Open Access Article
Open Access ArticleDNA aptamer-based dual-responsive nanoplatform for targeted MRI and combination therapy for cancer†
Mingming Zhaoab,
Xiaoxi Songab,
Jiahui Lub,
Siwen Liub,
Xuan Shab,
Qi Wangab,
Xu Caob,
Kai Xu*ab and
Jingjing Li *ab
*ab
aSchool of Medical Imaging, Xuzhou Medical University, Xuzhou 221004, P. R. China. E-mail: qingchao0124@163.com
bDepartment of Radiology, Affiliated Hospital of Xuzhou Medical University, Xuzhou 221006, P. R. China
First published on 31st January 2022
Abstract
Accurate drug delivery is a common topic, and it has always been an aim that scientists strive to achieve. To address this need, multifunctional and stimulus-sensitive nanoplatforms have attracted significant attention. Here we fabricated a glutathione (GSH) and adenosine-5′-triphosphate (ATP) dual-sensitive nanoplatform for controlled drug release and activatable MRI of tumors based on DNA aptamer and manganese dioxide (MnO2) nanosheets. Cleverly utilizing the DNA tunability, AS1411 aptamer which binds nucleolin, a protein specifically expressed on tumor-associated endothelial cells, was designed with ATP aptamer and its cDNA to load the anticancer drug, doxorubicin (Dox). The formed DNA-Dox complex was delivered to the tumor region with the help of MnO2 nanosheets and AS1411 aptamer. Then, the on-demand drug release in tumor cells was realized with the co-effect of the ATP aptamer and GSH reduction. It was found that without the structure of the MnO2 nanosheets being broken by GSH, Dox almost could not be released even in the presence of ATP. Similarly, without ATP, Dox was still maintained in the duplex even with GSH. Further combining the MRI ability and chemodynamic therapy of the produced Mn2+, an improved effect of the inhibition of tumor growth and imaging was achieved. Our designed DNA aptamer-based dual-responsive nanoplatform can realize the targeted drug delivery and MRI of breast tumor cells both in vitro and in vivo.
1. Introduction
Accurate drug delivery plays an important role in improving the therapeutic effect and reducing side effects. Particularly, with the good development of nanotechnology, a series of nanomaterial-based drug delivery systems have emerged. Dendrimers with micellar solubilization, good encapsulation ability, biodegradable nature and excellent biocompatibility have been widely used for drug delivery.1 Biopolymeric nanoparticles such as chitosan and dextran, liposomes,2,3 mesoporous silica,4 gold nanomaterials,5 carbon nanomaterials,6 proteins7 and even cells8 have been employed for drug delivery. The introductions of aptamers as targeting ligands to the above drug delivery systems further favor the accurate treating and diagnosing of disease. The characteristics of aptamers with high affinity and specificity for their targets, small size for little influence on the drug delivery device, and synthesized chemically for easy conjugation with nanocarrier or imaging molecules enable their applications in the therapy and diagnosis of disease, especially tumors.9 AS1411 is a 26-nucleotide guanosine-rich DNA aptamer that can bind to the nucleolin proteins on tumor cells.10–12 It can be used for targeted drug delivery and imaging. Dai et al. constructed multifunctional nanoparticles through AS1411 aptamer-conjugated mesoporous polydopamine for targeted and synergistic chemotherapy/photothermal therapy of prostate cancer.13 Chen et al. reported an aptamer-dendrimer functionalized magnetic nano-octahedrons for AS1411 aptamer-targeted, NIR/MR dual-modality imaging guided, and HSP70/HSP90 silencing sensitized magnetochemotherapy.14 To realize the on-demand release, a series of tumor related stimulus have been employed to design stimuli-triggered drug delivery systems. A series of endotrigger stimulies, such as pH,15 hypoxia,16 enzyme,17 temperature,18 redox,19,20 glucose21,22 and other exo-trigger stimuli such as light,23 electric,24 magnetic,25 and ultrasound26 responsive drug delivery systems have been well documented.27 For the design of these stimuli-triggered drug delivery systems, DNA molecules have been shown to be excellent substrates for the design and construction of intelligent vehicles for imaging and cargo delivery. Zhao et al. fabricated an autonomous DNA robot with a DNA aptamer binding with nucleolin and the blood coagulation protease thrombin. The nucleolin-targeting aptamer triggered the mechanical opening of the DNA nanorobot and exposed the thrombin inside to activate coagulation at the tumor site.28 Gu et al. developed a nanogel containing ATP aptamer and hyaluronic acid for active tumor-targeting and ATP triggered drug release.29 Fan et al. reviewed the DNA nanotechnology-enabled drug delivery systems, showing a lot of state-of-the-art strategies for the construction of DNA nanostructures and drug payloads delivered by DNA nanovehicles.30 Inspired by these successful applications of DNA-based nanovehicles, we tried to design strategy for the possibility of combining more than one stimuli for much greater targeted imaging and more efficient drug delivery as well as the integration of the imaging and combination therapy to further improve the theranostic effect of tumor.To realize this aim, in this work we designed a dual-responsive nanoplatform comprised of DNA scaffold and MnO2 nanosheets. The DNA scaffold was designed to contain AS1411 aptamer as targeting molecule, ATP aptamer and its cDNA for Dox loading and ATP triggered drug release (Scheme 1). Due to the different concentrations in the extracellular environment (<0.4 mM) and in the intracellular cytosol (1–10 mM), ATP has been applied as a trigger for drug release control.29,31 Specifically, the GC pairs of the duplex of ATP aptamer and its cDNA provided loading sites for Dox and ATP-triggered Dox release. MnO2 nanosheets were employed as the activatable MRI nano contrast agent, chemodynamic therapy (CDT) agent, and the GSH triggered drug delivery nanoplatform to load the functional DNA scaffold with Dox through coordination interaction between phosphate backbone of DNA and MnO2 nanosheets. In tumor, nucleolin-medicated endocytosis by AS1411 aptamer realized the internalization of our fabricated nanoplatform to tumor cells. Then, the overexpressed GSH reduced MnO2 nanosheets to Mn2+ and the broken up of the nanosheet structure favored the release of loaded functional DNA scaffold. The produced Mn2+ can further light up MRI signal and mediated Fenton-like reactions for CDT. Later, a significantly higher ATP level in the cytosol compared with the extracellular fluid brought the binding between ATP and its aptamer to cause the structure change of DNA scaffold and the release of Dox eventually. The on-demand intracellular drug release significantly increased the amount of subsequently accumulated Dox in the nuclei beyond the threshold. Finally, the dual-response mediated drug delivery and combination of CT and CDT were realized with MRI monitoring. The co-existence of GSH and ATP could produce the maximum therapeutic effect.
