Open Access Article
Open Access ArticleObservation of macrophage autophagy in the healing of diabetic ulcers via a lysosome-targeting polarity-specific two-photon probe†
Lina Wang‡
a,
Taotao Qiang‡ *a,
Longfang Rena,
Fei Chenga,
Wei Huab and
Renyu Qu
*a,
Longfang Rena,
Fei Chenga,
Wei Huab and
Renyu Qu *c
*c
aCollege of Bioresources and Materials Engineering, Shaanxi Collaborative Innovation Center of Industrial Auxiliary Chemistry & Technology, Shaanxi University of Science & Technology, Xi'an, 710021, China. E-mail: qiangtt515@163.com
bShaanxi Collaborative Innovation Center of Industrial Auxiliary Chemistry & Technology, Shaanxi University of Science & Technology, Xi'an, 710021, China
cJiangsu Sevencontinent Green Chemical Co., Ltd., Zhangjiagang, 215600, China. E-mail: Renyu199107@163.com
First published on 27th January 2022
Abstract
As a disease with high incidence, mutilation, and fatality rates, diabetic ulcers (DUs) have become a difficult and complicated disease of widely concern in recent years due to the unclear healing mechanism. The main reason for the delayed healing in DU patients is the unduly long chronic inflammation window, and the polarization state of macrophages plays a key role in this process. Since autophagy is believed to be closely related to the polarization trend of macrophages, recent studies have shown that autophagy is closely related to the healing of DU. To this end, a lysosome-targeting polarity-sensitive probe, XZTU-VIS, was developed to monitor the changes in lysosomal polarity, thereby assessing the autophagy of macrophages in mice suffering from DU. The experimental results showed that under two-photon fluorescence microscopy, the green channel fluorescence signal of XZTU-VIS decreased significantly during autophagy. In the meantime, DU models established using BV-2 cells and mice showed a process that could cause inflammation and the release of ROS, thereby inducing autophagy.
Introduction
As one of the complications of diabetes (a common public health concern in modern society), diabetic ulcers (DUs) account for about 36% of all chronic skin ulcers.1 Meanwhile, as many as 19.03% of the DU patients end up receiving amputation.2 Unfortunately, the mechanism of the healing of diabetic ulcers is still unclear. The main reason is that the healing mechanism of DU involves interactions between inflammatory cells and biochemical mediators stimulated by various factors.3–5 Specifically, macrophage polarization determines the length of inflammation, and the main reason for the delayed healing of DU is the unduly long chronic inflammation window.6 As an important protective mechanism in eukaryotic cells, autophagy plays a vital role in maintaining homeostasis and repairing damaged cells in the intracellular environment.7 In addition, macrophages are the main cells undergoing autophagy in DU wounds.8 Therefore, fully understanding and accurately detecting the degree of autophagy in the process of DU is the key to its treatment.Traditional methods for monitoring autophagy include transmission electron microscopy (TEM), western blotting, and plasmid transfection.9 However, TEM and western blotting cannot reflect the degree of autophagy in real-time. Although plasmid transfection facilitates direct visual observation of autophagy, the LC3 protein used in transfection is prone to fluorescence quenching in acidic environments (e.g., lysosomes), thus not suitable for long-term detection of autophagy in living cells.10 Therefore, a real-time and efficient method to detect the occurrence of autophagy is urgently needed. In comparison, fluorometric analysis is non-invasive, easy to operate, and highly sensitive, which makes it a powerful tool for the real-time sensing and tracking of biological species and events.11 In particular, two-photon confocal microscopy is a promising fluorescence imaging method. Firstly, its longer excitation wavelength can reduce biological background fluorescence and improve the signal-to-noise ratio of the images.12 Secondly, its near-infrared excitation has less damage to the biological samples, making it a suitable method for long-term imaging and observation of the biological samples.13 Thirdly, photobleaching outside the focus is avoided due to its focus excitation property, which improves the spatial resolution. Finally, the deep penetration of near-infrared excitation makes it a good tool for deep tissue imaging.14 Recent studies have shown that the lysosomal membrane fuses with the autophagosome membrane during the autophagy process, which leads to an increase in its polarity.15 Therefore, polarity-sensitive fluorescence probes with lysosomal targeting ability are becoming popular autophagy level assessment tools.
