 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Insights into the binding and covalent inhibition mechanism of PF-07321332 to SARS-CoV-2 Mpro†
Son Tung Ngo *ab,
Trung Hai Nguyenab,
Nguyen Thanh Tung
*ab,
Trung Hai Nguyenab,
Nguyen Thanh Tung cd and
Binh Khanh Mai
cd and
Binh Khanh Mai *e
*e
aLaboratory of Theoretical and Computational Biophysics, Ton Duc Thang University, Ho Chi Minh City, Vietnam. E-mail: ngosontung@tdtu.edu.vn
bFaculty of Applied Sciences, Ton Duc Thang University, Ho Chi Minh City, Vietnam
cInstitute of Materials Science, Vietnam Academy of Science and Technology, Hanoi, 11307, Vietnam
dGraduate University of Science and Technology, Vietnam Academy of Science and Technology, Hanoi, 11307, Vietnam
eDepartment of Chemistry, University of Pittsburgh, Pittsburgh, PA 15260, USA. E-mail: binh.mai@pitt.edu
First published on 28th January 2022
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been causing the COVID-19 pandemic, resulting in several million deaths being reported. Numerous investigations have been carried out to discover a compound that can inhibit the biological activity of the SARS-CoV-2 main protease, which is an enzyme related to the viral replication. Among these, PF-07321332 (Nirmatrelvir) is currently under clinical trials for COVID-19 therapy. Therefore, in this work, atomistic and electronic simulations were performed to unravel the binding and covalent inhibition mechanism of the compound to Mpro. Initially, 5 μs of steered-molecular dynamics simulations were carried out to evaluate the ligand-binding process to SARS-CoV-2 Mpro. The successfully generated bound state between the two molecules showed the important role of the PF-07321332 pyrrolidinyl group and the residues Glu166 and Gln189 in the ligand-binding process. Moreover, from the MD-refined structure, quantum mechanics/molecular mechanics (QM/MM) calculations were carried out to unravel the reaction mechanism for the formation of the thioimidate product from SARS-CoV-2 Mpro and the PF-07321332 inhibitor. We found that the catalytic triad Cys145–His41–Asp187 of SARS-CoV-2 Mpro plays an important role in the activation of the PF-07321332 covalent inhibitor, which renders the deprotonation of Cys145 and, thus, facilitates further reaction. Our results are definitely beneficial for a better understanding of the inhibition mechanism and designing new effective inhibitors for SARS-CoV-2 Mpro.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a β-coronavirus belonging to the Coronaviridae virus family, has caused the global pandemic named coronavirus disease 2019 (COVID-19).1–3 SARS-CoV-2, which was thought to originate from bats, can rapidly transfect between humans and humans.4 The virus can be rapidly spread among the community via aerosol transmission.5,6 Despite international exertions to restrict the rate of the virus spreading, the number of infected cases has increased.7 Moreover, although three vaccines, the Pfizer-BioNTech, Moderna, and Janssen COVID-19 vaccines, have been approved by the FDA for emergency use,8 the pandemic is still causing numerous issues to community health. In particular, recent work has suggested that long-term health problems of fully recovered patients with no or minor symptoms are increasingly being recorded.9 Furthermore, a growing number of variants escaping from the neutralizing antibodies have been observed.10,11 These variants contain mutations in the piece of the genome encoding the spike protein, which has been used by the vaccines to generate immunity.12,13 The vaccine effectiveness in the near future will likely be decreased. Therefore, developing an appropriate treatment for COVID-19 is accordingly of great urgency.The viral genome, with a length of 29.2 kb, encodes more than 20 nonstructural (nsp) and structural proteins.1,14 Those of SARS-CoV and SARS-CoV-2 are more than 82% similar to each other.15 In particular, SARS-CoV-2 consists of two proteases, the SARS-CoV-2 main protease (Mpro or 3CLpro) and papain-like protease (PLpro), which correspond to nsp5 and nsp3, respectively. The main protease (Mpro) of SARS-CoV-2 virus has >96% sequence identity to the one of SARS-CoV,16,17 while the SARS-CoV-2 PLpro shares 83% sequence identity to the SARS-CoV PLpro.18 The protease first self-cleaves from the product of the messenger ribonucleic acid (mRNA) translation, and polyproteins are then cleaved to polypeptides. Because the polypeptides are required for viral replication and encapsulation, the proteases are directly associated with viral replication and proliferation.16,17 In more detail, PLpro responds to the formation of nsp1-3 and Mpro is required for the establishment of the nsp4-16.19 Therefore, the SARS-CoV-2 Mpro has become a high-profile target for antiviral drug design, since inhibiting the biological activity of the SARS-CoV-2 Mpro is able to prevent the replication of a new virus.
Although there are already positive signs in the development of COVID-19 therapies,20 the race for antiviral drugs to prevent COVID-19 continues to be urgent.21 Numerous investigations have thus been performed to characterize a potential inhibitor for the SARS-CoV-2 Mpro.22–36 Several compounds have been suggested to be able to inhibit the biological activity of Mpro. In this context, a compound, named PF-07321332, has emerged as one of the most potent candidates for an oral antiviral therapeutic factor. The compound is currently under clinical trials as an antiviral agent against SARS-CoV-2 and was shown to be a potential inhibitor for Mpro in in vitro studies37,38 and phase I clinical trials.39 Moreover, a clinical phase III study in non-hospitalized high-risk adults with COVID-19 has also started.40 Understanding the binding and covalent inhibition mechanism of PF-07321332 to the SARS-CoV-2 Mpro would be beneficial in the design of antivirus drugs. Therefore, in this work, we tried to reveal physical insights into the binding and covalent inhibition mechanism of PF-07321332 to the SARS-CoV-2 Mpro. The work was supported by steered-molecular dynamics (SMD) and quantum mechanics/molecular mechanics (QM/MM) simulations. In particular, SMD simulations were first employed to preliminarily evaluate the binding pose of the ligand to Mpro. QM/MM calculations then probed the covalent inhibition mechanism. The obtained results are believed to enhance COVID-19 therapy.
