Open Access Article
Open Access ArticlePreparation and characterization of stable core/shell Fe3O4@Au decorated with an amine group for immobilization of lipase by covalent attachment
Marziyeh Aghamolaei,
Amir Landarani-Isfahani,
Mehrnaz Bahadori ,
Zahra Zamani Nori,
Saghar Rezaei,
Majid Moghadam
,
Zahra Zamani Nori,
Saghar Rezaei,
Majid Moghadam *,
Shahram Tangestaninejad
*,
Shahram Tangestaninejad *,
Valiollah Mirkhani and
Iraj Mohammadpoor-Baltork
*,
Valiollah Mirkhani and
Iraj Mohammadpoor-Baltork
Catalysis Division, Department of Chemistry, University of Isfahan, Isfahan 81746-73441, Iran. E-mail: moghadamm@sci.ui.ac.ir; stanges@sci.ui.ac.ir
First published on 18th February 2022
Abstract
The self-assembly approach was used for amine decoration of core/shell Fe3O4@Au with 4-aminothiophenol. This structure was used for covalent immobilization of lipase using a Ugi 4-component reaction. The amine group on the structure and carboxylic group from lipase can react in the Ugi reaction and a firm and stable covalent bond is created between enzyme and support. The synthesized structure was fully characterized and its activity was explored in different situations. The results showed the pH and temperature stability of immobilized lipase compared to free lipase in a wide range of pH and temperature. Also after 60 days, it showed excellent activity while residual activity for the free enzyme was only 10%. The synthesized structure was conveniently separated using an external magnetic field and reused 6 times without losing the activity of the immobilized enzyme.
Introduction
Undoubtedly, one of the important branches in green and sustainable chemistry is using enzyme catalysis that effectively has a wide range of applications in food manufacturing, pharmaceuticals, and catalysis.1–3 As one of the most useful enzymes, lipase can be introduced because of its unique ability in the hydrolysis of carboxylic ester bonds in fatty acids with high enantio- and regio-selectivity4,5 and polymerization reactions.6 The lack of thermal stability and pH resistance restrict the performance of free lipase in industrial processes or some reaction media. Besides, difficulty in separation and loss of activity after recovery have remained as major limiting factors.7,8 To overcome these restrictions, researchers have presented immobilization of enzyme on supports through physical or chemical adsorption,9 encapsulation,10 diffusion,11 etc. Immobilization can improve the stability of the enzyme under hard reaction conditions and leads to easy recovery and reusability of the enzyme.12 The method of immobilization and the kind of support are key factors for achieving bio-structures that are stable under acidic and basic conditions, have a high surface area for raising enzyme loading, and minimize enzyme leaching.13,14 Due to high enzyme loading, high mechanical strength, and reduction of diffusion problems,15 various nanoparticles and nanoporous structures such as magnetic nanoparticles,16 nanoporous silicates,13,17 nanofibers,18 graphene oxide,19 and gold nanoparticles20 were used for effective immobilization of enzymes. Because of individual bio-compatible properties,15,21 gold nanoparticles have been considered as an excellent candidate for immobilizing the enzymes. Considering the high cost of Au, the design and synthesis of its composite with other useful structures such as cellulose nanocrystal/gold nanoparticle,20 gold nanoparticle/polyurethane,22 gold nanoparticle/amine-functionalized Na–Y zeolite23 and gold/magnetite nanoparticles24 have been implemented. It is worth mentioning that by using gold/magnetic nanoparticles as support, besides the low cost of composite, the difficulty in the separation of the nanoparticles from reaction media is solved. The immobilized substrates on the magnetic nanoparticle (Fe3O4) can be easily separated from solution in the magnetic field and this property makes them reusable. This unique character inspired researchers for using Fe3O4@Au (core/shell) nanoparticle as a support for immobilization of enzymes.Anchoring of enzyme on the surface of Au in advantageous way that does not reduce the activity, increase the stability of the enzyme and prevents its leaching, is next challenge and guides the scientists to use linkers which can connect enzyme and nanoparticles by strong covalent bond. Based on several evidences for strong interaction between Au and S,25,26 creation of covalent gold–sulfur and synthesis of amine-decorated gold nanoparticles can be introduced for modifying the Au surface.
