Open Access Article
Open Access ArticleUnravelling the antifungal and antiprotozoal activities and LC-MS/MS quantification of steroidal saponins isolated from Panicum turgidum†
Ahmed A. Zaki *ab,
Mohamed M. Y. Kaddah
*ab,
Mohamed M. Y. Kaddah c,
Hamada S. Abulkhair
c,
Hamada S. Abulkhair *de and
Ahmed Ashour
*de and
Ahmed Ashour ab
ab
aDepartment of Pharmacognosy, Faculty of Pharmacy, Mansoura University, Mansoura 35516, Egypt. E-mail: ahmed.awad@fulbrightmail.org
bDepartment of Pharmacognosy, Faculty of Pharmacy, Horus University-Egypt, International Coastal Road, New Damietta 34518, Egypt
cPharmaceutical and Fermentation Industries Development Center, City of Scientific Research and Technological Applications, New Borg El-Arab 21934, Alexandria, Egypt
dPharmaceutical Organic Chemistry Department, Faculty of Pharmacy, Al-Azhar University, Nasr City 11884, Cairo, Egypt. E-mail: hamadaorganic@azhar.edu.eg
ePharmaceutical Chemistry Department, Faculty of Pharmacy, Horus University-Egypt, International Coastal Road, New Damietta 34518, Egypt
First published on 21st January 2022
Abstract
Bioassay-guided investigation of Panicum turgidum extract resulted in the identification of seven steroidal saponins (Turgidosterones 1–7). They were evaluated for their in vitro antifungal, antileishmanial, and antitrypanosomal activities. Turgidosterone 6 was the most active antifungal against Candida albicans and Candida neoformans (IC50 values of 2.84 and 1.08 μg mL−1, respectively). Turgidosterones 4–7 displayed antileishmanial activity against Leishmania donovani promastigotes with IC50 values ranging from 4.95 to 8.03 μg mL−1 and against Leishmania donovani amastigote/THP with IC50 values range of 4.50–9.29 μg mL−1. Activity against Trypanosoma brucei was also observed for Turgidosterones 4–7 with an IC50 values range of 1.26–3.77 μg mL−1. Turgidosterones 1–3 did not display any activity against the tested pathogens. The study of structure–activity relationships of the isolated saponins indicated that the antifungal, antileishmanial, and antitrypanosomal activities are markedly affected by the presence of spirostane-type saponins and the elongation of the sugar residue at C-3. To quantitatively determine the most abundant active ingredient in Panicum turgidum extract, a single run, sensitive, and highly selective liquid chromatography-tandem mass spectrometry (LC-MS/MS) method has been applied under positive and negative modes. The obtained results showed that compound 5 was the most abundant (95.93 ± 1.10 mg per gram of dry Panicum turgidum extract), followed by 6 (52.51 ± 1.05 mg gm−1), 4 (32.71 ± 0.48 mg gm−1), and 7 (13.19 ± 0.50 mg gm−1). Docking of these saponins against the Candida albicans oxidoreductases and Leishmania infantum trypanothione reductase active sites revealed their potential to effectively bind with a number of key residues in both receptor targets.
1. Introduction
Panicum (panic grass) is one of the important genera belonging to the family Poaceae.1 Traditionally, it has been reported to treat various ailments. The genus Panicum includes 450 species native to the tropical regions, with a few species extending into the northern temperate zone.2 P. maximum was reported to have antimalarial, antirheumatic, anti-inflammatory, antidiabetic, antiplasmodial, and analgesic activities.3,4 It was reported that the consumption of P. miliaceum grains protects against hypercholesterolemia, cardiovascular disease, liver injury, and type II diabetes.5,6 Panicum turgidum Forssk. is a widely distributed grass in the Egyptian desert. In previous work, we have reported the anti-inflammatory activity of extracts and isolated compounds from P. turgidum.1,7 Moreover, in our recent publication, the isolation of steroidal saponins and their antiproliferative activity was studied.5 However, the antifungal and antiprotozoal activities of P. turgidum have not yet been investigated.Microbes and protozoa are the leading causes of infectious diseases, death, and many global health issues.8–10 The treatment of most pathogens is difficult because of their resistance to traditional medications. Additionally, antifungal drugs such as amphotericin B which is effective against candida infections showed severe side effects, including renal toxicity.11 Therefore, there is an urgent unmet need to develop new safe antifungal agents. On one hand, antimony (pentavalent antimonials) is the most effective treatment for Leishmania infections.12 Alternative drugs commonly used for leishmaniasis are amphotericin B and pentamidine, which are also considered ineffective.13 On the other hand, nifurtimox and benznidazole are the front-line antitrypanosomal against acute Chagas' disease.14 They are trypanocidal agents, affecting mainly the circulating trypomastigotes.15 However, the parasites' resistance together with the high toxicity of current antiprotozoal drugs has proved to be an inefficient treatment.12 These drugs can induce serious unwanted effects including gastrointestinal intolerance, neurological symptoms, nausea, vomiting, and abdominal pain.15 Serious problems caused by a continuous increase in drug-resistant pathogens have become a leading goal for the discovery of novel antiprotozoal drugs.