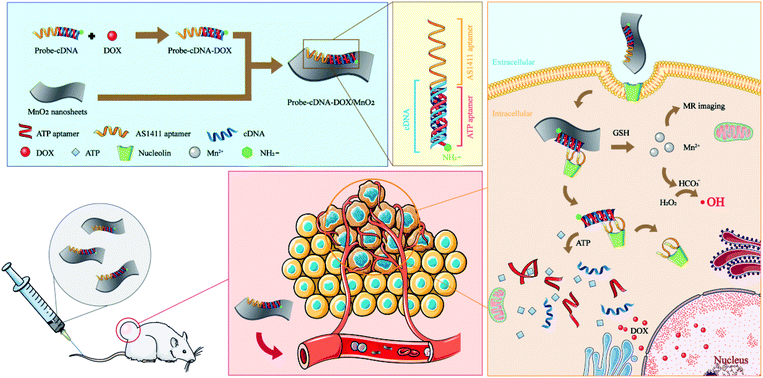 | ||
| Scheme 1 Schematic illustration of the construction of NH2-Probe 1-cDNA-Dox@MnO2 nanoplatform and their application for MRI and combination therapy of tumor. | ||
2. Experimental section
2.1 Materials, cell line and animals
Manganese chloride tetrahydrate (MnCl2·4H2O), tetra-methylammonium hydroxide (TMAH), hydrogen peroxide (H2O2) and ethanol were purchased from Aladdin Biochemical Polytron Technologies, Inc. (Shanghai, China). DNA oligos were obtained from Sangon Biotech Co. Ltd (Shanghai, China). Doxorubicin was obtained from Kangmei Biotechnology Co., Ltd (Xuzhou, China). All chemicals involved in this work were analytical grade. All aqueous solutions were prepared with ultrapure water (≥18 MΩ, Milli-Q, Millipore). The MDA-MB-231 breast cancer cells and mouse NIH-3T3 fibroblasts cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Incomplete medium, fetal bovine serum and penicillin–streptomycin solution were added to a sterile culture flask in a ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and formulated into a fully medium and stored in a 4 °C refrigerator. Cells were cultured at 37 °C, 5% CO2 incubator (Thermo, USA). The BALB/c mice were purchased from Xuzhou Medical University Animal Experiment Center (Xuzhou, China). Animal experiments were performed in accordance with the National Institutes of Health guidelines on the use of animals in research, and all animal experiments were approved by the Animal Care Committee of the Xuzhou Medical University (no. 202007W001).
1, and formulated into a fully medium and stored in a 4 °C refrigerator. Cells were cultured at 37 °C, 5% CO2 incubator (Thermo, USA). The BALB/c mice were purchased from Xuzhou Medical University Animal Experiment Center (Xuzhou, China). Animal experiments were performed in accordance with the National Institutes of Health guidelines on the use of animals in research, and all animal experiments were approved by the Animal Care Committee of the Xuzhou Medical University (no. 202007W001).
2.2 Apparatus and characterization
KS-180EI medical ultrasonic cleaning machine was used to redisperse MnO2 powder (Ningbo, China). Atomic force microscopy (AFM) (SP13800, Japan) and transmission electron microscopy (TEM) (TEC-NAI G2, USA) were used to get size and morphology characterizations. Nano ZS90 (Malvern, England) was employed to determine the zeta potential. The UV-vis absorption spectra were observed by the UH4150 spectrophotometer (Techcomp, China). LS 55 Fluorescence Spectrometer (PerkinEimer, USA) was used to get fluorescence spectra. The Mn2+ concentration was measured by inductively coupled plasma-mass spectrometry (ICP-MS) (PerkinElmer, USA). In vitro and in vivo MRI scans were performed on a 3.0 T MR imaging system (GE Discovery 750w) using a small animal coil. An inverted fluorescence microscope (Olympus bx 51, Japan) was used to observed histological changes of related organs of mice and cellular morphology.2.3 Preparation of MnO2 nanosheets
MnO2 nanosheets were prepared according to previous reports.32 Briefly, 0.5937 g MnCl2·4H2O was dissolved in 10 mL H2O. Then 20 mL 3% H2O2, and 2.172 g hydroxylated TMAH were added. The solution immediately turned dark brown and stirred violently overnight at room temperature in the open air. The overnight reactants were centrifuged, and the supernatant was removed. The obtained precipitate was washed twice with ultra-pure water and methanol respectively. The precipitate was then placed in a vacuum constant temperature drying box and dried at 40 °C. The dried powder was dissolved in ultra-pure water with 40 mL per 10 mg and treated with ultrasonic for 12 hours. Finally, the liquid was centrifuged and filtered with a filter head with a pore diameter of 0.2 μm to collect MnO2 nanosheets solution.2.4 Preparation of NH2-Probe 1-cDNA-Dox@MnO2
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 30 min. Then, they were incubated with MnO2 (20 μL, 15.3 mM Mn). The final volume of NH2-Probe 1-cDNA-Dox@MnO2 was fixed to be 300 μL with ultrapure water.
1 for 30 min. Then, they were incubated with MnO2 (20 μL, 15.3 mM Mn). The final volume of NH2-Probe 1-cDNA-Dox@MnO2 was fixed to be 300 μL with ultrapure water.2.5 The release of Dox triggered by ATP
The release of Dox from NH2-Probe 1-cDNA-Dox triggered by ATP was monitored by the fluorescence changes of Dox. Briefly, NH2-Probe 1-cDNA-Dox was incubated with different ATP concentrations (0, 0.2, 0.4, 1, 2, 4, 6, 8, 10 mM) for 30 min. Then, the changes of Dox fluorescence were recorded. The released Dox from ATP-triggered NH2-Probe 2-cDNA-Dox were also determined to evaluate the influence of the existence of AS1411 aptamer on the Dox release.2.6 The dual-responsive release of Dox from NH2-Probe 1-cDNA-Dox@MnO2
To evaluate the effect of the presence of GSH and ATP on the release of Dox, the fluorescence emissions of NH2-Probe 1-cDNA-Dox@MnO2 in the presence of ATP, GSH or ATP + GSH were detected. Briefly, 16 μL 25 μM NH2-Probe 1 was hybridized with 16 μL 25 μM cDNA for 30 minutes. Then, 10 μL 40 μM Dox was added and incubated for 30 minutes to obtain NH2-Probe 1-cDNA-Dox. Different amount of MnO2 (0, 10, 20, 50 μL Mn 15.3 mM) was introduced and incubated for 30 minutes, respectively. The fluorescence changes of Dox were measured after the above solution was mixed with 10 mM ATP, 10 mM GSH or 10 mM ATP + 10 mM GSH for 30 minutes.2.7 Relaxivity calculation of MnO2 and NH2-Probe 1-cDNA-Dox@MnO2 with GSH
The Mn concentration was detected according to ICP-MS. In the presence of GSH, the solution with manganese ion concentration of 0–1.2 mM was prepared respectively. The corresponding linear regression equation graph was made with the concentration of manganese ion as the horizontal coordinate and the reciprocal of T1 relaxation time of samples with different concentrations as the vertical coordinate (Fig. 6B). The T1 relaxation rates of the MnO2 and NH2-Probe 1-cDNA-Dox@MnO2 were compared. The scanning parameters were setted as follows: repetition time (RT) = 425 ms, echo time (TE) = min full, matrix size = 384 × 224, field of view (FOV) = 18 cm × 18 cm, slice thickness = 3.0 mm, spacing = 1.5 mm.332.8 Evaluation of drug release
The NH2-Probe 1-cDNA-Dox@MnO2 solution was packaged in a dialysis bag with a molecular weight cut-off of 3500–5000 Da. It was placed in a test tube fitted with a 3 mL release buffer solution, containing 5 mM HEPES, 137 mM NaCl, 10 mM MgCl2, 10 mM ATP and 10 mM GSH. Magnetical stirring was performed at 37 °C. At each scheduled time (0, 15, 30, 40, 50, 70, 90, 120, 150, 200, 300 min), 300 μL release medium was taken out for fluorescence test. The buffer volume was maintained at 3 mL all the time.2.9 MRI stability
The NH2-Probe 1-cDNA-Dox@MnO2 solution was stored in a refrigerator at 4 °C. At 0 day, 1 day, 5 day, 10 day, 15 day, 20 day, 30 day, the change of T1 signal intensity in the presence of GSH was measured.2.10 In vitro and in vivo toxicity test
The in vitro cytotoxicity tests were performed using MTT assay34 on MDA-MB-231 (breast cancer cell line) and NIH-3T3 (mouse fibroblasts cells). Briefly, cells in the logarithmic growth phase were evenly inoculated on the 96-well culture plate with a total number of 0.5 × 105 cells per well. The cells were cultured for 24–36 h to become overgrown. Subsequently, NH2-Probe 1-cDNA-Dox, NH2-Probe 1-cDNA@MnO2, and NH2-Probe 1-cDNA-Dox@MnO2 with various NH2-Probe 1-cDNA concentrations were introduced. After incubation for 24 h, the cells were washed with PBS for 3 times. Then, 100 μL 1 mg mL−1 MTT was added and incubated for another 4 h. After discarding the supernatant, 100 μL DMSO was added to each well. The absorbance value of each well at 490 nm was measured with a microplate analyzer, and the average absorbance value of each group was calculated.The experiment of live and dead cell observations was also introduced to evaluate the toxicity and in vitro therapy effect. In brief, the MDA-MB-231 cells were co-cultured with NH2-Probe 1-cDNA@MnO2 and NH2-Probe 1-cDNA-Dox@MnO2 at the equivalent concentration of NH2-Probe 1-cDNA for 2 h. After that, the cells were washed and incubated with calcein acetoxymethyl ester (CA) (4 μM) and propidium iodide (PI) (4 μM) staining reagents for 10 min and then observed using an inverted fluorescence microscope.
Intravenous injection of NH2-Probe 1-cDNA-Dox@MnO2 was used to observe the histological changes of the heart, liver, spleen, kidney, lung of mice, and to evaluate the toxicity of NH2-Probe 1-cDNA-Dox@MnO2 in vivo. 200 μL NH2-Probe 1-cDNA-Dox@MnO2 (3.06 mM Mn) was injected into the mice via the tail vein. The mice were sacrificed at day 1, 7 and 21 post-injection and the heart, liver, spleen, lung, kidney and other organs were taken out, fixed with 10% formalin for 3 days and stained with hematoxylin and eosin (H & E). The tissue structure was observed under a microscope. Untreated mice were dissected as controls.
Hematological and biochemical analyses were also used to evaluate the in vivo toxicity of NH2-Probe 1-cDNA-Dox@MnO2. Twenty-four Kunming mice were divided into 4 groups. Six mice in group 0 day were injected with normal saline and eighteen mice in group 1 day, 7 day and 21 day were injected with NH2-Probe 1-cDNA-Dox@MnO2 (200 μL of 3.06 mM Mn) through tail vein, respectively. At the time point (0 day, 1 day, 7 day and 21 day), the blood samples from mice in the corresponding group were collected by eyeball extirpating for blood routine and biochemical analyses (n = 3).
2.11 Blood half-life determination
The NH2-Probe 1-cDNA-Dox@MnO2 solution (200 μL, 3.06 mM Mn) was injected into the mice through a tail vein. The blood samples (40 μL) were collected at different time periods (15 min, 30 min, 45 min, 1 h, 2 h, 4 h, 6 h, 12 h) post-injection. The collected blood was thoroughly mixed with nitric acid solution. After digestion at 80 °C for 30 min, the concentrations of Mn in each sample were determined by ICP-MS. Fitting half-life curve and half-life (T1/2) was calculated with Origin 8.0 software.2.12 In vitro and in vivo specific MRI
For in vitro MRI, MDA-MB-231 breast cancer cells were first cultured overnight on a 6-well plate at a density of 2 × 105 cells per well. Then, different concentrations of NH2-Probe 1-cDNA-Dox@MnO2 or NH2-Probe 2-cDNA-Dox@MnO2 (0.26 mM, 0.51 mM, 1.02 mM Mn) were co-incubated with cells for 2 h. Subsequently, the cells were washed twice with fresh PBS, and digested with EDTA-trypsin. The collected cells were dispersed in a 1% agarose gel for MRI scanning.A 100 μL cell suspension containing 2 × 106 MDA-MB-231 breast cancer cells was injected into the subcutaneous soft tissue on the right dorsum axillary of BALB/c female nude mice. When the tumor diameter reached about 5 mm, MDA-MB-231 nude mice were used for MRI. In vivo MRI was performed on the MDA-MB-231 nude mice before (0 min) and post-injection (10 min, 30 min, 1 h, 2 h, 3 h, 6 h, 12 h, 24 h) of NH2-Probe 1-cDNA-Dox/MnO2 or NH2-Probe 2-cDNA-Dox/MnO2 (200 μL, 3.06 mM Mn) using an animal T/R coil on a 3.0T magnetic resonance imaging system (GE Discovery 750W, USA). The change of signal intensity in the tumor with the time passing by was recorded accordingly. The T1-weighted MR images were obtained using a gradient echo (GRE) sequence with the following parameters, repetition times (TR) = 16 ms, echo time (TE) = 3.2 ms, field of view (FOV) = 8 cm × 8 cm, matrix size = 320 × 288, slice thickness = 0.4 mm, and spacing = 0.5 mm.35
2.13 ·OH generation of NH2-Probe 1-cDNA-Dox@MnO2 in the presence of GSH