In addition to stabilizing gold nanoparticles by the thiol groups in RSH, the R group can be designed containing different functional groups for special targets. Regard to the presence of carboxylic acid group in lipase as selected enzyme in this work, formation of N–C bond between amine and carboxylic acid though Ugi four components reaction was chosen as the best accessible way to anchoring the lipase on nanoparticle surface. So 4-aminothiophenol was considered as superb linker because of amine group for the connection to carboxylic acid in lipase and thiol group for the connection to Au in gold nanoparticle. The created covalent bond leads to the strong attachment of the enzyme to the support and decreases the leaching of the enzyme during the reactions. This excellent performance in reusability is the superiority of covalent bonds compared to electrostatic bands. In this work, immobilization of the enzyme using covalent bonds will carry out on the magnetic nanoparticle coated by gold. Less leaching of the enzyme through firm covalent bond and easy separation via magnetic nanoparticle core are the main advantages of this structure.
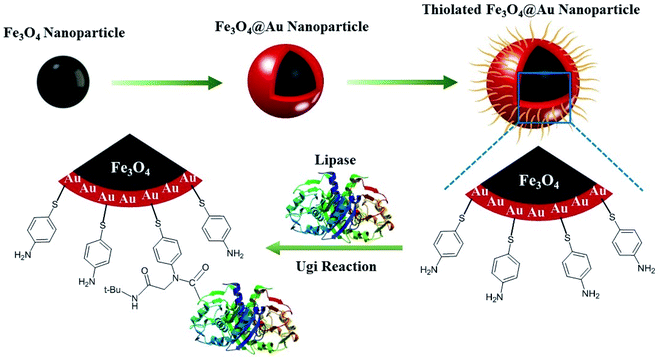
Subsequently, the synthesis of Fe3O4@Au and its decoration with 4-aminothiophenol as support for lipase was defined as a goal in this project (Scheme 1). The obtained bio-structure through immobilization of lipase on amine-decorated Fe3O4@Au nanoparticles exhibits easy separation, bio-compatibility, the lack of leaching, and stabilization of enzyme in reaction media.
Results
Characterizations
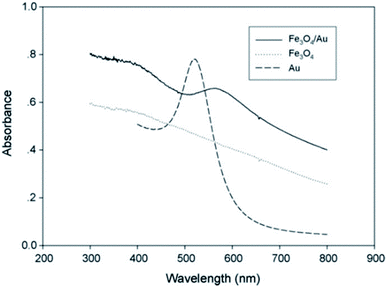
Fe3O4 nanoparticles were synthesized and coated by an Au layer. Based on Au–S interactions, 4-aminothiophenol was embedded on the nanoparticles, and by Ugi's four-component reaction, the lipase enzyme was immobilized on these gold-coated magnetic nanoparticles. The UV-Vis absorption spectra of the as-synthesized Fe3O4, Au, and core/shell Fe3O4@Au nanoparticles showed different adsorption properties as illustrated in Fig. 1. A decrease in UV absorption was observed in the range of 300–800 nm for Fe3O4 NPs which was in agreement with previous literature.27The UV-Vis absorption spectrum of Au nanoparticles displayed an absorption band at 520 nm which was shifted to 570 nm at core/shell Fe3O4@Au NP. This red-shift is because of the surface plasmon resonance property of the Au shell which has been reported by previous researches.28,29
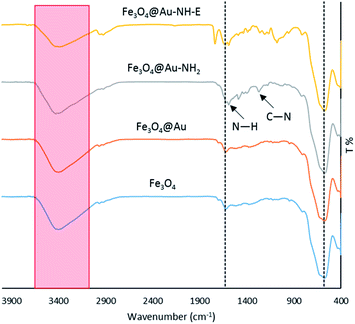
The FT-IR spectra of Fe3O4, Fe3O4@Au, Fe3O4@Au-NH2, and Fe3O4@Au-NH-E were recorded to confirm the successful synthesis of each compound (Fig. 2). The characteristic bands for Fe3O4 NPs in 580, 1830, and around 3400 cm−1 are attributed to Fe–O, H–O–H bending and O–H stretching vibrations, respectively.30 These bands exist in Fe3O4@Au, Fe3O4@Au-NH2, Fe3O4@Au-NH-E spectrum that indicates the stability of nano-magnetic particles during the syntheses steps. After functionalization of Fe3O4@Au with 4-aminothiophenol, two bands at 1262 and 1589 cm−1 corresponds to N–H bending and C–N stretching, respectively, were observed in the FT-IR spectrum of Fe3O4@Au-NH2. The appearance of the band at 1736 cm−1 after Ugi reaction in the FT-IR spectrum of Fe3O4@Au-NH-E is attributed to the carbonyl group of the lipase enzyme.
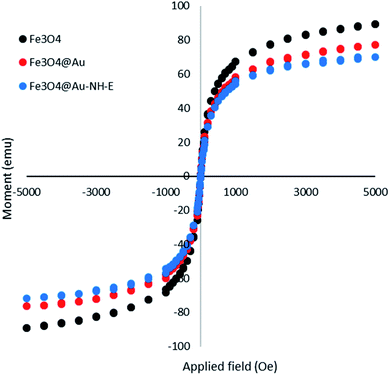
The magnetic property of the biocatalyst was investigated using a vibrating sample magnetometer (VSM) (Fig. 3). A slight decrease in magnetic property was observed in Fe3O4@Au and Fe3O4@Au-NH-E in comparison with Fe3O4 that is due to the diamagnetic layer of Au and lipase on the nano-magnetic nanoparticles. However, it should be noted that the magnetic property in biocatalyst was completely enough for the separation of biocatalyst from reaction media.
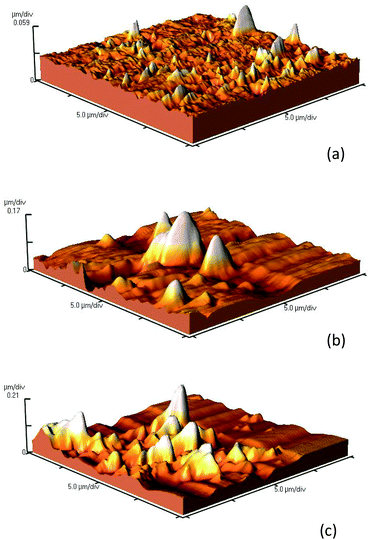
The change of surface morphology upon immobilizing of lipase was surveyed using atomic force microscopy (AFM) (Fig. 4, Table 1). The results in Table 1 and the images of the surface (Fig. 4) verified the higher roughness in Fe3O4@Au-NH-E compared to Fe3O4@Au and Fe3O4@Au-NH2 which confirmed the successful immobilization of enzyme on the structure. These results are in accordance with the previously reported results.31
| Entry | pH stability, activity | Temperature stability, activity | Storage stability, activity | Recyclability, activity | Ref. |
|---|---|---|---|---|---|
| 1 | pH = 4, 10% | T = 75 °C, 40% | 7 cycles, 20% | 32 | |
| pH = 10, 10% | |||||
| 2 | pH = 5, 10% | — | — | 7 cycles, 70% | 33 |
| pH = 11, 5% | |||||
| 3 | — | — | 75 days, 90% | 10 cycles, 70% | 34 |
| 4 | pH = 5, 70% | T = 80 °C, 30% | 30 days, 90% | 35 | |
| pH = 9, 60% | |||||
| 5 | pH = 4, 20% | T = 90 °C, 85% | 60 days, 80% | 5 cycles, >90% | This work |
| pH = 9, 85% |
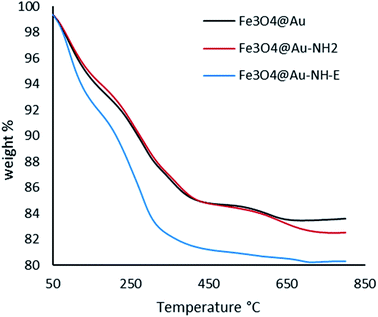
The thermograms of Fe3O4@Au, Fe3O4@Au-NH2, and Fe3O4@Au-NH-E are presented in Fig. 5. As can be seen, up to 330 °C, the solvent and unreacted organic compounds adsorbed on the surface have been removed. In Fe3O4@Au-NH-E, weight loss attributed to the decomposition of loaded enzyme. The residual part in the curves illustrates that for Fe3O4@Au we have the maximum amount and for Fe3O4@Au-NH-E. There isn't any specific weight loss after 450 °C.