Computational methods
Structures of the receptor and ligand
The three-dimensional conformation of the SARS-CoV-2 Mpro was downloaded from the Protein Data Bank (PDB) with the identity 7JYC.41 The protonation states of the Mpro catalytic dyad, including His41 and Cys145, were assigned, as shown in Fig. 1A, since it plays an important role in the protease activity and ligand effectiveness.42 The three-dimensional structure of PF-07321332 was generated using MarvinSketch, a package of ChemAxon.43 The ligand structure was then optimized via density-functional theory (DFT) calculations with the B3LYP functional at the 6-31G(d,p) level of theory. During QM calculations to prepare the ligand for MD simulations, the implicit solvent environment, ε = 78.4, was implemented. | ||
| Fig. 1 (A) The protonation states of the catalytic dyad. The distance between the Cys145-Sγ and His41-Nε atoms from the X-ray diffraction structure is also presented. (B) Three-dimensional structure of the SARS-CoV-2 Mpro (PDB ID 7JYC) + PF-07321332. (C) and (D) System configuration. In particular, the minimum distance between SARS-CoV-2 Mpro Cys145-Sγ and the ligand is ca. 28 Å. A constant force with a spring constant cantilever of 1 kcal mol−1 Å−2 was put on SARS-CoV-2 Mpro Cys145-Sγ and the nitrile group of the ligand. The solvent was hidden to clarify the view. | ||
Atomistic simulations
Atomistic simulations were performed to obtain the binding pose between SARS-CoV-2 Mpro + PF-07321332 using GROMACS version 2019.44 The protease and ions were presented using the Amber14SB force field.45 The TIP3P water model was employed for water molecules.46 PF-07321332 was topologized via the general Amber force field47 with the support of the AmberTools18 and ACPYPE packages.48,49 In particular, the QM calculations using the B3LYP functional at the 6-31G(d,p) level of theory with the implicit solvent were performed to obtain the ligand geometrical information and atomic charges. The restrained electrostatic potential scheme was employed to estimate the ligand atomic charges.47 In particular, the ligand was placed on the position having a minimum distance from the Cys145-Sγ atom of 28 Å, as shown in Fig. 1. The complex was then inserted into a rectangular periodic boundary condition (PBC) box with a size of 9.40 × 5.65 × 8.51 nm3, as shown in Fig. 1. The soluble complex contained 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 789 atoms, involving the protease, PF-07321332, 13
789 atoms, involving the protease, PF-07321332, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 012 water molecules, and 4 Na+ ions.
012 water molecules, and 4 Na+ ions.
Atomistic simulations were carried out with the parameters referred to in the previous studies.50,51 The simulations were executed at 310 K. A non-bonded contact between two atoms is available when the pair distance is smaller than 0.9 nm. The electrostatic (cou) and van der Waals (vdW) interactions were computed using the fast particle-mesh Ewald electrostatics and cut-off approaches, respectively.52 The solvated system was first minimized using the steepest descent method. The minimized system was then relaxed using NVT and NPT simulations with a length of 0.1 ns each. During these simulations, the SARS-CoV-2 Mpro Cα atoms and the ligand atoms were positionally restrained via a small harmonic force having ca. 24 kcal mol−1 nm−2 spring constant.
Steered-molecular dynamics (SMD) simulations
The last conformation of NPT simulations was then used as the starting shape for further SMD simulations. During the SMD simulating process, a small constant force with a spring constant of 1 kcal mol−1 nm−2 was employed to pull the nitrile group of PF-07321332 and the sulfur atom of the residue Cys145 together because a covalent bond is able to form between the two groups.53 During the SMD simulations, the SARS-CoV-2 Mpro reorientation and translation were prevented via a small restraining force applied on the Cα atoms. The ligand was slowly mobilized from the unbound to the bound state under the effects of a small constant force, with a schematic representation of the simulations shown in Fig. 1. It should be noted that using a stronger pulling force can make the ligand bind quicker, but at the same time may cause distortion to the binding pocket and result in the ligand adopting the wrong binding pose. We found that the chosen pulling strength is a good trade off between computational efficiency and accuracy. Moreover, because the pulling force is very weak, the ligand would mobilize very slowly and probably fail to bind to the active site of the protease. SMD simulations were thus carried out with a length of 50 ns and repeated 81 times independently to produce the binding conformation of SARS-CoV-2 Mpro + PF-07321332. In addition, a restraining force with ca. 24 kcal mol−1 nm−2 spring constant was also applied on SARS-CoV-2 Mpro Cα atoms to avoid system reorientation. Furthermore, the trajectory was extended to 1.0 μs of MD simulations to allow the system enough time to reach the “native” binding pose. There were 5 μs of MD simulations in total, which were produced to assess the binding process of PF-07321332 to the SARS-CoV-2 Mpro. The coordinates of the complex were recorded every 1 ps.QM/MM calculations
The covalent inhibition mechanism for the reaction between PF-07321332 and the SARS-CoV-2 Mpro to give the thioimidate product was investigated using the ONIOM algorithm54 implemented in Gaussian 16.55 The MolUP package56 was used to support input preparations. A representative snapshot of the SARS-CoV-2 Mpro + PF-07321332 complex in the minimum region of the free energy landscape was used as the starting structure for the ONIOM calculations (see the Results and discussion section). Only waters and counter-ions within a distance of 7 Å from the protein were kept, giving a system with a total of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 327 atoms and a neutral charge. The MM parameters and atomic charges were extracted from the parameter from MD simulations (vide supra). The QM region for the ONIOM calculations was defined as shown in Scheme 1, including Cys145, His41, and Asp187 residues. To facilitate the calculations, only the part of the PF-07321332 that is close to Cys145 is included in the QM region (Scheme 1). Hydrogen-link (H-link) atoms were added to the QM atoms at the boundary. The QM region has 49 atoms, including H-link atoms, with a total charge of −1.