1.1 Aim of the work
Previously, our research group has evaluated the cytotoxic activity for the total extract of Panicum turgidum in bio guided isolation of its active components. It resulted in the isolation of seven steroidal saponins and the activity of these compounds as cytotoxic and anti-inflammatory were evaluated.5 In continuation of our recent works for the bioassay-guided isolation and biological evaluation of natural bioactive molecules,16–20 herein, we have selected other activities than selected before to be used for the bio guided isolation, antifungal and antiprotozoal effects are selected especially that total extract exhibited good activity in both assays. Also, we report bioassay-guided fractionation and the first quantitative and fully validated LC-MS/MS for the analysis of the most abundant biologically active constituents, NMR and HR-ESI-MS spectral characterization, and biological evaluation of these steroidal saponins.2. Results and discussion
2.1 Isolation of compounds
Total extract of Panicum turgidum showed antifungal activity against C. albicans, C. glabrata and C. neoformans at a low range of the IC50. It also exhibited good antiprotozoal activity against L. donovani Promastigote, L. donovani Amastigote/THP, and T. brucei. The extract was partitioned into n-hexane, methylene chloride, and remaining aqueous fractions. Both n-hexane and methylene chloride extracts showed no activity against fungi and leishmania strains used at the highest tested concentrations. However, the aqueous fraction was the most active against both fungi and protozoa. The bioactive remaining aqueous extract was subjected to fractionation in a vacuum liquid chromatography (VLC) over C-18 silica gel and the mixture of water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
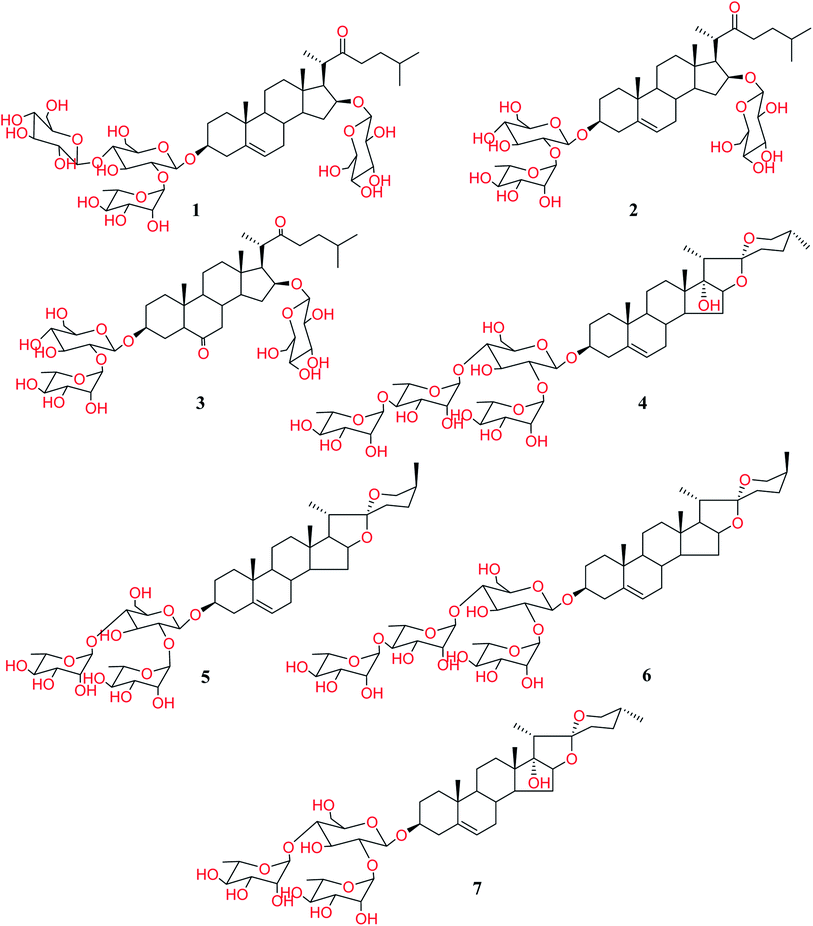
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (different proportions) was used to obtain fifteen fractions. The re-investigation of antifungal and anti-parasitic activities for the resulted fractions revealed that the fractions 14 and 15 are the only active ones. Therefore, these latter fractions were chromatographed over silica gel columns to isolate the constituting compounds. Seven steroidal saponins were isolated from these fractions and identified based on the analysis of the NMR spectra and HR-ESI-MS. Upon identification, it was found that they are the same as those we have isolated and reported before (Fig. 1).5
methanol (different proportions) was used to obtain fifteen fractions. The re-investigation of antifungal and anti-parasitic activities for the resulted fractions revealed that the fractions 14 and 15 are the only active ones. Therefore, these latter fractions were chromatographed over silica gel columns to isolate the constituting compounds. Seven steroidal saponins were isolated from these fractions and identified based on the analysis of the NMR spectra and HR-ESI-MS. Upon identification, it was found that they are the same as those we have isolated and reported before (Fig. 1).5
2.2 Quantitative analysis of active Turgidosterones 4–7 in Panicum extract
It has been shown that Turgidosterones 4–7 are the compounds responsible for the activity of P. turgidum extract. Therefore, we have decided to determine their naturally occurring concentration required to give the biological effect. The Turgidosterones 4–7 and vancomycin as an internal standard (IS) were individually prepared in methanol and directly injected into the mass spectrometer using a gas-tight syringe at a flow rate of 7 μL min−1 to implement automatic compound optimization using quadrupole full MS/MS scans in positive and negative ion modes to obtain the maximal sensitivity. Table 1 shows the molecular ions used to establish the multiple reaction monitoring (MRM) scanning method with its optimum mass spectrometry parameters for compounds 4–7 and IS. The ESI-MS/MS spectral data of these compounds and the IS are shown in Fig. S1–S6 in ESI.† Many papers have been reported with proposals for pharmaceutical compounds' MS/MS fragmentation pattern.21,22 Here, we further used the Competitive Fragmentation Modeling for Metabolite Identification CFM-ID23 to develop a proposal for the fragmentation pattern for each compound (Fig. S1–S6†).| Compounds | Mode of analysis | Precursor ion (m/z) | Product ion (m/z) | Declustering potential (V) | Collision energy (V) | Collision cell exit potential (V) | Dwell time (ms) |
|---|---|---|---|---|---|---|---|
| a Quantifier ion.b Qualifier ion. | |||||||
| 4 | Negative | 1029.36 | 737.4a | −25 | −64 | −37 | 50 |
| 1029.36 | 59b | −25 | −130 | −7 | 50 | ||
| 5 | Positive | 869.48 | 70.9a | 1 | 95 | 10 | 100 |
| 869.48 | 84.8b | 1 | 75 | 14 | 100 | ||
| 6 | Negative | 1013.34 | 721.4a | −20 | −60 | −55 | 100 |
| 1013.34 | 59b | −20 | −130 | −19 | 100 | ||
| 7 | Positive | 885.39 | 293.2a | 1 | 23 | 8 | 50 |
| 885.39 | 413.3b | 1 | 29 | 20 | 50 | ||
| Vancomycin (IS) | Negative | 722.81 | 700.7 | −190 | −20 | −27 | 50 |
| Vancomycin (IS) | Positive | 724.73 | 83 | 141 | 67 | 8 | 50 |