Also, the amount of Au loaded on the surface of Fe3O4 was determined by ICP analysis and showed a value of about 0.096 mmolAu g−1 for Fe3O4@Au.
TEM images and particle size distribution curve for Fe3O4@Au and Fe3O4@Au-NH-E are presented in Fig. 6. The images don't display remarkable change in the morphology and sphere structure of the particles indicating the stability of the particles and the lack of agglomeration during the reaction. The size of the particles showed a slight increase after functionalization of the magnetic nanoparticle with lipase which related to functional groups and the presence of lipase on the structure.
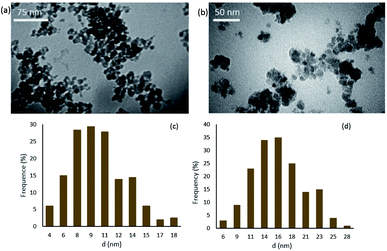 | ||
| Fig. 6 TEM images and particle size distribution curve for (a and c) Fe3O4@Au and (b and d) Fe3O4@Au-NH-E and. | ||
Optimization of lipase immobilization through Ugi reaction
The Ugi reaction is a four-component reaction including amine group (from Fe3O4@Au-NH2), carboxylic acid group (from lipase), formaldehyde, and tert-butyl isocyanide. The amount of every component can affect the amount of enzyme loading. To obtain the maximum loading, different amounts o0066 each component were applied in the Ugi reaction (Table 2), and the amount of unloaded enzyme was measured by Bradford assay in each reaction. The loaded amount was calculated by subtracting the amount of unloaded enzyme from the initial enzyme value. Based on the obtained results, using 15 mg formaldehyde, 25 μL tert-butyl isocyanide, and 1 mL lipase (5 mg mL−1) led to 82.3% enzyme loading on 10 mg of Fe3O4@Au-NH2 and which were chosen as optimized conditions for the synthesis of Fe3O4@Au-NH-E.| Vmax (mM min−1) | Km (mM) | |
|---|---|---|
| Free lipase | 3.15 | 3.01 |
| Immobilized lipase | 3.04 | 3.82 |
Stability of Fe3O4@Au-NH-E in different conditions
By considering the deactivation of the enzyme under different conditions, the effect of several factors including temperature, pH, and storage time on the stability of the enzyme was explored (Fig. 7). The results of the temperature stability in the range of 40–90 °C are presented in Fig. 7a and the best activity was observed for Fe3O4@Au-NH-E at 60 °C. In the whole investigated range, the immobilized lipase showed a better performance ratio than free enzyme which refers to the lack of movement of lipase and improvement of stability. The covalent bonds between support and lipase protect it from denaturation in high temperatures. Furthermore, pH checking at the range of 4–9 demonstrated that in the range of 4–6.5 the activity for Fe3O4@Au-NH-E is higher than that of the free enzyme (Fig. 7b). The better performance of the immobilized enzyme in acidic pH makes it so suitable for the reactions carried out in acidic conditions. The storage stability of immobilized enzyme was also investigated (Fig. 7c). After 30 days, a significant loss in activity was not observed and after 60 days, the immobilized enzyme lost just 20% of its initial activity, while the free enzyme lost 90% of its activity, indicating that the constructed covalent bonds created a stable environment for lipase. Besides these excellent stabilities in temperature, pH, and storage, the important factor in enzyme immobilization is the reusability of the obtained structure. In this view, considering the presence of magnetic nanoparticles in the core of the structure, Fe3O4@Au-NH-E was easily and efficiently separated by an external magnet and reused 6 times. As it is shown in Fig. 7d, even after the fifth cycle, the lipase activity in the sixth cycle is higher than 95% indicating the lack of leaching for lipase based on firm covalent bonds.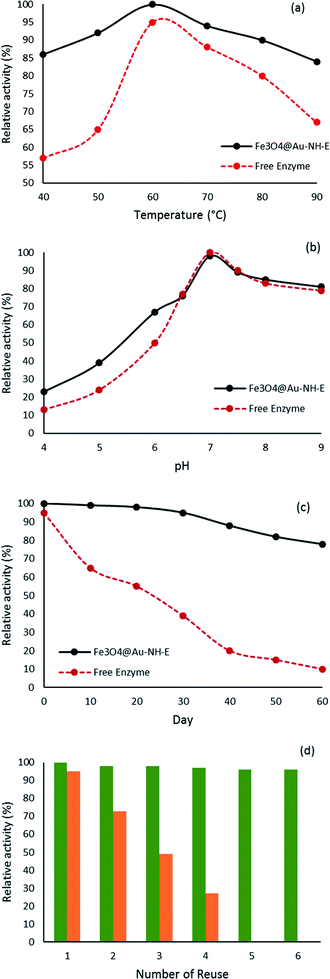 | ||
| Fig. 7 Thermal stability (a), pH stability (b), storage ability (c) of free enzyme (red) and Fe3O4@Au-NH-E (black), and number of reusability Fe3O4@Au-NH-E (green) and Fe3O4@Au-NH-E (orange) (d). | ||