327 atoms and a neutral charge. The MM parameters and atomic charges were extracted from the parameter from MD simulations (vide supra). The QM region for the ONIOM calculations was defined as shown in Scheme 1, including Cys145, His41, and Asp187 residues. To facilitate the calculations, only the part of the PF-07321332 that is close to Cys145 is included in the QM region (Scheme 1). Hydrogen-link (H-link) atoms were added to the QM atoms at the boundary. The QM region has 49 atoms, including H-link atoms, with a total charge of −1.
All intermediates and transition states were fully optimized using the quadratic coupled algorithm57 with the dispersion-corrected58 B3LYP functional,59,60 i.e., the B3LYP-D3(BJ), and 6-31G(d) basis sets. All residues in the MM region which were not within 10 Å of the QM region were constrained in all calculations. Vibrational frequency calculations at the same level of theory as the optimization were performed to confirm if each structure was a local minimum (no imaginary frequency) or a transition state (one imaginary frequency). Single-point calculations were carried out using the M06-2X functional61 and 6-311+G(2d,2p) basis set. The free energy profile was constructed by the Gibbs free energy differences between stationary points, i.e., intermediates and transition states, and the SARS-CoV-2 Mpro–PF-07321332 complex.
Analysis tools
The collective variable free energy landscape (FEL) was constructed using “gmx sham”, a tool of GROMACS. The two variables were the non-hydrogen atom root-mean-square deviation (RMSD) of the complex and the distance between the sulfur atom of the Cys145 residue and the nitrile group of PF-07321332. All of the snapshots locating the minima were used as the initials of the clustering analysis. The clustering method was applied with a non-hydrogen RMSD of nine critical residues of the SARS-CoV-2 Mpro and the ligand PF-07321332. Specifically, the nine critical residues were Thr26, His41, Ser46, Asn142, Gly143, Cys145, His164, Glu166, and Gln189, which play an important role in the ligand-binding process of SARS-CoV-2 Mpro.62 The clustering cutoff was 1 Å. The protonation states of PF-07321332 were predicted using Chemicalize.63Results and discussion
In this work, SMD simulations were employed to search for the binding position between PF-07321332 and the SARS-CoV-2 Mpro. In particular, the nitrile group of PF-07321332 and the sulfur atom of the Cys145 residue were pulled together using a small constant force. 81 SMD trajectories with a length of 50 ns each were produced to investigate the diffusion of PF-07321332 around the SARS-CoV-2 Mpro active site. Among these, 21 trajectories (26%) successfully reached the binding pocket, since the distances between Cys145-Sγ and the non-hydrogen atoms of PF-0321332 were less than 4.5 Å (Fig. S1–S9 of the ESI).† However, there were only 13 trajectories (16%), where the ligand was stable in the binding pocket until the trajectories were completed. Some trajectories reached the binding pocket just after ca. 4 ns of simulation, whereas some required more than ca. 40 ns.Among the 21 trajectories mentioned above, 10 of them (12%) successfully generated the bound states by forming a contact between the Cys145-Sγ atom and the nitrile group of PF-07321332, in which the distance (dSγ–CN) was less than 4.5 Å (Fig. 2A and S9–S17 of the ESI).† It should be noted that a short contact between two groups would allow the nitrile group and the catalytic cysteine to be able to adopt a covalent bond between them.53 PF-07321332 required more time (at least ca. 4.5 ns) to reach the bound state after entering the binding pocket of the SARS-CoV-2 Mpro. However, there were only 2 trajectories where the dSγ–CN stayed below 4.5 Å until the simulations were completed (Movie S1† describes a representative binding process). Moreover, although the binding mechanism of PF-07321332 probably is a complex pathway instead of a simple mobilization of the ligand to the pocket,64 two successfully generated bound states trajectories preliminarily suggested that the residues Glu166 and Gln189 play an important role during the binding process of the inhibitor (Fig. S18 and S19 of the ESI).† Furthermore, analyzing these two trajectories indicated that the pyrrolidinyl group of the ligand first inserted itself into the space between the residues Glu166 and Gln189, before the whole of PF-07321332 fully inserted into the binding pocket of the SARS-CoV-2 Mpro.
Because the nitrile group of PF-07321332 likely forms a covalent bond with the Cys145-Sγ atom, in which the force constant is much larger than 1 kcal mol−1 nm−2, one of the successfully generated bound state trajectories was extended to 1 μs to assess the stability of the complex. The non-hydrogen RMSD of the complex reaches a stable state after ca. 100 ns of SMD simulations (Fig. 2B). 900![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 snapshots over the interval 100–1000 ns were collected to be inputs for the collective-variable FEL analysis. The non-hydrogen RMSD of the complex and the distance dSγ–CN were calculated over these conformations to use as the two reaction coordinates of FEL. The obtained FEL are shown in Fig. 2C, with one minimum denoted as S1. The binding pose of PF-0721332 to the SARS-CoV-2 Mpro in the representative structure S1 is shown in Fig. 2D. In particular, the ligand nitrile group formed a contact with the Cys145-Sγ atom at a distance of dSγ–CN = 3.73 Å. The ligand nitrile group also adopted a contact with the His-Nε atom with dNε–CN = 3.86 Å. The presence of contacts between the PF-0721332 nitrile group increases the distance between the Cys145-Sγ and His41-Eε atoms, dNε–Sγ, from 3.99 to 4.17 Å (Fig. 1A and 2D). Therefore, the catalytic dyad Cys145–His41 is probably disturbed. In addition, the ligand also formed HBs to three residues, Asn142, Glu166, and Gln189 (Fig. 2D).