The chromatographic conditions were optimized to achieve high sensitivity, a good symmetric chromatographic peak shape, and a shorter retention time for the chosen compounds. HILIC chromatography is a fast, reliable, and highly selective HPLC technique that may be utilized to analyze polar substances quantitatively. The high polarity of the studied substances instructed that the chromatographic conditions be optimized utilizing the Luna® 3 μm HILIC column (100 × 4.6 mm) in this analysis. The mobile phase composition influenced the chromatographic separation and ionization of the compounds 4–7. Several trials were executed to determine the optimal chromatographic conditions by changing the elution type and flow rate. Gradient elution in various mobile phase ratios was investigated using different time programming elution systems. Methanol was used as the organic component of the mobile phase in this investigation instead of acetonitrile because it produced the best chromatographic peak shape. In this study, using 0.01 percent formic acid instead of 0.1 percent in both the aqueous and organic phases improved the peak shape, sensitivity, and retention behavior of the analytes substantially.24 The detailed gradient elution program is illustrated in Table S1 in ESI.† A stable isotopically labelled version of the analyte you want to measure is the perfect internal standard. It is considered to yield higher technique accuracy and precision since it acts in the same way as the analyte in ESI ion source, extraction recovery, and comparable chromatography.25 However, it necessitates a particular form of synthesis, which is both costly and time-consuming. In many situations, compound analogs to the analytes may be utilized as an IS.26–28 Vancomycin was chosen as the IS in this investigation because it demonstrated similar responses in both positive and negative modes. The MRM extracted chromatograms of the IS and Turgidosterones 4–7 are given in Fig. S7 and S8.†
| a y = a + xb where y is the analyte/IS peak area ratios; x is the concentration of the analyte; a is the intercept and b is the slope. | ||||
|---|---|---|---|---|
| Linear regression | 4 | 5 | 6 | 7 |
| Linearity range (ng mL−1) | 4–1200 | 4–1200 | 4–1200 | 4–1200 |
| LOD (ng mL−1) | 0.96 | 1.03 | 0.74 | 1.05 |
| LOQ (ng mL−1) | 2.89 | 3.12 | 2.26 | 3.19 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Linear regressiona | ||||
| Correlation coefficient (r) | 0.998 | 0.998 | 0.999 | 0.998 |
| Slope | 2.04 × 10−3 | 2.17 × 10−3 | 1.01 × 10−3 | 4.95 × 10−4 |
| Intercept | 3.18 × 10−3 | − 10.60 × 10−3 | 1.05 × 10−3 | 3.08 × 10−3 |
The contents of the chosen compounds in P. turgidum extract were 95.93 ± 1.10 mg per gram of dry Panicum turgidum extract, 52.51 ± 1.05 mg gm−1, 32.71 ± 0.48 mg gm−1 and 13.19 ± 0.50 mg gm−1 for compound 5, 6, 4, and 7, respectively. The estimated extraction efficiencies for compounds 4, 5, 6, and 7 were 79.29, 68.17, 74.93, and 64.30%, respectively.
2.3 Biological evaluation
The chemical investigation of the biologically active fractions in P. turgidum extract revealed the presence of Turgidosterones 1–7. With the guidance of the appropriate in silico approach, researchers can determine the accurate biological target and to what extent the activity could be attained. Also, the cost and implementation time of the experimental work could be reduced to the optimum. Accordingly, to decipher the proposed biological effect of new steroidal saponins, an in silico PASS online tool was used.29 Results of online PASS-prediction are shown in Table S3 (see ESI).† The tabulated results surprisingly showed that the antifungal and antileishmanial potencies were among the highest predicted actions of the newly isolated saponins. All the isolated Turgidosterones revealed probabilities of being active as antifungal and as antileishmanial over 71.30%. Consequently, encouraged by these promising results, we decided to experimentally investigate these predicted actions in vitro. The biological activities of 1–7 are discussed in the following sections.| Comp. # | IC50 μg mL−1 | ||||
|---|---|---|---|---|---|
| C. albicans | C. glabrata | C. krusei | A. fumigatus | C. neoformans | |
| Plant-total | 106.88 ± 5.97 | 22.2 ± 4.20 | >200 | >200 | 50.01 ± 1.01 |
| Remaining aq | 8.28 ± 0.01 | — | — | — | 21.03 ± 2.00 |
| 1 | >20 | NA | NA | >20 | >20 |
| 2 | >20 | NA | NA | >20 | >20 |
| 3 | >20 | NA | NA | >20 | >20 |
| 4 | 13.83 ± 2.50 | NA | NA | >20 | >20 |
| 5 | 10.83 ± 1.97 | NA | NA | >20 | 14.51 ± 2.01 |
| 6 | 2.84 ± 0.55 | NA | NA | >20 | 1.08 ± 0.002 |
| 7 | 16.04 ± 3.20 | NA | NA | >20 | >20 |
| Amphotericin B | 0.2 ± 0.002 | 0.27 ± 0.001 | 0.61 ± 0.001 | 1.21 ± 0.003 | 0.33 ± 0.0003 |
| Comp. # | IC50 (μg mL−1) | |||
|---|---|---|---|---|
| L. donovani Promastigote | L. Donovan Amastigote | L. donovani Amastigote/THP | T. brucei | |
| Plant -total | 19.11 ± 0.90 | >20 | 16.84 ± 3.16 | 7.79 ± 0.12 |
| Remaining aq. | 13.17 ± 1.79 | 14.82 ± 1.93 | 16.14 ± 1.09 | 4.64 ± 2.74 |
| 1 | >10 | >10 | >10 | >10 |
| 2 | >10 | >10 | >10 | >10 |
| 3 | >10 | >10 | >10 | >10 |
| 4 | 7.72 ± 1.58 | >10 | 9.03 ± 0.98 | 1.26 ± 0.25 |
| 5 | 8.04 ± 1.25 | >10 | 4.51 ± 0.66 | 2.87 ± 0.34 |
| 6 | 4.92 ± 2.16 | >10 | 5.15 ± 0.14 | 3.78 ± 0.31 |
| 7 | 7.80 ± 1.71 | >10 | 9.29 ± 0.65 | 1.42 ± 0.16 |
| Pentamidine | 1.78 ± 0.59 | 6.77 ± 0.004 | 5.48 ± 3.06 | 0.002 ± 0.001 |
As shown in Table 4, all the cholestane-type steroidal saponins 1–3 were totally inactive. The obtained results revealed that all the active compounds are of yamogenin-type glycosides (5 and 6) and pennogenine-type glycosides (4 and 7).