Clearly, less sensitivity and more stability for Fe3O4@Au-NH-E compared to the free enzyme in the wide range of pH, temperature, and storage has been observed, which can be explained by strong covalent bonds and a stable environment for lipase on the support. Putting these individual performances in harsh situations together with so effortless separation in the magnetic field without loose of activity provided a structure which can be so useful in industry part.
To survey the role of covalent bond in Fe3O4@Au-NH-E, a different biostructure using electrostatic interactions between enzyme and support was synthesized (Fe3O4@Au-E) and its activity in the same conditions was tested. Based on the results, Fe3O4@Au-E showed good activity ratio than Fe3O4@Au-NH-E but it didn't display good performance in the next cycles (Fig. 7d). Testing of buffer solution after separation of Fe3O4@Au-E using Bradford assay revealed the presence of the loaded enzyme in the solution. It seems the release of the enzyme from the support in Fe3O4@Au-E through electrostatic interaction is the reason for losing activity.
Comparison between this work and previous work which used the covalent bond between support and enzyme was presented in Table 1. The results confirmed the good performance of synthesized bio-composite compared to similar systems in pH, temperature, storage stability, and recyclability.
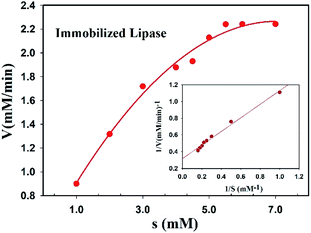
The Michaelis constant (Km) and the maximum reaction velocity (Vmax) of free and immobilized lipase were measured and compared together. The Michaelis–Menten and Lineweaver–Burk plots were displayed in Fig. 8 and the values of Km and Vmax were summarized in Table 2. According to the obtained results, immobilized lipase showed a little increase and decrease values of Km and Vmax, respectively. Generally, this phenomenon is observed due to strong enzyme immobilization to support which lead to cover some active sites and diffusional limitation of the substrates to them.36
Experimental
Methods
The UV-Vis diffuse reflectance spectra of the samples were recorded by a JASCO V-670 spectrophotometer. Transmission electron microscopy (TEM) was carried out on a Philips CM10 analyzer operating at 100 kV. Fourier transform infrared (FT-IR) spectroscopy was recorded in the range 4000–400 cm−1 on a JASCO FT-IR 6300 spectrometer. Vibrating sample magnetometer (VSM) measurement was measured using a SQUID magnetometer at 298 K (Quantum Design MPMS XL) in the magnetic field intensity (H) range of −8500 to +8500 Oe.Synthesis of Fe3O4 nanoparticles
Fe3O4 nanoparticles were synthesized through the chemical co-precipitation method.37,38 FeCl2·4H2O (2 g, 0.01 mol) and FeCl3·6H2O (5.4 g, 0.02 mol) were completely dissolved in HCl 10 mM (25 mL) which was deoxygenated with argon beforehand. The mixture was added to deoxygenated NH3 1.5 M (250 mL) and kept at 80 °C for 30 min under vigorous mechanical stirring and argon gas protection continuously. Completely the reactions were carried out under an ultrasonic bath. The obtained black Fe3O4 NPs were easily separated from reaction media by a magnetic field and washed with double distilled water.Synthesis of core/shell Fe3O4@Au nanoparticles
The obtained Fe3O4 NPs were dispersed using an ultrasonic bath in tetramethylammonium hydroxide (TMAOH) 0.1 M (250 mL) to prepare a stock solution. A 7.5 mL portion of the stock solution was added to 140 mL deionized water and after the addition of sodium citrate 0.1 M (7.5 mL), the mixture was stirred for 15 min. Then, NH2OH·HCl 0.2 M (750 μL) and HAuCl4 1% (v/w) (625 μL) were added to the mixture and the mixture was stirred for 3 h. Changing the color of the mixture from black to red confirmed the covering of Fe3O4 NPs by Au nanoparticles. The synthesized core/shell Fe3O4@Au nanoparticles were separated using a magnetic field and rinsed with double distilled water three times.39Functionalization of core/shell Fe3O4@Au nanoparticles with 4-aminothiophenol (Fe3O4@Au-NH2)