000 snapshots over the interval 100–1000 ns were collected to be inputs for the collective-variable FEL analysis. The non-hydrogen RMSD of the complex and the distance dSγ–CN were calculated over these conformations to use as the two reaction coordinates of FEL. The obtained FEL are shown in Fig. 2C, with one minimum denoted as S1. The binding pose of PF-0721332 to the SARS-CoV-2 Mpro in the representative structure S1 is shown in Fig. 2D. In particular, the ligand nitrile group formed a contact with the Cys145-Sγ atom at a distance of dSγ–CN = 3.73 Å. The ligand nitrile group also adopted a contact with the His-Nε atom with dNε–CN = 3.86 Å. The presence of contacts between the PF-0721332 nitrile group increases the distance between the Cys145-Sγ and His41-Eε atoms, dNε–Sγ, from 3.99 to 4.17 Å (Fig. 1A and 2D). Therefore, the catalytic dyad Cys145–His41 is probably disturbed. In addition, the ligand also formed HBs to three residues, Asn142, Glu166, and Gln189 (Fig. 2D).
In recent theoretical studies,42,65–68 using the adaptive string method, Moliner and Tuñón et al. proposed that the catalytic dyad Cys145H–His41 plays an important role in the reactivity of the SARS-CoV-2 Mpro, where a proton transfer from Cys145H to His41 takes place, first giving the ion pair Cys145−–His41H+, which is followed by a nucleophilic addition, producing a covalent bond with the inhibitor. In our initial ONIOM calculations, only the residues Cys145 and His41, and the PF-0721332 inhibitor were included in the QM region. However, we were unsuccessful in locating the ion pair Cys145−–His41H+. During the optimization, the proton automatically transferred from His41H+ to Cys145− (see Fig. S20 of the ESI).† We then performed a constrained optimization by fixing the N–H distances to 1.01 Å. Interestingly, the ion pair structure was calculated to be 31.6 kcal mol−1 higher in energy than the neutral form (Scheme 2). Our DFT calculations suggest that the imidazole ring of His41 is not basic enough to abstract a proton from the thiol group of Cys145. Our QM/MM calculations are consistent with a recent study, where the ion pair of the catalytic dyad (Cys145−–His41H+) was suggested to be a transient intermediate and was very high in energy compared to the neutral form.69
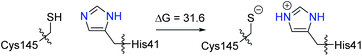 | ||
| Scheme 2 Computed free energy difference (kcal mol−1) for the ion pair formation of the catalytic dyad (Cys145−–His41H+) from the neutral form (Cys145H–His41). | ||
Searching for alternative mechanisms for this reaction, we found that the residue Asp187 was close to the residue His41, which could form a catalytic triad,70 Cys145–His41–Asp187, which facilitated the deprotonation of Cys145. Therefore, the Asp187 residue was then included in our ONIOM calculations. The computed free energy profile for the covalent inhibition mechanism between the SARS-CoV-2 Mpro and PF-0721332 is shown in Fig. 3. At Int-1, a strong hydrogen bond with a distance of 1.64 Å between His41 and Asp187 is found. A proton transfer via TS-1 between His41 and Asp187 can easily take place, giving His41−, which is followed by another proton transfer via TS-2 from Cys145 to His41−, generating Cys145−. The activation barriers of the two proton transfer steps, TS-1 and TS-2, are very low, amounting to 0.7 and 3.4 kcal mol−1, respectively, relative to Int-1. It should be noted that although the electronic energy of TS-1 is calculated to be 0.5 kcal mol−1 higher than that of Int-2, because of the entropic effect, the Gibbs free energy value of TS-1 is slightly lower than that of Int-2 (Fig. 3).
From Int-2, the nucleophilic addition of Cys145− to the nitrile group of PF-07321332 via TS-3 can then take place. Interestingly, at TS-3, a water molecule is crucial to stabilize the developed negative charge on the nitrogen atom of the nitrile group resulting from the nucleophilic addition of Cys145−. The small size of the nitrile group allows for the water molecule to participate in the reaction and transfer proton from His47 to nitrogen atom. In the nucleophilic addition TS-3, water played an essential role, where it stabilized the developed negative charge on the nitrogen atom of the nitrile group and, thus, facilitated this step.42,65–68 We also tried to optimize the nucleophilic addition without a water molecule. However, no TS could be located. The activation barrier of TS-3 was calculated to be 19.1 kcal mol−1 relative to Int-1, which is consistent with the previous study by Tuñón and co-workers.42 Our reaction mechanism is in good agreement with the covalent inhibition mechanism of antidiabetic drugs in dipeptidyl peptidase-4.71 Furthermore, the activation barrier of 19.1 kcal mol−1 of this reaction is also consistent with a previous study, where energy barriers of enzymatic reactions were found to be in a range of 14–20 kcal mol−1.72
It should be mentioned that from Int-4, the hydrolysis of the S–C bond could also occur by the nucleophilic addition of water to form an amide product. (Fig. S21 of the ESI† for the optimized transition state). However, the activation barrier of this transition state was calculated to be 39.6 kcal mol−1 relative to Int-3, ruling out this possibility and validating the inhibition ability of PF-07321332 to the SARS-CoV-2 Mpro. Furthermore, we also optimized the alternate mechanism where the proton transfer and nucleophilic addition occur synchronously. However, no transition state for this mechanism could be located because of the instability of the high negative partial charge in the nitrogen atom of the nitrile group of PF-07321332.