2.4 Structure–activity relationships (SARs)
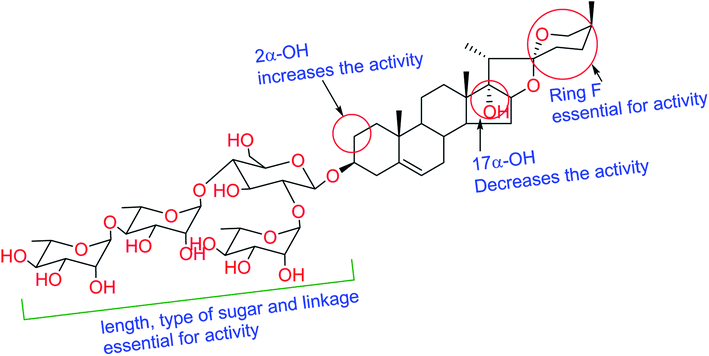
Literature survey revealed that saponins have activity against a broad spectrum of microorganisms, mainly fungi but showed weak activity against bacteria.30 Antifungal and antiprotozoal activities of saponins are affected by both the type of sapogenin and the sugar residue.31 The number of sugar residues and presence of 3β-linked glucose is crucial for activity, and branched sugar chains containing derivatives exhibited higher activities than straight chains.32 Compound 6 showed better activity than that of 5, which has fewer sugar residues. For antifungal and antiprotozoal activities, the closed F-ring is vital for this kind of activity.33 Accordingly, compounds lacking F-ring (1–3) presented almost no activity. Our results fully comply with previous reports; glycosides of spirostane-type have better activity than cholestane- and furostane-type. Cammarata et al. reported the MIC of series of steroidal saponins of spirostane- and cholestane-type against C. albicans, C. glabrata, and C. neoformans and they found that the antifungal activity of both spirostane- and cholestane-type is enhanced by the presence of hydroxyl group in 2α-position.34 Furthermore, the presence of the C-17 hydroxyl group caused a reduction of fungicidal activity confirmed by the moderate IC50 values exhibited by compounds 4 and 7 compared to 5 and 6. The structure requirements for steroidal saponins to be active as antifungal and/or antiprotozoal are concluded in Fig. 2.2.5 Molecular docking study
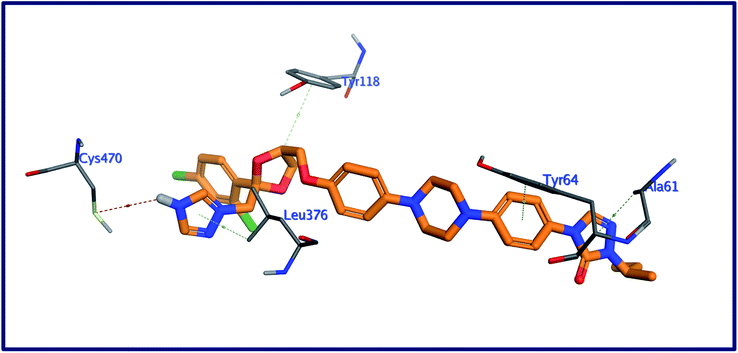
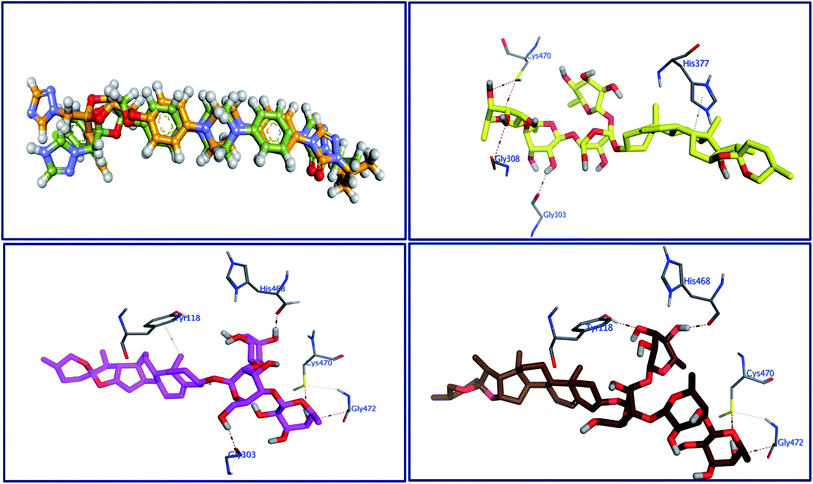
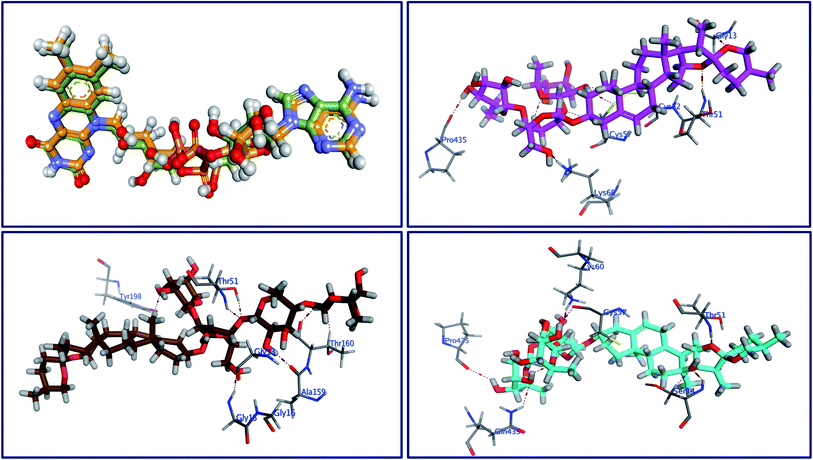
Molecular docking as a bioinformatic tool is applied at different steps of drug development for three vital purposes: (i) suggesting the binding mode of the structurally interrelated compound; (ii) expecting a new ligand using the technique of virtual screening; (iii) predicting the binding pattern of a known ligand.35–39 To unravel the possible underlying mode of action of new steroidal saponins as antifungal and antitrypanosomal, we employed molecular docking studies against the C. albicans oxidoreductases and L. infantum trypanothione reductase active sites. To date, only three main classes of FDA-approved antifungal drugs are available, polyenes (like amphotericin B & nystatin), azoles (like miconazole & ketoconazole), and candins (like caspofungin).40 The first two classes exert their antifungal activity through the inhibition of ergosterol biosynthesis. The third class acts on inhibiting the synthesis of β-1,3-D-glucan.40 On the other hand, antimonial antiprotozoals exert their effect through the inhibition of DNA topoisomerase while the mechanism of pentamidine has not been yet clearly identified.41 The mitochondrion is a suggested target of pentamidine action.42 Although none of these antifungals and antileishmanial agents target the oxidoreductase or trypanothione reductase, both proteins remain novel promising biological targets for the development of fungicidal and antileishmanial medications.43 Accordingly, we used the already FDA-approved, potent, and clinically useable antifungal and antileishmanial drugs (amphotericin B & pentamidine) as reference drugs to compare results of in vitro studies while oxidoreductases and trypanothione reductase were used to conduct the docking studies as predicted by the in silico PASS tools. Based on PDB search for C. albicans oxidoreductases and trypanothione reductase, more than one hundred protein codes were obtained for each. Based on the obtained docking score values and nature of the internal co-crystallized ligand, 5V5Z and 2JK6 complexes were selected to conduct molecular docking studies and consequently understand the virtual binding modes of the newly defined saponins with these receptor targets.44The behavior of newly isolated steroidal saponins inside the C. albicans oxidoreductase active pocket is summarized in Fig. 4 and S9 in ESI.† With low value of root mean square deviation (RMSD), the binding mode of Turgidosterone 4 exhibited an affinity value of −11.32 kcal mol−1 and obeyed a similar interaction pattern with that of 1YN. Turgidosterone 4 interacted with the active pocket through a set of four hydrogen bonds and one hydrophobic interaction. This new isolated saponin played as a hydrogen bond donor to form two hydrogen bonds with the amino acid residues Gly303 and Cys470. One more hydrogen bond arose between one alcoholic OH group attached with the terminal sugar moiety and the carboxylate oxygen of Gly308. The last interaction is in the form of hydrophobic stacking between ring C of Turgidosterone 4 steroidal nucleus and His377. This set of desirable covalent and noncovalent interactions of compound 4 might explain the good activity of this compound as antifungal agent.