Considering the self-assembly of the Au and thiol group, 4-aminothiophenol (1.84 g) was dissolved in deoxygenated ethanol (1 mL) and then, the core/shell Fe3O4@Au (1.32 g) nanoparticles dispersed in H2O was added to this solution. The mixture was stirred for 30 min under argon gas and kept stable for 24 h at room temperature. The modified nanoparticles were separated by a magnetic field, washed with ethanol three times, and dried in a vacuum oven at 100 °C for 24 h.Preparation of bio-catalyst (Fe3O4@Au-NH-E)
Immobilization of lipase on functionalized Fe3O4@Au-NH2 nanoparticles was carried out using Ugi's four components reaction as a perfect way. The Fe3O4@Au-NH2 nanoparticles (300 mg) and formaldehyde (45 μL) were mixed in a 15 mL phosphate buffer and stirred for 30 min. After adding the lipase (15 mg mL−1) and stirring for 30 min, tert-butyl isocyanide (75 μL) was added and the mixture was stirred for 24 h at room temperature. The obtained solid was separated with a magnet, washed with buffer several times, and dried in a vacuum oven at 25 °C for 24 h.Preparation of Fe3O4@Au-E
300 mg Fe3O4@Au nanoparticle was reacted with lipase (15 mg mL−1) in 15 mL phosphate buffer and stirred for 24 h at room temperature. The resulting solid was washed with phosphate buffer and dried in vacuum conditions for 24 h.Lipase activity assay
The hydrolysis of ester catalyzed by free lipase and immobilized one was selected to evaluate the activity of the enzyme.40 Two stock solutions were prepared: (a) 90 mg p-nitrophenyl palmitate (p-NPP) in 30 mL 2-propanol was dissolved thoroughly using an ultrasonic bath, (b) 0.4 mL Triton X-100 and 0.1 g Arabic gum were added to 90 mL HCl buffer (50 mM, pH = 8). 5 mL solution a, 45 mL solution (b) and 1 mL enzyme solution (0.1 mg mL−1) were mixed at 37 °C. the solution was monitored every 5 min at 405 nm. For bio-composite, the loaded enzyme was determined using the Bradford method and an appropriate amount of composite was used for investigating enzyme activity.Stability investigation
For pH stability, every sample has been kept in HCl buffer with different pH for 1 hour before activity assay. Temperature stability was carried out by keeping the samples in different temperatures for 1 hour before the activity assay.Conclusions
In summary, a new magnetic nanoparticle-based support was designed and decorated by an amine group for the creation of covalent bonds between lipase and support using a Ugi reaction. The magnetic nanoparticles were covered by Au and based on a firm Au–S bond, 4-aminothiophenol embedded on the surface and Au shell mobilized by an amine group. Considering the carboxylic acid in the lipase structure, the Ugi-4 component was used for grafting the enzyme on the amine group from the Core/Shell Fe3O4@Au nanoparticles. The immobilized lipase presented high activity ratio than free enzyme in a wide range of temperature and pH and was completely stable after 60 days. The separation of this bio-structure was carried out conveniently in the external magnetic field without loss of activity for 6 cycles. The mentioned advantages and stability of this structure make it proper for use in the industry.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thankfully acknowledge the financial support of the Iran National Science Foundation (INSF) [research project 94001249].Notes and references
- O. L. Tavano, A. Berenguer-Murcia, F. Secundo and R. Fernandez-Lafuente, Compr. Rev. Food Sci. Food Saf., 2018, 17, 412–436 CrossRef PubMed.
- R. Das, M. Talat, O. Srivastava and A. M. Kayastha, Food Chem., 2018, 245, 488–499 CrossRef CAS PubMed.
- R. A. Sheldon and J. M. Woodley, Chem. Rev., 2018, 118, 801–838 CrossRef CAS PubMed.
- A. Rao, A. Bankar, A. Shinde, A. R. Kumar, S. Gosavi and S. Zinjarde, ACS Appl. Mater. Interfaces, 2012, 4, 871–877 CrossRef CAS PubMed.
- D. A. Sánchez, G. M. Tonetto and M. L. Ferreira, Biotechnol. Bioeng., 2018, 115, 6–24 CrossRef PubMed.
- P. Ye, Z.-K. Xu, J. Wu, C. Innocent and P. Seta, Macromolecules, 2006, 39, 1041–1045 CrossRef CAS.
- S. K. Patel, S. H. Choi, Y. C. Kang and J.-K. Lee, Nanoscale, 2016, 8, 6728–6738 RSC.
- S. K. Patel, S. H. Choi, Y. C. Kang and J.-K. Lee, ACS Appl. Mater. Interfaces, 2017, 9, 2213–2222 CrossRef CAS PubMed.
- R. C. Rodrigues, C. Ortiz, Á. Berenguer-Murcia, R. Torres and R. Fernández-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- S. Rafiei, S. Tangestaninejad, P. Horcajada, M. Moghadam, V. Mirkhani, I. Mohammadpoor-Baltork, R. Kardanpour and F. Zadehahmadi, Chem. Eng. J., 2018, 334, 1233–1241 CrossRef CAS.
- H. Gumz, S. Boye, B. Iyisan, V. Krönert, P. Formanek, B. Voit, A. Lederer and D. Appelhans, Adv. Sci., 2019, 6, 1801299 CrossRef PubMed.
- N. M. Thomson, S. Sangiambut, K. Ushimaru, E. Sivaniah and T. Tsuge, ACS Biomater. Sci. Eng., 2017, 3, 3076–3082 CrossRef CAS PubMed.
- S. Mei, J. Shi, S. Zhang, Y. Wang, Y. Wu, Z. Jiang and H. Wu, ACS Appl. Bio Mater., 2019, 2, 777–786 CrossRef CAS PubMed.
- J. C. S. d. Santos, O. Barbosa, C. Ortiz, A. Berenguer-Murcia, R. C. Rodrigues and R. Fernandez-Lafuente, ChemCatChem, 2015, 7, 2413–2432 CrossRef.
- E. P. Cipolatti, M. J. A. Silva, M. Klein, V. Feddern, M. M. C. Feltes, J. V. Oliveira, J. L. Ninow and D. de Oliveira, J. Mol. Catal. B: Enzym., 2014, 99, 56–67 CrossRef CAS.
- S. Rezaei, A. Landarani-Isfahani, M. Moghadam, S. Tangestaninejad, V. Mirkhani and I. Mohammadpoor-Baltork, ACS Appl. Bio Mater., 2020, 3, 8414–8426 CrossRef CAS PubMed.
- K. Kumar and P. Paik, ACS Biomater. Sci. Eng., 2021, 7, 4847–4858 CrossRef CAS PubMed.
- C. Tang, C. D. Saquing, S. W. Morton, B. N. Glatz, R. M. Kelly and S. A. Khan, ACS Appl. Mater. Interfaces, 2014, 6, 11899–11906 CrossRef CAS PubMed.
- J. Zhang, F. Zhang, H. Yang, X. Huang, H. Liu, J. Zhang and S. Guo, Langmuir, 2010, 26, 6083–6085 CrossRef CAS PubMed.