Based on our ONIOM calculations, we found that the catalytic triad Cys145–His41–Asp187 plays an important role in the covalent inhibition of the SARS-CoV-2 Mpro, which enables the deprotonation of Cys145 and, thus, facilitates further reaction. This finding is consistent with a previous study73 which demonstrated that Asp187 favors proton transfer from Cys145 to His41.
Conclusions
In this work, 5 μs of SMD simulations were first generated to preliminarily estimate the binding pose of PF-07321332 to the SARS-CoV-2 Mpro. In particular, the ligand reached the binding pocket of SARS-CoV-2 Mpro in 26% of the trajectories. Among these, the nitrile group of PF-07321332 successfully adopted a contact with the Cys145-Sγ atom in 12% of the trajectories. However, the contact only stabilized over 2 trajectories until the simulations were completed. Moreover, the residues Glu166 and Gln189 were suggested to play an important role during the binding process of the inhibitor. The pyrrolidinyl group of PF-07321332 is probably key in leading the compound into a successful bound state.A representative structure of SARS-CoV-2 Mpro + PF-07321332 was obtained by using a combined calculation of FEL and clustering analyses. In this state, the distance between the Cys145-Sγ and His41-Eε atoms dNε–Sγ was increased from 3.99 to 4.17 Å when the PF-07321332 nitrile group adopted a contact with the Cys145-Sγ atom (dSγ–CN = 3.73 Å). The catalytic dyad Cys145–His41 is probably disturbed. In addition, three residues, Asn142, Glu166, and Gln189, play a crucial role in the ligand-binding process by forming HBs with the inhibitor.
From the representative structure of the complex, quantum mechanics/molecular mechanics (QM/MM) calculations were performed to unravel the reaction mechanism for the formation of the thioimidate product from the SARS-CoV-2 Mpro and the PF-07321332 inhibitor. We found that the catalytic triad Cys145–His41–Asp187 of the SARS-CoV-2 Mpro plays an important role in the activation of the PF-07321332 covalent inhibitor, which leads to the deprotonation of Cys145 and, thus, facilitates further reaction. The outcome is in good agreement with a previous study73 which found that Asp187 favors proton transfer from Cys145 to His41. Our results are definitely beneficial for a better understanding of the inhibition mechanism and designing new effective inhibitors for the SARS-CoV-2 Mpro.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Vietnam National Foundation for Science & Technology Development (NAFOSTED) grant no. 104.99-2019.57.References
- F. Wu, S. Zhao, B. Yu, Y.-M. Chen, W. Wang, Z.-G. Song, Y. Hu, Z.-W. Tao, J.-H. Tian, Y.-Y. Pei, M.-L. Yuan, Y.-L. Zhang, F.-H. Dai, Y. Liu, Q.-M. Wang, J.-J. Zheng, L. Xu, E. C. Holmes and Y.-Z. Zhang, Nature, 2020, 579, 265–269 CrossRef CAS PubMed.
- A. E. Gorbalenya, S. C. Baker, R. S. Baric, R. J. de Groot, C. Drosten, A. A. Gulyaeva, B. L. Haagmans, C. Lauber, A. M. Leontovich, B. W. Neuman, D. Penzar, S. Perlman, L. L. M. Poon, D. V. Samborskiy, I. A. Sidorov, I. Sola and J. Ziebuhr, Coronaviridae Study Group of the International Committee on Taxonomy of, Nat. Microbiol., 2020, 5, 536–544 CrossRef PubMed.
- WHO Director-General’s opening remarks at the media briefing on COVID-19 – 11 March 2020, https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19-11-march-2020 Search PubMed.
- J. F. W. Chan, S. F. Yuan, K. H. Kok, K. K. W. To, H. Chu, J. Yang, F. F. Xing, J. L. Liu, C. C. Y. Yip, R. W. S. Poon, H. W. Tsoi, S. K. F. Lo, K. H. Chan, V. K. M. Poon, W. M. Chan, J. D. Ip, J. P. Cai, V. C. C. Cheng, H. L. Chen, C. K. M. Hui and K. Y. Yuen, Lancet, 2020, 395, 514–523 CrossRef CAS.
- T. Greenhalgh, J. L. Jimenez, K. A. Prather, Z. Tufekci, D. Fisman and R. Schooley, Lancet, 2021, 397, 1603–1605 CrossRef CAS.
- N. van Doremalen, T. Bushmaker, D. H. Morris, M. G. Holbrook, A. Gamble, B. N. Williamson, A. Tamin, J. L. Harcourt, N. J. Thornburg, S. I. Gerber, J. O. Lloyd-Smith, E. de Wit and V. J. Munster, N. Engl. J. Med., 2020, 382, 1564–1567 CrossRef PubMed.
- Worldometers, COVID-19 Coronavirus Pandemic, https://www.worldometers.info/coronavirus/ Search PubMed.
- COVID-19 Vaccines, https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccines, accessed Jan 08, 2021 Search PubMed.
- M. Augustin, P. Schommers, M. Stecher, F. Dewald, L. Gieselmann, H. Gruell, C. Horn, K. Vanshylla, V. D. Cristanziano, L. Osebold, M. Roventa, T. Riaz, N. Tschernoster, J. Altmueller, L. Rose, S. Salomon, V. Priesner, J. C. Luers, C. Albus, S. Rosenkranz, B. Gathof, G. Fätkenheuer, M. Hallek, F. Klein, I. Suárez and C. Lehmann, Lancet Reg. Health Eur., 2021, 6, 100122 CrossRef PubMed.