Turgidosterone 5 revealed RMSD value of 1.59, exhibited an affinity value of −9.89 kcal mol−1 and exhibited a little different virtual binding mode with the C. albicans oxidoreductase enzyme. Turgidosterone 5 played as a HB donor to form three hydrogen bonds with Gly303, His468, Cys470, and Gly472. Also, the latter saponin exerted one hydrophobic interaction with the amino acid residue Tyr118.
The best effective steroidal saponin as antifungal (6) revealed a much better affinity value than that of the co-crystallized ligand, (−13.43 kcal mol−1), and showed four covalent interactions within a distance range of 2.89–3.29 Å. Two alcoholic OH groups of this ligand acted as hydrogen bond donors to create two hydrogen bonds with the His468 and Cys470 residues of the C. albicans oxidoreductase receptor backbone. Two additional hydrogen bonds also formed between the ligand's OHs and the hydroxyl groups of Tyr118 and Gly472. This set of covalent bindings may describe the potent activity of this compound as antifungal against both C. albicans and C. neoformans. Free energy of bindings, RMSD values, hydrophobic interactions, H-bonding interactions, and ligand positioning of the co-crystallized ligand and the best effective antifungal saponins (4, 5, 6, and 7) are presented in Table S4 (see ESI).† In order to validate the above results further, molecular docking experiments were also conducted on saponins that revealed the lower activity in the in vitro studies (Turgidosterones 1–3). Results of these experiments showed notable lower affinities and fewer numbers of covalent and non-covalent interactions. All results of these experiments are available in the ESI.† In particular, Turgidosterones 1 gives a number of poses with very high values of RMSD. The nearest acceptable pose exhibited a binding energy of −1.80 kcal mol−1 with C. albicans oxidoreductase (compared with −11.32, −9.89, −13.43, and −9.90 for compounds # 4, 5, 6, and 7 respectively). As well, this steroidal saponin interacted with the C. albicans oxidoreductase via three hydrogen bonds only (Cys470, Met508, and His377).
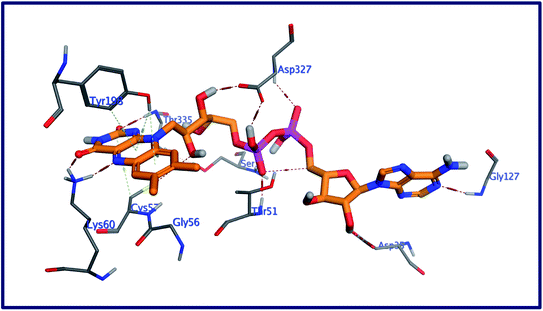
The behavior of new saponins in the L. infantum trypanothione reductase active pocket is summarized in Fig. 6 & S10 (in ESI).† With five hydrogen bonding interactions, the binding mode of compound 5 exhibited an affinity value of −0.78 kcal mol−1. Two alcoholic OHs played as hydrogen bond donors to create three hydrogen bonds with the essential amino acid residues Cys52, Cys57, and Pro435. Additionally two more hydrogen bonds formed between the new ligand and the residues Thr51 and Lys60, in which the ligand's primary alcoholic OH and oxolane oxygen played as HB acceptors. Obeying almost the same binding mode and occupying identical pocket, the binding mode of Turgidosterone 6 exhibited an affinity value of −3.72 kcal mol−1. The etherial oxygen linking the first two sugar moieties played as a hydrogen bond acceptor to form a bidirectional hydrogen bond with OH and NH2 groups of Thr51. Three of the ligand's alcoholic functionalities played as HB donors to form three additional hydrogen bonds with Ala159, Thr160, and Gly15 residues. As well, another alcoholic OH played as a hydrogen bond acceptor to form the last interaction between this ligand and the phenolic group of Tyr198. This set of strong covalent interactions may justify the superior activity of this compound as antitrypanosomal against both L. donovani Amastigote/THP and L. donovani Promastigote. With much lower affinity (4.61 kcal mol−1), the last compound formed six hydrogen bonds with the active site of the target trypanothione reductase. The oxolane and OH functionalities of this ligand played as HB acceptors to form three hydrogen bonds with Lys60, Gln439, and Ser14 and as hydrogen bond donors to form three more covalent interactions with Cys57, Pro435, and Thr51. An outline of free energy of bindings, RMSD values, H-bonding interactions, hydrophobic interactions, and ligand positioning of selected saponins and that of the internal co-crystallized ligand is shown in Table S5 (ESI).† Results of molecular docking experiments of Turgidosterones with low in vitro antitrypanosomal activity (1–3) on trypanothione reductase showed notable lower affinities and fewer numbers of effective interactions. For instance, Turgidosterone 1 formed only one hydrogen bond with the amino acid residue Asp327 and exhibited a very low affinity (22.17 kcal mol−1). Also, Turgidosterone 2 revealed a low binding affinity (13.73 kcal mol−1) and interacted with only five amino acid residues (Asp327, Ala365, Cys364, Gly13, and Cys364).
3. Conclusion
In our ongoing study to find biologically active compounds from medicinal plants, a bio-guided chemical investigation of methanolic extract of Panicum turgidum against fungi and protozoa was adopted. Seven steroidal saponins were isolated from active fractions. The biological evaluation of the isolated compounds against these two pathogens revealed that the yamogenin glycoside (yamogenin 3β-O-α-L-rhamnopyranosyl-(1→2)-O-[α-L-rhamnopyranosyl-(1→4)-O-α-L-rhamnopyranosyl-(1→4)]-O-β-D-glucopyranoside, 6) is the most active saponin. The structure–activity relationships of the active compounds were also studied, it revealed that spirostane-type saponins are active compounds with some structure requirements. These two activities were assessed before for the steroidal saponins and our results found comply with the those reported earlier. However, this is the first time for these two activities to be investigated for Panicum turgidum. A new LC-MS/MS method was developed to quantify the bioactive saponins 4–7 to determine their concentration in total extract responsible for the tested activities. Computational docking studies were conducted to determine the binding modes of the most potent steroidal saponins to identify their potential interactions with the active sites of C. albicans oxidoreductase and L. infantum trypanothione reductase. In line with the results of the in vitro experiments, the bioactive saponins 4–7 demonstrated the best virtual affinities and binding interactions.4. Material and methods
4.1 Plant material
Panicum turgidum Forssk. aerial parts were obtained from Egypt, and the plant identity was confirmed in the Botany Department, Faculty of Science, Mansoura University, Mansoura 35516, Egypt. In addition, a plant specimen was kept for reference in the Department of Pharmacognosy, Faculty of Pharmacy, Mansoura University.4.2 Isolation of compounds
The P. turgidum Forssk. aerial parts (450 g) were dried under shade, ground into a fine powder, and repeatedly extracted with methanol at 27 °C (3 L × 3 × 24 h). The combined extract was dried by using a rotary evaporator at 40 °C to afford crude residue (22.16 g). The total amount of the crude extract was re-dissolved in distilled water: methanol (9.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) and defatted by shaking vigorously with n-hexane, and the process was repeated three times. The residual aqueous phase was extracted by CHCl3. The remaining aqueous layer was dried to yield (7.2 g) then applied to a VLC packed with RP18 silica (30 cm × 3.5 cm), and the following polarity mixtures were used for elution of H2O–MeOH (1
0.5) and defatted by shaking vigorously with n-hexane, and the process was repeated three times. The residual aqueous phase was extracted by CHCl3. The remaining aqueous layer was dried to yield (7.2 g) then applied to a VLC packed with RP18 silica (30 cm × 3.5 cm), and the following polarity mixtures were used for elution of H2O–MeOH (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0, v/v), (4
0, v/v), (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), (7
1), (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), (3
3), (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), (1
2), (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), (2
1), (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), (3
3), (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), (1
7), (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), and (0
4), and (0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 15 fractions. Fractions 15 (1.98 g) and 14 (1.99 g) were subjected to repeated silica gel column and eluted gradually within EtOAc
1) to give 15 fractions. Fractions 15 (1.98 g) and 14 (1.99 g) were subjected to repeated silica gel column and eluted gradually within EtOAc![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CHCl3
CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH
MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O (10
H2O (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6
6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 then 6
1 then 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v/v/v) to get the steroidal saponin 1–7.
1, v/v/v/v) to get the steroidal saponin 1–7.
4.3 Biological evaluation
4.4 Chemicals and reagents for quantitative analysis
Sigma Pharmaceutical Industries in Egypt kindly provided the internal standard (IS) vancomycin hydrochloride (99.50%). The LC-MS grade acetic acid, formic acid, ammonium acetate, and ammonium formate were bought from Fisher Scientific, UK. Acetonitrile and methanol of UHPLC-MS grade were bought from Sigma-Aldrich (Steinheim, Germany). The water used in all procedures was purified using the Milli-Q water purification system (Millipore, Bedford, MA, USA).4.5 Preparation of calibration standards and quality control samples
A separate stock solution of 4–7 compounds were prepared in methanol (1.0 mg mL−1) while the IS was prepared in Mill-Q water (1.0 mg mL−1) and kept in refrigerator for one month. A methanol–water mixture (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) was used as a diluent to prepare a combined intermediate stock solution of 2000 ng mL−1. A serial dilution was applied to the intermediate stock solution to generate a standard working solution in the range of 4 ng mL−1 to 1200 ng mL−1 in which each concentration level contained a fixed amount of the IS (50 ng mL−1). 20 μL of these working standard solutions were injected into the LC-MS/MS. In the same way, the quality control samples were prepared in methanol–water mixture (50
50, v/v) was used as a diluent to prepare a combined intermediate stock solution of 2000 ng mL−1. A serial dilution was applied to the intermediate stock solution to generate a standard working solution in the range of 4 ng mL−1 to 1200 ng mL−1 in which each concentration level contained a fixed amount of the IS (50 ng mL−1). 20 μL of these working standard solutions were injected into the LC-MS/MS. In the same way, the quality control samples were prepared in methanol–water mixture (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v): low-quality control (LQC, 20.0 ng mL−1), medium-quality control (MQC, 600.0 ng mL−1), and high-quality control (HQC, 800.0 ng mL−1). To determine the compounds 4–7 in the plant extract, a stock solution of the plant extract was prepared in methanol (1.0 mg mL−1), sonicated for 5.0 minutes, and further centrifuged at 10
50, v/v): low-quality control (LQC, 20.0 ng mL−1), medium-quality control (MQC, 600.0 ng mL−1), and high-quality control (HQC, 800.0 ng mL−1). To determine the compounds 4–7 in the plant extract, a stock solution of the plant extract was prepared in methanol (1.0 mg mL−1), sonicated for 5.0 minutes, and further centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 minutes. The stock solution supernatant was further serially diluted by methanol–water mixture (50
000 rpm for 15 minutes. The stock solution supernatant was further serially diluted by methanol–water mixture (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, v/v) to produce a working solution of 10 μg mL−1 that contained a fixed amount of IS (50 ng mL−1).
50, v/v) to produce a working solution of 10 μg mL−1 that contained a fixed amount of IS (50 ng mL−1).
4.6 Instrumentation and software
The UPLC analysis was carried out using Eksigent ekspert™ ultraLC 100 system (Dublin-California-USA). Mass spectrometric detection was implemented on SCIEX QTRAP® 5500 (SCIEX instruments, Foster City, Canada) in multiple reaction monitoring (MRM). A Turbo V™ electrospray ionization (ESI) interface under positive and negative ionization modes was used. Analyst® software v. 1.6.2 was utilized to control both the UHPLC and the mass spectrometer. Data acquisition, analysis, and post-run data processing were implemented with Analyst® 1.6.2 and MultiQuant 3.0.1 software.4.7 Quantitative MRM method and chromatographic conditions
Positive and negative MRM scanning modes were implemented for the quantitative estimation of 4–7 compounds. A direct injection of a methanolic solution of 4–7 compound and IS at 20 ng mL−1 was employed to adjust the mass spectral data. The best mass spectrometer's main operating parameters are illustrated in Table 1.The quantitative estimation of 4–7 was executed on Luna® 3 μm HILIC column (100 × 4.6 mm), Phenomenex, USA. The column was maintained at 40 °C and run under gradient elution program with a flow rate of 0.5 mL min−1 and 20 μL injection volume. Table S1 in ESI† demonstrates the full chromatographic elution programs for quantitative assay.
4.8 Method validation
The LC-MS/MS method's validation was executed as described under the current Food and Drug Administration (FDA) guideline on bioanalytical method validation45 and ICH guideline on validation of analytical procedure.46| LOD = 3 Cs/(S/N) & LOQ = 10 Cs/(S/N) |
| Extraction efficiency (%) = (spiked extract − unspiked extract)/concentration of the analyte added to the spiked portion |
4.9 Molecular docking study
Docking experiments were conducted using the Molecular Operating Environment (MOE) software (MOE2014, https://www.chemcomp.com/Products.htm). Crystal structures of C. albicans oxidoreductases and L. infantum trypanothione reductase were retrieved from the protein data bank (PDB IDs: 5V5Z, resolution: 2.20 Å, https://www.rcsb.org/structure/5V5Z; 2JK6: resolution: 2.10 Å, https://www.rcsb.org/structure/2JK6) and considered as target for docking simulation. Molecular docking studies were conducted following our previously reported procedures.48–53Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
The authors would like to say thanks to the website of https://biorender.com/for providing clip art materials that have been used to design the graphical abstract.References
- G. Volpato, P. Kourková and V. Zelený, J. Ethnobiol. Ethnomed., 2012, 8, 49 CrossRef PubMed.
- M. Hedayetullah and P. Zaman, Forage Crops of the World, Volume I: Major Forage Crops, Apple Academic Press, 2018, DOI:10.1201/9781351167369.
- J. E. Okokon, P. A. Nwafor and U. E. Andrew, Asian Pac. J. Trop. Med., 2011, 4, 442–446 CrossRef PubMed.
- E. J. Akpan, J. E. Okokon and E. Offong, Asian Pac. J. Trop. Biomed., 2012, 2, 523–527 CrossRef PubMed.
- A. A. Zaki, Z. Ali, Y.-H. Wang, Y. A. El-Amier, S. I. Khan and I. A. Khan, Steroids, 2017, 125, 14–19 CrossRef CAS PubMed.
- L. Zhang, R. Liu and W. Niu, PLoS One, 2014, 9, e104058 CrossRef PubMed.
- A. A. Zakia, Z. Qiub, L. Alib, S. I. Khanb and I. A. Khanb, J. Agric. Basic Sci., 2016, 1, 1–6 Search PubMed.
- H. M. Elsheikha, Front. Vet. Sci., 2014, 1(25) DOI:10.3389/fvets.2014.00025.
- R. Capela, R. Moreira and F. Lopes, Int. J. Mol. Sci., 2019, 20(22), 5748 CrossRef CAS PubMed.
- A. Abo Elmaaty, M. I. A. Hamed, M. I. Ismail, E. B. Elkaeed, H. S. Abulkhair, M. Khattab and A. A. Al-Karmalawy, Molecules, 2021, 26, 3772 CrossRef PubMed.
- R. Laniado-Laborín and M. N. Cabrales-Vargas, Rev. Iberoam. Micol., 2009, 26, 223–227 CrossRef PubMed.
- J. Chakravarty and S. Sundar, J. Global Infect. Dis., 2010, 2, 167 CrossRef PubMed.
- D. O. Santos, C. E. R. Coutinho, M. F. Madeira, C. G. Bottino, R. T. Vieira, S. B. Nascimento, A. Bernardino, S. C. Bourguignon, S. Corte-Real, R. T. Pinho, C. R. Rodrigues and H. C. Castro, Parasitol. Res., 2008, 103, 1–10 CrossRef PubMed.
- B. J. Maguire, P. Dahal, S. Rashan, R. Ngu, A. Boon, C. Forsyth, N. Strub-Wourgaft, E. Chatelain, F. Barreira, S. Sosa-Estani and P. J. Guérin, PLoS Neglected Trop. Dis., 2021, 15, e0009697 CrossRef CAS PubMed.
- Imported Infectious Diseases, Elsevier, 2014, DOI:10.1016/C2013-0-18223-2.
- R. M. Samra, A. F. Soliman, A. A. Zaki, A. Ashour, A. A. Al-Karmalawy, M. A. Hassan and A. M. Zaghloul, S. Afr. J. Bot., 2021, 139, 210–216 CrossRef CAS.
- A. A. Zaki, A. A. Al-Karmalawy, Y. A. El-Amier and A. Ashour, New J. Chem., 2020, 44, 16752–16758 RSC.
- R. M. Samra, A. F. Soliman, A. A. Zaki, A. N. El-Gendy, M. A. Hassan and A. M. Zaghloul, J. Essent. Oil-Bear. Plants, 2020, 23, 648–659 CrossRef CAS.
- A. A. Zaki and L. Qiu, Nat. Prod. Res., 2020, 34, 2938–2944 CrossRef CAS PubMed.
- A. A. Zaki, A. A. Ashour and L. Qiu, Nat. Prod. Res., 2020, 1–5 CrossRef PubMed.
- M. M. Y. Kaddah, S. Billig, R. Oehme and C. Birkemeyer, J. Chromatogr. A, 2021, 1645, 462095 CrossRef CAS PubMed.
- M. M. Y. Kaddah, W. Talaat and M. A. El Demellawy, Biomed. Chromatogr., 2021, 35(9) DOI:10.1002/bmc.5146.
- Competitive Fragmentation Modeling for Metabolite Identification, http://cfmid2.wishartlab.com, accessed 7 January 2020 Search PubMed.
- O. Núñez and P. Lucci, Analysis, 2014, 71–86 Search PubMed.
- D. H. Chace, M. W. Duncan, D. Matern, M. R. Morris, D. E. Palmer-Toy, A. Rockwood, J. Siuzdak, A. Urbani, A. I. Yergi and Y. M. Han, Mass Spectrometry in the Clinical Laboratory: General Principles and Guidance, Chace, 1st edn, 2007 Search PubMed.
- J. W. Honour, Ann. Clin. Biochem., 2011, 48, 97–111 CrossRef CAS PubMed.
- M. M. K. Sharaf El-Din, M. W. I. Nassar, K. A. M. Attia, M. A. E. Demellawy and M. M. Y. Kaddah, J. Pharm. Biomed. Anal., 2016, 125, 236–244 CrossRef CAS PubMed.
- K. A. M. Attia, M. W. I. Nassar, M. M. K. Sharaf El-Din, A. A. A. Mohamad and M. M. Y. Kaddah, Anal. Methods, 2016, 8, 1836–1851 RSC.
- PASS online, http://www.way2drug.com/PASSOnline/predict.php, accessed 22 September 2021 Search PubMed.
- J. J. Magadula and P. Erasto, Nat. Prod. Rep., 2009, 26, 1535–1554 RSC.
- E. Ramos-Morales, L. Lyons, G. de la Fuente, R. Braganca and C. J. Newbold, FEMS Microbiol. Lett., 2019, 366(13) DOI:10.1093/femsle/fnz144.
- A. K. Patra and J. Saxena, Nutr. Res. Rev., 2009, 22, 204–219 CrossRef CAS PubMed.
- M. González, A. Zamilpa, S. Marquina, V. Navarro and L. Alvarez, J. Nat. Prod., 2004, 67, 938–941 CrossRef PubMed.
- A. Cammarata, S. K. Upadhyay, B. S. Jursic and D. M. Neumann, Bioorg. Med. Chem. Lett., 2011, 21, 7379–7386 CrossRef CAS PubMed.
- A. Turky, A. H. Bayoumi, F. F. Sherbiny, K. El-Adl and H. S. Abulkhair, Mol. Diversity, 2021, 25, 403–420 CrossRef CAS PubMed.
- H. G. Ezzat, A. H. Bayoumi, F. F. Sherbiny, A. M. El-Morsy, A. Ghiaty, M. Alswah and H. S. Abulkhair, Mol. Diversity, 2021, 25, 291–306 CrossRef CAS PubMed.
- H. Abul-Khair, S. Elmeligie, A. Bayoumi, A. Ghiaty, A. El-Morsy and M. H. Hassan, J. Heterocycl. Chem., 2013, 50, 1202–1208 CAS.
- M. H. El-Shershaby, K. M. El-Gamal, A. H. Bayoumi, K. El-Adl, H. E. A. Ahmed and H. S. Abulkhair, Arch. Pharm., 2021, 354(2), e2000277 CrossRef PubMed.
- K. El-Adl, M. K. Ibrahim, F. Khedr, H. S. Abulkhair and I. H. Eissa, Arch. Pharm., 2021, 354(3), e202000219 CrossRef PubMed.
- T. K. Mazu, B. A. Bricker, H. Flores-Rozas and S. Y. Ablordeppey, Mini-Rev. Med. Chem., 2016, 16, 555–578 CrossRef CAS PubMed.
- A. E. Kip, J. H. M. Schellens, J. H. Beijnen and T. P. C. Dorlo, Clin. Pharmacokinet., 2018, 57, 151–176 CrossRef CAS PubMed.
- A. C. Coelho, N. Messier, M. Ouellette and P. C. Cotrim, Antimicrob. Agents Chemother., 2007, 51, 3030–3032 CrossRef CAS PubMed.
- R. K. Verma, V. K. Prajapati, G. K. Verma, D. Chakraborty, S. Sundar, M. Rai, V. K. Dubey and M. S. Singh, ACS Med. Chem. Lett., 2012, 3, 243–247 CrossRef CAS PubMed.
- M. V. Keniya, M. Sabherwal, R. K. Wilson, M. A. Woods, A. A. Sagatova, J. D. A. Tyndall and B. C. Monk, Antimicrob. Agents Chemother., 2018, 62(11) DOI:10.1128/AAC.01134-18.
- FDA, Safety Testing of Drug Metabolites, https://www.fda.gov/media/72279/download, accessed 24 September 2021 Search PubMed.
- P. Borman and D. Elder, in ICH Quality Guidelines, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2017, pp. 127–166 Search PubMed.
- Y. V. Kazakevich and R. LoBrutto, HPLC for Pharmaceutical Scientists, John Wiley & Sons, Inc., 2nd edn, 2007 Search PubMed.
- H. S. Abulkhair, S. Elmeligie, A. Ghiaty, A. El-Morsy, A. H. Bayoumi, H. E. A. Ahmed, K. El-Adl, M. F. Zayed, M. H. Hassan, E. N. Akl and M. S. El-Zoghbi, Arch. Pharm., 2021, 354(5), 2000449 CrossRef CAS PubMed.
- M. H. El-Shershaby, A. Ghiaty, A. H. Bayoumi, A. A. Al-Karmalawy, E. M. Husseiny, M. S. El-Zoghbi and H. S. Abulkhair, Bioorg. Med. Chem., 2021, 42, 116266 CrossRef CAS PubMed.
- M. H. El-Shershaby, A. Ghiaty, A. H. Bayoumi, H. E. A. Ahmed, M. S. El-Zoghbi, K. El-Adl and H. S. Abulkhair, New J. Chem., 2021, 45(25), 11136–11152 RSC.
- A. M. Omar, S. Ihmaid, E.-S. E. Habib, S. S. Althagfan, S. Ahmed, H. S. Abulkhair and H. E. A. Ahmed, Bioorg. Chem., 2020, 99, 103781 CrossRef CAS PubMed.
- K. El-Adl, A. G. A. El-Helby, H. Sakr, R. R. Ayyad, H. A. Mahdy, M. Nasser, H. S. Abulkhair and S. S. A. El-Hddad, Arch. Pharm., 2021, 354(2), 2000279 CrossRef CAS PubMed.
- M. H. El-Shershaby, K. M. El-Gamal, A. H. Bayoumi, K. El-Adl, M. Alswah, H. E. A. Ahmed, A. A. Al-Karmalamy and H. S. Abulkhair, New J. Chem., 2021, 45(31), 13986–14004 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08532h |
| This journal is © The Royal Society of Chemistry 2022 |