Chemodynamic therapy of NH2-Probe 1-cDNA-Dox/MnO2 was dependent on their ability to generate ·OH in tumor microenvironment. ·OH degrades methylene blue (MB), and ·OH induced degradation of MB is monitored by absorbance change at 665 nm. Briefly, 25 mM NaHCO3 containing NH2-Probe 1-cDNA-Dox/MnO2 (15.3 mM Mn) and 1 mM GSH was first shaken for 15 min. Then, 10 μg mL−1 MB and 8 mM H2O2 were added to the above solution with a final volume of 300 μL. After incubated at 37 °C for 30 min, the absorbance of MB at 665 nm was determined.2.14 In vivo therapy
The inhibitory effect of MDA-MB-231 on tumor growth in BALB/c nude mice was evaluated. NH2-Probe 1-cDNA-Dox@MnO2 or NH2-Probe 2-cDNA-Dox/MnO2 were injected intravenously (i.v.) 7 times at a dose of 200 μL 3.06 mM Mn with a 3 day-interval, respectively. PBS was used as the control. Tumor length and width were recorded with a vernier caliper every 3 days for 21 days. The volume of the tumor was calculated by the formula: volume = width2 × length/2. On the last day, MRI scans were performed on the tumor-bearing nude mice. The body weight of the nude mice during the above treatment was recorded every three days.2.15 Statistical analysis
SPSS 22.0 software package was used for statistical analysis. One-way ANOVA and Dunnettt-tests were used to compare the statistical significance among different groups. Quantitative data conform to normal distribution and are expressed as mean ± SD. According to the test level α = 0.05, p < 0.05 was considered statistically significant.3. Results and discussion
3.1 DNA scaffold design
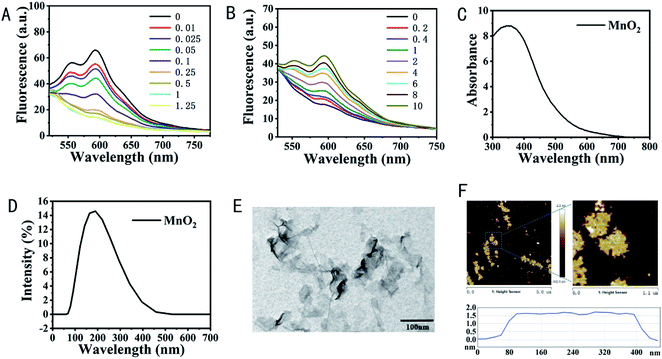
In this work, we aim to fabricate the tumor microenvironment dual-responsive nanoplatform for MRI and combination therapy of tumor based on DNA scaffold and MnO2 nanosheets. To realize this aim, a DNA sequence named Probe 1 containing ATP aptamer and AS411 aptamer sequence was first designed (Table S1†). ATP aptamer still maintained the second structure with the addition of AS1411 aptamer (Fig. S1A†). To load Dox, a partial complementarity cDNA sequence was introduced to hybridize with Probe 1. The formed duplex had a 27-base pair with GC-rich motif which could be used for anthracycline-contained Dox loading.36 To evaluate the amount of Dox molecules loaded into the Probe 1-cDNA duplex, the changes of Dox fluorescence were monitored before and after intercalation. As shown in Fig. 1A, when the concentration of Dox was fixed, the fluorescence intensity of Dox was decreased with an increasing molar ratio of the Probe 1-cDNA duplex, indicating more Dox molecules were loaded onto the duplex due to the initiation of forster resonance energy transfer (FRET) between Dox molecules and the DNA duplex.37 A maximum quenching efficacy was achieved when the molar ratio of duplex to Dox was 1. Furthermore, the existence of ATP could trigger the release of Dox and the release amount was increased with the ATP concentration raise (Fig. 1B), showing the possibility of the ATP triggered drug release. We also compared the Dox loading and ATP triggered drug release without AS1411 aptamer. As shown in Fig. S1B and C,† such binding and releasing property was not influenced by the existence of AS1411 aptamer, favoring the specific targeting and drug delivery of the nanoplatform to tumor.3.2 Characterization of MnO2 nanosheets and NH2-Probe 1-cDNA-Dox@MnO2
The MnO2 nanosheets were employed as nanocarrier and activatable MRI contrast agent as well as the chemodynamic therapy agent in this work. The MnO2 nanosheets have a wide UV-vis absorption peak at 300–800 nm, and the characteristic absorption center is located at 360 nm (Fig. 1C).32 Their hydrodynamic diameters were 161.2 nm (Fig. 1D). TEM images revealed that MnO2 nanosheets exhibited a typical two-dimensional morphology with occasional folds and wrinkles (Fig. 1E) and a brown, transparent liquid under the white light (Fig. S2A†) with a thickness of approximately 1.5 nm learnt from atomic force microscopy (AFM) analysis (Fig. 1F). MnO2 maintained excellent stability without obvious precipitation even at 4 °C for a month (Fig. S2B†).To testify the feasibility of MnO2 nanosheets as nanocarrier to load DNA scaffold, fluorophore fluorescein, FAM labeled Probes 1 (FAM-Probe 1) were incubated with MnO2 nanosheets. Considering the Mn–N coordinate bonding might further increase the binding of DNA scaffold on the surface of MnO2 nanosheets, FAM-NH2-Probe 1 were also introduced as comparision. Utilizing the fluorescence quenching ability of MnO2 nanosheets to FAM in a close range, the loading ability of MnO2 nanosheets to DNA scaffold was evaluated. As shown in Fig. S3A,† with the increase of the ratio between FAM-Probe 1 and MnO2 nanosheets, the signal-to-noise ratio (SNR) determined by the changes of Dox fluorescence became larger, indicating more FAM-Probe 1 were loaded on MnO2 nanosheets and when the concentration of FAM-Probe 1 was 1.67 μM, the most optimized loading ability was obtained. The same tendency was obtained in the case of FAM-NH2-Probe 1 (Fig. S3A†). But it should be noticed that the existence of amino group helped the loading of DNA scaffold on the MnO2 nanosheets. We then further investigated the influence of the presence of cDNA and Dox on the loading of fabricated DNA scaffold on MnO2 nanosheets. As shown in Fig. S3B,† the hybridization between FAM-NH2-Probe 1 and cDNA released only a small amount of FAM-NH2-Probe 1 from MnO2 nanosheets and most of the FAM-NH2-Probe 1-cDNA could adsorb on the surface of MnO2 nanosheets, which provided the good basis for their following applications. The fabrication of NH2-Probe 1-cDNA-Dox@MnO2 was also proved by the changes of zeta potential from −29.13 mV for MnO2 to −5.28 mV for NH2-Probe 1-cDNA-Dox and −30.43 mV for NH2-Probe 1-cDNA-Dox@MnO2 (Fig. S4A†). AFM analysis showed the thickness of MnO2 was changed from 1.62 nm to 2.37 nm in local area for NH2-Probe 1-cDNA-Dox@MnO2 (Fig. S4B†).
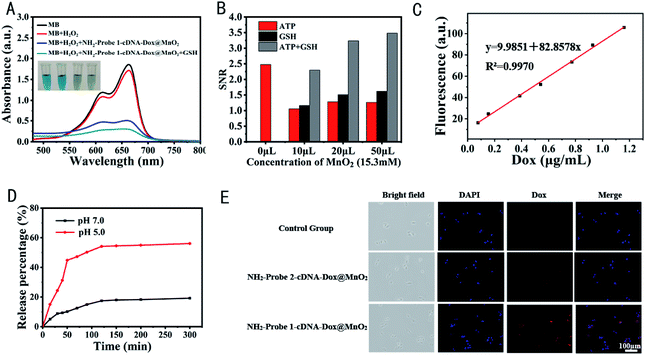
In addition, MnO2 nanosheets were proved to be used as nanoenzyme for chemodynamic therapy of tumor because Mn2+ can produce ·OH through a Fenton like reaction with the assistance of HCO3−, which raises oxygen levels and depletes the antioxidants.38,39 Thus, we evaluated the capacity of NH2-Probe 1-cDNA-Dox@MnO2 to produce ·OH in the presence of H2O2, HCO3− and GSH using methylene blue (MB) as an indicator. ·OH induced degradation of MB was monitored by the change of absorbance at 665 nm. As shown in Fig. 2A, the absorbance of MB was significantly decreased when NH2-Probe 1-cDNA-Dox@MnO2 were incubated with H2O2, showing the production of ·OH. Such absorbance of MB was further decreased with the addition of GSH, which might come from the more Mn2+ formation and more ·OH production, presenting the potential application of NH2-Probe 1-cDNA-Dox@MnO2 nanocomplex for the combined therapy of CT and CDT.
3.3 GSH and ATP dual-responsive Dox release from NH2-Probe 1-cDNA-Dox/MnO2
To evaluate the activatable Dox release and MRI signal, the corresponding roles of GSH and ATP were studied. It is reported that the GSH concentration inside the cells is about 1 × 10−3 to 10 × 10−3 M,40 as compared with 2 × 10−6 to 40 × 10−6 M in blood and other body fluids.41 The concentration of GSH in cancer cells is 100- to 1000-fold higher than that in the blood, and in a tumor mass. It is also markedly (100-fold) higher than the extracellular level, which has been designed for the delivery of drugs and as imaging agents.42 Firstly, to evaluate the ATP-responsive Dox release characteristics, NH2-Probe 1-cDNA-Dox was incubated at different concentrations of ATP (0, 0.2, 0.4, 1, 2, 4, 6, 8, 10 mM), respectively. Among them, 0.4 mM and 4 mM ATP represented the typical ATP level in the extracellular fluid and intracellular cytosol.43 The binding between ATP and its aptamer would change the structure of ATP aptamer and destroy the duplex, leading to the release of Dox from NH2-Probe 1-cDNA-Dox. Such changes could be monitored by the Dox fluorescence. As mentioned in Fig. 1B, a remarkable fluorescence recovery was observed with the increase of ATP concentration, indicating an ATP-triggered Dox release of our designed DNA scaffold. However, in the case of NH2-Probe 1-cDNA-Dox@MnO2, only the presence of ATP could only release a small amount of Dox, which might come from the interference of MnO2 nanosheets on the binding between ATP and its aptamer (Fig. 2B). Similarly, the existence of GSH could only destroy the structure of MnO2 and release NH2-Probe 1-cDNA-Dox, but no released Dox could be observed. Dox could only be released from NH2-Probe 1-cDNA-Dox@MnO2 with GSH and ATP co-existence (Fig. 2B).To further investigate the in vitro release profiles of NH2-Probe 1-cDNA-Dox@MnO2, the releasing buffer solutions containing 5 mM HEPES, 137 mM NaCl, 10 mM MgCl2, 10 mM ATP and 10 mM GSH (pH 7.0 or 5.0) were prepared. The Dox release was determined based on the linear fitting curves of Dox (Fig. 2C). As shown in Fig. 2D, with a releasing buffer of pH 7.0, only 10.2% of Dox was released from NH2-Probe 1-cDNA-Dox@MnO2 in the first 50 min and 19.25% was released within 300 min. When pH of the buffer was adjusted to 5.0, the release of Dox was dramatically accelerated. It was showed that about 44.83% of Dox was released from NH2-Probe 1-cDNA-Dox@MnO2 in the first 50 min and more than 56.1% was released within 300 min.
The intracellular delivery of NH2-Probe 1-cDNA-Dox@MnO2 in MDA-MB-231 cells was also explored. As shown in Fig. 2E, after incubation for two hours, the red fluorescence of Dox could be observed in the MDA-MB-231 cells, which well overlapped with the blue fluorescence of the nucleus, showing the accumulation of Dox into the nuclei. Additionally, the presence of AS1411 aptamer favored the Dox delivery to tumor cells benefitting from the nucleolin-mediated endocytosis with the help of AS1411 aptamer.
Taken together, our fabricated NH2-Probe 1-cDNA-Dox@MnO2 nanocomplex presented tumor microenvironment related multicomponent response, showing their promising potential for the specific MRI and combination therapy of CT and CDT of tumor.
3.4 Antitumor efficacy in vitro and in vivo
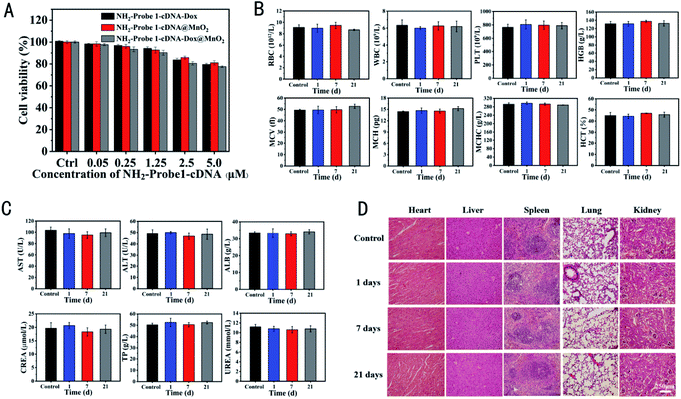
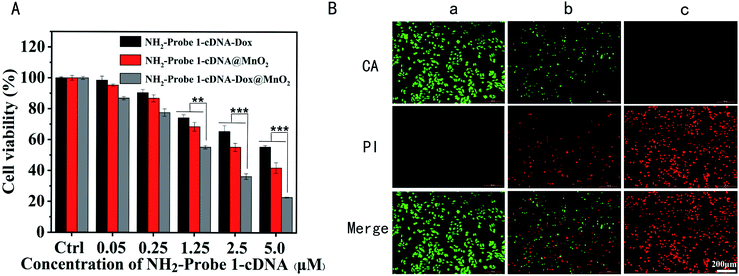
To explore their potential for the application of the tumor cell growth inhibition, MDA-MB-231 and NIH-3T3 cells were treated with different concentrations of NH2-Probe 1-cDNA-Dox, NH2-Probe 1-cDNA@MnO2 and NH2-Probe 1-cDNA-Dox@MnO2, respectively. The cell viabilities were determined by MTT assay. As expected, no obvious cytotoxicity was observed for the above complex to NIH-3T3 cells, showing their good biocompatibility to normal cells (Fig. 3A). Furthermore, the hematological and biochemical analyses as well as pathological section analysis also testified the excellent biocompatibility of our fabricated NH2-Probe 1-cDNA-Dox@MnO2. The results of blood routine (Fig. 3B) and blood biochemical (Fig. 3C) tests showed no obvious abnormality compared with the normal control group, showing no obvious acute toxicity of NH2-Probe 1-cDNA-Dox@MnO2 to liver and kidney. What's more, we compared the histological structures of the heart, liver, spleen, kidneys and lungs dissected from the mice without and post-injection of NH2-Probe 1-cDNA-Dox@MnO2 for 1, 7, and 21 day. As shown in Fig. 3D, the structures of the myocardium, hepatic lobules, splenic sinuses, glomeruli, and alveoli were clearly visible, suggesting no acute or chronic tissue damage after the NH2-Probe 1-cDNA-Dox@MnO2 injection.But for MDA-MB-231 breast cancer cells, all of NH2-Probe 1-cDNA-Dox, NH2-Probe 1-cDNA@MnO2 and NH2-Probe 1-cDNA-Dox@MnO2 presented a concentration-dependent cytotoxicity (Fig. 4A). Furthermore, the lowest cell viability in NH2-Probe 1-cDNA-Dox@MnO2 group indicated the excellent effect of the combination therapy of CT and CDT. In order to visualize living and dead cells, the cells after different treatments were co-stained with CA and PI solution. Living cells are dyed green and dead cells are dyed red, respectively. The methyl acetate of CA has high lipophilicity, which makes it permeable through the cell membrane. Although CA itself is not a fluorescent molecule, CA can remove the acetyl methoxymethyl ester (AM) group through the action of esterase in living cells, and the resulting calcein can emit strong green fluorescence. Therefore, living cells can be stained by CA with green emission. On the other hand, PI, as a nuclear dye, can not pass through the living cell membrane, but it can pass through the disordered region of the dead cell membrane and reach the nucleus, and embed the DNA double helix of the cell to produce red fluorescence.44 Learnt from Fig. 4B, green fluorescence emission and almost no red fluorescence emission showed the excellent growth situation in control group. But with the addition of NH2-Probe 1-cDNA@MnO2 or NH2-Probe 1-cDNA-Dox@MnO2, less green fluorescence emission and more red fluorescence emission were observed, showing their cytotoxicity to tumor cells. Furthermore, almost no green fluorescence emission could be found with the treatment of NH2-Probe 1-cDNA-Dox@MnO2, displaying their higher cytotoxic activity towards the tumor cells.
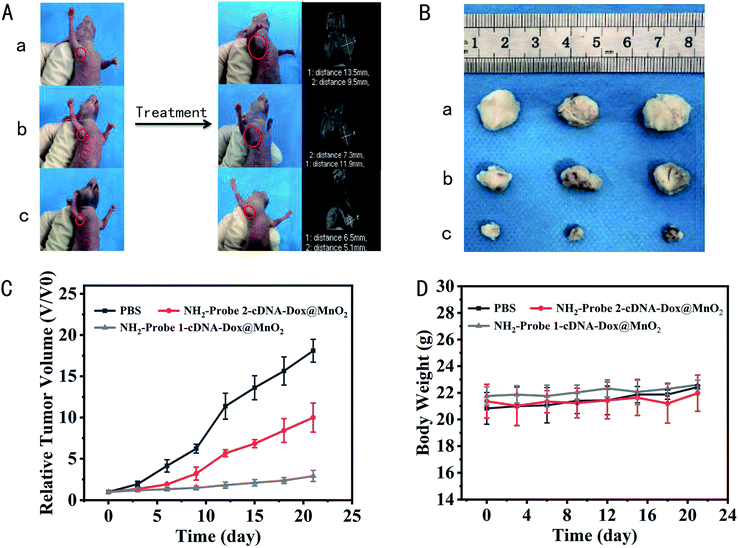
The excellent tumor cell inhibition of the fabricated nanocomplex in vitro promoted us to explore their antitumor efficacy in vivo. After treatment, tumor growth was almost completely suppressed in mice treated with NH2-Probe 1-cDNA-Dox@MnO2, and more significantly than the mice treated with PBS and NH2-Probe 2-cDNA-Dox@MnO2. MRI scans taken on the last day further confirmed these changes (Fig. 5A). The mice were killed at the 21st day and the tumor sizes in the different groups were compared. The tumors in the NH2-Probe 1-cDNA-Dox@MnO2 group showed almost no significant enlargement. In contrast, tumor growth in the NH2-Probe 2-cDNA-Dox@MnO2 group was relatively faster, suggesting that more chemotherapy drugs were concentrated in the tumor region due to antibody target recognition (Fig. 5B). In addition, tumors in mice treated with PBS and NH2-Probe 2-cDNA-Dox@MnO2 showed rapid growth during subsequent treatment (Fig. 5C). More importantly, there was no significant weight loss or animal death in all three groups during the entire monitoring period, indicating that the prepared nanomedicine showed excellent therapeutic efficacy without significant side effects on the tested animals (Fig. 5D).
3.5 In vitro and in vivo enhanced MR imaging
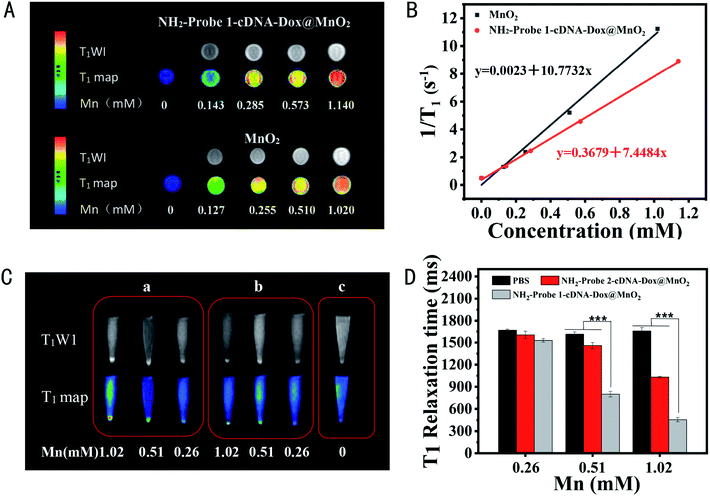
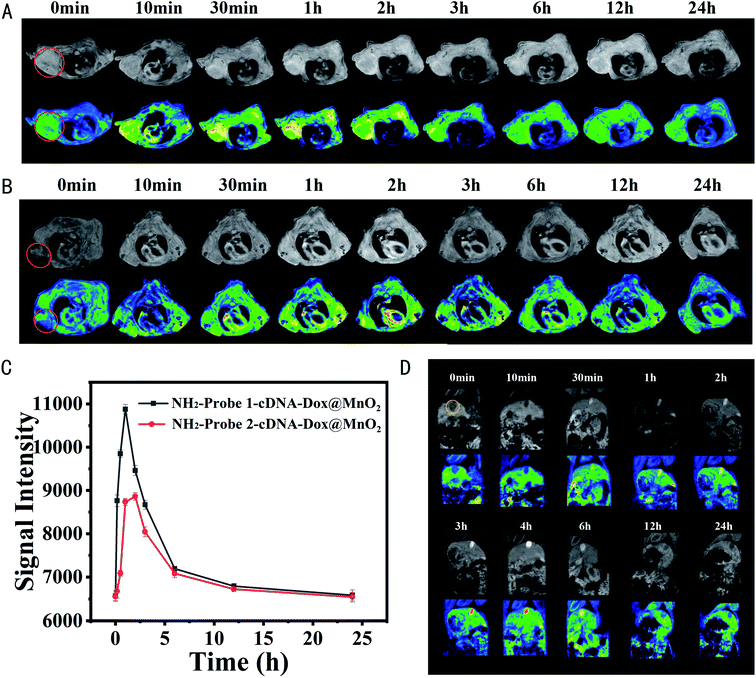
To explore the specific MRI ability of NH2-Probe 1-cDNA-Dox@MnO2 to tumor cells in vitro and in vivo, its r1 relaxivity in the presence of 10 mM GSH was first determined. As shown in Fig. 6A and B, the r1 MRI relaxivity values of MnO2 and NH2-Probe 1-cDNA-Dox@MnO2 were 10.77 mM−1 s−1 and 7.45 mM−1 s−1 Mn, respectively. The MRI signal intensity of NH2-Probe 1-cDNA-Dox@MnO2 within 30 day period presented unnoticeable changes, providing excellent basis for their use as MRI contrast agent (Fig. S5†). Then, their performance for the specific MRI of MDA-MB-231 cells was evaluated. It was showed that the designed NH2-Probe 1-cDNA-Dox@MnO2 nanocomplex could target MDA-MB-231 cells visualized by MR imaging with the help of AS1411 aptamer and GSH-activated MRI signal (Fig. 6C). At the same MnO2 concentration, the signal intensity of the targeted group (NH2-Probe 1-cDNA-Dox@MnO2) was significantly higher than that of the non-targeted group (NH2-Probe 2-cDNA-Dox@MnO2) and the control group (PBS), indicating the higher accumulation amount of nanocomplex on tumor cells with the help of AS1411 aptamer (Fig. 6D).Later, such specific MRI of tumor cells was expanded to breast carcinoma bearing BALB/c nude mice. NH2-Probe 1-cDNA-Dox@MnO2 or NH2-Probe 2-cDNA-Dox@MnO2 was injected into the nude mouse model via tail vein and the T1-weighted MRI scans were conducted at scheduled time points. As shown in Fig. 7A and B, the tumors both exhibited contrast-enhanced regions after post-injection of NH2-Probe 1-cDNA-Dox@MnO2 or NH2-Probe 2-cDNA-Dox@MnO2. The T1 MR signal in the tumor gradually brightens over the first 3 hours and then weakens over time. However, it should be noted that the T1 signal of NH2-Probe 1-cDNA-Dox@MnO2 was significantly brighter than that observed after injection of NH2-Probe 2-cDNA-Dox@MnO2 at the same time. Benefit from the targeting ability of the AS1411 aptamer (Fig. 7C). Meanwhile, we found a strong enhanced signal from the gallbladder, indicating the excretion of the reduced Mn2+ ions might mainly via hepatobiliary system (Fig. 7D), which was consistent with our previous report.32 The blood half-life time was determined to be 1.65 h (Fig. S6†).
4. Conclusions
Here, we propose a dual-sensitive nanoplatform for the controlled drug delivery and activatable MRI and CT & CDT combined therapy of tumor based on DNA aptamers and MnO2 nanosheets. The cleaver DNA design of the integration of targeting molecule and drug loading molecule simplified the preparation procedure. The MnO2 nanosheets as nanocarrier and AS1411 aptamer as targeting molecule favored the efficient drug delivery to breast cancer cells. Then, the Dox release triggered by the co-existence of ATP and GSH further increase the on-demand drug delivery accurately and therapeutic efficacy. In addition, Mn2+ mediated Fenton-like reactions for CDT, which synergistically enhanced the therapy effect. More importantly, MRI from Mn2+ realized the real-time diagnosis and monitoring of tumor changes after the drug intervention. This strategy might provide a new idea for the design of theranostic nanoplatform for the more accurate diagnosis and treatment of tumor. In the future study, we will try to evaluate the therapy effect of our fabricated nanocomposites on advanced tumor model, which is more meaningful for their later clinical transformation.Author contributions
Mingming Zhao: data curation, formal analysis, methodology, investigation, writing – original draft. Xiaoxi Song: investigation. Jiahui Lu: methodology, investigation. Siwen Liu: validation. Xuan Sha: methodology, investigation. Qi Wang: software, formal analysis. Xu Cao: scheme drawing. Kai Xu: funding acquisition, supervision. Jingjing Li: conceptualization, supervision, resources, funding acquisition, writing – review & editing. All the authors have approved the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81771904), the Natural Science Foundation of Jiangsu Province for the Excellent Young Scholars (BK20170054), Qing Lan Project, the Peak of Six Talents of Jiangsu Province (WSN-112), Jiangsu Provincial Medical Youth Talent (QNRC2016776), Six one project of Jiangsu Province (LGY2018083), Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX21_2737, KYCX21_2724).Notes and references
- R. Barman, A. Nain and S. Jain, J. Mater. Chem. B, 2018, 6, 2368–2384 RSC.
- R. Cheng, L. Liu, Y. Xiang, L. Deng, H. Zhang, H. A. Santos and W. Cui, Biomaterials, 2020, 232, 119706 CrossRef CAS PubMed.
- X. Nguyen, L. Huang, M. Gauthier, G. Yang and Q. Wang, Nanomedicine, 2016, 11, 1169–1185 CrossRef PubMed.
- E. Bagheri, L. Ansari, K. Abnous, S. M. Taghdisi, F. Charbgoo, M. Ramezani and M. Alibolandi, J. Controlled Release, 2018, 277, 57–76 CrossRef CAS PubMed.
- J. Amina and B. Guo, Int. J. Nanomed., 2020, 15, 9823–9857 CrossRef PubMed.
- J. Liu, J. Dong, T. Zhang and Q. Peng, J. Controlled Release, 2018, 286, 64–73 CrossRef CAS PubMed.
- S. Er, U. Laraib, R. Arshad, S. Sargazi, A. Rahdar, S. Pandey, V. Thakur and A. M. Díez-Pascual, Nanomaterials, 2021, 11(11), 3002–3036 CrossRef CAS PubMed.
- S. Timin, M. M. Litvak, D. A. Gorin, E. N. AtochinaVasserman, D. N. Atochin and G. B. Sukhorukov, Adv. Healthcare Mater., 2017, 1700818 Search PubMed.
- E. Levy-Nissenbaum, A. F. Radovic-Moreno, A. Z. Wang, R. Langer and O. C. Farokhzad, Trends Biotechnol., 2018, 26(8), 442–449 CrossRef PubMed.
- X. Liao, E. Y. Chuang, C. C. Lin, Y. C. Ho, K. Ju. Lin, P. Y. Cheng, K. J. Chen, H. J. Wei and H. W. Sung, J. Controlled Release, 2015, 208, 42–51 CrossRef PubMed.
- J. Bates, D. A. Laber, D. M. Miller, S. D. Thomas and J. O. Trent, Exp. Mol. Pathol., 2009, 86, 151–164 CrossRef PubMed.
- C. Mi, S. D. Thomas, X. H. Xu, L. K. Casson, D. M. Miller and P. J. Bates, J. Biol. Chem., 2003, 278, 8572–8579 CrossRef PubMed.
- L. Dai, D. Wei, J. Zhang, T. Shen, Y. Zhao, J. Liang, W. Ma, L. Zhang, Q. Liu and Y. Zheng, Cell Proliferation, 2021, e13130 CAS.
- Z. Chen, Y. Peng, Y. Li, X. Xie, X. Wei, G. Yang, H. Zhang, N. Li, T. Li, X. Qin, S. Li, C. Wu, F. You, H. Yang and Y. Liu, ACS Nano, 2021, 15, 16683–16696 CrossRef CAS PubMed.
- K. Han, W. Zhang, J. Zhang, Z. Ma and H. Han, Adv. Healthcare Mater., 2017, 6, 2192–2640 Search PubMed.
- Y. Li, J. Jeon and J. H. Park, Cancer Lett., 2020, 490, 31–43 CrossRef CAS PubMed.
- M. Shahriari, M. Zahiri, K. Abnous, S. Taghdisi, M. Ramezani and M. Alibolandi, J. Controlled Release, 2019, 308, 172–189 CrossRef CAS PubMed.
- M. Sponchioni, P. U. Capasso and D. Moscatelli, Mater. Sci. Eng., C, 2019, 102, 589–605 CrossRef CAS PubMed.
- X. Guo, Y. Cheng, X. Zhao, Y. Luo, J. Chen and W. Yuan, J. Nanobiotechnol., 2018, 16, 74 CrossRef PubMed.
- W. Q. Lim, S. Phua and Y. Zhao, ACS Appl. Mater. Interfaces, 2019, 11, 31638–31648 CrossRef CAS PubMed.
- J. Wang, Z. Wang, J. Yu, A. R. Kahkoska, T. B. Buse and Z. Gu, Adv. Mater., 2020, 32, e1902004 CrossRef PubMed.
- R. Volpatti, M. A. Matranga, A. B. Cortinas, D. Delcassian, K. B. Daniel, R. Langer and D. G. Andersonet, ACS Nano, 2020, 14, 488–497 CrossRef PubMed.
- W. Zhao, Y. Zhao, Q. Wang, T. Liu, J. Sun and R. Zhang, Small, 2019, 15, e1903060 CrossRef PubMed.
- J. Qu, Y. Liang, M. Shi, B. Guo, Y. Gao and Z. Yin, Int. J. Biol. Macromol., 2019, 140, 255–264 CrossRef CAS PubMed.
- W. Chen, C. Cheng and J. I. Zink, ACS Nano, 2019, 13, 1292–1308 CAS.
- D. Huang, M. Sun, Y. Bu, F. Luo, C. Lin, Z. Lin, Z. Weng, F. Yang and D. Wu, J. Mater. Chem. B, 2019, 7(14), 2330–2337 RSC.
- S. El-Sawy, A. M. Al-Abd, T. A. Ahmed, K. M. El-Sayet and V. P. Torchilin, ACS Nano, 2018, 12, 10636–10664 CrossRef PubMed.
- S. Li, Q. Jiang, S. Liu, Y. Zhang, Y. Tian, C. Song, J. Wang, Y. Zou, G.-J. Anderson, J. Han, Y. Chang, Y. Liu, C. Zhang, L. Chen, G. Zhou, G. Nie, H. Yan, B. Ding and Y. A. Zhao, Nat. Biotechnol., 2018, 36, 258–264 CrossRef CAS PubMed.
- R. Mo, T. Jiang, R. DiSanto, W. Tai and Z. Gu, Nat. Commun., 2014, 5, 3364 CrossRef PubMed.
- Q. Hu, H. Li, L. Wang, H. Gu and C. Fan, Chem. Rev., 2019, 119, 6459–6506 CrossRef CAS PubMed.
- X. Yang, Q. Tang, Y. Jiang, M. Zhang, M. Wang and L. Mao, J. Am. Chem. Soc., 2019, 141, 3782–3786 CrossRef CAS PubMed.
- M. Shi, S. Wang, S. Zheng, P. Hou, L. Dong, M. He, C. Wu, X. Zhang, F. Zuo, K. Xu and J. Li, Colloids Surf., B, 2020, 185, 110625 CrossRef CAS PubMed.
- S. Zheng, M. Zhang, H. Bai, M. He, L. Dong, L. Cai, M. Zhao, Q. Wang, K. Xu and J. Li, Int. J. Nanomed., 2019, 14, 9513–9524 CrossRef CAS PubMed.
- F.-R. Balkwill, M. Capasso and T. Hagemann, J. Cell Sci., 2012, 125, 5591–5596 CrossRef CAS PubMed.
- S. Zheng, N. Yu, C. Han, T. Xie, B. Dou, Y. Kong, F. Zuo and M. Shi, Biochem. Biophys. Res. Commun., 2019, 511, 207–213 CrossRef CAS PubMed.
- B. Chaires, J. E. Herrera and M. J. Waring, Biochemistry, 1990, 29, 6145–6153 CrossRef PubMed.
- Z. Xiao, C. Ji, J. Shi, E. M. Pridgen, J. Frieder, J. Wu and O. C. Farokhzad, Angew. Chem., Int. Ed. Engl., 2012, 51, 11853–11857 CrossRef CAS PubMed.
- Z. Qing, A. Bai, L. Chen, S. Xing, Z. Zou, Y. Lei, J. Li, J. Liu and R. Yang, CCS Chem., 2020, 2, 1217–1230 Search PubMed.
- L. Lin, J. Song, L. Song, K. Ke, Y. Liu, Z. Zhou, Z. Shen, J. Li, Z. Yang, W. Tang, G. Niu, H. Yang and X. Chen, Angew. Chem., Int. Ed. Engl., 2018, 57, 4902–4906 CrossRef CAS PubMed.
- C. Hwang, A. J. Sinskey and H. F. Lodish, Science, 1992, 257, 1496–1502 CrossRef CAS PubMed.
- R. Hong, G. Han, J. M. Fernández, B. J. Kim, N. S. Forbes and V. M. Rotello, J. Am. Chem. Soc., 2006, 128, 1078–1079 CrossRef CAS PubMed.
- P. Torchilin, Nat. Rev. Drug Discovery, 2014, 13, 813–827 CrossRef PubMed.
- W. Gorman, E. O. Feigl and C. W. Buffington, Clin. Chem., 2007, 53, 318–325 CrossRef PubMed.
- Y. Wang, J. Chen, X. Liang, H. Han, H. Wang, Y. Yang and Q. Li, Mol. Pharm., 2017, 14, 2323–2332 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra08373b |
| This journal is © The Royal Society of Chemistry 2022 |