In this work, a D–π–A two-photon fluorescence probe XZTU-VIS was designed and synthesized with the classic two-photon fluorophore quinoline derivative as the core because of its large two-photon active cross-section and quantum yield, as well as high water solubility, easy modification, and low toxicity. In the meantime, literature has shown that the D–π–A polarity structure is considered conducive to the polarity response and can reflect the subtle polarity differences in the environment.16 In addition, the morpholine group is very easy to be protonated and bound in the lysosome due to its weakly alkaline nature (pKa = 5–6), thereby achieving specific anchoring of lysosomes (pH = 4–5) in the cell.17 Therefore, we believe that XZTU-VIS is very suitable for two-photon fluorescence imaging of lysosomal polarity changes in living cells and achieving long-term imaging analysis of autophagy. In vitro experiments showed that XZTU-VIS exhibited high sensitivity to environmental viscosity and could achieve long-term in situ imaging analysis of autophagy. A DU mouse model was established in this work, and in situ detection of autophagy in cells and mice was successfully achieved. Studies have found that DU can cause autophagy, and inhibiting inflammation significantly reduces the degree of autophagy, indicating that DU induces autophagy through inflammation.
Results and discussion
Synthesis and characterization of XZTU-VIS
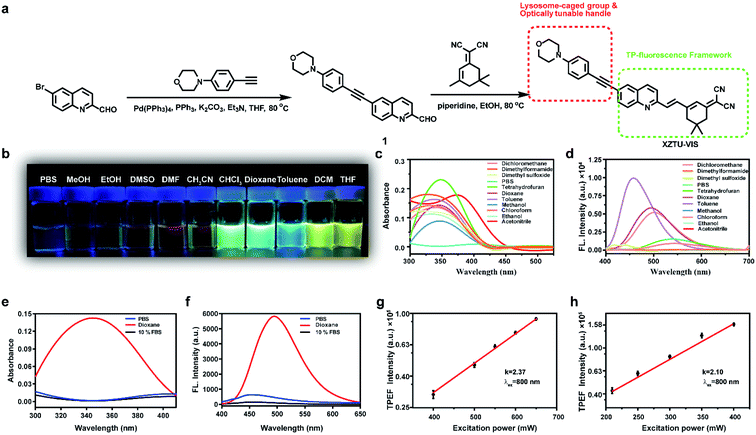
In this study, a two-photon fluorescence probe XZTU-VIS was designed and synthesized. The synthesized probe uses quinoline as the fluorescent matrix (π), 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile as the electron acceptor (A), and 4-(4-ethynylphenyl)morpholine as the electron donor (D) as well as served as a solvent polarity-dependent group enabling the probe to indicate polarity changes in the environment. In addition, the organic base morpholine enriches in the lysosomes with a strong acid environment, thus ensuring that the probe was effectively targeted to lysosomes in the cell. The synthetic route is shown in Fig. 1a. The 4-(4-ethynylphenyl)morpholine, 6-iodoquinoline-2-carbaldehyde, and 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile were synthesized in accordance with the procedures reported in the literature.18–20 Briefly, 4-(4-ethynylphenyl)morpholine and 6-iodoquinoline-2-carbaldehyde underwent a Sonogashira coupling reaction under the action of CuI and Pd(PPh3)Cl2 to generate compound 1. Specifically, under the action of piperidine, compound 1 underwent a condensation reaction with 2-(3,5,5-trimethylcyclohex-2-en-1-ylidene)malononitrile to generate the target probe XZTU-VIS. The detailed characterization data for XZTU-VIS was presented in Fig. S1–S4.†Sensing performance of XZTU-VIS
The optical properties of XZTU-VIS were measured first in solvents with different polarities. As shown in Fig. 1b, XZTU-VIS exhibits different brightness under excitation with 365 nm excitation in different solvents. The brightness in solvents with small polarity, including toluene, chloroform, dioxane, and dichloromethane, is significantly higher than that in solvents with large polarity such as PBS, MeOH and EtOH, etc. Fig. 1c and d show the absorption and emission spectra of XZTU-VIS in different solvents, and the photophysical parameters of XZTU-VIS, including maximum absorbance, maximum emission, quantum yield, and Stokes shift, are listed in Table S1.† Meanwhile, the relationship between absorption and the dielectric constant as well as fluorescence intensity and the dielectric constant of the different solvents is shown in Fig. S5.