- P. Wang, M. S. Nair, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P. D. Kwong, B. S. Graham, J. R. Mascola, J. Y. Chang, M. T. Yin, M. Sobieszczyk, C. A. Kyratsous, L. Shapiro, Z. Sheng, Y. Huang and D. D. Ho, Nature, 2021, 593, 130–135 CrossRef CAS PubMed.
- M. Hoffmann, P. Arora, R. Groß, A. Seidel, B. F. Hörnich, A. S. Hahn, N. Krüger, L. Graichen, H. Hofmann-Winkler, A. Kempf, M. S. Winkler, S. Schulz, H.-M. Jäck, B. Jahrsdörfer, H. Schrezenmeier, M. Müller, A. Kleger, J. Münch and S. Pöhlmann, Cell, 2021, 184, 2384–2393 CrossRef CAS PubMed.
- South African SARS-CoV-2 Variant Alarms Scientists, https://www.the-scientist.com/news-opinion/south-african-sars-cov-2-variant-alarms-scientists-68317, accessed Jan 08, 2021 Search PubMed.
- H. Tu, M. R. Avenarius, L. Kubatko, M. Hunt, X. Pan, P. Ru, J. Garee, K. Thomas, P. Mohler, P. Pancholi and D. Jones, bioRxiv, 2021 DOI:10.1101/2021.01.12.426407.
- A. Francés-Monerris, C. Hognon, T. Miclot, C. García-Iriepa, I. Iriepa, A. Terenzi, S. Grandemange, G. Barone, M. Marazzi and A. Monari, J. Proteome Res., 2020, 19, 4291–4315 CrossRef PubMed.
- WHO, Coronavirus disease 2019 (COVID-19) Situation Report – 52 Search PubMed.
- C. M. Fauquet and D. Fargette, Virology, 2005, 2, 64 CrossRef CAS PubMed.
- Z. Alex, A. Vladimir, Z. Alexander, Z. Bogdan, T. Victor, B. S. Dmitry, P. Daniil, S. Rim, F. Andrey, O. Philipp, Y. Yilin, P. Olga, V. Quentin, A. Alex and I. Yan, Potential COVID-2019 3C-like Protease Inhibitors Designed Using Generative Deep Learning Approaches, 2020 Search PubMed.
- D. Shin, R. Mukherjee, D. Grewe, D. Bojkova, K. Baek, A. Bhattacharya, L. Schulz, M. Widera, A. R. Mehdipour, G. Tascher, P. P. Geurink, A. Wilhelm, G. J. van der Heden van Noort, H. Ovaa, S. Müller, K.-P. Knobeloch, K. Rajalingam, B. A. Schulman, J. Cinatl, G. Hummer, S. Ciesek and I. Dikic, Nature, 2020, 587, 657–662 CrossRef CAS PubMed.
- B. T. Freitas, I. A. Durie, J. Murray, J. E. Longo, H. C. Miller, D. Crich, R. J. Hogan, R. A. Tripp and S. D. Pegan, ACS Infect. Dis., 2020, 6, 2099–2109 CrossRef CAS PubMed.
- FDA Approves First Treatment for COVID-19, https://www.fda.gov/news-events/press-announcements/fda-approves-first-treatment-covid-19?fbclid=IwAR3jElh3p4H0YrLtL0o92OE931R6ygixc2edh3zPX4E6SL4AbmMFNu19q8U, accessed Oct 27, 2020 Search PubMed.
- E. Dolgin, Nature, 2021, 592, 340–343 CrossRef CAS PubMed.
- T. S. Komatsu, N. Okimoto, Y. M. Koyama, Y. Hirano, G. Morimoto, Y. Ohno and M. Taiji, Sci. Rep., 2020, 10, 16986 CrossRef CAS PubMed.
- M. D. Sacco, C. Ma, P. Lagarias, A. Gao, J. A. Townsend, X. Meng, P. Dube, X. Zhang, Y. Hu, N. Kitamura, B. Hurst, B. Tarbet, M. T. Marty, A. Kolocouris, Y. Xiang, Y. Chen and J. Wang, Sci. Adv., 2020, 6, eabe0751 CrossRef CAS PubMed.
- W. Vuong, M. B. Khan, C. Fischer, E. Arutyunova, T. Lamer, J. Shields, H. A. Saffran, R. T. McKay, M. J. van Belkum, M. A. Joyce, H. S. Young, D. L. Tyrrell, J. C. Vederas and M. J. Lemieux, Nat. Commun., 2020, 11, 4282 CrossRef CAS PubMed.
- Z. Li, X. Li, Y.-Y. Huang, Y. Wu, R. Liu, L. Zhou, Y. Lin, D. Wu, L. Zhang, H. Liu, X. Xu, K. Yu, Y. Zhang, J. Cui, C.-G. Zhan, X. Wang and H.-B. Luo, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 27381–27387 CrossRef CAS PubMed.
- C. Ma, M. D. Sacco, B. Hurst, J. A. Townsend, Y. Hu, T. Szeto, X. Zhang, B. Tarbet, M. T. Marty, Y. Chen and J. Wang, Cell Res., 2020, 30, 678–692 CrossRef CAS PubMed.
- W. Dai, B. Zhang, H. Su, J. Li, Y. Zhao, X. Xie, Z. Jin, F. Liu, C. Li, Y. Li, F. Bai, H. Wang, X. Cheng, X. Cen, S. Hu, X. Yang, J. Wang, X. Liu, G. Xiao, H. Jiang, Z. Rao, L.-K. Zhang, Y. Xu, H. Yang and H. Liu, Science, 2020, 368, 1331–1335 CrossRef CAS PubMed.
- M. Q. Pham, K. B. Vu, T. N. Han Pham, L. T. Thuy Huong, L. H. Tran, N. T. Tung, V. V. Vu, T. H. Nguyen and S. T. Ngo, RSC Adv., 2020, 10, 31991–31996 RSC.
- S. T. Ngo, N. Hung Minh, H. Le Thi Thuy, Q. Pham Minh, T. Vi Khanh, T. Nguyen Thanh and V. Van, RSC Adv., 2020, 10, 40284–40290 RSC.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang, C. Peng, Y. Duan, J. Yu, L. Wang, K. Yang, F. Liu, R. Jiang, X. Yang, T. You, X. Liu, X. Yang, F. Bai, H. Liu, X. Liu, L. W. Guddat, W. Xu, G. Xiao, C. Qin, Z. Shi, H. Jiang, Z. Rao and H. Yang, Nature, 2020, 582, 289–293 CrossRef CAS PubMed.
- Z. Jin, Y. Zhao, Y. Sun, B. Zhang, H. Wang, Y. Wu, Y. Zhu, C. Zhu, T. Hu, X. Du, Y. Duan, J. Yu, X. Yang, X. Yang, K. Yang, X. Liu, L. W. Guddat, G. Xiao, L. Zhang, H. Yang and Z. Rao, Nat. Struct. Mol. Biol., 2020, 27, 529–532 CrossRef CAS PubMed.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed.
- N. M. Tam, M. Q. Pham, N. X. Ha, P. C. Nam and H. T. T. Phung, RSC Adv., 2021, 11, 17478–17486 RSC.
- S. T. Ngo, N. Quynh Anh Pham, L. Thi Le, D.-H. Pham and V. V. Vu, J. Chem. Inf. Model., 2020, 60, 5771–5780 CrossRef CAS PubMed.
- O. Yañez, M. I. Osorio, C. Areche, A. Vasquez-Espinal, J. Bravo, A. Sandoval-Aldana, J. M. Pérez-Donoso, F. González-Nilo, M. J. Matos, E. Osorio, O. García-Beltrán and W. Tiznado, Biomed. Pharmacother., 2021, 140, 111764 CrossRef PubMed.
- O. Yañez, M. I. Osorio, E. Uriarte, C. Areche, W. Tiznado, J. M. Pérez-Donoso, O. García-Beltrán and F. González-Nilo, Front. Chem., 2021, 8 Search PubMed.
- Study of PF-07321332 in Healthy Participants, https://clinicaltrials.gov/ct2/show/NCT04756531, accessed Jun 17, 2021 Search PubMed.
- Pfizer Initiates Phase 1 Study of Novel Oral Antiviral Therapeutic Agent Against SARS-CoV-2, https://www.pfizer.com/news/press-release/press-release-detail/pfizer-initiates-phase-1-study-novel-oral-antiviral, accessed Jun 17, 2021 Search PubMed.
- D. R. Owen, C. M. N. Allerton, A. S. Anderson, L. Aschenbrenner, M. Avery, S. Berritt, B. Boras, R. D. Cardin, A. Carlo, K. J. Coffman, A. Dantonio, L. Di, H. Eng, R. Ferre, K. S. Gajiwala, S. A. Gibson, S. E. Greasley, B. L. Hurst, E. P. Kadar, A. S. Kalgutkar, J. C. Lee, J. Lee, W. Liu, S. W. Mason, S. Noell, J. J. Novak, R. S. Obach, K. Ogilvie, N. C. Patel, M. Pettersson, D. K. Rai, M. R. Reese, M. F. Sammons, J. G. Sathish, R. S. P. Singh, C. M. Steppan, A. E. Stewart, J. B. Tuttle, L. Updyke, P. R. Verhoest, L. Wei, Q. Yang and Y. Zhu, Science, 2021, 374, 1586–1593 CrossRef CAS PubMed.
- A Study of PF-07321332/Ritonavir in Nonhospitalized High Risk Adult Participants With COVID-19, https://clinicaltrials.gov/ct2/show/NCT04960202, accessed Sep 03, 2021 Search PubMed.
- B. Andi, D. Kumaran, D. F. Kreitler, A. S. Soares, W. Shi, J. Jakoncic, M. R. Fuchs, J. Keereetaweep, J. Shanklin and S. McSweeney, Hepatitis C Virus NSP3/NSP4A Inhibitors as Promising Lead Compounds for the Design of New Covalent Inhibitors for SARS-CoV-2 3CLpro/Mpro Protease, accessed Oct 04, 2020 Search PubMed.
- C. A. Ramos-Guzmán, J. J. Ruiz-Pernía and I. Tuñón, Chem. Commun., 2021, 57, 9096–9099 RSC.
- Marvin was Used for Drawing, Displaying and Characterizing Chemical Structures, Substructures and Reactions, Marvin 20.8.0, ChemAxon, 2020, http://www.chemaxon.com Search PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- D. A. Case, H. M. Aktulga, K. Belfon, I. Y. Ben-Shalom, S. R. Brozell, D. S. Cerutti, T. E. Cheatham III, G. A. Cisneros, V. W. D. Cruzeiro, T. A. Darden, R. E. Duke, G. Giambasu, M. K. Gilson, H. Gohlke, A. W. Goetz, R. Harris, S. Izadi, S. A. Izmailov, C. Jin, K. Kasavajhala, M. C. Kaymak, E. King, A. Kovalenko, T. Kurtzman, T. S. Lee, S. LeGrand, P. Li, C. Lin, J. Liu, T. Luchko, R. Luo, M. Machado, V. Man, M. Manathunga, K. M. Merz, Y. Miao, O. Mikhailovskii, G. Monard, H. Nguyen, K. A. O'Hearn, A. Onufriev, F. Pan, S. Pantano, R. Qi, A. Rahnamoun, D. R. Roe, A. Roitberg, C. Sagui, S. Schott-Verdugo, J. Shen, C. L. Simmerling, N. R. Skrynnikov, J. Smith, J. Swails, R. C. Walker, J. Wang, H. Wei, R. M. Wolf, X. Wu, Y. Xue, D. M. York, S. Zhao and P. A. Kollman, AMBER 2018, University of California, San Francisco, 2018 Search PubMed.
- A. W. Sousa da Silva and W. F. Vranken, BMC Res. Notes, 2012, 5, 1–8 CrossRef PubMed.
- S. T. Ngo, T. H. Nguyen, N. T. Tung, P. C. Nam, K. B. Vu and V. V. Vu, J. Comput. Chem., 2020, 41, 611–618 CrossRef CAS PubMed.
- S. T. Ngo, K. B. Vu, L. M. Bui and V. V. Vu, ACS Omega, 2019, 4, 3887–3893 CrossRef CAS PubMed.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089–10092 CrossRef CAS.
- R. Löser, K. Schilling, E. Dimmig and M. Gütschow, J. Med. Chem., 2005, 48, 7688–7707 CrossRef PubMed.
- L. W. Chung, W. M. C. Sameera, R. Ramozzi, A. J. Page, M. Hatanaka, G. P. Petrova, T. V. Harris, X. Li, Z. Ke, F. Liu, H.-B. Li, L. Ding and K. Morokuma, Chem. Rev., 2015, 115, 5678–5796 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, Williams, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavacha-ri, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian Inc., Wallingford CT, 2016.
- H. S. Fernandes, M. J. Ramos and N. M. F. S. A. Cerqueira, J. Comput. Chem., 2018, 39, 1344–1353 CrossRef CAS PubMed.
- T. Vreven, M. J. Frisch, K. N. Kudin, H. B. Schlegel and K. Morokuma, Mol. Phys., 2006, 104, 701–714 CrossRef CAS.
- S. Grimme, S. Ehrlich and L. Goerigk, J. Comput. Chem., 2011, 32, 1456–1465 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785–789 CrossRef CAS PubMed.
- A. D. Becke, J. Chem. Phys., 1993, 98, 5648–5652 CrossRef CAS.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- S. T. Ngo, N. M. Tam, M. Q. Pham and T. H. Nguyen, J. Chem. Inf. Model., 2021, 61, 2302–2312 CrossRef CAS PubMed.
- Chemicalize was used for prediction of chemical properties, https://chemicalize.com/, developed by ChemAxon Search PubMed.
- I. Buch, T. Giorgino and G. De Fabritiis, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10184–10189 CrossRef CAS PubMed.
- K. Świderek and V. Moliner, Chem. Sci., 2020, 11, 10626–10630 RSC.
- C. A. Ramos-Guzmán, J. J. Ruiz-Pernía and I. Tuñón, ACS Catal., 2020, 10, 12544–12554 CrossRef PubMed.
- C. A. Ramos-Guzmán, J. J. Ruiz-Pernía and I. Tuñón, ACSCatal., 2021, 11, 4157–4168 Search PubMed.
- C. A. Ramos-Guzmán, J. J. Ruiz-Pernía and I. Tuñón, Chem. Sci., 2021, 12, 3489–3496 RSC.
- H. T. H. Chan, M. A. Moesser, R. K. Walters, T. R. Malla, R. M. Twidale, T. John, H. M. Deeks, T. Johnston-Wood, V. Mikhailov, R. B. Sessions, W. Dawson, E. Salah, P. Lukacik, C. Strain-Damerell, C. D. Owen, T. Nakajima, K. Świderek, A. Lodola, V. Moliner, D. R. Glowacki, J. Spencer, M. A. Walsh, C. J. Schofield, L. Genovese, D. K. Shoemark, A. J. Mulholland, F. Duarte and G. M. Morris, Chem. Sci., 2021, 12, 13686–13703 RSC.
- A. R. Buller and C. A. Townsend, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, E653–E661 CrossRef CAS PubMed.
- Y.-H. Wang, F. Zhang, H. Diao and R. Wu, ACS Catal., 2019, 9, 2292–2302 CrossRef CAS.
- J. P. M. Sousa, R. P. P. Neves, S. F. Sousa, M. J. Ramos and P. A. Fernandes, ACS Catal., 2020, 10, 9545–9554 CrossRef CAS.
- L. Zanetti-Polzi, M. D. Smith, C. Chipot, J. C. Gumbart, D. L. Lynch, A. Pavlova, J. C. Smith and I. Daidone, J. Phys. Chem. Lett., 2021, 12, 4195–4202 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Includes a movie describing a representative binding process, the minimum distance between the non-hydrogen atoms of PF-07321332 and the non-hydrogen atoms of the catalytic dyad, the minimum distance dSγ–CN between the nitrile group of PF-07321332 and the sulfur atom of the Cys145 residue, the binding pathway of PF-07321332 to SARS-CoV-2 Mpro over the trajectories 0 and 20, the optimization starting from the ion pair Cys145−–His41H+, the optimized transition state for the hydrolysis of Int-3, and energy and Cartesian coordinates. See DOI: 10.1039/d1ra08752e |
| This journal is © The Royal Society of Chemistry 2022 |



