.
- N. Sriplai, R. Mangayil, A. Pammo, V. Santala, S. Tuukkanen and S. Pinitsoontorn, Enhancing piezoelectric properties of bacterial cellulose films by incorporation of MnFe2O4 nanoparticles, Carbohydr. Polym., 2020, 231, 115730 CrossRef CAS PubMed.
- D. T. Luat, D. Van Thom, T. T. Thanh, P. Van Minh, T. Van Ke and P. Van Vinh, Mechanical analysis of bi-functionally graded sandwich nanobeams, Adv. Nano Res., 2021, 11(1), 55–71 Search PubMed.
- B. Paczosa-Bator, L. Cabaj, M. Raś, B. Baś and R. Piech, Potentiometric sensor platform based on a carbon black modified electrodes, Int. J. Electrochem. Sci., 2014, 9, 2816–2823 Search PubMed.
- E. l. Santos, M. C. B. Montenegro, C. Couto, A. N. Araújo, M. F. Pimentel and V. L. da Silva, Sequential injection analysis of chloride and nitrate in waters with improved accuracy using potentiometric detection, Talanta, 2004, 63(3), 721–727 CrossRef CAS PubMed.
- S. S. Hassan, H. Sayour and S. S. Al-Mehrezi, A novel planar miniaturized potentiometric sensor for flow injection analysis of nitrates in wastewaters, fertilizers and pharmaceuticals, Anal. Chim. Acta, 2007, 581(1), 13–18 CrossRef CAS PubMed.
- H. Karimi-Maleh, F. Karimi, S. Malekmohammadi, N. Zakariae, R. Esmaeili, S. Rostamnia, M. L. Yola, N. Atar, S. Movaghgharnezhad and S. Rajendran, An amplified voltammetric sensor based on platinum nanoparticle/polyoxometalate/two-dimensional hexagonal boron nitride nanosheets composite and ionic liquid for determination of N-hydroxysuccinimide in water samples, J. Mol. Liq., 2020, 310, 113185 CrossRef CAS.
- S. Menon, S. Jesny and K. G. Kumar, A voltammetric sensor for acetaminophen based on electropolymerized-molecularly imprinted poly(o-aminophenol) modified gold electrode, Talanta, 2018, 179, 668–675 CrossRef CAS PubMed.
- R. Arshad, A. Rhouati, A. Hayat, M. H. Nawaz, M. A. Yameen, A. Mujahid and U. Latif, MIP-based impedimetric sensor for detecting dengue fever biomarker, Appl. Biochem. Biotechnol., 2020, 191(4), 1384–1394 CrossRef CAS PubMed.
- S. Xiao, L. Chen, X. Xiong, Q. Zhang, J. Feng, S. Deng and L. Zhou, A new impedimetric sensor based on anionic intercalator for detection of lead ions with low cost and high sensitivity, J. Electroanal. Chem., 2018, 827, 175–180 CrossRef CAS.
- C. Chi, X. Sun, N. Xue, T. Li and C. Liu, Recent progress in technologies for tactile sensors, Sensors, 2018, 18(4), 948 CrossRef PubMed.
- C. Chen and J. Wang, Optical biosensors: an exhaustive and comprehensive review, Analyst, 2020, 145(5), 1605–1628 RSC.
- X. Fan, I. M. White, S. I. Shopova, H. Zhu, J. D. Suter and Y. Sun, Sensitive optical biosensors for unlabeled targets: a review, Anal. Chim. Acta, 2008, 620(1–2), 8–26 CrossRef CAS PubMed.
- J. Zhang, I. Khan, Q. Zhang, X. Liu, J. Dostalek, B. Liedberg and Y. Wang, Lipopolysaccharides detection on a grating-coupled surface plasmon resonance smartphone biosensor, Biosens. Bioelectron., 2018, 99, 312–317 CrossRef CAS PubMed.
- W. Xu, J. Tian, X. Shao, L. Zhu, K. Huang and Y. Luo, A rapid and visual aptasensor for lipopolysaccharides detection based on the bulb-like triplex turn-on switch coupled with HCR-HRP nanostructures, Biosens. Bioelectron., 2017, 89, 795–801 CrossRef CAS PubMed.
- X. Fan, Z. Li, S. Wang, Y. Wang, L. Yu and X. Fan, An electrochemiluminescence sandwich biosensor for the detection of lipopolysaccharide, Quim. Nova, 2020, 43, 747–751 CAS.
- L. Gloag, M. Mehdipour, D. Chen, R. D. Tilley and J. J. Gooding, Advances in the application of magnetic nanoparticles for sensing, Adv. Mater., 2019, 31(48), 1904385 CrossRef CAS PubMed.
- J. Kudr, Y. Haddad, L. Richtera, Z. Heger, M. Cernak, V. Adam and O. Zitka, Magnetic nanoparticles: from design and synthesis to real world applications, Nanomaterials, 2017, 7(9), 243 CrossRef PubMed.
- C. Xu, O. U. Akakuru, J. Zheng and A. Wu, Applications of iron oxide-based magnetic nanoparticles in the diagnosis and treatment of bacterial infections, Front. Bioeng. Biotechnol., 2019, 7, 141 CrossRef PubMed.
- C.-M. Tîlmaciu and M. C. Morris, Carbon nanotube biosensors, Front. Chem., 2015, 3, 59 Search PubMed.
- X.-M. Liu, Z. Dong Huang, S. Woon Oh, B. Zhang, P.-C. Ma, M. M. Yuen and J.-K. Kim, Carbon nanotube (CNT)-based composites as electrode material for rechargeable Li-ion batteries: a review, Compos. Sci. Technol., 2012, 72(2), 121–144 CrossRef CAS.
- M. Hussain, M. N. Naeem, M. Taj and A. Tounsi, Simulating vibration of single-walled carbon nanotube using Rayleigh-Ritz's method, Adv. Nano Res., 2020, 8(3), 215–228 Search PubMed.
- D. C. Ferrier, Carbon Nanotube (CNT)-Based Biosensors, Biosensors, 2021, 11, 486 CrossRef CAS PubMed.
- V. Biju, Chemical modifications and bioconjugate reactions of nanomaterials for sensing, imaging, drug delivery and therapy, Chem. Soc. Rev., 2014, 43(3), 744–764 RSC.
- H.-K. Choi, J. Lee, M.-K. Park and J.-H. Oh, Development of Single-Walled Carbon Nanotube-Based Biosensor for the Detection of Staphylococcus aureus, J. Food Qual., 2017, 2017, 5239487 CrossRef.
- A. Sobhan, J. Lee, M.-K. Park and J.-H. Oh, Rapid detection of Yersinia enterocolitica using a single-walled carbon nanotube-based biosensor for Kimchi product, LWT, 2019, 108, 48–54 CrossRef CAS.
- B. E. Stoica, A.-M. Gavrila, A. Sarbu, H. Iovu, H. Brisset, A. Miron and T.-V. Iordache, Uncovering the behavior of screen-printed carbon electrodes modified with polymers molecularly imprinted with lipopolysaccharide, Electrochem. Commun., 2021, 124, 106965 CrossRef CAS.
- R. M. Mayall, M. Renaud-Young, N. W. C. Chan and V. I. Birss, An electrochemical lipopolysaccharide sensor based on an immobilized Toll-Like Receptor-4, Biosens. Bioelectron., 2017, 87, 794–801 CrossRef CAS PubMed.
- E. Brzozowska, M. Śmietana, M. Koba, S. Górska, K. Pawlik, A. Gamian and W. J. Bock, Recognition of bacterial lipopolysaccharide using bacteriophage-adhesin-coated long-period gratings, Biosens. Bioelectron., 2015, 67, 93–99 CrossRef CAS PubMed.
- Y. Tang, A. Kang, X. Yang, L. Hu, Y. Tang, S. Li, Y. Xie, Q. Miao, Y. Pan and D. Zhu, A robust OFF-ON fluorescent biosensor for detection and clearance of bacterial endotoxin by specific peptide based aggregation induced emission, Sens. Actuators, B, 2020, 304, 127300 CrossRef CAS.
- Y. Huang, L. Wang, L. Sha, Y. Wang, X. Duan and G. Li, Highly sensitive detection of lipopolysaccharide based on collaborative amplification of dual enzymes, Anal. Chim. Acta, 2020, 1126, 31–37 CrossRef CAS PubMed.
- Z. Zhao, J. W. Y. Lam and B. Zhong Tang, Aggregation-induced emission of tetraarylethene luminogens, Curr. Org. Chem., 2010, 14(18), 2109–2132 CrossRef CAS.
- F. Würthner, Aggregation-induced emission (AIE): a historical perspective, Angew. Chem., Int. Ed., 2020, 59(34), 14192–14196 CrossRef PubMed.
- G. Feng, G.-Q. Zhang and D. Ding, Design of superior phototheranostic agents guided by Jablonski diagrams, Chem. Soc. Rev., 2020, 49(22), 8179–8234 RSC.
- G. Feng and B. Liu, Aggregation-induced emission (AIE) dots: emerging theranostic nanolights, Acc. Chem. Res., 2018, 51(6), 1404–1414 CrossRef CAS PubMed.
- X. Liu, Z. Yang, W. Xu, Y. Chu, J. Yang, Y. Yan, Y. Hu, Y. Wang and J. Hua, Fine tuning of pyridinium-functionalized dibenzo[a, c]phenazine near-infrared AIE fluorescent biosensors for the detection of lipopolysaccharide, bacterial imaging and photodynamic antibacterial therapy, J. Mater. Chem. C, 2019, 7(40), 12509–12517 RSC.
- C. N. R. Rao, A. K. Sood, K. S. Subrahmanyam and A. Govindaraj, Graphene: the new two-dimensional nanomaterial, Angew. Chem., Int. Ed., 2009, 48(42), 7752–7777 CrossRef CAS PubMed.
- F. Sha'bani and S. Rash-Ahmadi, Molecular dynamics investigation of pull-in instability in graphene sheet under electrostatic and van der Waals forces, Adv. Nano Res., 2021, 11(2), 173–181 Search PubMed.
- T. Kuila, S. Bose, P. Khanra, A. K. Mishra, N. H. Kim and J. H. Lee, Recent advances in graphene-based biosensors, Biosens. Bioelectron., 2011, 26(12), 4637–4648 CrossRef CAS PubMed.
- C. S. Park, H. Yoon and O. S. Kwon, Graphene-based nanoelectronic biosensors, J. Ind. Eng. Chem., 2016, 38, 13–22 CrossRef CAS.
- X. Wu, Y. Xing, K. Zeng, K. Huber and J. X. Zhao, Study of fluorescence quenching ability of graphene oxide with a layer of rigid and tunable silica spacer, Langmuir, 2018, 34(2), 603–611 CrossRef CAS PubMed.
- T. Kuila, S. Bose, P. Khanra, A. K. Mishra, N. H. Kim and J. H. Lee, Recent advances in graphene-based biosensors, Biosens. Bioelectron., 2011, 26(12), 4637–4648 CrossRef CAS PubMed.
- C. S. Park, H. Yoon and O. S. Kwon, Graphene-based nanoelectronic biosensors, J. Ind. Eng. Chem., 2016, 38, 13–22 CrossRef CAS.
- S. K. Lim, P. Chen, F. L. Lee, S. Moochhala and B. Liedberg, Peptide-assembled graphene oxide as a fluorescent turn-on sensor for lipopolysaccharide (endotoxin) detection, Anal. Chem., 2015, 87(18), 9408–9412 CrossRef CAS PubMed.
- S. Ebrahim, M. Reda, A. Hussien and D. Zayed, CdTe quantum dots as a novel biosensor for Serratia marcescens and Lipopolysaccharide, Spectrochim. Acta, Part A, 2015, 150, 212–219 CrossRef CAS PubMed.
- A. T. Bayraç and S. I. Donmez, Selection of DNA aptamers to Streptococcus pneumonia and fabrication of graphene oxide based fluorescent assay, Anal. Biochem., 2018, 556, 91–98 CrossRef PubMed.
- W. Su, M. Lin, H. Lee, M. Cho, W.-S. Choe and Y. Lee, Determination of endotoxin through an aptamer-based impedance biosensor, Biosens. Bioelectron., 2012, 32(1), 32–36 CrossRef CAS PubMed.
- H. Jiang, D. Jiang, J. Shao, X. Sun and J. Wang, High-throughput living cell-based optical biosensor for detection of bacterial lipopolysaccharide (LPS) using a red fluorescent protein reporter system, Sci. Rep., 2016, 6(1), 1–12 CrossRef PubMed.
- P. Xie, L. Zhu, X. Shao, K. Huang, J. Tian and W. Xu, Highly sensitive detection of lipopolysaccharides using an aptasensor based on hybridization chain reaction, Sci. Rep., 2016, 6(1), 29524 CrossRef CAS PubMed.
- H. Kwon, Y. Yang, S. Kumar, D. W. Lee, P. Bajracharya, T. L. Calkins, Y. Kim and P. V. Pietrantonio, Characterization of the first insect prostaglandin (PGE(2)) receptor: MansePGE(2)R is expressed in oenocytoids and lipoteichoic acid (LTA) increases transcript expression, Insect Biochem. Mol. Biol., 2020, 117, 103290 CrossRef CAS PubMed.
- J. Z. Kubicek-Sutherland, D. M. Vu, A. Noormohamed, H. M. Mendez, L. R. Stromberg, C. A. Pedersen, A. C. Hengartner, K. E. Klosterman, H. A. Bridgewater, V. Otieno, Q. Cheng, S. B. Anyona, C. Ouma, E. Raballah, D. J. Perkins, B. H. McMahon and H. Mukundan, Direct detection of bacteremia by exploiting host-pathogen interactions of lipoteichoic acid and lipopolysaccharide, Sci. Rep., 2019, 9(1), 6203 CrossRef PubMed.
- J.-N. Yih, Y.-M. Chu, Y.-C. Mao, W.-H. Wang, F.-C. Chien, C.-Y. Lin, K.-L. Lee, P.-K. Wei and S.-J. Chen, Optical waveguide biosensors constructed with subwavelength gratings, Appl. Opt., 2006, 45(9), 1938–1942 CrossRef CAS PubMed.
- H. Hosseini-Toudeshky and F. A. Amjad, Sensor placement optimization for guided wave-based structural health monitoring, Struct. Monit. Maint., 2021, 8(2), 125–150 Search PubMed.
- A. Noormohamed, L. R. Stromberg, A. S. Anderson, Z. Karim, P. Dighe, P. Kempaiah, J. M. Ong'echa, D. J. Perkins, N. Doggett and B. McMahon, Detection of lipopolysaccharides in serum using a waveguide-based optical biosensor, Optical Diagnostics and Sensing XVII: Toward Point-of-Care Diagnostics, SPIE, 2017, pp. 30–37 Search PubMed.
- G. Naberezhnykh, V. Gorbach, E. Kalmykova and T. Solov'eva, Determination of the parameters of binding between lipopolysaccharide and chitosan and its N-acetylated derivative using a gravimetric piezoquartz biosensor, Biophys. Chem., 2015, 198, 9–13 CrossRef CAS PubMed.
- M. Moskovits, Surface-enhanced spectroscopy, Rev. Mod. Phys., 1985, 57(3), 783–826 CrossRef CAS.
- N. Valley, N. Greeneltch, R. P. Van Duyne and G. C. Schatz, A look at the origin and magnitude of the chemical contribution to the enhancement mechanism of surface-enhanced Raman spectroscopy (SERS): theory and experiment, J. Phys. Chem. Lett., 2013, 4(16), 2599–2604 CrossRef CAS.
- F. N. Kenry, L. Clark, S. R. Panikkanvalappil, B. Andreiuk and C. Andreou, Advances in Surface Enhanced Raman Spectroscopy for in Vivo Imaging in Oncology, Nanotheranostics, 2022, 6(1), 31 CrossRef PubMed.
- S. Xiang, C. Ge, S. Li, L. Chen, L. Wang and Y. Xu, In Situ Detection of Endotoxin in Bacteriostatic Process by SERS Chip Integrated Array Microchambers within Bioscaffold Nanostructures and SERS Tags, ACS Appl. Mater. Interfaces, 2020, 12(26), 28985–28992 CAS.
- A. Verde, M. Mangini, S. Managò, C. Tramontano, I. Rea, D. Boraschi, P. Italiani and A. C. De Luca, SERS Sensing of Bacterial Endotoxin on Gold Nanoparticles, Front. Immunol., 2021, 12, 758410 CrossRef CAS PubMed.
- Z. Luo, Q. Qi, L. Zhang, R. Zeng, L. Su and D. Tang, Branched polyethylenimine-modified upconversion nanohybrid-mediated photoelectrochemical immunoassay with synergistic effect of dual-purpose copper ions, Anal. Chem., 2019, 91(6), 4149–4156 CrossRef CAS PubMed.
- L. Liu, F. He, Y. Yu and Y. Wang, Application of FRET biosensors in mechanobiology and mechanopharmacological screening, Front. Bioeng. Biotechnol., 2020, 8, 1299 Search PubMed.
- J. Wu, A. Zawistowski, M. Ehrmann, T. Yi and C. Schmuck, Peptide functionalized polydiacetylene liposomes act as a fluorescent turn-on sensor for bacterial lipopolysaccharide, J. Am. Chem. Soc., 2011, 133(25), 9720–9723 CrossRef CAS PubMed.
- J. Sun, P. Zhu, X. Wang, J. Ji, J. d. D. Habimana, J. Shao, H. Lei, Y. Zhang and X. Sun, Cell based-green fluorescent biosensor using cytotoxic pathway for bacterial lipopolysaccharide recognition, J. Agric. Food Chem., 2018, 66(26), 6869–6876 CrossRef CAS PubMed.
- P. Xie, L. Zhu, X. Shao, K. Huang, J. Tian and W. Xu, Highly sensitive detection of lipopolysaccharides using an aptasensor based on hybridization chain reaction, Sci. Rep., 2016, 6(1), 1–8 CrossRef PubMed.
- J. Z. Kubicek-Sutherland, D. M. Vu, A. Noormohamed, H. M. Mendez, L. R. Stromberg, C. A. Pedersen, A. C. Hengartner, K. E. Klosterman, H. A. Bridgewater and V. Otieno, Direct detection of bacteremia by exploiting host-pathogen interactions of lipoteichoic acid and lipopolysaccharide, Sci. Rep., 2019, 9(1), 1–14 CAS.
- M. A. Khadimallah, M. Hussain, K. M. Khedher, S. M. Bouzgarrou, A. F. Al Naim, M. N. Naeem, M. Taj, Z. Iqbal and A. Tounsi, Vibration of SWCNTs: Consistency and behavior of polynomial law index with Galerkin's model, Adv. Nano Res., 2020, 9(4), 251–261 Search PubMed.
- T. Chen, A. Sheng, Y. Hu, D. Mao, L. Ning and J. Zhang, Modularization of three-dimensional gold nanoparticles/ferrocene/liposome cluster for electrochemical biosensor, Biosens. Bioelectron., 2019, 124, 115–121 CrossRef PubMed.
- J. Zhang, R. Oueslati, C. Cheng, L. Zhao, J. Chen, R. Almeida and J. Wu, Rapid, highly sensitive detection of Gram-negative bacteria with lipopolysaccharide based disposable aptasensor, Biosens. Bioelectron., 2018, 112, 48–53 CrossRef CAS PubMed.
- S. Sannigrahi, S. K. Arumugasamy, J. Mathiyarasu and K. Suthindhiran, Magnetosome-anti-Salmonella antibody complex based biosensor for the detection of Salmonella typhimurium, Mater. Sci. Eng., C, 2020, 114, 111071 CrossRef CAS PubMed.
- M. Cho, L. Chun, M. Lin, W. Choe, J. Nam and Y. Lee, Sensitive electrochemical sensor for detection of lipopolysaccharide on metal complex immobilized gold electrode, Sens. Actuators, B, 2012, 174, 490–494 CrossRef CAS.
- H. Jiang, J. Yang, K. Wan, D. Jiang and C. Jin, Miniaturized paper-supported 3D cell-based electrochemical sensor for bacterial lipopolysaccharide detection, ACS Sens., 2020, 5(5), 1325–1335 CrossRef CAS PubMed.
- J. Hicks, N. Silman, S. Jackson, J. Aylott and F. Rawson, Mass transport of lipopolysaccharide induced H2O2 detected by an intracellular carbon nanoelectrode sensor, Bioelectrochemistry, 2020, 135, 107547 CrossRef CAS PubMed.
- X. Fan, Z. Li, S. Wang, L. Liu, P. Liu, F. Chen and X. Zheng, Electrochemical Impedance biosensor for the determination of lipopolysaccharide using peptide as the recognition molecule, J. Braz. Chem. Soc., 2019, 30, 1762–1768 CAS.
- F. Ma, A. Rehman, M. Sims and X. Zeng, Antimicrobial susceptibility assays based on the quantification of bacterial lipopolysaccharides via a label free lectin biosensor, Anal. Chem., 2015, 87(8), 4385–4393 CrossRef CAS PubMed.
- J. Qin, H. Sun, H. Hao, L. Jia, C. Yao, Q. Wang and H. Yang, Impedimetric Biosensor for the Detection of Bacterial Lipopolysaccharide Based on Lectin-functionalized Gold Nanoparticle–Graphene Composites, Sens. Mater., 2019, 31(9), 2917–2929 CAS.
- W.-J. Shen, Y. Zhuo, Y.-Q. Chai and R. Yuan, Ce-based metal-organic frameworks and DNAzyme-assisted recycling as dual signal amplifiers for sensitive electrochemical detection of lipopolysaccharide, Biosens. Bioelectron., 2016, 83, 287–292 CrossRef CAS PubMed.
- M. Pourmadadi, J. S. Shayeh, M. Omidi, F. Yazdian, M. Alebouyeh and L. Tayebi, A glassy carbon electrode modified with reduced graphene oxide and gold nanoparticles for electrochemical aptasensing of lipopolysaccharides from Escherichia coli bacteria, Microchim. Acta, 2019, 186(12), 1–8 CrossRef PubMed.
- M. D. Oliveira, C. A. Andrade, M. T. Correia, L. C. Coelho, P. R. Singh and X. Zeng, Impedimetric biosensor based on self-assembled hybrid cystein-gold nanoparticles and CramoLL lectin for bacterial lipopolysaccharide recognition, J. Colloid Interface Sci., 2011, 362(1), 194–201 CrossRef CAS PubMed.
- B. Posha, S. R. Nambiar and N. Sandhyarani, Gold atomic cluster mediated electrochemical aptasensor for the detection of lipopolysaccharide, Biosens. Bioelectron., 2018, 101, 199–205 CrossRef CAS PubMed.
- S. Chen, X. Chen, L. Zhang, J. Gao and Q. Ma, Electrochemiluminescence Detection of Escherichia coli O157:H7 Based on a Novel Polydopamine Surface Imprinted Polymer Biosensor, ACS Appl. Mater. Interfaces, 2017, 9(6), 5430–5436 CrossRef CAS PubMed.
- V. Gaudin, Chapter 11 – Receptor-based electrochemical biosensors for the detection of contaminants in food products, in Electrochemical Biosensors, ed. A. A. Ensafi, Elsevier, 2019, pp. 307–365 Search PubMed.
- L.-X. You, N.-J. Chen, L. Wang, J. Chen, S.-F. Qin, C. Rensing, Z.-Y. Lin and S.-G. Zhou, Electrochemiluminescence for the identification of electrochemically active bacteria, Biosens. Bioelectron., 2019, 137, 222–228 CrossRef CAS PubMed.
- H. Sun, P. Miao, Y. Tang, B. Wang, J. Qian and D. Wang, An elastography analytical method for the rapid detection of endotoxin, Analyst, 2015, 140(13), 4374–4378 RSC.
- Z. Yang, Y. Wang and D. Zhang, An integrated multifunctional photoelectrochemical platform for simultaneous capture, detection, and inactivation of pathogenic bacteria, Sens. Actuators, B, 2018, 274, 228–234 CrossRef CAS.
- Y. Hou, L. Zhu, H. Hao, Z. Zhang, C. Ding, G. Zhang, J. Bi, S. Yan, G. Liu and H. Hou, A novel photoelectrochemical aptamer sensor based on rare-earth doped Bi2WO6 and Ag2S for the rapid detection of Vibrio parahaemolyticus, Microchem. J., 2021, 165, 106132 CrossRef CAS.
- S. Prolongo, R. Moriche, A. Jiménez-Suárez, M. Sánchez and A. Ureña, Advantages and disadvantages of the addition of graphene nanoplatelets to epoxy resins, Eur. Polym. J., 2014, 61, 206–214 CrossRef CAS.
- E. Frackowiak, K. Metenier, V. Bertagna and F. Beguin, Supercapacitor electrodes from multiwalled carbon nanotubes, Appl. Phys. Lett., 2000, 77(15), 2421–2423 CrossRef CAS.
- Y. Yim, H. Shin, S. M. Ahn and D.-H. Min, Graphene oxide-based fluorescent biosensors and their biomedical applications in diagnosis and drug discovery, Chem. Commun., 2021, 57(77), 9820–9833 RSC.
- Y. Yim, H. Shin, S. M. Ahn and D.-H. Min, Graphene oxide-based fluorescent biosensors and their biomedical applications in diagnosis and drug discovery, Chem. Commun., 2021, 57(77), 9820–9833 RSC.
- I. M. Obaidat, B. Issa and Y. Haik, Magnetic properties of magnetic nanoparticles for efficient hyperthermia, Nanomaterials, 2015, 5(1), 63–89 CrossRef PubMed.
- A. G. Kolhatkar, A. C. Jamison, D. Litvinov, R. C. Willson and T. R. Lee, Tuning the magnetic properties of nanoparticles, Int. J. Mol. Sci., 2013, 14(8), 15977–16009 CrossRef PubMed.
- S. Höfinger, M. Melle-Franco, T. Gallo, A. Cantelli, M. Calvaresi, J. A. Gomes and F. Zerbetto, A computational analysis of the insertion of carbon nanotubes into cellular membranes, Biomaterials, 2011, 32(29), 7079–7085 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |