Open Access Article
Open Access ArticleFacile and wide-range size tuning of conjugated polymer nanoparticles for biomedical applications as a fluorescent probe†
Noriko Nakamura a,
Nobuaki Tanakab and
Seiichi Ohta
a,
Nobuaki Tanakab and
Seiichi Ohta *abcd
*abcd
aInstitute of Engineering Innovation, The University of Tokyo, 2-11-16 Yayoi, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: s-ohta@sogo.t.u-tokyo.ac.jp
bDepartment of Chemical System Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
cDepartment of Bioengineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
dPrecursory Research for Embryonic Science and Technology (PRESTO), Japan Science and Technology Agency (JST), 4-2-8 Honcho, Kawaguchi, Saitama 332-0012, Japan
First published on 14th April 2022
Abstract
Conjugated polymer nanoparticles (Pdots) are expected to be novel bioimaging and sensing probes. However, the size tuning required to control biological interactions has not been well established. Herein, we achieved a size-tunable synthesis of Pdots ranging from 30 to 200 nm by controlling the hydrolysis rate of the stabilising agent and evaluated their cellular imaging properties.
1. Introduction
Nanoparticles (NPs) play a key role in biomedical applications. A variety of nanoparticulated materials, such as lipid nanoparticles, polymer nanoassemblies, and inorganic nanocrystals, have demonstrated their enormous potential for biomedical applications, such as drug delivery, bioimaging, and sensing. The physicochemical properties of NPs, including size, shape, and surface chemistry, significantly affect their fate in biological systems, thereby determining their therapeutic or diagnostic activity. In particular, the correlation between the size of NPs and their interaction with cells and tissue has recently attracted considerable research interest.1–3 In addition to their physicochemical properties, the precise design and synthesis of NPs further render their specific characteristics, such as fluorescent,4 plasmonic,5 and magnetic properties.6Fluorescent NPs have attracted significant attention as effective bioimaging and sensing agents. Since small molecular fluorescent dyes were first reported in the late nineteenth century, they have been representative imaging probes for monitoring biological phenomena.7 The emergence of fluorescent NPs has accelerated the development of numerous imaging methodologies based on their brightness and controllable optical properties. A representative example is quantum dots (QDs), which are semiconductor nanocrystals showing size-dependent fluorescence due to the quantum confinement effect.4,8–10 They show excellent optical properties, including high photo-stability and tunable emission wavelengths, but the toxicity of QDs caused by their heavy metal components (e.g. cadmium) has been a potential concern.11,12 Recently, conjugated polymer NPs, often called polymer dots (Pdots), have been investigated as a new class of fluorescent NPs without heavy metal-related toxicity.13,14 While QDs show visible photoluminescence only when their size is within the single-nm range, Pdots maintain their fluorescent property with a size up to several hundred nanometres. This is advantageous for optimising their biological interaction via size. Additionally, Pdots can incorporate photo-functional molecules, such as fluorescent dyes15,16 or photosensitizers,17,18 leading to tunable photo-functions, such as efficient intra-particle fluorescent resonance energy transfer. A wide range of applications of Pdots has been proposed not only as a substitute for QDs but also as novel diagnostic/therapeutic agents.19,20
In general, Pdots are prepared using a post-polymerisation approach, such as the nanoprecipitation method using commercially available conjugated polymers. The nanoprecipitation method has demonstrated Pdot synthesis with a reliable size, usually up to 70 nm.7,21 NPs containing fluorescent conjugated polymers have also been synthesized by loading them into matrix polymer nanoparticles, such as poly(lactic-co-glycolic acid),22,23 phospholipid,24 as well as hyperbranched polyglycerol25 via the mini-emulsion method. For the application of Pdots for bioimaging and sensing, the size of Pdots needs to be carefully tuned by considering their behaviour in the biological system. The size-controlled synthesis of Pdot was firstly explored based on the nanoprecipitation method without stabilizing agent by changing the conjugated polymer concentration in organic phase. Wu et al. succeeded in synthesizing Pdot with different size ranging from 8 nm to 50–70 nm using five different conjugated polymers and demonstrating the labeling of macrophages in vitro.7 After that, there have been many literatures which demonstrated Pdot synthesis utilizing stabilizing agents such as polystyrene grafted with ethylene oxide functionalized with carboxyl groups26 as well as poly(styrene-co-maleic anhydride) (PSMAnh).21 These amphiphilic or hydrophilic stabilizing agents not only contributed to the colloidal stabilization of larger Pdots but also to the enhancement of biocompatibility as well as the ability for surface functionalization with biomolecules.27,28 Additionally, the size-tunable synthesis of Pdots with the stabilizing agents have been demonstrated by adjusting the synthetic condition. For instance, the concentration of fluorescent conjugated polymer, the water content in the organic solvent, and the temperature were reported to be the effective parameter to control the size of Pdots ranging from 50 nm to 200 nm via nanoprecipitation.29 The feed volume of the oil phase and the concentration of the surfactant were reported as the parameters to tune the size of Pdots from 40 nm to 210 nm via mini-emulsion method.25 However, although there have been demonstrated synthetic methods of Pdot with wide-range size tuning by controlling multiple parameters at the same time,30,31 a facile synthesis methodology just adjusting one parameter to achieve wide-range size-tuning of Pdot has not been explored yet. Furthermore, the size effect on the performance of Pdots in the biological environment has not been investigated before.
Herein, we propose a facile, size-controllable Pdot synthesis strategy for controlling biological interactions. In the nanoprecipitation method for Pdot synthesis, PMSAnh is widely used for stabilising hydrophobic conjugated polymers. During nucleation and growth via condensation/coagulation of conjugated polymers, hydrophobic PMSAnh is initially hydrolysed into a hydrophilic maleic acid copolymer to stabilise the particle surface. The particle growth is assumed to be terminated when the hydrolysis reaction proceeds to a certain threshold, upon which the particle surface becomes sufficiently stable to prevent further growth. This hypothetical formation mechanism has motivated us to alter the hydrolysis rate and subsequent electrostatic repulsion of PSMAnh via pH and ionic strength to control the size of Pdots, as shown in Fig. 1. The surface and optical properties of the synthesised Pdots, as well as their size-dependent cellular imaging functions, were investigated.
2. Experimental
2.1. Materials
Poly(9,9-duictylfluorene-alt-benzothiazole) (F8BT) and PSMAnh were purchased from Sigma-Aldrich (St. Louis, MO, USA). Tetrahydrofuran (THF), acetic acid, sodium acetate, sodium chloride (NaCl), N-hydroxysuccinimide (NHS), and dimethyl sulfoxide (DMSO) were purchased from FIJIFILM Wako Chemicals (Tokyo, Japan). 1-(3-Dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) and 4-(4,5-dimethyltheiazol-2,5-diphenyltetrazoliu) bromide (MTT) were purchased from Tokyo Chemical Industry Co., Ltd (Tokyo, Japan). Amine-terminated polyethylene glycol (PEG) was purchased from Yuka Sangyo Co., Ltd (Tokyo, Japan). LysoTraker™ Deep Red was purchased from Thermo Fisher Scientific, Inc. (Waltham, MA, USA).2.2. Synthesis of Pdot with different diameters
Pdots were synthesised using a nanoprecipitation method. F8BT and PMSAnh were dissolved in THF separately to prepare each solution at a concentration of 500 μg mL−1 and then the F8BT and PSMAnh solutions were mixed to prepare a solution mixture with a F8BT concentration of 400 μg mL−1 and a PSMAnh concentration of 100 μg mL−1. A 1 mL aliquot of the mixture solution was added to 20 mL of 10 mM acetate buffer with varying pH and NaCl concentrations (pH: 5.4, 5.6, 5.8, 6.0, or 6.2, NaCl concentration: 0, 50, 80, or 100 mM) under sonication and vigorous stirring to form a homogenous Pdot solution. After the nanoparticles were formed, the THF was removed by evaporation at room temperature. The obtained Pdots were purified by ultracentrifugation three times and then dispersed in deionised water.2.3. Morphology and size characterization of Pdots
The size and morphology of the obtained Pdots were evaluated by transmission electron microscopy (TEM, JEM-1400; JEOL, Tokyo, Japan). A copper grid surface with 400 mesh was subjected to plasma hydrophilic treatment. Next, 2 μL of Pdot dispersed in deionised water was applied to the grid, followed by staining with 2 μL of 2 wt% uranyl acetate. The TEM observations were conducted at an acceleration voltage of 120 kV.The hydrodynamic diameter of the Pdots was characterised by dynamic light scattering (DLS) in deionised water at 25 °C using a Zetasizer Pro (Marvern Instruments Ltd, Worcestershire, UK) with a He–Ne laser (wavelength = 633 nm). The intensity-averaged hydrodynamic diameter and polydispersity index (PDI) were derived using the cumulant method.
2.4. Optical properties of Pdots
UV-vis absorption spectra of Pdots dispersed in deionised water were obtained with a UV-1900i spectrophotometre (Shimadzu Co., Kyoto, Japan) using a 1 cm quartz cuvette at room temperature. Fluorescence spectra were recorded using a fluorophotometer RF-6000 (SHIMADZU Co., Kyoto, Japan).The quantum yield (QY) of the synthesised Pdots was evaluated using Rhodamine 6G, whose QY was 0.95,32 as a reference. The QY of the Pdot samples was calculated using the following equation:
2.5. Surface functionalization of Pdots
PEG conjugation to Pdots was performed by utilising carbodiimide cross-linking reaction between carboxyl groups on the surface of Pdots and terminal amines of PEG molecules. The Pdot solution in 20 mM HEPES buffer (pH 7.3) was reacted with 20 equivalents of amine-terminated PEG, EDC, and NHS for the surface carboxyl groups on Pdot. After the reaction solution was stirred overnight at room temperature, excess PEG, EDC, and NHS molecules were removed by ultracentrifugation three times.2.6. Characterization of PEGylated Pdots
The surface zeta potentials of the Pdots and PEGylated Pdots (PEG-Pdots) dispersed in deionised water were evaluated by conducting electrophoretic light scattering (ELS) measurements using a Zetasizer Pro at 25 °C.Fourier transform infrared spectroscopy (FT-IR) measurements of the Pdots and PEGylated Pdots were conducted using an IRSpirit (SHIMADZU Co., Kyoto, Japan) equipped with QATR-S. FT-IR spectra were obtained with a 4 cm−1 spectral resolution and 128 scans.
2.7. Cellular uptake of PEGylated Pdots
Mouse fibroblast NIH/3T3 cells were cultured and maintained in D-MEM medium with 10% fetal bovine serum and 1% penicillin streptomycin at 37 °C and 5% CO2. Cells were seeded on a 96-well plate with a density of 5000 cells per well, and then incubated for 6 h. These cells were subsequently treated with cell culture medium containing 5, 10 and 15 μg mL−1 of PEG-Pdot60, 80, 110, 170 or 220 for 18 h. Cells were then treated with 10 μL per well of MTT (5 mg mL−1 in PBS) and incubated for 2 h. After removing the medium, the produced formazan was lysed by adding 100 μL per well of DMSO. The absorbance of formazan in each well was measured at 560 nm using a multilabel plate reader ARVO X2 (ParkinElmer, Inc., Waltham, MA, USA).For confocal observation, cells were seeded on a 35 mm-glass bottom dish with a cell density of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2, and then incubated for 6 h. These cells were subsequently treated with culture medium containing 30 μg mL−1 PEG-Pdot60, 80, 110, 170, or 220 for 3 h. These cells were thoroughly treated with 50 nM of LysoTracker in the culture medium for 1 h subsequently after treating with PEG-Pdots. The LysoTracker and PEG-Pdots were observed after replacing culture medium to the fresh one. PEG-Pdots internalised into NIH/3T3 cells were observed using a confocal laser scanning microscopy LSM800 (Carl Zeiss Co. Ltd, Jena, Germany) with a 40× lens. A diode laser was used for LysoTracker, and an Ar laser was used for the PEG-Pdots.
000 cells per cm2, and then incubated for 6 h. These cells were subsequently treated with culture medium containing 30 μg mL−1 PEG-Pdot60, 80, 110, 170, or 220 for 3 h. These cells were thoroughly treated with 50 nM of LysoTracker in the culture medium for 1 h subsequently after treating with PEG-Pdots. The LysoTracker and PEG-Pdots were observed after replacing culture medium to the fresh one. PEG-Pdots internalised into NIH/3T3 cells were observed using a confocal laser scanning microscopy LSM800 (Carl Zeiss Co. Ltd, Jena, Germany) with a 40× lens. A diode laser was used for LysoTracker, and an Ar laser was used for the PEG-Pdots.
Quantification of Pdot uptake by NIH/3T3 cells was performed using flow cytometry. NIH/3T3 cells were maintained in D-MEM supplemented with 10% fetal bovine serum and 1% penicillin streptomycin at 37 °C and 5% CO2. Cells were seeded at 3 × 105 cells per well onto 12-well plates and allowed to attach for 24 h. These cells were incubated with a culture medium containing 5 μg mL−1 PEG-Pdots of different diameters for 18 h. The treated cells were washed with D-PBS (−) and harvested by trypsinisation. The fluorescent signal from PEG-Pdots internalised into each cell was quantified and analysed using FACSAria II and FlowJo software (BD Biosciences, Franklin Lakes, NJ, USA).
3. Results and discussion
3.1. Pdots with different diameters
F8BT was used as a fluorescent conjugated polymer in this study. F8BT and PSMAnh were dissolved in THF, followed by addition to the water phase with varying pH values and ionic strengths under sonication and stirring to synthesise Pdots. Fig. 2a shows the representative TEM images of Pdots synthesised at various pH, from 5.4 to 6.2. Acetate buffer (10 mM) with 80 mM NaCl was used as the water phase. The size distribution of the Pdots evaluated from the TEM images is shown in Fig. 2b. Pdots were of spherical morphology for all pH conditions. The size of Pdots decreased from ca. 220 to 60 nm with increasing pH. The decrease in Pdot size can be attributed to the accelerated hydrolysis rate of PSMAnh in the basic condition.33 The hydrodynamic size distribution of Pdots determined by DLS was also consistent with the TEM observation (Table S1†), thereby confirming the successful control of Pdot size by controlling the pH of the water phase. Additionally, once the Pdots formed, their size was not affected by the environmental condition such as salt concentration and pH of phosphate buffered saline.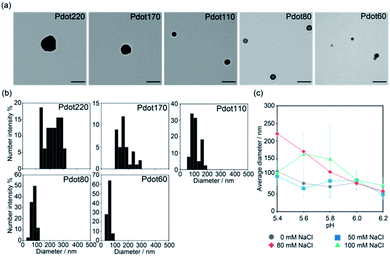 | ||
| Fig. 2 Pdot with different diameters obtained by the nanoprecipitation method with different conditions. (a) Typical TEM images of Pdot with an average diameter of 220 nm, 170 nm, 110 nm, 80 nm, and 60 nm which were obtained by the nanoprecipitation method with different pH at 80 mM NaCl. Scale bars: 200 nm. Lower magnification images are also shown in Fig. S1 (see ESI†). Other TEM images of Pdots obtained by nanoprecipitation with different NaCl concentration are also shown in Fig. S2 (ESI†). (b) Size distribution of Pdot with an average diameter of 220 nm, 170 nm, 110 nm, 80 nm, and 60 nm evaluated by TEM image analysis using ImageJ FIJI. More than 30 Pdots were analyzed for each experimental condition. (c) Effect of pH and salt concentration on the average diameter of obtained Pdots. Each value was calculated from the analysis of TEM image shown in (a) and S2 (ESI†). More than 30 Pdots were analyzed for each experimental condition. Data are shown as means ± SD. | ||
The effects of pH and salt concentration on the average diameter of the formed Pdots are summarised in Fig. 2c. As shown in Fig. 2a, the size of Pdots decreased with increasing pH in the presence of 80 mM NaCl in the water phase. However, the lower salt concentrations (0 and 50 mM) mitigated the pH-responsive size increase of Pdots (Fig. 2c, S2a and b, ESI†). It was suggested that a lower ionic strength facilitated the stabilising effect of PSMAnh even at a low hydrolysis degree via decreased electrostatic shielding, thereby suppressing the effect of the hydrolysis rate. Furthermore, it was found that Pdots prepared with the presence of a higher NaCl concentration of 100 mM showed an increase in size when the pH decreased from 6.2 (74 nm) to 5.4 (163 nm), followed by a decrease in size with a further decrease in pH to 5.2 (110 nm) (Fig. 2c and S2c, ESI†). In addition to the hydrolysis rate of PSMAnh, pH also affected the colloidal stability of the nanoparticles formed during the synthesis of Pdots via nanoprecipitation. Pdots become more unstable with decreasing pH because of the lower degree of carboxylic group ionisation. This effect becomes more prominent under high salt concentration conditions that exert severe electrostatic shielding, leading to the different pH dependence of the formed Pdot size observed at 100 mM NaCl. At a pH higher than 5.8, the pH-dependency of the Pdot size under 100 mM NaCl followed a similar trend to that of other NaCl concentrations. However, when the pH was lower than 5.8, the decreased colloidal stability caused the aggregation of the formed Pdots during the synthesis, which prevented their further growth to form small particles. Sedimentation was observed in the reaction solution after particle formation (Fig. S3, ESI†), which could be due to the formation of large aggregates. Since the growth period of each particle would vary depending on the timing of its collision with other particles to form aggregates, the formed Pdots showed wider size distribution than other conditions. The size distribution in Fig. 2c and the TEM images in Fig. S1 and S2 (ESI†) of the Pdots were obtained after re-dispersing them in pure water; thus, the above-mentioned aggregation is considered to disappear upon evaluation. These results suggest that fine-tuning the Pdot size can be achieved by choosing the appropriate value of salt concentration and pH in the water phase during nanoprecipitation.
3.2. Optical and surface properties of Pdots
The optical properties of Pdots are crucial for their performance as fluorescent probes for bioimaging and sensing. The absorption, excitation, and emission spectra of the obtained Pdots were measured to determine whether the synthetic conditions affected these properties and are shown in Fig. 3a, b and S5 (ESI†). The absorption peak of Pdots with different diameters appeared in the wavelength range of 480–500 nm. This peak was slightly red-shifted with increasing particle size, which could be due to the extension of the delocalisation of π-electron conjugation. Conversely, the fluorescence emission spectra of Pdots with different diameters showed a minor difference. Quantum yield also did not change significantly, regardless of the size (Table S2, ESI†).The surface properties of nanoparticles affect their interaction with the biological environment, which is another crucial parameter for determining their performance as a fluorescent probe. The zeta potentials of the Pdots with various diameters are shown in Fig. 3c. They represented a negative zeta potential of −50 to −44 mV, irrespective of the particle size. These results demonstrated that our proposed synthetic method alters the size of Pdots without changing other characteristics, such as optical and surface properties, as a fluorescent probe for bioimaging.
 | ||
| Fig. 3 Optical and surface properties of Pdot with different diameters. (a) Absorbance spectra and (b) fluorescent emission spectra of Pdot with different diameters. Excitation wavelength was 450 nm. Red, green, blue, yellow, and gray lines indicate Pdot220, Pdot170, Pdot110, Pdot80, and Pdot60, respectively. Inset shows Pdot60 dispersion irradiated with UV. Excitation spectra are also shown in Fig. S4 (ESI†). (c) Surface zeta potential of Pdot with five different diameters before and after PEG functionalization, measured by electrophoresis light scattering. Black and white columns indicate the zeta potential of Pdots before and after the PEGylation, respectively. n = 3, means ± SD. | ||
As we succeeded in synthesising Pdots under precise size control, the size effect of Pdots on their performance as a bioimaging probe was further investigated. The surface of the Pdots was first modified with polyethylene glycol (PEG) to minimise non-specific interactions with serum proteins. The obtained Pdots with five different diameters (Pdot60, Pdot80, Pdot110, Pdot170 and Pdot220) were functionalised with amine-terminated PEG via a coupling reaction catalysed by EDC. The surface zeta potential of the Pdots before PEGylation was −50 to −44 mV, which increased to approximately −31 to −29 mV after PEGylation (Fig. 3c). FT-IR spectroscopic spectra of Pdots before and after PEG conjugation are shown in Fig. S5 (ESI†). It was confirmed that the surface functionalisation of Pdot with PEG was achieved, and the negatively charged carboxyl groups were well covered by PEG chains.
3.3. Pdot properties as a bioimaging probe in vitro
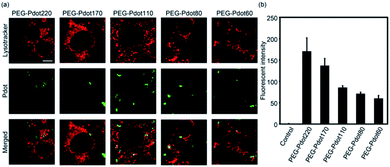
We then examined the performance of Pdots with different diameters as a bioimaging probe using mouse fibroblasts (NIH/3T3) cells in vitro. PEGylated Pdots (30 μg mL−1) with different diameters (60, 80, 110, 170 and 220 nm) were incubated with NIH/3T3 cells at 37 °C under 5% CO2 for 4 h, followed by the observation via confocal laser microscopy, as shown in Fig. 4a. Cellular uptake of fluorescent nanoparticles was successfully observed in Pdots of all sizes. Furthermore, fluorescence from Pdots was colocalized with endosomes and/or lysosomes stained with LysoTracker, suggesting their internalization into the cells via endocytosis. Further investigation on the detailed endocytic pathway would deepen the understanding of the biological interaction of Pdots in the future. To further quantify the cellular uptake, the fluorescent intensity of Pdots internalised into each cell was measured using flow cytometry (Fig. 4b). NIH/3T3 cells were incubated with Pdots of different sizes under the same conditions as above, harvested via 0.05 wt% trypsin/EDTA, and then analysed using a flow cytometer with a 488 nm Ar laser. The raw data of flow cytometry is provided in Fig. S6 (ESI†). The obtained mean fluorescence was used as an index of cellular uptake. It was found that the PEG-Pdots showed higher fluorescent intensity in the cells depending on their increasing size (Fig. 4b), which was consistent with previous reports.34,35 Therefore, it is confirmed that our size-tuned Pdots can demonstrate the size effect of nanoparticles in a biological environment. Note that PEG-Pdots showed no cytotoxicity under these experimental conditions (Fig. S7, ESI†). This is consistent with previous reports that indicated low cytotoxicity of Pdots functionalized with PEG.21,364. Conclusions
In this study, we demonstrated the size-controllable synthesis of Pdots based on a nanoprecipitation method focusing on the hydrolysis rate of PSMAnh as a stabilising agent. Previously, the size of nanoparticles synthesised by nanoprecipitation was controlled by polymer concentration, temperature, and water content.37 However, these methods have challenges. For example, it is difficult to obtain sub-100 nm particles and the wide size distribution of the obtained particles. However, the introduction of acid anhydrides, such as maleic anhydride, as a stabilising agent enables well-defined termination of particle growth at an early stage, leading to the formation of smaller nanoparticles and thus has been widely used in the synthesis of Pdots in previous studies.21,29 However, since the formation mechanism is different, the above-mentioned parameters often used for particle size control in conventional nanoprecipitation methods (such as polymer concentration and temperature) were not sufficiently effective to achieve the wide-range tuning of Pdot size. In this study, we found that the size of Pdot was successfully regulated by the pH and salt concentration of the water phase during nanoprecipitation because these parameters determined the hydrolysis rate of maleic anhydride segments of PSMAnh. Our strategy works when the acid anhydrides are used during nanoparticle formation, and it is expected to be adapted not only to conjugated polymers but also to other hydrophobic polymer materials.We consider that the novel size-tuning method of Pdots proposed herein has the potential to dramatically broaden the application of Pdots in biomedical applications. Cellular imaging applications of Pdots, such as cellular labelling through endocytosis7 and immunofluorescent labelling for membrane proteins,21,26 have been widely investigated. Furthermore, in vivo imaging38 and drug delivery39–41 based on Pdots have been extensively studied in recent years. The size tuning of Pdots between several tens to hundreds of nanometres is expected to provide precise tuning of their biodistribution and targeting efficiency for disease sites, thereby expanding their potential in the field of biomedical engineering. We envision size-tunable Pdots as a strong tool to understand the size effect of nanoparticles on their performance in biological conditions. Furthermore, this Pdot synthesis methodology is expected to impact biomedical applications of Pdots, such as tumour imaging,38,41 detection of disease-related physiological environments, such as reactive oxygen species,42 and photodynamic therapy20,43 with the optimisation of their functions.
In summary, we succeeded in size-tunable synthesis of Pdots by controlling the hydrolysis rate of the stabilising agent during nanoprecipitation and evaluated their functions using cultured cells. The novel synthetic method demonstrated precise and on-demand size control of Pdots, which is crucial for achieving high performance as a bioimaging and sensing probe. Our findings are not limited to the specific conjugated polymer F8BT and are expected to apply to other conjugated polymers and other types of hydrophobic polymers. We expect that size-tunable Pdots will significantly broaden the application of Pdots as fluorescent materials in the biomedical field.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We appreciate Prof. Taichi Ito for the helpful discussion. TEM observations were conducted at the Advanced Characterisation Nanotechnology Platform of the University of Tokyo, supported by MEXT. This work was supported by JST PRESTO (grant number: 30801), JSPS KAKENHI (grant numbers: 21H01722, 21H05092, and 21K20478), AMED (grant number: 21ck0106518h0003), and Takeda Science Foundation.References
- H. Cabral, Y. Matsumoto, K. Mizuno, Q. Chen, M. Murakami, M. Kimura, Y. Terada, M. R. Kano, K. Miyazono, M. Uesaka, N. Nishiyama and K. Kataoka, Nat. Nanotechnol., 2011, 6, 815–823 CrossRef CAS PubMed
.
- S. Wilhelm, A. J. Tavares, Q. Dai, S. Ohta, J. Audet, H. F. Dvorak and W. C. W. Chan, Nat. Rev. Mater., 2016, 1, 16014 CrossRef CAS
.
- S. Ohta, E. Kikuchi, A. Ishijima, T. Azuma, I. Sakuma and T. Ito, Sci. Rep., 2020, 10, 18220 CrossRef CAS
.
- X. Wu, H. Liu, J. Liu, K. N. Haley, J. A. Treadway, J. P. Larson, N. Ge, F. Peale and M. P. Bruchez, Nat. Biotechnol., 2003, 21, 41–46 CrossRef CAS
.
- I. H. El-Sayed, X. Huang and M. A. El-Sayed, Nano Lett., 2005, 5, 829–834 CrossRef CAS PubMed
.
- L. Babes, B. t. Denizot, G. Tanguy, J. J. Le Jeune and P. Jallet, J. Colloid Interface Sci., 1999, 212, 474–482 CrossRef CAS PubMed
.
- C. Wu, B. Bull, C. Szymanski, K. Christensen and J. McNeill, ACS Nano, 2008, 2, 2415–2423 CrossRef CAS PubMed
.
- W. C. Chan and S. Nie, Science, 1998, 281, 2016–2018 CrossRef CAS
.
- M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013–2016 CrossRef CAS PubMed
.
- I. L. Medintz, H. T. Uyeda, E. R. Goldman and H. Mattoussi, Nat. Mater., 2005, 4, 435–446 CrossRef CAS PubMed
.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS PubMed
.
- C. Kirchner, T. Liedl, S. Kudera, T. Pellegrino, A. Muñoz Javier, H. E. Gaub, S. Stölzle, N. Fertig and W. J. Parak, Nano Lett., 2005, 5, 331–338 CrossRef CAS PubMed
.
- C. Wu and D. Chiu, Angew. Chem., Int. Ed. Engl., 2013, 52, 3086–3109 CrossRef CAS PubMed
.
- X. Xu, R. Liu and L. Li, Chem. Commun., 2015, 51, 16733–16749 RSC
.
- C. Cordovilla and T. M. Swager, J. Am. Chem. Soc., 2012, 134, 6932–6935 CrossRef CAS PubMed
.
- X. Zhang, C. F. Chamberlayne, A. Kurimoto, N. L. Frank and E. J. Harbron, Chem. Commun., 2016, 52, 4144–4147 RSC
.
- J. L. Grimland, C. Wu, R. R. Ramoutar, J. L. Brumaghim and J. McNeill, Nanoscale, 2011, 3, 1451–1455 RSC
.
- X. Shen, F. He, J. Wu, G. Q. Xu, S. Q. Yao and Q.-H. Xu, Langmuir, 2011, 27, 1739–1744 CrossRef CAS
.
- J. H. Moon, P. MacLean, W. McDaniel and L. F. Hancock, Chem. Commun., 2007, 4910–4912, 10.1039/B710807A
.
- M. Zhao, E. Leggett, S. Bourke, S. Poursanidou, S. Carter-Searjeant, S. Po, M. Palma do Carmo, L. A. Dailey, P. Manning, S. G. Ryan, L. Urbano, M. A. Green and A. Rakovich, ACS Nano, 2021, 15, 8790–8802 CrossRef CAS
.
- C. Wu, Y. Jin, T. Schneider, D. R. Burnham, P. B. Smith and D. T. Chiu, Angew. Chem., Int. Ed. Engl., 2010, 49, 9436–9440 CrossRef CAS PubMed
.
- K. Li, J. Pan, S. S. Feng, A. Wu, y. Kan, Y. Liu and B. Liu, Adv. Funct. Mater., 2009, 19, 3535–3542 CrossRef CAS
.
- K. Li, Y. Liu, K. Pu, S.-s. Feng, R. Zhan and B. Liu, Adv. Funct. Mater., 2011, 21, 287–294 CrossRef CAS
.
- P. Howes, M. Green, J. Levitt, K. Suhling and M. Hughes, J. Am. Chem. Soc., 2010, 132, 3989–3996 CrossRef CAS
.
- L. Zhou, J. Geng, G. Wang, J. Liu and B. Liu, ACS Macro Lett., 2012, 1, 927–932 CrossRef CAS
.
- C. Wu, T. Schneider, M. Zeigler, J. Yu, P. G. Schiro, D. R. Burnham, J. D. McNeill and D. T. Chiu, J. Am. Chem. Soc., 2010, 132, 15410–15417 CrossRef CAS PubMed
.
- F. Ye, C. Wu, Y. Jin, M. Wang, Y.-H. Chan, J. Yu, W. Sun, S. Hayden and D. T. Chiu, Chem. Commun., 2012, 48, 1778–1780 RSC
.
- X. Zhang, J. Yu, C. Wu, Y. Jin, Y. Rong, F. Ye and D. T. Chiu, ACS Nano, 2012, 6, 5429–5439 CrossRef CAS
.
- Y. Lin, C. Dong, F. Cao, L. Xiong, H. Gu and H. Xu, RSC Adv., 2017, 7, 55957–55965 RSC
.
- Y. Braeken, S. Cheruku, A. Ethirajan and W. Maes, Materials, 2017, 10, 1420 CrossRef
.
- P. Palani and S. Karpagam, New J. Chem., 2021, 45, 19182–19209 RSC
.
- M. Fischer and J. Georges, Chem. Phys. Lett., 1996, 260, 115–118 CrossRef CAS
.
- J. M. Dorr, S. Scheidelaar, M. C. Koorengevel, J. J. Dominguez, M. Schafer, C. A. van Walree and J. A. Killian, Eur. Biophys. J., 2016, 45, 3–21 CrossRef
.
- H. J. Je, E. S. Kim, J.-S. Lee and H. G. Lee, J. Agric. Food Chem., 2017, 65, 10899–10906 CrossRef CAS
.
- M. Wu, H. Guo, L. Liu, Y. Liu and L. Xie, Int. J. Nanomed., 2019, 14, 4247–4259 CrossRef CAS
.
- F. Wang, H. Chen, Z. Liu, F. Mi, X. Fang, J. Liu, M. Wang, P. K. Lo and Q. Li, New J. Chem., 2019, 43, 14443–14449 RSC
.
- W. Huang and C. Zhang, Biotechnol. J., 2018, 13, 1700203 CrossRef
.
- S. Kim, C.-K. Lim, J. Na, Y.-D. Lee, K. Kim, K. Choi, J. F. Leary and I. C. Kwon, Chem. Commun., 2010, 46, 1617–1619 RSC
.
- J. H. Moon, E. Mendez, Y. Kim and A. Kaur, Chem. Commun., 2011, 47, 8370–8372 RSC
.
- X. Feng, Y. Tang, X. Duan, L. Liu and S. Wang, J. Mater. Chem., 2010, 20, 1312–1316 RSC
.
- X. Feng, F. Lv, L. Liu, H. Tang, C. Xing, Q. Yang and S. Wang, ACS Appl. Mater. Interfaces, 2010, 2, 2429–2435 CrossRef CAS
.
- K. Pu, A. J. Shuhendler and J. Rao, Angew. Chem., Int. Ed. Engl., 2013, 52, 10325–10329 CrossRef CAS PubMed
.
- M. Doshi, M. Krienke, S. Khederzadeh, H. Sanchez, A. Copik, J. Oyer and A. J. Gesquiere, RSC Adv., 2015, 5, 37943–37956 RSC
.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d1ra09101h |
| This journal is © The Royal Society of Chemistry 2022 |