† These data showed that XZTU-VIS was very sensitive to solvent polarity and met the design requirements of polarity probes. The results showed that although the absorption of XZTU-VIS did not change significantly in different solvents except in PBS, but the fluorescence intensity changed significantly. In order to further explore the relationship between XZTU-VIS and polarity, the spectral response of XZTU-VIS to polarity changes was studied using the dioxane/PBS system. As shown in Fig. 1e and f, the absorption intensity of XZTU-VIS in PBS and PBS contain 10% foetal bovine serum (FBS, physiological environment simulation system) is negligible, while the maximum absorption wavelength in dioxane reaches 345 nm (ε = 1.43 × 104, in which ε is the molar absorptivity). The fluorescence spectrum shows that XZTU-VIS has almost no fluorescence release in PBS and PBS contain 10% FBS, while the fluorescence intensity in dioxane increases by about 80-fold (Φ = 0.173, in which Φ is the quantum yield), and the Stocks shift reaches 149 nm. What's more, in order to prove the two-photon excitation of XZTU-VIS in both PBS and dioxane, the relationship between two-photon emission intensity and excitation light power was investigated. The results showed that the square of the two-photon excitation light power and the slope of the two-photon fluorescence intensity of XZTU-VIS in ethanol and dioxane were both close to 2.0 (2.04 and 2.29, respectively), indicating two-photon absorption processes of XZTU-VIS under different polarities21 (Fig. 1g and h).In addition, fluorescence titration experiments showed that under one-photon excitation and two-photon excitation, the fluorescence intensity of XZTU-VIS in dioxane was about 80-fold and 628-fold higher than that in PBS, respectively (Fig. 2a and c). The relationship between fluorescence intensity and the dielectric constant of the solvents is shown in Fig. 2b. These data showed that XZTU-VIS was very sensitive to solvent polarity and met the design requirements of polarity probes. In order to evaluate whether XZTU-VIS can be used for two-photon confocal imaging, the two-photon active cross-section of the probe in ethanol and dioxane was explored. According to the results shown in Fig. 2d, under an excitation of 810 nm, the maximum two-photon activity cross-sections of XZTU-VIS in PBS and dioxane reached 8 GM and 36 GM (1 GM = 1 × 10−50 cm4 s per photon), respectively, indicating that the probe can be applied to two-photon confocal imaging. Finally, the influence of potentially interfering intracellular substances on the fluorescence of XZTU-VIS was investigated. According to the results shown in Fig. 2e, regardless of whether the probe is in dioxane or PBS, the potential interferers (e.g., 1 mM Fe2+, Fe3+, K+, Na+, Ca2+, Ag+, Zn2+, Cys, GSH, Sec, H2S, and SO2 or 100 μM H2O2, ·OH, HClO, ·O2−, NO, NO2, and ONOO−) cannot change the fluorescence intensity of XZTU-VIS, indicating that XZTU-VIS has good selectivity and can accurately achieve the in situ imaging of cell polarity. To further investigate the selectivity of XZTU-VIS to indicate cell polarity in physiological environment, we next explored the effect of serum and protein (bovine serum albumin as an example) on the fluorescence intensity of XZTU-VIS. The results clearly showed that serum and different concentrations of bovine serum albumin did not interfere with the fluorescence intensity of XZTU-VIS, indicating that XZTU-VIS can accurately detect cell polarity in a physiological environment (Fig. S6†).