- K. A. Mahmoud, K. B. Male, S. Hrapovic and J. H. Luong, ACS Appl. Mater. Interfaces, 2009, 1, 1383–1386 CrossRef CAS PubMed.
- Y. Lv, Z. Lin, T. Tan and F. Svec, Biotechnol. Bioeng., 2014, 111, 50–58 CrossRef CAS PubMed.
- S. Phadtare, A. Kumar, V. Vinod, C. Dash, D. V. Palaskar, M. Rao, P. G. Shukla, S. Sivaram and M. Sastry, Chem. Mater., 2003, 15, 1944–1949 CrossRef CAS.
- K. Mukhopadhyay, S. Phadtare, V. Vinod, A. Kumar, M. Rao, R. V. Chaudhari and M. Sastry, Langmuir, 2003, 19, 3858–3863 CrossRef CAS.
- K. A. Mahmoud, E. Lam, S. Hrapovic and J. H. Luong, ACS Appl. Mater. Interfaces, 2013, 5, 4978–4985 CrossRef CAS PubMed.
- M. Tambasco, S. K. Kumar and I. Szleifer, Langmuir, 2008, 24, 8448–8451 CrossRef CAS PubMed.
- J. E. Matthiesen, D. Jose, C. M. Sorensen and K. J. Klabunde, J. Am. Chem. Soc., 2012, 134, 9376–9379 CrossRef CAS PubMed.
- D. Tang, R. Yuan and Y. Chai, J. Phys. Chem. B, 2006, 110, 11640–11646 CrossRef CAS PubMed.
- J. L. Lyon, D. A. Fleming, M. B. Stone, P. Schiffer and M. E. Williams, Nano Lett., 2004, 4, 719–723 CrossRef CAS.
- L. Wang, L. Wang, J. Luo, Q. Fan, M. Suzuki, I. S. Suzuki, M. H. Engelhard, Y. Lin, N. Kim and J. Q. Wang, J. Phys. Chem. B, 2005, 109, 21593–21601 CrossRef CAS PubMed.
- M. Baghayeri, R. Ansari, M. Nodehi, I. Razavipanah and H. Veisi, Microchim. Acta, 2018, 185, 320 CrossRef PubMed.
- S. Hirsh, M. Bilek, N. Nosworthy, A. Kondyurin, C. Dos Remedios and D. McKenzie, Langmuir, 2010, 26, 14380–14388 CrossRef CAS PubMed.
- X. Liu, Y. Fang, X. Yang, Y. Li and C. Wang, Chem. Eng. J., 2018, 336, 456–464 CrossRef CAS.
- N. S. Rios, D. M. A. Neto, J. C. S. Dos Santos, P. B. A. Fechine, R. Fernández-Lafuente and L. R. B. Gonçalves, Int. J. Biol. Macromol., 2019, 134, 936–945 CrossRef CAS PubMed.
- R. M. Bezerra, R. R. Monteiro, D. M. A. Neto, F. F. da Silva, R. C. de Paula, T. L. de Lemos, P. B. Fechine, M. A. Correa, F. Bohn and L. R. Gonçalves, Enzyme Microb. Technol., 2020, 138, 109560 CrossRef CAS PubMed.
- H. Aghaei, A. Yasinian and A. Taghizadeh, Int. J. Biol. Macromol., 2021, 178, 569–579 CrossRef CAS PubMed.
- T. Du, B. Liu, X. Hou, B. Zhang and C. Du, Appl. Surf. Sci., 2009, 255, 7937–7941 CrossRef CAS.
- X. Zhao, Y. Shi, T. Wang, Y. Cai and G. Jiang, J. Chromatogr. A, 2008, 1188, 140–147 CrossRef CAS PubMed.
- X. Zhao, Y. Shi, Y. Cai and S. Mou, Environ. Sci. Technol., 2008, 42, 1201–1206 CrossRef CAS PubMed.
- X. Zhao, Y. Cai, T. Wang, Y. Shi and G. Jiang, Anal. Chem., 2008, 80, 9091–9096 CrossRef CAS PubMed.
- U. K. Winkler and M. Stuckmann, J. Bacteriol., 1979, 138, 663–670 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |