Open Access Article
Open Access ArticleNovel urea derivative-loaded PLGA nanoparticles to inhibit caries-associated Streptococcus mutans
Mengyun Zhang†
 a,
Ying Liao†b,
Xin Tong*c and
Fuhua Yan
a,
Ying Liao†b,
Xin Tong*c and
Fuhua Yan *d
*d
aNanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, Jiangsu 210008, China
bDepartment of Pediatric Dentistry, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, Jiangsu 210008, China
cDepartment of Oral Implantology, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, Jiangsu 210008, China. E-mail: 419311196@qq.com
dDepartment of Periodontology, Nanjing Stomatological Hospital, Medical School of Nanjing University, Nanjing, Jiangsu 210008, China. E-mail: yanfh@nju.edu.cn
First published on 2nd February 2022
Abstract
Caries is the most common chronic infectious disease in the human oral cavity and the existing anti-caries agents may lead to drug resistance and microecological imbalance. A novel urea derivative, 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea, has a potentially prominent antibacterial effect on cariogenic bacterial strain Streptococcus mutans UA159. In this study, we encapsulated the water-insoluble urea derivative in poly(lactic-co-glycolic acid) (PLGA) nanoparticles, performed physicochemical characterizations and explored its potential as a caries-preventive agent. The results showed that the drug-loaded PLGA nanoparticles exhibited satisfying surface morphology, particle size, size distribution and stability. With an optimized theoretical drug loading (10%), the drug-loaded PLGA nanoparticles exhibited negligible cytotoxicity against human oral squamous cell carcinoma cells. We noticed a biphasic drug release in vitro and the rate and cumulative release was higher in an acidic environment (pH 4.5) compared to a neutral environment (pH 7.4). The drug-loaded PLGA nanoparticles significantly inhibited the growth and lactic acid production of planktonic S. mutans as well as S. mutans biofilms. Our results indicate that the novel urea derivative-loaded PLGA nanoparticles serve as a promising anti-caries agent with remarkable pharmaceutical characteristics, low cytotoxicity, and satisfying antimicrobial effect.
Introduction
Dental caries is the most prevalent chronic infectious disease in the oral cavity. Antimicrobial agents have been used for decades to prevent, slow down and, hopefully, cease the process of dental caries.1 Systematic antibiotics including penicillin and tetracyclines can lead to antibiotic resistance which significantly reduces the efficiency of the drugs. Multiple topical antimicrobials have been developed in recent years to target the local micro-environment. Among them, fluoride, chlorhexidine and quaternary ammonium salts have been of interest.2 However, studies have also shown that the application of these antimicrobials is associated with unfavorable events, including fluoride resistance, systematic toxicity, genotoxicity and gastrointestinal symptoms.3–5 It is still challenging to develop novel antimicrobials to inhibit caries.In recent years, researchers have screened a series of small molecules which can potentially target cariogenic bacteria.6,7 Nelson et al. managed to locate a potential fluoride agonist, 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea (Fig. 1, CAS number: 3824-74-6), which exhibited a strong ability to increase fluoride toxicity on a cariogenic bacterial species, Streptococcus mutans (S. mutans).7 An investigation of the molecular structure showed that this chemical did not only interfere with fluoride transport but also had strong potential to exhibit antimicrobial effects alone.8 Previous studies have confirmed that similar urea derivatives can significantly inhibit the growth of Staphylococcus aureus and Pseudomonas aeruginosa.9 The diaryl rings contributed as hydrogen-bond donors and had the potential to bind and inhibit bacterial proteins such as DNA gyrase B and penicillin binding protein 1a (PBP 1a), which were known targets for several antibacterial agents.10,11 Moreover, the trifluoromethyl groups on the diaryl rings can enhance membrane permeation of the drug and increase its lipophilicity and metabolic stability.12
While 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea has great potentials as a locally administered antimicrobial, its insolubility in water and difficulty of staying in oral cavity remains obstacles. Over the years, poly(lactic-co-glycolic acid) (PLGA) nanoparticles, a FDA- and EMA-approved biodegradable drug delivery system, have been used as drug carriers for cancer, cardiovascular diseases, infections, and other health disorders.13,14 PLGA nanoparticulate systems can encapsulate various hydrophobic drugs and they have shown good stability with sustained drug release in vivo.15 The rate of polymer degradation has been found to be related to external pH conditions. Previous studies have reported that PLGA microspheres had an accelerated rate of degradation at lower pH compared to neutral state,16–18 indicating that drug release from PLGA systems could be pH-responsive. It has been widely accepted that dental caries only occurs and progresses in acidic environments.19 A pH-responsive drug carrier has long been favored in the development of anti-caries drugs. Therefore, PLGA is selected in the current study as an ideal carrier for 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea.
In this study, we loaded the novel urea derivative 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea in PLGA nanoparticles and explored its antimicrobial effects against cariogenic S. mutans. Nanoprecipitation was applied to obtain PLGA and drug-loaded PLGA nanoparticles, followed by physicochemical characterizations of nanoparticles, including examination of surface morphology, particle size, size distribution, polydispersity index (PDI), zeta potential, encapsulation efficiency, as well as loading efficiency. The stability and drug release profiles were evaluated under different pH conditions. Cytotoxicity of PLGA and drug-loaded PLGA nanoparticles was examined. Finally, the antibacterial efficacy of drug-loaded PLGA nanoparticles against planktonic S. mutans and S. mutans biofilms was determined.
Materials and methods
Chemicals, test cells and bacteria, and growth conditions
1,3-Bis[3,5-bis(trifluoromethyl)phenyl]urea was obtained from Sigma-Aldrich (St Louis, MO, USA). Poly(D,L-lactide-co-glycolide) (PLGA, lactide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glycolide 50
glycolide 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, molecular weight 24
50, molecular weight 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–38
000–38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) were obtained from Aladdin Biochemical Technology Co., Ltd (Shanghai, China). Chemicals including polysorbate 80 and acetonitrile which were used to prepare PLGA nanoparticles were purchased from Sigma-Aldrich and Titan Scientific Co., Ltd (Shanghai, China).
000 Da) were obtained from Aladdin Biochemical Technology Co., Ltd (Shanghai, China). Chemicals including polysorbate 80 and acetonitrile which were used to prepare PLGA nanoparticles were purchased from Sigma-Aldrich and Titan Scientific Co., Ltd (Shanghai, China).
Oral squamous cell carcinoma (OSCC) cell line CAL27, provided by the American Type Culture Collection (ATCC; Manassas, VA, USA), was used for cytotoxicity test. Cells were grown in Dulbecco's modified Eagle medium, high glucose (DMEM-high, Gibco, NY, USA) containing 10% fetal bovine serum (FBS, Sigma-Aldrich) and 1% penicillin–streptomycin solution (HyClone, Logan, USA) at 37 °C with 5% CO2.
S. mutans UA159 was used in this study for antimicrobial efficacy test. Bacterial strain was a gift from the State Key Laboratory of Oral Diseases (Sichuan University, Chengdu, China). For planktonic status, bacteria were cultured in Brain Heart Infusion (BHI, Becton, Dickinson and Company, USA) media and grown on BHI agar plates at 37 °C anaerobically (10% H2, 5% CO2, and 85% N2). For S. mutans biofilms, BHIS (BHI medium containing 1% sucrose) was used. Biofilms were formed on the surface of clarified saliva-coated hydroxyapatite (sHA) discs. Clarified saliva was prepared by centrifuging human whole unstimulated saliva at 4000 rpm for 20 min at 4 °C, followed by filtration through 0.22 μm polyethersulfone membrane.20 HA discs (8 mm in diameter and 2 mm in thickness) were purchased from Baiameng Bioactive Materials Co., Ltd (Chengdu, China).
Preparation of PLGA and drug-loaded PLGA nanoparticles
The nanoprecipitation technology was used to prepare the PLGA and drug-loaded PLGA nanoparticles. Polysorbate 80 (w/v 0.1%) was added in the aqueous phase. The organic phase was obtained by accurately weighing and dissolving 10 mg PLGA and specified amount of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea in 2 mL acetonitrile. The organic phase was added dropwise to 2 mL of the aqueous phase at a constant rate of 1 mL min−1 and thoroughly stirred at 1200 rpm with a magnetic stirrer (big squid white, IKA, Germany). The theoretical drug loading of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea in the PLGA was 100% w/w, 50% w/w, 20% w/w, 10% w/w and 5% w/w, respectively. Acetonitrile was removed by evaporation in a ventilator cabinet at room temperature overnight. The next day, the suspension was filtered with 0.45 μm filters (Titan Scientific, Shanghai, China) to remove the free drug. Finally, the resulting PLGA and drug-loaded PLGA nanoparticles suspension was directly used for further physicochemical characterization or stored at 4 °C until used for antimicrobial tests.Physicochemical characterization of nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and then placed in a water bath at 37 °C. At 0, 3, 9, 24 and 48 h, samples were taken and examined with TEM and DLS to investigate the changes in particle distribution and size.
1 and then placed in a water bath at 37 °C. At 0, 3, 9, 24 and 48 h, samples were taken and examined with TEM and DLS to investigate the changes in particle distribution and size.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (85
water (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 v/v) mixture and the isocratic flow rate was 10 mL min−1. Injection volume was 10 μL and detection was performed at 260 nm wavelength. Briefly, a specific amount of drug-loaded PLGA nanoparticles were completely dissolved in HPLC-grade acetonitrile (Merck, USA) under magnetic stirring at a uniform speed of 1200 rpm for 10 min to destroy the polymer network fully. The content of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea was then quantified with HPLC and the encapsulation and loading efficiency were determined using the following calculation formulas:
15 v/v) mixture and the isocratic flow rate was 10 mL min−1. Injection volume was 10 μL and detection was performed at 260 nm wavelength. Briefly, a specific amount of drug-loaded PLGA nanoparticles were completely dissolved in HPLC-grade acetonitrile (Merck, USA) under magnetic stirring at a uniform speed of 1200 rpm for 10 min to destroy the polymer network fully. The content of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea was then quantified with HPLC and the encapsulation and loading efficiency were determined using the following calculation formulas:| Encapsulation efficiency (% EE) = amount of drug in nanoparticles/total amount of drug added × 100%. |
| Loading efficiency (% LE) = amount of drug in nanoparticles/total weight of nanoparticles × 100%. |
Antimicrobial activity of drug-loaded nanoparticles on S. mutans
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 and incubated at 37 °C anaerobically until OD600 nm reached 0.2. Cells were then centrifuged and resuspended in fresh BHI medium. Twenty μL of PLGA nanoparticles (5 mg mL−1) or drug-loaded nanoparticles was added to 1.98 mL bacterial suspension and incubated at 37 °C. Sterile water was added in the control group. Bacteria growth was monitored with Spectra MaxM3 (Molecular Devices, San Jose, CA, USA) by measuring OD600 nm every hour.
20 and incubated at 37 °C anaerobically until OD600 nm reached 0.2. Cells were then centrifuged and resuspended in fresh BHI medium. Twenty μL of PLGA nanoparticles (5 mg mL−1) or drug-loaded nanoparticles was added to 1.98 mL bacterial suspension and incubated at 37 °C. Sterile water was added in the control group. Bacteria growth was monitored with Spectra MaxM3 (Molecular Devices, San Jose, CA, USA) by measuring OD600 nm every hour.Lactic acid production of planktonic S. mutans cells in the presence of drug-loaded nanoparticles was measured as previously described.21 Briefly, S. mutans cells were incubated until mid-exponential phase (OD600 nm ∼ 0.5), harvested with centrifugation and resuspended in potassium phosphate buffer (pH 7.0, 90 mM). Either PLGA or drug-loaded PLGA nanoparticles were added and the suspensions were incubated at 37 °C for 1 h. The same amount of sterile water was added as control. At the end of the incubation, the suspensions were centrifuged, and the supernatant was used to determine the amount of lactic acid enzymatically.22
Preparation of the saliva coated-hydroxyapatite discs (s-HA). Hydroxyapatite (HA) discs were sonicated in deionized water for 10 min to remove free debris on the surface and then put in a glass Petri dish with deionized water for sterilization at 121 °C for 15 min. The sterile HA discs were randomly placed into the wells of 24-well plates. A volume of 2 mL of clarified saliva (sterile saliva containing biologically active substances such as various proteins, inorganic ions and trace elements) was added to each well and incubated at 37 °C for 2 h to form saliva-coated hydroxyapatite discs (s-HA) as described previously.20
SEM images, CFU counts and pH measurement. Overnight cultures of S. mutans UA159 were transferred into fresh BHI medium until they reached the mid-exponential phase (OD600 nm ∼ 0.5) and were diluted 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
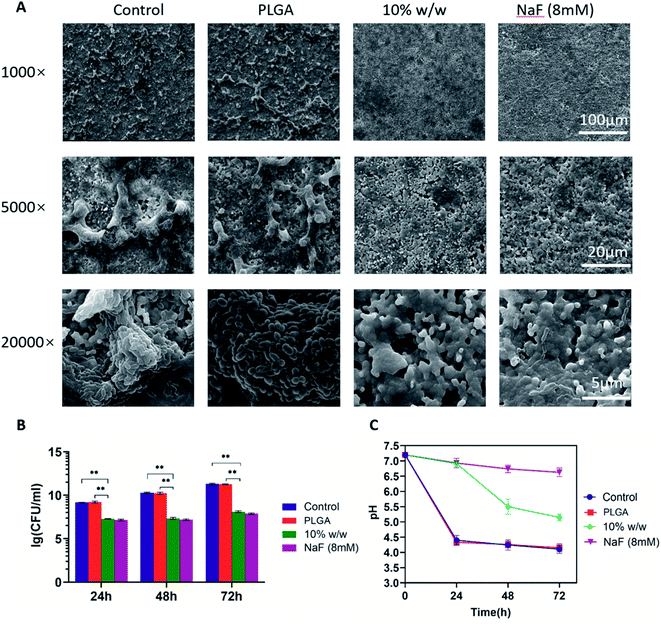
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in fresh BHI with 1% sucrose (BHIS). Next, 1.98 mL of culture was transferred into wells of 24-well plates with an sHA disc in each well. Twenty μL suspensions of PLGA nanoparticles (5 mg mL−1), drug-loaded nanoparticles (theoretical drug loading 10% w/w), sodium fluoride (final concentration 8 mM) or sterile water (control) was added in each well. Cultures were then incubated at 37 °C anaerobically and media were changed every 24 h. Biofilms formed at 24 h were examined with scanning electron microscopy (SEM) (Quanta 400 FEG, FEI, USA). Samples were taken and washed twice with CPW and then fixed in 2.5% glutaraldehyde at 4 °C overnight. They were then serially dehydrated with ethanol (50%, 60%, 70%, 80%, 90%, and 100%). Photos were examined at ×1000, ×5000, and ×20
100 in fresh BHI with 1% sucrose (BHIS). Next, 1.98 mL of culture was transferred into wells of 24-well plates with an sHA disc in each well. Twenty μL suspensions of PLGA nanoparticles (5 mg mL−1), drug-loaded nanoparticles (theoretical drug loading 10% w/w), sodium fluoride (final concentration 8 mM) or sterile water (control) was added in each well. Cultures were then incubated at 37 °C anaerobically and media were changed every 24 h. Biofilms formed at 24 h were examined with scanning electron microscopy (SEM) (Quanta 400 FEG, FEI, USA). Samples were taken and washed twice with CPW and then fixed in 2.5% glutaraldehyde at 4 °C overnight. They were then serially dehydrated with ethanol (50%, 60%, 70%, 80%, 90%, and 100%). Photos were examined at ×1000, ×5000, and ×20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 magnification under the microscope with an accelerating voltage of 20 kV. The images displayed are randomly selected from at least three fields.
000 magnification under the microscope with an accelerating voltage of 20 kV. The images displayed are randomly selected from at least three fields.After 24, 48 and 72 h incubation at 37 °C anaerobically, the HA discs were washed twice with sterile cysteine peptone water (CPW, containing yeast extract, peptone, sodium chloride, and cysteine HCl, pH 7.2) to remove loosely attached bacteria. Then, the discs were transferred to a 5 mL tube with 2 mL sterile CPW, and the S. mutans biofilms were dispersed by cell scrapers and sonicated on ice for 10 min at an amplitude of 40 W. The bacterial suspension was serially diluted with sterile deionized water, and 100 μL of the diluted solution was plated on BHI agar plates, incubating at 37 °C under anaerobic conditions for 48 h before the colony forming unit (CFU) was counted.
The supernatant (centrifuged at 5000 rpm for 10 min at 4 °C) of biofilms at 24, 48 and 72 h were taken and pH was measured with pH meter (FiveEasy Plus, FE28-standard, Mettler Toledo, Switzerland).
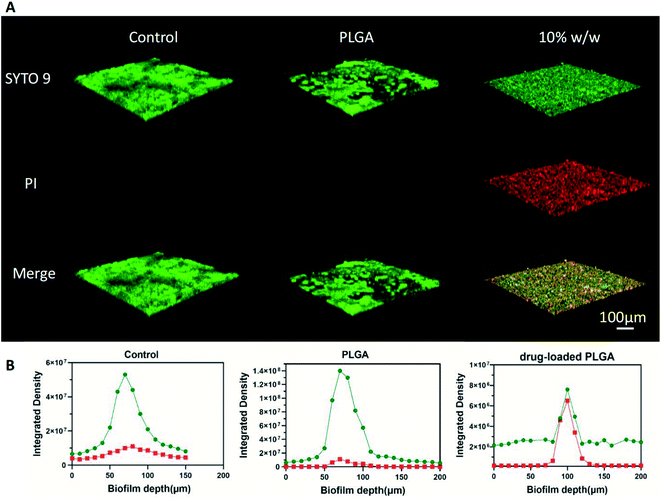
Bacteria live/dead staining assay. Confocal laser scanning microscopy (CLSM, Nikon A1; Nikon Inc., Tokyo, Japan) was used to visualize live and dead bacterial cells in 24 h biofilms. Samples were first washed with 0.9% sodium chloride to remove loosely adhered cells. LIVE/DEAD BacLight Bacterial Viability Kit (Life Technologies Corporation, New York, USA) was used according to the manufacturer's instructions. SYTO 9 (excitation, 480 nm; emission, 500 nm) and propidium iodide (PI, excitation, 490 nm; emission, 635 nm) were used to stain viable and dead cells, respectively. Biofilm images were captured with CLSM at magnifications of 10×. The biofilm depth was examined by generating the optical sections of 10 μm.23 ImageJ software (v1.48, National Institutes of Health, USA) was used to quantify the integrated density. The quantity of viable and dead cells was represented by the integrated density of green and red fluorescence, respectively.
Cytotoxicity assay
The cytotoxicity of PLGA and drug-load PLGA nanoparticles was detected using OSCC CAL27 cell line with CCK-8 assay kit (Meilun Biotechnology, Dalian, China). In brief, 5000 cells per well were seeded in 100 μL of growth medium (high-glucose Dulbecco's modified Eagle's medium (DMEM) with 10% (v/v) fetal bovine serum and 1% (v/v) penicillin–streptomycin solution.) in 96-well polypropylene microtiter plates and then cultured in an incubator at 37 °C with 5% CO2 for 24 h. After the cells adhered to the wall, the culture medium was replaced by a fresh medium containing nanoparticles of different concentrations. Control groups (with deionized water) and blank groups (cell-free medium) were set. Five replicates were placed for each concentration. After incubation for 24, 48 and 72 h, 10 μL CCK-8 solution was added to each well and incubated at 37 °C for 2 h. Finally, the light absorption value of each well was detected by Spectra Max M3 at 450 nm.Results
The morphology and size of nanoparticles
The PLGA and drug-loaded PLGA nanoparticles were successfully prepared by nanoprecipitation technology. TEM images showed that the nanoparticles were almost spherical in shape and had a smooth surface without pore. They were dispersed with each other (Fig. 2A and B). The size of all nanoparticles was less than 200 nm, which was confirmed by DLS analysis (Fig. 2C and D). Also, DLS showed that the nanoparticles had a narrow and normalized size distribution.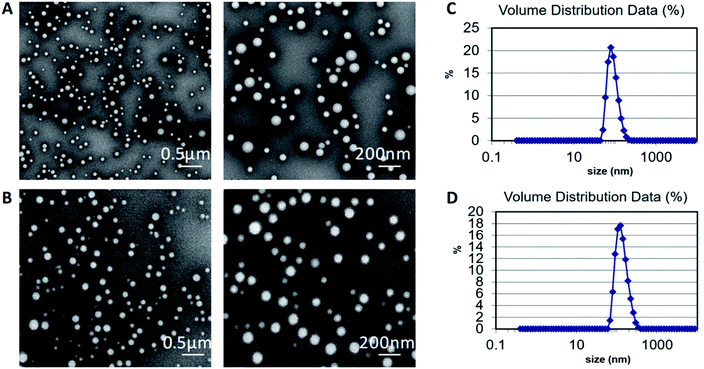 | ||
| Fig. 2 Representative TEM images of (A) PLGA nanoparticles and (B) drug-loaded PLGA nanoparticles. DLS size distribution of (C) PLGA nanoparticles and (D) drug-loaded PLGA nanoparticles. | ||
Encapsulation and loading efficiency of drug-loaded PLGA nanoparticles
Encapsulation efficiency (% EE) and loading efficiency (% LE) of drug-loaded PLGA nanoparticles were evaluated with different theoretical drug loadings. Particle size, PDI, and zeta potential were also measured. Table 1 showed that the average particle size of PLGA nanoparticles was 102.63 ± 1.46 nm, which was significantly smaller than that of drug-loaded PLGA nanoparticles (ranging from 112.6 nm to 140.6 nm). Particle sizes increased with the rise of theoretical drug loading. Zeta potentials of nanoparticles also increased in negativity with the increase of theoretical drug loading (from −13.1 mV to −27.3 mV). PDIs of all groups were less than 0.2, indicating a narrow size distribution. Based on % EE and % LE of different theoretical drug loading, 10% w/w was selected for further experiments as it exhibited a satisfying % EE (94.8 ± 2.8%) and % LE (8.7 ± 0.2%).| Theoretical drug loading (%, w/w) | Particle size (nm) | PDI | Zeta potential (mV) | % EE | % LE |
|---|---|---|---|---|---|
| 0 | 102.6 ± 1.5 | 0.07 ± 0.04 | −13.1 ± 0.4 | — | — |
| 100 | 140.7 ± 0.6 | 0.08 ± 0.04 | −13.3 ± 0.8 | 11.5 ± 0.7 | 10.3 ± 0.6 |
| 50 | 127.9 ± 2.2 | 0.11 ± 0.02 | −14.2 ± 1.1 | 40.9 ± 9.6 | 16.8 ± 2.9 |
| 20 | 120.2 ± 0.6 | 0.15 ± 0.06 | −24.9 ± 1.0 | 64.6 ± 14.5 | 11.4 ± 2.4 |
| 10 | 113.5 ± 2.1 | 0.04 ± 0.01 | −25.6 ± 0.5 | 94.8 ± 2.8 | 8.7 ± 0.2 |
| 5 | 112.6 ± 2.0 | 0.05 ± 0.02 | −27.3 ± 0.7 | 95.6 ± 1.5 | 4.6 ± 0.0 |
Stability of PLGA and drug-loaded PLGA nanoparticles
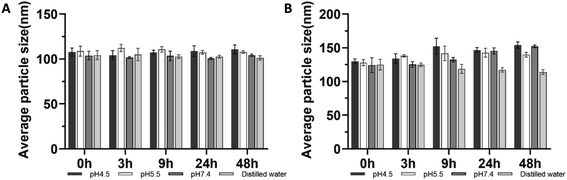
Fig. 3 shows the change of particle sizes of PLGA and drug-loaded PLGA nanoparticles at 37 °C under different pH conditions during 48 h incubation. Changes were insignificant, while we noticed the size of drug-loaded particles had a slight trend of increasing under acidic conditions. Otherwise, particle sizes remained around 100 nm for PLGA nanoparticles and 130 nm for drug-loaded nanoparticles throughout the incubation period.In vitro drug release behavior of drug-loaded PLGA nanoparticles
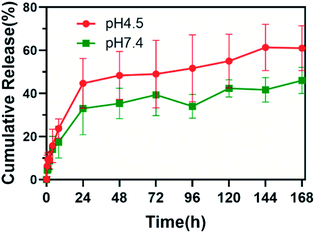
The release behavior of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea from drug-loaded PLGA was evaluated at different pH values (7.4 and 4.5). The result is presented in Fig. 4. At pH 4.5, approximately 45% of the total loaded content was released from drug-loaded PLGA during the first 24 h. After the initial rapid release, drug release continued and finally reached 61% of the total loaded content after 7 days. In neutral environments, a relatively smaller amount of drug (33% of total loaded content) was released within the first 24 h, and the cumulative release was about 46% of total loaded content after 7 days.Inhibition on growth and lactic acid production in planktonic S. mutans
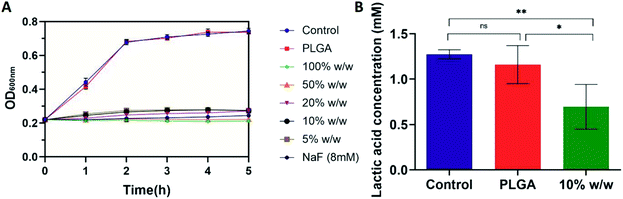
The growth profiles of S. mutans UA159 in the presence of PLGA and drug-loaded nanoparticles with different theoretical loadings were shown in Fig. 5A. PLGA nanoparticles did not inhibit bacterial growth. All drug-loaded nanoparticles showed almost complete growth inhibition. The inhibitive effect was similar to that of 8 mM NaF.Fig. 5B exhibited lactic acid production profiles of S. mutans UA159 treated with PLGA nanoparticles for 1 h. The co-incubation of bacterial cells with PLGA nanoparticles did not affect the ability of lactic acid production (1.16 ± 0.21 mM). Drug-loaded nanoparticles with 10% theoretical loading led to significantly less production of lactic acid (0.70 ± 0.25 mM).
Inhibition on formation and acid production of S. mutans biofilm
SEM images confirmed that drug-loaded PLGA nanoparticles inhibited the formation and development of biofilms on sHA discs (Fig. 6). While we noticed a thick layer of biofilms as well as a vast amount of extracellular matrix in control and PLGA groups, biofilms appeared thinner and less matrix was visible in drug-loaded PLGA group and sodium fluoride group (Fig. 6A).CFU counts confirmed that the presence of drug-loaded PLGA nanoparticles significantly decreased the number of viable cells in 24, 48 and 72 h biofilms (**p < 0.01, Fig. 6B). In control and PLGA groups, CFU counts reached 1.6 × 109, 1.8 × 1010 and 1.9 × 1011 CFU mL−1 at 24, 48 and 72 h respectively. In drug-loaded PLGA group, CFU counts was 1.7 × 107, 2.0 × 107 and 1.1 × 108 CFU mL−1 at 24, 48 and 72 h respectively. No significant difference was observed between drug-loaded PLGA and sodium fluoride groups.
At different time points (24, 48 and 72 h), pH of supernatant of biofilms was determined to reflect the ability of the biofilms to produce acid. As shown in Fig. 6C, biofilms in control and PLGA groups had similar supernatant pH, which dropped to pH 4.5 in the first 24 h. Drug-loaded PLGA nanoparticles significantly alleviated the pH drop, especially in the first 24 h. During the first 24 h, pH dropped from 7.2 to around 6.9. The decrease was similar to the NaF group during this period. The pH drop became more obvious afterwards. By the end of 72 h, the pH of the drug-loaded PLGA group was about 5.2 (Fig. 6C).
Live/dead staining of biofilms
As illustrated in Fig. 7A, CLSM images showed that the treatment of drug-loaded PLGA nanoparticles significantly increased fluorescence of dead bacterial cells in 24 h biofilms. The quantified integrated intensity of green and red fluorescence showed that the calculated live/dead bacterial ratio of 24 h S. mutans biofilms exposed to the drug-loaded PLGA nanoparticles was remarkably lower than that of the control group and PLGA group. Furthermore, differences were noticed in the thickness of 24 h S. mutans biofilms. The thickness of biofilms treated by sterile water and PLGA nanoparticles for 24 h was similar (approximately 90 μm), which was thicker than that of the biofilms treated by the drug-loaded PLGA nanoparticles samples (approximately 50 μm, Fig. 7B).Cytotoxicity assay
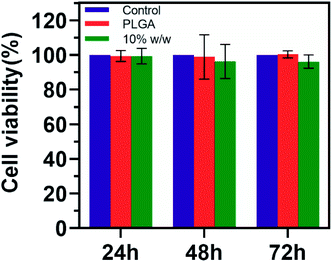
The cell cytotoxicity was evaluated with OSCC CAL-27. The viability of cells was tested 24, 48 and 72 h after the cells were treated with negative control, PLGA and drug-loaded PLGA (theoretical drug loading 10% w/w). Fig. 8 showed that both PLGA and drug-loaded PLGA nanoparticles did not affect the viability of cells significantly. The cell viability of CAL27 cells after co-incubation with drug-loaded PLGA nanoparticles for 24 (99.3% ± 4.5%), 48 (96.2% ± 9.8%) and 72 (96.2% ± 3.8%) h was higher than 90%.Discussion
The emergence of antimicrobial resistance in oral bacteria has been a rising problem when developing caries-preventive agents. In order to avoid drug resistance and promote effectiveness in local environment, small molecule antimicrobials have been of interest. This current study encapsulated a novel urea derivative, 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea, in biodegradable PLGA nanoparticles. The drug-loaded PLGA nanoparticles exhibited satisfying physicochemical characterizations and acceptable cytotoxicity. Moreover, the antimicrobial effect of the drug-loaded PLGA nanoparticles was confirmed in planktonic S. mutans as well as S. mutans biofilms. This novel urea derivative-loaded PLGA nanoparticles provide as a promising agent to be used in caries prevention and treatment.The application of urea derivatives as antimicrobials could date back to the 1940s when the antimicrobial activity of urea derivatives against Gram-positive bacteria was first reported by Weinstein L. et al.24 In the past several years, different urea derivates have been developed to target bacteria including Escherichia coli, Staphylococcus aureus, and Pseudomonas aeruginosa. 1,3-Bis[3,5-bis(trifluoromethyl)phenyl]urea was first identified as a fluoride toxicity agonist due to its strong ability to bind F− mediated by the diaryl rings.7,25 We identified several functional groups in its chemical structure and realized that it had the potential to exert antimicrobial effect alone (as described in Introduction). In order to deliver and confirm the antimicrobial effect, PLGA nanoparticles were employed.
The drug-loaded PLGA nanoparticles exhibited satisfying pharmaceutical characters. The particle size is a crucial parameter as it directly affects the physical stability, cellular uptake, biodistribution, and drug release.26 The average size of the prepared nanoparticles was 100–140 nm, which was within the ideal range of nanoparticles for drug delivery.27 The nanoparticle size increased with the rise of theoretical drug loading, which could be the result of the increased number of active ingredients encapsulated in nanoparticles. Also, the PDI values of all prepared nanoparticles (0.04–0.15) indicated that the nanoparticles were monodispersed,28 as observed in TEM images (Fig. 2). Zeta potentials of prepared nanoparticles represented carboxyl groups on the surface of the PLGA polymers and have been previously reported to be associated with the stability of nanoparticles in colloidal systems.29 All nanoparticles prepared in this study had negative zeta potentials, ranging from −13.1 to −27.3 mV, indicating that they were relatively stable. This was further confirmed with the stability test (Fig. 3). By optimizing theoretical loading, we reached a high % EE and % LE (94.8% and 8.7% respectively).
The in vitro release profiles of 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea from the drug-loaded nanoparticles in different pH environments showed a biphasic configuration, consisting of an initial burst followed by a steady, sustained release (Fig. 4). This was in line with previous studies which also used PLGA as drug carriers.30 During the first 24 hours, the burst release could be related to the free drug which was absorbed on the surface of the PLGA nanoparticles. After the initial burst release, the profile showed a steady sustained-release manner for over 168 h, probably due to the drug encapsulated at the core compartment retained firmly inside the nanoparticle.31 There are also reports that the encapsulated drug released slowly because of the relatively dense polymer or the few existing pores during the degradation and hydration proceed of PLGA nanoparticles.32,33 It is worth mentioning that the release profile of the drug from PLGA nanoparticles was pH-responsive. The rate and cumulative release of drug were significantly higher in acidic environment compared to neutral environment. This was closely related to the pH-responsiveness of PLGA polymer. Low pH condition catalyzes the breakage of ester bond of polymerization and enhances erosion of PLGA, thus accelerating drug release.34 Specifically, the pH-dependent release is highly appreciated for caries prevention, as only under acidic conditions does the disease initiate and progress.
In vitro antimicrobial experiments showed that drug-loaded PLGA nanoparticles inhibited the growth and acid production of both planktonic S. mutans and S. mutans biofilms. The ability to produce acid and form biofilms are the major virulence factors of S. mutans.35 It is somewhat surprising that the novel urea derivative used in this study can inhibit both processes. As described previously, in silico studies have predicted that similar urea derivatives can bind and inhibit key enzymes such as DNA gyrase B and PBP.36,37 The binding seems to be mediated by hydrogen bond between enzymes and the diaryl rings in the drug. It is speculated that 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea could also bind to other enzymes and act as an inhibitor, especially enzymes associated with metabolic pathways and biofilm formation. For example, we noticed that the extracellular matrix in biofilms treated with drug-loaded PLGA nanoparticles was significantly less with SEM examination (Fig. 6A). The activity of glucosyltransferase, which is responsible for the production of extracellular polysaccharides (EPS), was highly likely to be inhibited. Molecular docking and in vitro examinations will be performed to further confirm the binding of specific proteins with 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea. In addition, the urea derivative could be hydrolyzed by urease to produce NH3, which is an important alkalic substance in oral cavity and can be related to the limited pH drop in biofilms.38
Conclusion
In conclusion, by encapsulating 1,3-bis[3,5-bis(trifluoromethyl)phenyl]urea in PLGA nanoparticles, we aimed to develop a novel anti-caries agent. The drug is released from the nanoparticles in a pH-dependent and slow released manner. Both acid production and biofilm formation of S. mutans are inhibited by the drug-loaded nanoparticles. Furthermore, the mechanism of the novel anti-caries agent to inhibit S. mutans still requires further research.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Funds (grant number 82001035); the Jiangsu Natural Science Funds (grant number SBK2020041847); and the Nanjing Clinical Research Center for Oral Diseases (grant number 2019060009).References
- J. Y. Niu, I. X. Yin, W. K. K. Wu, Q. L. Li, M. L. Mei and C. H. Chu, Arch. Oral Biol., 2021, 122, 105022 CrossRef CAS PubMed.
- W. Qiu, Y. Zhou, Z. Li, T. Huang, Y. Xiao, L. Cheng, X. Peng, L. Zhang and B. Ren, BioMed Res. Int., 2020, 2020, 5658212 Search PubMed.
- Y. Liao, J. Chen, B. W. Brandt, Y. Zhu, J. Li, C. van Loveren and D. M. Deng, PLoS One, 2015, 10, e0122630 CrossRef PubMed.
- T. M. Karpinski and A. K. Szkaradkiewicz, Eur. Rev. Med. Pharmacol. Sci., 2015, 19, 1321–1326 CAS.
- L. Drew, Nature, 2016, 537, S60–S62 CrossRef CAS PubMed.
- Q. Zhang, B. Nijampatnam, Z. Hua, T. Nguyen, J. Zou, X. Cai, S. M. Michalek, S. E. Velu and H. Wu, Sci. Rep., 2017, 7, 5974 CrossRef PubMed.
- J. W. Nelson, M. S. Plummer, K. F. Blount, T. D. Ames and R. R. Breaker, Chem. Biol., 2015, 22, 527–534 CrossRef CAS PubMed.
- Y. Fan, C. Payne and S. R. Kass, J. Org. Chem., 2018, 83, 10855–10863 CrossRef CAS PubMed.
- Y. L. Wu, Yaoxue Xuebao, 1965, 12, 523–532 CAS.
- C. Limban, M. C. Chifiriuc, M. T. Caproiu, F. Dumitrascu, M. Ferbinteanu, L. Pintilie, A. Stefaniu, I. M. Vlad, C. Bleotu, L. G. Marutescu and D. C. Nuta, Molecules, 2020, 25 DOI:10.3390/molecules25071478.
- F. M. Sroor, A. M. Othman, M. A. Tantawy, K. F. Mahrous and M. E. El-Naggar, Bioorg. Chem., 2021, 112, 104953 CrossRef CAS PubMed.
- J. A. Ma and D. Cahard, Chem. Rev., 2008, 108, PR1–PR43 CrossRef CAS PubMed.
- M. Mir, N. Ahmed and A. U. Rehman, Colloids Surf., B, 2017, 159, 217–231 CrossRef CAS PubMed.
- F. Danhier, E. Ansorena, J. M. Silva, R. Coco, A. Le Breton and V. Preat, J. Controlled Release, 2012, 161, 505–522 CrossRef CAS PubMed.
- M. Dandamudi, P. McLoughlin, G. Behl, S. Rani, L. Coffey, A. Chauhan, D. Kent and L. Fitzhenry, Pharmaceutics, 2021, 13 DOI:10.3390/pharmaceutics13101590.
- B. S. Zolnik and D. J. Burgess, J. Controlled Release, 2007, 122, 338–344 CrossRef CAS PubMed.
- Y. Hua, Y. Su, H. Zhang, N. Liu, Z. Wang, X. Gao, J. Gao and A. Zheng, Drug Delivery, 2021, 28, 1342–1355 CrossRef CAS PubMed.
- Y. Liu, A. H. Ghassemi, W. E. Hennink and S. P. Schwendeman, Biomaterials, 2012, 33, 7584–7593 CrossRef CAS PubMed.
- W. Krzysciak, A. Jurczak, D. Koscielniak, B. Bystrowska and A. Skalniak, Eur. J. Clin. Microbiol. Infect. Dis., 2014, 33, 499–515 CrossRef CAS PubMed.
- X. Wang, L. Liu, X. Zhou, Y. Huo, J. Gao and H. Gu, BMC Oral Health, 2020, 20, 169 CrossRef CAS PubMed.
- Y. Liao, B. W. Brandt, M. Zhang, J. Li, W. Crielaard, C. van Loveren and D. M. Deng, Antimicrob. Agents Chemother., 2016, 60, 7509–7512 CrossRef CAS PubMed.
- W. A. Belli, D. H. Buckley and R. E. Marquis, Can. J. Microbiol., 1995, 41, 785–791 CrossRef CAS PubMed.
- M. Vaknin, D. Steinberg, J. D. Featherstone and O. Feuerstein, Laser Med. Sci., 2020, 35, 709–718 CrossRef PubMed.
- L. Weinstein and A. McDonald, Science, 1945, 101, 44–45 CrossRef CAS PubMed.
- N. Busschaert and P. A. Gale, Angew. Chem., Int. Ed., 2013, 52, 1374–1382 CrossRef CAS PubMed.
- G. Mittal, D. K. Sahana, V. Bhardwaj and M. N. Ravi Kumar, J. Controlled Release, 2007, 119, 77–85 CrossRef CAS PubMed.
- W. Huang and C. Zhang, Biotechnol. J., 2018, 13 DOI:10.1002/biot.201700203.
- A. A. Öztürk, L. M. Banderas, M. D. C. Otero, E. Yenilmez, B. Şenel and Y. Yazan, Trop. J. Pharm. Res., 2019, 18 DOI:10.4314/tjpr.v18i1.1.
- Z. Rahman, A. S. Zidan, M. J. Habib and M. A. Khan, Int. J. Pharm., 2010, 389, 186–194 CrossRef CAS PubMed.
- Y. Wang and Y. Tan, J. Drug Delivery Sci. Technol., 2016, 36, 183–191 CrossRef CAS.
- C. J. Martinez Rivas, M. Tarhini, W. Badri, K. Miladi, H. Greige-Gerges, Q. A. Nazari, S. A. Galindo Rodriguez, R. A. Roman, H. Fessi and A. Elaissari, Int. J. Pharm., 2017, 532, 66–81 CrossRef CAS PubMed.
- S. Fredenberg, M. Wahlgren, M. Reslow and A. Axelsson, Int. J. Pharm., 2011, 415, 34–52 CrossRef CAS PubMed.
- J. Wang, B. M. Wang and S. P. Schwendeman, J. Controlled Release, 2002, 82, 289–307 CrossRef CAS PubMed.
- N. Faisant, J. Siepmann and J. P. Benoit, Eur. J. Pharm. Sci., 2002, 15, 355–366 CrossRef CAS PubMed.
- L. Jia, S. Tao, J. Yang, K. Liang, Z. Yu, Y. Gao, M. Fan, M. Zhang, L. He and J. Li, Colloids Surf., B, 2021, 197, 111409 CrossRef CAS PubMed.
- K. W. Pugh, Z. Zhang, J. Wang, X. Xu, V. Munthali, A. Zuo and B. S. J. Blagg, ACS Med. Chem. Lett., 2020, 11, 1535–1538 CrossRef CAS PubMed.
- Z. T. Wen, J. P. Bitoun and S. Liao, PLoS One, 2015, 10, e0124319 CrossRef PubMed.
- Y. L. Liu, M. Nascimento and R. A. Burne, Int. J. Oral Sci., 2012, 4, 135–140 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this study. |
| This journal is © The Royal Society of Chemistry 2022 |