Bioimaging application of XZTU-VIS in living cells
Before being adopted in fluorescence imaging, the biocompatibility and photostability of XZTU-VIS were investigated. As shown in Fig. S7,† after 24 h incubation with different concentrations of XZTU-VIS (0, 5, 10, 15, 20 μM), BV-2 cells still had a survival rate above 90% according to the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. In the meantime, the probe-loaded cells were subjected to 810 nm two-photon excitation, and data were collected every 5 min. According to the results shown in Fig. S8,† the fluorescence intensity of the green channel (500–550 nm) remained almost unchanged within 60 min. The above experiments showed that XZTU-VIS had good biocompatibility and photostability and was suitable for real-time imaging monitoring and analysis of the autophagy process in living cells. Then, the distribution of XZTU-VIS in cells was investigated to verify whether XZTU-VIS was capable of indicating the degree of autophagy. Commercial colocalization dyes (Lyso-Tracker Green for lysosomes, Mito-Tracker Green for mitochondria, and ER-Tracker Green for the endoplasmic reticulum) were used along XZTU-VIS to stain the cells and explore the localization ability of the probe to different subcellular organelles. The probe-loaded cells were incubated with 60 nM Lyso-Tracker-Red, Mito-Tracker-Red, and ER-Tracker-Red for 30 min before being subjected to confocal imaging. The results are shown in Fig. S9.† The fluorescence of XZTU-VIS overlaps well with that of Lyso-Tracker-Red. The Pearson coefficient reaches 0.94, while the Pearson coefficients in the mitochondria and endoplasmic reticulum are 0.24 and 0.28, respectively, indicating that XZTU-VIS can specifically target lysosomes. These features provide a theoretical basis for in situ detection of autophagy.In order to explore whether XZTU-VIS can be interfered with by viscosity in living cells, it was incubated with BV-2 cells at different temperatures (37 °C, 25 °C, and 5 °C, where the viscosity increases with decreasing temperature22), and the changes in cell viscosity were measured. As shown in Fig. S10,† under two-photon excitation, the fluorescence intensity of probes incubated at 4 °C and 25 °C showed no significant changes, thus excluding the response of XZTU-VIS to intracellular viscosity. In addition, the cells were incubated with sucrose to increase their polarity successively.15 The results showed significantly reduced fluorescence intensity, indicating that the probe was capable of in situ imaging analysis of cell polarity fluctuations. Subsequently, autophagy inducer rapamycin (100 nm, mTORC1 complex inhibitor) and starvation were adopted to induce autophagy and prove the probe's autophagy detecting ability.23 As shown in Fig. S9,† under 810 nm two-photon excitation, the green channel (500–550 nm) fluorescence intensity of the cells treated with rapamycin and starvation is negligible. It is worth noting that the green channel fluorescence of the cells treated with starvation recovered after continued incubation with 3-methyladenine15 (3-MA, 10 μM). The above experiments showed that XZTU-VIS could respond to intracellular autophagy and achieve in situ imaging analysis of autophagy. Finally, LPS/IFN-γ was used to stimulate inflammatory responses in the cells,24 and the results showed that the green channel (500–550 nm) fluorescence intensity was significantly reduced. The fact that 3-MA could significantly enhance the green channel (500–550 nm) fluorescence intensity of inflammatory cells indicated that XZTU-VIS could achieve in situ imaging analysis of the autophagy degree during cellular inflammation.
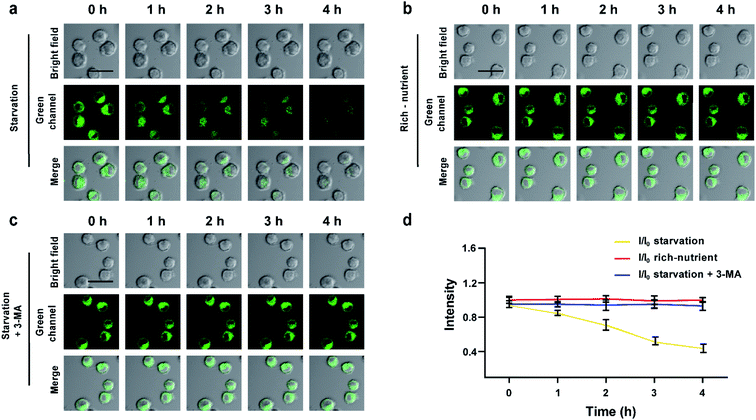
To further evaluate the real-time autophagy monitoring ability of XZTU-VIS, BV-2 cells stained with XZTU-VIS were cultured under different cell culture conditions (starvation, nutrient-rich, and starvation + 3-MA), and real-time two-photon confocal imaging analysis was performed at different times (0, 1, 2, 3, 4 h). As shown in Fig. 3, the green channel (500–550 nm) fluorescence intensity of the cells in starvation decreases continuously with time. However, the fluorescence intensity of cells pre-treated with autophagy inhibitor 3-MA or with normal nutrients remained constant over time, indicating that XZTU-VIS could achieve real-time imaging and analysis of autophagy. Next, inflammation inducer LPS was used to trigger an inflammatory response in BV-2 cells and determine whether XZTU-VIS could effectively respond to inflammation-induced autophagy.
Encouraged by the above experiments, LPS/IFN-γ was then used to stimulate inflammatory responses in the cells. Extensive literature indicated that autophagy was usually caused by the increase of intracellular ROS levels. In the meantime, autophagy consumes and degrades excess intracellular ROS to ensure cell stability. On the other hand, inflammation is usually caused by the release of large amounts of ROS induced by oxidative stress. Therefore, the DU-induced inflammation, in turn, induces autophagy by regulating the degree of cellular oxidative stress. The results are shown in Fig. 4a and b. The green channel (500–550 nm) fluorescence intensity of the LPS/IFN-γ treatment group was almost negligible compared to the sham group (untreated cells, p < 0.001). After being subjected to inflammation inhibitors NS-398 (10 mM, cyclooxygenase-2 inhibitor25), NOX-2 inhibitors apocynin (APO, 10 mM, NADPH oxidase 2 inhibitor26), and NOX-2 gene silencing (NOX2KD, operational approach according to the reported method27), the degree of autophagy was significantly reduced, indicating that LPS/IFN-γ causes inflammation by inducing oxidative stress in the cells. In the meantime, green channel (500–550 nm) fluorescence intensity of the negative control group (negative group, operational approach according to the reported method27) was significantly higher than that of the NOX-2KD group (p < 0.001). The above experiment fully demonstrated that the NOX-2 protein could regulate oxidative stress in cells and cause inflammation. The conclusion was also proved by ELISA (Fig. 4a–e). The trends of inflammatory factors (TNF-α and IL-1β) and hydrogen peroxide (H2O2, cellular oxidative stress marker28) content were consistent with that of the ratio signal, indicating that inhibiting oxidative stress could significantly inhibit cellular inflammation. More importantly, the CCK-8 experiment showed that both oxidative stress inhibitors and inflammation inhibitors alleviated the apoptosis induced by cell inflammation (Fig. 4f).
Based on the above findings, fluorescence imaging of inflammatory wound tissue of XZTU-VIS was investigated. The diabetic mice were divided into three groups: the first group is the control group; the second group received a cut in the right foot; the third group received a cut in the right foot and a subcutaneous injection of NS-398 on the inner thigh. After 12 h, the three groups of mice were injected subcutaneously with 200 μL of 100 μM XZTU-VIS on the inner thigh. After another 1 h, the wound sections of the three groups were subjected to imaging. As shown in Fig. 5a and b, the results were similar to the cell model. Compared with normal mice, the green channel (500–550 nm) fluorescence intensity of the skin tissue from mice with cuts was significantly reduced (p < 0.001). The fluorescence signal of the group pre-injected with NS-398 was significantly higher than that of the group with cuts (p < 0.001), indicating that the wound could induce inflammation in mice. H&E staining showed that the skin tissue of the mice in the control group had clear structures and normal cellular morphology. The skin tissue of the mice in the group with cuts showed obviously abnormal structures, pyknotic cell nuclei, and a significant increase in inflammatory cells. However, the tissue structure of mice injected with NS-398 was less damaged and even recovered, with reduced nuclear pyknosis and inflammatory cells (Fig. 5c). In the meantime, the expression of IL-6 measured by immunohistochemical staining and the ROS levels measured by DHE staining was consistent with the cell experiment results (Fig. 5c–f). The above experimental results showed that the wound caused inflammation in mice, and NS-398 protected the mice from skin damage by the inflammatory response.
Conclusions
In this work, we developed a two-photon fluorescence probe with lysosomal targeting properties for real-time, efficient, and accurate monitoring of autophagy. The probe XZTU-VIS showed high selectivity for changes in lysosomal polarity and lysosomes, which could help achieve high accuracy and fidelity in the fluorescence imaging of inflammation-induced autophagy. With good two-photon photophysical properties, low cytotoxicity, and sensitive response to polarity changes, the probe XZTU-VIS could help monitor the lysosomal polarity changes in BV-2 cells and skin ulcers in diabetic mice and achieve autophagy level assessment. In addition, the probe was the first molecular tool used to visualize the development of inflammation induced by skin ulcers in diabetic mice. This work proved that the lysosome-targeting two-photon fluorescence probe had great potential in studying inflammation and autophagy induced by inflammation.Ethical statement
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of South-Central University for Nationalities and experiments were approved by the Animal Ethics Committee of College of Biology (South-Central University for Nationalities). Wild-type C57BL/6J mice (n = 300; 25–30 g) were purchased from Hubei Experimental Animal Research Center. (Hubei, China; No. 43004700018817, 43004700020932). All animal experimental protocols were approved by the Animal Experimentation Ethics Committee of South-Central University for Nationalities (No. 2020-scuec-043) and were conducted according to the Animal Care and Use Committee guidelines of South-Central University for Nationalities.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This work was funded by the Open Project of Shaanxi Collaborative Innovation Center of Industrial Auxiliary Chemistry & Technology (No. XTKF-2020-05); the Key Scientific Research Group of Shaanxi Province (2020TD-009), Key Scientific Research Program of Shaanxi Provincial Education Department (Collaborative Innovation Center project) (20JY003), Science and Technology Plan Project of Xi'an Weiyang District (No. 201907) and the Youth Innovation Team of Shaanxi Universities.References
- M. M. Iversen, K. Midthjell, G. S. Tell, T. Moum, T. Østbye, M. W. Nortvedt, S. Uhlving and B. R. Hanestad, BMC Endocr. Disord., 2009, 9, 18 CrossRef PubMed.
- Y. Jiang, X. Ran, L. Jia, C. Yang, P. Wang, J. Ma, B. Chen, Y. Yu, B. Feng, L. Chen, H. Yin, Z. Cheng, Z. Yan, Y. Yang, F. Liu and Z. Xu, Int. J. Lower Extremity Wounds, 2015, 14, 19–27 CrossRef PubMed.
- R. E. Mirza and T. J. Koh, Cytokine, 2015, 71, 409–412 CrossRef CAS PubMed.
- L. Pradhan, X. Cai, S. Wu, N. D. Andersen, M. Martin, J. Malek, P. Guthrie, A. Veves and F. W. LoGerfo, J. Surg. Res., 2011, 167, 336–342 CrossRef CAS PubMed.
- P. Bannon, S. Wood, T. Restivo, L. Campbell, M. J. Hardman and K. A. Mace, Dis. Models Mech., 2013, 6, 1434–1447 CAS.
- M. Gooyit, Z. Peng, W. R. Wolter, H. Pi, D. Ding, D. Hesek, M. Lee, B. Boggess, M. M. Champion, M. A. Suckow, S. Mobashery and M. Chang, ACS Chem. Biol., 2014, 9, 105–110 CrossRef CAS PubMed.
- S. K. Biswas and A. Mantovani, Cell Metab., 2012, 15, 432–437 CrossRef CAS PubMed.
- S. Willenborg and S. A. Eming, J. Dtsch. Dermatol. Ges., 2014, 12, 214–221 Search PubMed.
- J. M. Swanlund, K. C. Kregel and T. D. Oberley, Autophagy, 2010, 6, 270–277 CrossRef PubMed.
- I. Tanida, N. Minematsu-Ikeguchi, T. Ueno and E. Kominami, Autophagy, 2005, 1, 84–91 CrossRef CAS PubMed.
- S. Zhai, W. Hu, C. Fan, W. Feng and Z. Liu, Chem. Commun., 2021, 57, 5542–5545 RSC.
- J. Zhang, Q. An, W. Li, L. Chai, W. Hu, Y. Wang, S. Su, Y. He, C. Li and D. Sun, Sens. Actuators, B, 2021, 345, 130329 CrossRef CAS.
- F. Cheng, T. Qiang, L. Ren, T. Liang, X. Gao, B. Wang and W. Hu, Analyst, 2021, 146, 2632–2637 RSC.
- M. Ye, W. Hu, M. He, C. Li, S. Zhai, Z. Liu, Y. Wang, H. Zhang and C. Li, Chem. Commun., 2020, 56, 6233–6236 RSC.
- J. Diao, R. Liu, Y. Rong, M. Zhao, J. Zhang, Y. Lai, Q. Zhou, L. M. Wilz, J. Li, S. Vivona, R. A. Pfuetzner, A. T. Brunger and Q. Zhong, Nature, 2015, 520, 563–566 CrossRef CAS PubMed.
- T. Liang, D. Zhang, W. Hu, C. Tian, L. Zeng, T. Wu, D. Lei, T. Qiang, X. Yang and X. Sun, Talanta, 2021, 235, 122719 CrossRef CAS PubMed.
- M. Tian, J. Zhan and W. Lin, Coord. Chem. Rev., 2022, 451, 214266 CrossRef CAS.
- C. Xu, C. W. Muir, A. G. Leach, A. R. Kennedy and A. J. B. Watson, Angew. Chem., Int. Ed. Engl., 2018, 57, 11374–11377 CrossRef CAS PubMed.
- L.-L. Li, K. Li, M.-Y. Li, L. Shi, Y.-H. Liu, H. Zhang, S.-L. Pan, N. Wang, Q. Zhou and X.-Q. Yu, Anal. Chem., 2018, 90, 5873–5878 CrossRef CAS PubMed.
- L. Dell'Amico, G. Rassu, V. Zambrano, A. Sartori, C. Curti, L. Battistini, G. Pelosi, G. Casiraghi and F. Zanardi, J. Am. Chem. Soc., 2014, 136, 11107–11114 CrossRef PubMed.
- Y. Lou, C. Wang, S. Chi, S. Li, Z. Mao and Z. Liu, Chem. Commun., 2019, 55, 12912–12915 RSC.
- L. Hou, P. Ning, Y. Feng, Y. Ding, L. Bai, L. Li, H. Yu and X. Meng, Anal. Chem., 2018, 90, 7122–7126 CrossRef CAS PubMed.
- Y. Liu, J. Zhou, L. Wang, X. Hu, X. Liu, M. Liu, Z. Cao, D. Shangguan and W. Tan, J. Am. Chem. Soc., 2016, 138, 12368–12374 CrossRef CAS PubMed.
- Z. Mao, M. Ye, W. Hu, X. Ye, Y. Wang, H. Zhang, C. Li and Z. Liu, Chem. Sci., 2018, 9, 6035–6040 RSC.
- W. Hu, L. Zeng, S. Zhai, C. Li, W. Feng, Y. Feng and Z. Liu, Biomaterials, 2020, 241, 119910 CrossRef CAS PubMed.
- V. E. Kagan, N. V. Konduru, W. Feng, B. L. Allen, J. Conroy, Y. Volkov, I. I. Vlasova, N. A. Belikova, N. Yanamala, A. Kapralov, Y. Y. Tyurina, J. Shi, E. R. Kisin, A. R. Murray, J. Franks, D. Stolz, P. Gou, J. Klein-Seetharaman, B. Fadeel, A. Star and A. A. Shvedova, Nat. Nanotechnol., 2010, 5, 354–359 CrossRef CAS PubMed.
- Z. Wang, X. Wei, K. Liu, X. Zhang, F. Yang, H. Zhang, Y. He, T. Zhu, F. Li, W. Shi, Y. Zhang, H. Xu, J. Liu and F. Yi, Free Radicals Biol. Med., 2013, 65, 942–951 CrossRef CAS PubMed.
- B. Zhai, W. Hu, R. Hao, W. Ni and Z. Liu, Analyst, 2019, 144, 5965–5970 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details and characterization probe XZTU-VIS supplementary spectral and cells experiments results. See DOI: 10.1039/d1ra08417h |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |