Open Access Article
Open Access ArticleNanozymes with reductase-like activities: antioxidant properties and electrochemical behavior
Nataliya Stasyuk *ab,
Galina Gayda
*ab,
Galina Gayda a,
Taras Kavetskyybc and
Mykhailo Goncharab
a,
Taras Kavetskyybc and
Mykhailo Goncharab
aInstitute of Cell Biology, National Academy of Sciences of Ukraine, 79005 Lviv, Ukraine. E-mail: stasuk_natalia@ukr.net; galina.gayda@gmail.com; gonchar@cellbiol.lviv.ua
bDrohobych Ivan Franko State Pedagogical University, 82100 Drohobych, Ukraine. E-mail: kavetskyy@yahoo.com
cThe John Paul II Catholic University of Lublin, 20-950 Lublin, Poland
First published on 12th January 2022
Abstract
Nanozymes (NZs) as stable cost-effective mimics of natural enzymes may be promising catalysts in food and environmental biotechnology, biosensors, alternative energy and medicine. The majority of known NZs are mimetics of oxidoreductases, although there are only limited data regarding mimetics of reductases. In the present research, a number of metal-based NZs were synthesized via chemical methods and screened for their antioxidant ability in solution. The most effective reductase-like Zn/Cd/Cu NZ was characterized in detail. Its antioxidant properties in comparison with several food products and Trolox, as well as substrate specificity, size and composition were studied. Zn/Cd/Cu NZ was shown to mimic preferentially selenite reductase. The amperometric sensor was constructed possessing a high sensitivity (1700 A M−1 m−2) and a broad linear range (16–1000 μM) for selenite ions. The possibility to apply the fabricated sensor for selenite determination in commercial mineral water has been demonstrated.
Introduction
In recent years, nanomaterials with enzyme-mimetic properties (nanozymes) have attracted considerable attention.1–3 Nanozymes (NZs) as artificial enzymes (AEs) are promising alternatives to the natural ones.1–5 AEs have essential advantages over natural enzymes such as low preparation costs, stability, high surface area, self-assembling capability, size and composition-dependent activities, broad possibility for modification, and biocompatibility. They have wide potential practical applications as catalysts in biosensors, fuel-cell technology, environmental biotechnology, and medicine.5–12 It has been shown that AEs mimic the activity of peroxidases,5,13–15 oxidases,5,16 superoxide dismutases,5,17 and hydrolases,5,18 although only limited data were reported regarding reductase mimetics.19–25Natural reductases are the enzymes which belong to the E.C. 1 class of oxidoreductases and catalyze reduction reactions. Reductases are dehydrogenases which transfer hydrogen from the substrate to biological acceptors and to dyes.26,27 Reductases lower the activation energy needed for redox reactions to occur. The substrates of microbial reductases are inorganic compounds, including metal ions, metals, and non-metal and metalloid oxianions, as well as organic compounds, including aldehydes, glutathione, nucleotides, proteins, etc.26–42
Diverse enzymes of various microorganisms take part in the reduction processes, that possess abilities of the metalloreductase,28,29 carboxylic acid reductase,30,31 selenate32 and selenite reductases,33–36 hydrogenase,37 cromate reductase,38,39 nitrite40 and nitrate reductases41 etc. However, the problem of identification and classification for many reductases was not solved up to day. For example, so far no gene product or enzyme solely responsible for selenite reduction in aerobic bacteria has been identified in vivo.42
All known NZs usually utilize oxygen or hydrogen peroxide to produce active oxygen forms to oxidize substrate. There exist limited data on the nanomaterials simulating reductase, since oxygen, rather than other biomolecules, is preferable to be reduced on the surface of nanoparticles.19,22,43 Therefore, it is a great challenge to develop novel mimetics of reductase to simulate its natural counterparts with diverse functions.
Reduction of different substrates using reductase-like (R-like) NZs as catalysts have so far been reported.19–25 Cr(VI) may be reduced to Cr(III) via catalysis by magnetite-based nanocomposites25 or by cobalt oxide containing nanocomposite,24 peroxynitrite – via grapheme-hemin hybrid nanosheets,44 and Cyt c – via zeolitic imidazolate frameworks encapsulated with polyethylenimine.19 A number of NZs were reported to catalyze the sodium borohydride-induced 4-nitrophenol reduction to 4-aminophenol, namely silver and gold nanoparticles,23 platinum nanoparticles,45 reduced graphene oxide–cobalt oxide-based nanocomposite,24 iron oxide-loaded alginate-bentonite hydrogel beads20 and other materials.21,22
In this paper, we demonstrate that hybride Zn/Cd/Cu (core/shell) NZ obtained via the chemical bath deposition method (further – Zn/Cd/Cubd) possesses the ability to mimic reductase, in particular, coenzyme-dependent selenite reductase. The synthesized selenite R-like NZ has been used for construction of a highly sensitive amperometric biosensor for selenite detection in water. Based on our findings, we envision that Zn/Cd/Cubd NZ can also be promising for remediation of waste waters by reducing Se(IV) to elemental selenium (Se0) and for isolation of this element in pure form. The detailed study of effective R-like AEs, including Zn/Cd/Cubd NZ, may be useful in fundamental biological investigations, in particular, for elucidation of mechanisms of their interactions with living cells. R-like AEs may become potential tools in medicine not only for diagnostics and drug delivery, but also be used as pharmaceuticals (mimetics of antioxidant enzymes) in enzyme-therapy of different diseases caused by unbalance of redox processes in organism.9–12,46–49
Results and discussion
Obtaining and characterization of reductase mimetics
| No. | AE | Synthesis method | Specific activity, U mg−1 |
|---|---|---|---|
| 1 | Zn/Cu/Cdbd | Chemical bath deposition on plate | 32.7 ± 1.8 |
| 2 | Ce/Cubd | 14.3 ± 1.1 | |
| 3 | Pd/Cubd | 0.27 ± 0.03 | |
| 4 | Cd/Cucr | Chemical reduction | 3.60 ± 0.25 |
| 5 | Ce/Pdcr | 0.81 ± 0.02 | |
| 6 | Cu/Ptcr | 1.63 ± 0.15 | |
| 7 | Cecr | 1.50 ± 0.04 | |
| 8 | Pd/Cdcr | 3.24 ± 0.22 |
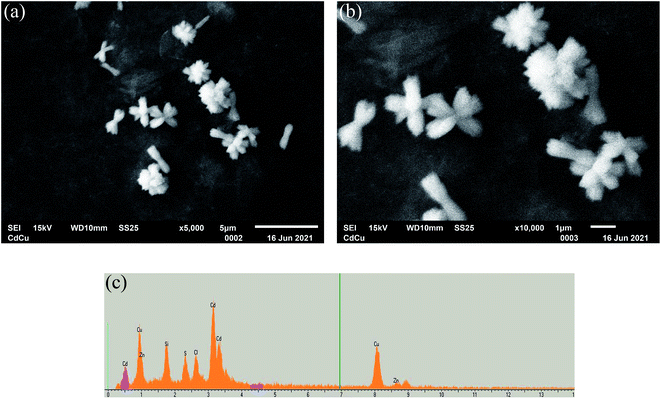
The Zn/Cd/Cubd-NZ with the highest R-like activity was chosen for further detailed investigation by SEM coupled with X-ray microanalysis (SEM-XRM). As shown in Fig. 1a–c, NZ has three dimensional flower-like structures with the size of near 1 μm. The XRM images show the characteristic peaks for Cd0, Cu0 and Zn0 (Fig. 1d).
Catalytic properties of Zn/Cu/Cdbd NZ
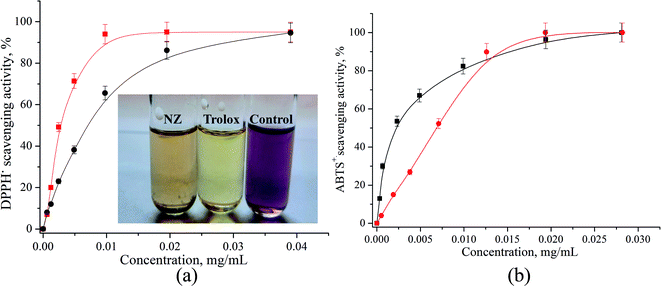
As demonstrated in Fig. 2a and b, Zn/Cd/Cubd NZ shows appreciable DPPH˙ and ABTS˙+ free radical scavenging activities at the selected range of concentrations. It was observed that the DPPH˙ and ABTS˙+ radical scavenging effect increases with the increase of the concentration of Zn/Cd/Cubd NZ. The results of the electrochemical ABTS˙+ method reported in Trolox equivalent units were in good agreement with a classic spectrophotometric DPPH˙ method. We have tested the samples of some plant foods (berries, vegetables, fruits) and beverages on their antioxidant capacity in comparison with Trolox as a standard. These activities were expressed as EC50 values and presented in Table 2.
| Species | EC50 (μg mL−1), that estimated with using | |
|---|---|---|
| DPPH˙ | ABTS˙+ | |
| a Not done. | ||
| Zn/Cd/Cubd NZ | 6.8 ± 0.2 | 6.9 ± 0.3 |
| Trolox | 2.7 ± 0.1 | 2.9 ± 0.2 |
| Red wine, dry | 2.9 ± 2.0 | NDa |
| Bilberry | 9.7 ± 0.1 | 10.5 ± 0.2 |
| Cognac | 12 ± 0.3 | ND |
| Yogurt with chokeberry | 13 ± 1.0 | ND |
| Garnet | 18 ± 2.5 | 19.3 ± 3.0 |
| Tomatoes | 26 ± 0.4 | ND |
| Lemon | 49 ± 0.3 | ND |
| Milk | 582 ± 3.4 | ND |
The EC50 values calculated from DPPH˙ and ABTS˙+ assays ranged between 9.7–49 mg mL−1 for food products and 2.9–12 mg mL−1 – for beverages. The values of antioxidant activities determined by two different methods are very similar. It should be noted that the lower EC50 value indicates a higher antioxidant activity for the product. The antioxidant activity values determined by these two different assays (Table 2) revealed that among food products and beverages, the Zn/Cd/Cubd NZ has a stronger antioxidant power when compared to garnet, lemon or milk and a lower power when compared to red wine and the standard Trolox. The obtained EC50 data of food products are in good correlation with the data described in literature.50
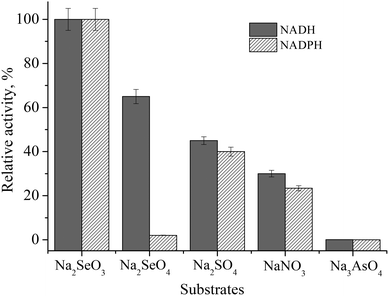 | ||
| Fig. 3 Substrate and co-factor specificity of Zn/Cd/Cubd NZ in acetate buffer, pH 4.5. Substrates (1 mM): Na2SeO3, Na2SeO4, Na2SO4 NaNO3 and Na3AsO4; cofactors (0.13 mM): NADH and NADPH. | ||
We have found that the formation of elemental selenium (Se0) in the presence of cofactors (NADH or NADPH), mediators of electron transfer (ABTS or DPPH), and Na2SeO3 as a substrate takes place even without the addition of NZ (Fig. 4a and b, lines 2). The addition of Zn/Cd/Cubd NZ activates this process (Fig. 4a and b, lines 1).
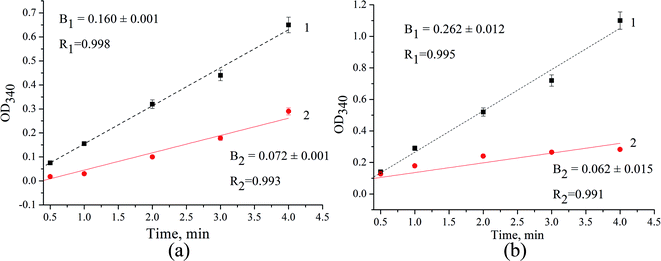 | ||
| Fig. 4 The kinetic curves for selenite reduction in the mixture of Na2SeO3, NADH, ABTS (a) or DPPH (b) in acetate buffer, pH 4.5 in the presence (line 1) and in the absence of Zn/Cd/Cubd NZ (line 2). | ||
The efficiency factor (F) of selenite reduction/Se0 formation for NZ was calculated as a relation of slope values (F = B1/B2) from the correspondent kinetic curves. In the presence of ABTS in reaction mixture (Fig. 4a), FABTS is 2.2, whereas in the presence of DPPH (Fig. 4b), the value of FDPPH is 1.8 fold higher and achieves 4.2.
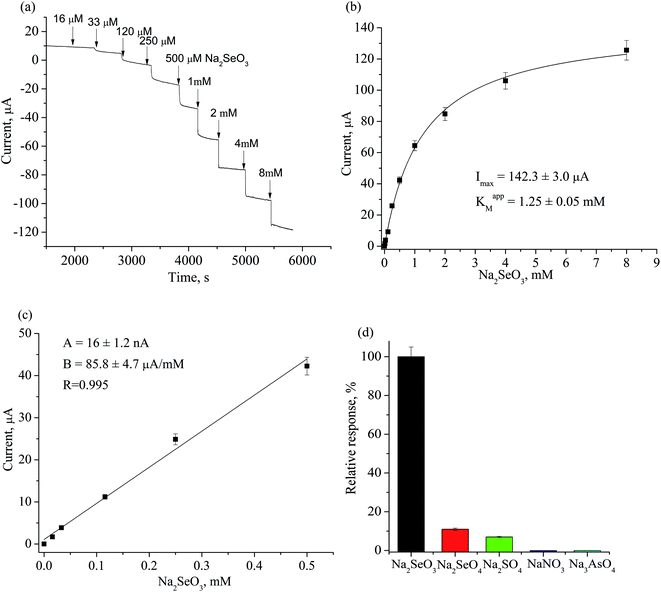
Construction, evaluation and application of amperometric sensor for selenite determination
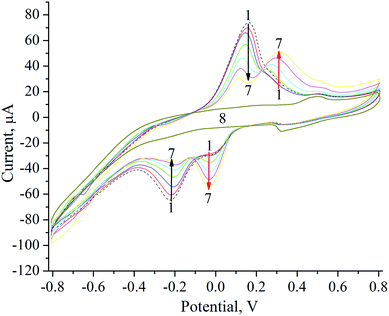 | ||
| Fig. 5 Cyclic voltammograms of NZ/GE (1–7) and unmodified GE (8) in acetate buffer, pH 4.5 upon subsequent additions of Na2SeO3, μM: 0 (1), 8 (2), 16 (3), 32 (4), 64 (5), 150 (6), 300 (7, 8). | ||
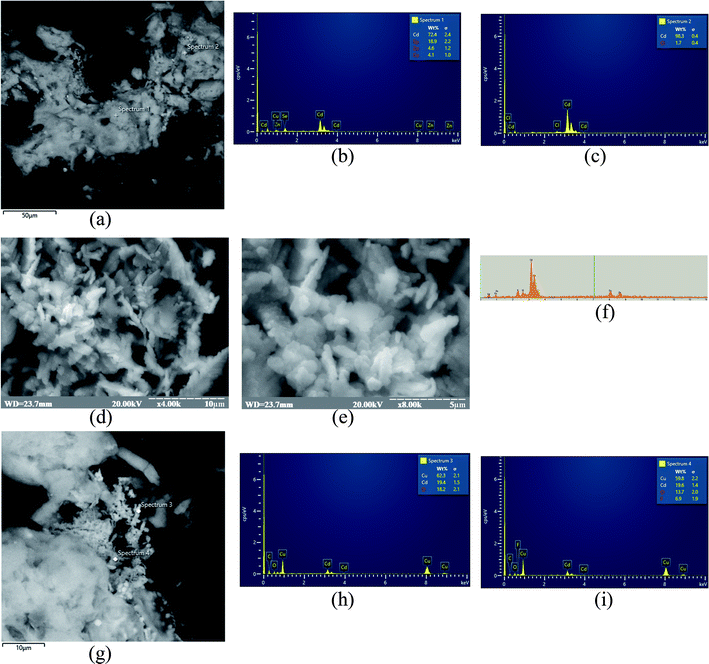
Formation of elemental selenium (Se0) on the surface of NZ/GE was visually observed and proved by the SEM and X-ray microanalysis (Fig. 6). According to SEM images, Se0 is generated as agglomerates with the sizes varying from 5 to 50 μm (Fig. 6a, d, e and g). Se0 formation has been proved by X-ray microanalysis, which showed emission peaks at 1.37, 11.22, and 12.49 keV (Fig. 6b, c, f, h and l) corresponding to the SeLα, SeKα, and SeKβ transitions, respectively.
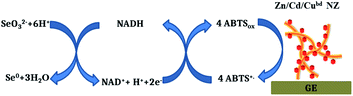
The general scheme of the reactions running on the electrode surface can be described as follows: SeO32− ions diffuse to the Zn/Cd/Cubd-modified GE and are reduced to Se0 in the presence of NADH and ABTS˙+. In turn, the NADH cofactor is oxidized to NAD+ and the ABTS˙+ radical cation is reduced to ABTS releasing 4 electrons and providing a reversible process (Fig. 7). It was shown that the peaks at +300 mV and −50 mV are corresponded to the oxidation of Se0 to SeO32− and to the reduction of SeO32− to Se0, respectively. The obtained oxidation/reduction peaks for the pair of SeO32−/Se0 are in good agreement with the literature data.53 It should be noted that the reduction SeO32− to Se0 on the Zn/Cd/Cubd/GE does not take place in the absence of NADH or ABTS (the data not shown).
The levels of Se in groundwater and surface water usually range from 0.06 μg L−1 to about 400 μg L−1, but in some areas, the Se concentration may to achieve 6000 μg L−1.54 Levels of Se in tap water samples from public water supplies around the world are usually much less than 10 μg L−1, although they may exceed 50 μg L−1.
The developed selenite specific sensor was tested on the model solution of water (after adding a known concentration of sodium selenite) as well as on the real sample of “Morshynska Antioxiwater, Se + Cr + Zn” mineral water. The results of estimation of SeO32− in tested waters are presented in Table 3.
| Sample | Concentration of selenite, mg L−1 | |
|---|---|---|
| Sensor analysis | Added to the model sample or declared by producer | |
| a CV – coefficient of variation. Values are expressed as mean ± SD.b Total Se in the form of SeO32− and Se2−. | ||
| Model solution, CV,a % | 46 ± 2.4, 5.4 | 43.3 ± 1.6 (added) |
| Mineral water “Morshynska”, CV, % | 0.0605 ± 0.003, 7.3 | 0.092b (declared) |
The reproducibility of the proposed analytical method is satisfactory: the coefficients of variation (CV) are less than 10%. It is worthwhile mentioning that the detected selenite contents in the water were close to those found by other authors (0.06–400 μg L−1).54 These results indicate that the developed Zn/Cd/Cubd NZ-based amperometric sensor can be utilized for a fast and simple assay of selenite in water.
Experimental
Materials and methods/reagents
Cerium(III) chloride, ascorbic acid, cadmium(II) chloride, chloroplatinic acid, copper(II) sulfate, zinc(II) sulfate, palladium chloride, sodium borohydride, 2,2′-azinobis-(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS), 2,2-diphenyl-1-picrylhydrazyl (DPPH), cetrimonium bromide (CTAB), nicotinamide adenine dinucleotide reduced (NADH), nicotinamide adenine dinucleotide phosphate reduced (NADPH), Nafion (5% solution in 90% low-chain aliphatic alcohols), potassium persulfate and all other reagents and solvents used in this work were purchased from Sigma-Aldrich (Steinheim, Germany). The antioxidant standard was Trolox from Merck KGaA (Darmstadt, Germany). All reagents were of analytical grade and were used without further purification. All solutions were prepared using ultra-pure water obtained with the Milli-Q® IQ 7000 Water Purification system (Merck KGaA, Darmstadt, Germany).Synthetic procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 40 min (Hettich Micro-22R centrifuge), washed twice with water, and precipitated by centrifugation. Pellets were suspended in 1.0 mL water and stored until use at +4 °C.
000g for 40 min (Hettich Micro-22R centrifuge), washed twice with water, and precipitated by centrifugation. Pellets were suspended in 1.0 mL water and stored until use at +4 °C.
| No. | NZ | Reaction mixture and conditions |
|---|---|---|
| 1 | Cd/Cu | 5 mL 100 mM CdCl2 + 10 mL 12.5 mM CTAB, vigorous stirring for 5 min at 20 °C followed by adding 0.2 mL 100 mM ascorbic acid and heating at 100 °C during 10 min; +5 mL 100 mM CuSO4 followed by adding 0.2 mL 100 mM ascorbic acid, heating at 100 °C and stirring for 15 min at 20 °C |
| 2 | Ce/Pd | 5 mL 100 mM CeCl3 + 10 mL 12.5 mM CTAB, vigorous stirring for 5 min at 20 °C followed by adding 0.2 mL 100 mM ascorbic acid and heating at 100 °C during 10 min; +5 mL 100 mM PdCl3 followed by adding 0.2 mL 100 mM ascorbic acid, heating at 100 °C and stirring for 15 min at 20 °C |
| 3 | Cu/Pt | 5 mL 100 mM H2PtCl6 + 10 mL 12.5 mM CTAB, vigorous stirring for 5 min at 20 °C followed by adding 0.2 mL 100 mM ascorbic acid and heating at 100 °C during 10 min; +5 mL 100 mM CuSO4 followed by adding 0.2 mL 100 mM ascorbic acid, heating at 100 °C and stirring for 15 min at 20 °C |
| 4 | Ce | 5 mL 100 mM CeCl3 + 10 mL 12.5 mM CTAB, vigorous stirring for 5 min at 20 °C followed by adding 0.2 mL 100 mM ascorbic acid and heating at 100 °C during 10 min |
| 5 | Cd/Pd | 5 mL 100 mM PdCl3 + 10 mL 12.5 mM CTAB, vigorous stirring for 5 min at 20 °C followed by adding 0.2 mL 100 mM ascorbic acid and heating at 100 °C during 10 min; + 5 mL 100 mM CdCl2 followed by adding 0.2 mL 100 mM ascorbic acid, heating at 100 °C and stirring for 15 min at 20 °C |
The synthesis of core–shell NZs on stainless steel substrate was performed by chemical bath deposition method (bd) similar as reported earlier,56,57 with some modifications. They include preparation of an aqueous bath containing 0.1 M solution of salt, namely, cadmium(II) chloride, palladium(II) chloride or cerium(II) chloride with the addition of aqueous NH3 solution (28%) under constant stirring at room temperature with resultant pH ∼ 12. Well cleaned Zn plate substrate was placed in reaction bath at room temperature for 20 h resulting in the direct growth of metallic nanowires on Zn substrate.
For the formation of core–shell Zn/Cd/Cubd NZs, the previously synthesized Cd/Zn thin film was vertically dipped into beaker containing 0.01 M copper sulphate solution making an angle of 45° with the wall of beaker. The reaction bath maintained at room temperature for 3 days resulting in the conversion of gray color cadmium film into brown color. The resulting Zn/Cd/Cubd NZs were washed several times with water, dried in air atmosphere and used for further characterization.
Determination of pseudo-enzymatic activities of nanomaterials in solution
The procedure of the assay was the following: 10 μL aqueous suspension of NZ was incubated in a glass tube with 1 mL of the chromogenic substrate (ABTS˙+ solution). Addition of NZ to the substrate stimulated the discoloration from dark blue to colorless over time, indicating an pseudo-enzymatic reaction. The reaction was monitored kinetically during 5 min. The analytical results were statistically processed using the OriginPro 8.5 software.
The specific R-like activity was expressed in μmol min−1 mg−1 of NZ and calculated using the following formula:
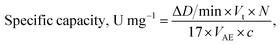 | (1) |
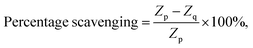 | (2) |
Electrochemical assay of the free radical scavenging activity was done using the ABTS˙+ based method.59
Amperometric measurements were carried out using a CHI 1200A potentiostat (IJ Cambria Scientific, Burry Port, UK) connected to a personal computer and performed in a batch mode under continuous stirring in an electrochemical cell with a 20 mL volume at 25 °C.
All experiments were carried out in triplicate trials. Analytical characteristics of the electrodes were statistically processed using the OriginPro 2021 software. The error bars represent the standard error derived from three independent measurements. Calculation of the apparent Michaelis–Menten constants (KappM) was performed automatically by this program according to the Lineweaver–Burk equation.
The electrochemical properties of the synthesized NZs were studied by cyclic voltammetry (CV) on the GEs in the range from −800 to +800 mV with the scan rate of 50 mV min−1; the profiles of amperometric signals in increasing concentrations of Na2SeO3 were compared.
The monitored signal was the cathodic current produced from reduction of the ABTS˙+ radical at the applied potential −0.10 V vs. Ag/AgCl on the GE working electrode. The responses were obtained as the current–time plot. The peak currents from antioxidant samples were calibrated using Trolox as a standard in concentration range 0–28 μg mL−1.
Conclusions
In the current research, a number of nano- and microzymes based on transition and noble metals were synthesized by the reduction and chemical bath deposition methods. The antioxidant and radical-scavenging properties of the synthesized Zn/Cd/Cubd NZ were studied by its ability to reduce the DPPH˙ or ABTS˙+ radicals in solution. The structure, size, morphology, composition, catalytic properties, and electrochemical activities of the chosen Zn/Cd/Cubd NZ in solution and in immobilized form on the electrode were characterized.The novelty of the work is the following: (1) obtaining for the first time the nanozymes with a high reductase activity; (2) the discovery of the nanozyme with selenite-specific reducing activity; (3) the construction of selenite-selective chemosensor based on metal hybrid nanocomposite. Being sensitive, valid, low-cost, highly selective, the proposed nanozyme-based chemosensor would be promising in different fields of fundamental and practical science, including environmental chemistry, plant and animal biochemistry, nutrition, and medicine.
Because of a high antioxidant activity of the synthesized nanozymes, we plan to study in future the influence of the nanozymes in vivo on eucaryotic cells to test possible consequences of nanozymes' uptake on cell viability and redox-activity of the modified living cells.
Funding
This research was partially supported by the National Research Foundation of Ukraine (projects no. 2020.02/0100 “Development of new nanozymes as catalytic elements for enzymatic kits and chemo/biosensors” and 2021.01/0010 “Development of an enzymatic kit and portable biosensors for express-analysis of creatinine, a marker of acute functional disorders of the kidneys”), National Academy of Sciences of Ukraine (The program “Smart sensor devices of a new generation based on modern materials and technologies”; grant “New dual casein kinase 2 inhibitors and histone deacetylase for targeted tumor chemotherapy” for research laboratories/groups of young scientists in priority areas of science and technology in 2021–2022), and Ministry of Education and Science of Ukraine (projects no. 0120U103398, 0121U109539 and 0121U109543).Author contributions
Nataliya Stasyuk and Galina Gayda: investigation, validation, writing – original draft, visualization. Taras Kavetskyy: methodology, formal analysis, visualization. Mykhailo Gonchar: conceptualization, supervision, writing – review & editing, project administration, funding acquisition, resources.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Dr I. Mat'ko (Institute of Physics, Slovak Academy of Sciences, Bratislava, Slovakia) and R. Serkiz (Institute of Cell Biology NAS of Ukraine, Lviv, Ukraine) for their help with SEM-XRM experiments.References
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- R. Tian, J. Xu, Q. Luo, C. Hou and J. Liu, Front. Chem., 2021, 8, 1–21, DOI:10.3389/fchem.2020.00831.
- J. Wu, X. Wang, Q. Wang, Z. Lou, S. Li, Y. Zhu, L. Qin and H. Wei, Chem. Soc. Rev., 2019, 48, 1004–1076 RSC.
- N. Stasyuk, O. Smutok, O. Demkiv, T. Prokopiv, G. Gayda, M. Nisnevitch and M. Gonchar, Sensors, 2020, 20, 4509–4550 CrossRef CAS PubMed.
- N. Alizadeh and A. Salimi, J. Nanobiotechnol., 2021, 19(1) DOI:10.1186/s12951-021-00771-1.
- Y. Huang, J. Ren and X. Qu, Chem. Rev., 2019, 119, 4357–4412 CrossRef CAS PubMed.
- A. A. Nayl, A. I. Abd-Elhamid, A. Y. El-Moghazy, M. Hussin, M. A. Abu-Saied, A. A. El-Shanshory and H. M. A. Soliman, Trends Environ. Anal. Chem., 2020, 26, e00087, DOI:10.1016/j.teac.2020.e00087.
- W. Wang and S. Gunasekaran, TrAC, Trends Anal. Chem., 2020, 115841, DOI:10.1016/j.trac.2020.115841.
- N. E. Castillo, E. M. Melchor-Martínez, J. S. Ochoa Sierra, N. M. Ramírez-Torres, J. E. Sosa-Hernández, H. M. N. Iqbal and R. Parra-Saldívar, Int. J. Biol. Macromol., 2021, 179, 80–89 CrossRef PubMed.
- R. Mahmudunnabi, F. Z. Farhana, N. Kashaninejad, S. H. Firoz, Y. B. Shim and M. J. A. Shiddiky, Analyst, 2020, 145, 4398–4420, 10.1039/d0an00558d.
- M. Kumawat, A. Umapathi, E. Lichtfouse and H. K. Daima, Environ. Chem. Lett., 2021, 19(6), 3951–3957 CrossRef PubMed.
- S. Khan, M. Sharifi, S. H. Bloukh, Z. Edis, R. Siddique and M. Falahati, Talanta, 2021, 224, 121805, DOI:10.1016/j.talanta.2020.121805.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- F. Attar, M. G. Shahpar, B. Rasti, M. Sharifi, A. A. Saboury, S. M. Rezayat and M. Falahati, J. Mol. Liq., 2019, 278, 130–144 CrossRef CAS.
- B. Neumann and U. Wollenberger, Sensors, 2020, 20, 3692, DOI:10.3390/s20133692.
- R. Fu, J. Zhou, Y. Wang, Y. Liu, H. Liu, Q. Yang, Q. Zhao, B. Jiao and Y. He, ACS Appl. Bio Mater., 2021, 4(4), 3539–3546 CrossRef CAS PubMed.
- H. Zhao, R. Zhang, X. Yan and K. Fan, J. Mater. Chem. B, 2021, 9, 6939–6957 RSC.
- F. Mancin, L. J. Prins, P. Pengo, L. Pasquato, P. Tecilla and P. Scrimin, Molecules, 2016, 21(8), 1014, DOI:10.3390/molecules21081014.
- J. Chen, Q. Ma, M. Li, W. Wu, L. Huang, L. Liu, Y. Fang and S. Dong, Nanoscale, 2020, 12, 23578–23585 RSC.
- S. Biswas and A. Pal, Mater. Today Commun., 2021, 28, 102588 CrossRef CAS.
- R. D. Neal, R. A. Hughes, P. Sapkota, S. Ptasinska and S. Neretina, ACS Catal., 2020, 10(17), 10040–10045 CrossRef CAS.
- L. Lu, S. Zou and B. Fang, ACS Catal., 2021, 11(10), 6020–6058 CrossRef CAS.
- D. Besold, S. Risse, Y. Lu, J. Dzubiella and M. Ballauff, Ind. Eng. Chem. Res., 2021, 60(10), 3922–3935 CrossRef CAS.
- A. A. Nafiey, A. Addad, B. Sieber, G. Chastanet, A. Barras, S. Szunerits and R. Boukherroub, Chem. Eng. J., 2017, 322, 375–384 CrossRef.
- R. Acharya, A. Lenka and K. Parida, J. Mol. Liq., 2021, 337, 116487 CrossRef CAS.
- H. Luck, in Method of Enzymatic Analysis, ed. H. U. Bergmeyer, Academic Press, New York and London, 1st edn, 1965, pp. 885–894 Search PubMed.
- F. Hollmann, D. J. Opperman and C. E. Paul, Angew. Chem., Int. Ed. Engl., 2021, 60(11), 5644–5665 CrossRef CAS PubMed.
- D. R. Lovley, Annu. Rev. Microbiol., 1993, 47, 263–290 CrossRef CAS PubMed.
- https://www.creative-biolabs.com/metalloreductase-family.html.
- N. Butler and A. M. Kunjapur, J. Biotechnol., 2020, 307, 1–14 CrossRef CAS PubMed.
- H. Stolterfoht, D. Schwendenwein, C. W. Sensen, F. Rudroff and M. Winkler, J. Biotechnol., 2017, 257, 222–232 CrossRef CAS PubMed.
- L. Li, B. Zhang, L. Li and A. G. L. Borthwick, J. Hazard. Mater., 2022, 422, 126932 CrossRef CAS PubMed.
- E. P. Painter, Chem. Rev., 1941, 28, 179–213 CrossRef CAS.
- M. M. Stenchuk, L. B. Chaban and M. V. Gonchar, Biopolym. Cell, 2006, 22(1), 3–17 CAS.
- W. J. Hunter, Curr. Microbiol., 2014, 69(1), 69–74 CrossRef CAS PubMed.
- M. Wells, J. McGarry, M. M. Gaye, P. Basu, R. S. Oremland and J. F. Stolz, J. Bacteriol., 2019, 201(7), e00614-18 CrossRef PubMed.
- L. J. Yanke, R. D. Bryant and E. J. Laishley, Anaerobe, 1995, 1(1), 61–67 CrossRef CAS PubMed.
- D. F. Ackerley, C. F. Gonzalez, M. Keyhan, R. Blake and A. Matin, Environ. Microbiol., 2004, 6, 851–860 CrossRef CAS PubMed.
- A. G. O’Neill, B. A. Beaupre, Y. Zheng, D. Liu and G. R. Moran, Appl. Environ. Microbiol., 2020, 86(22), e01758-20 CrossRef PubMed.
- J. C. Begara-Morales, M. Chaki, R. Valderrama, C. Mata-Pérez, M. N. Padilla-Serrano and J. B. Barroso, in Plant Life Under Changing Environment, ed. D. K. Tripathi, et al., Elsevier, Academic Press, 2020, ch. 29, pp. 735–754 Search PubMed.
- D. J. Richardson, R. van Spanning and S. J. Ferguson, in Biology of the Nitrogen Cycle, ed. H. Bothe, S. Ferguson and W. E. Newton, Elsevier, Elsevier Science, 1st edn, 2006, ch. 3 Search PubMed.
- M. Guiral, L. Prunetti, C. Aussignargues, A. Ciaccafava, P. Infossi, M. Ilbert, E. Lojou and M.-T. Giudici-Orticoni, in Advances in Microbial Physiology, ed. R. K. Poole, Academic Press, New York, 2012, vol. 61, ch. 4, pp. 125–194 Search PubMed.
- Z. Liang, H. Guo, G. Zhou, K. Guo, H. Lei, W. Zhang, H. Zheng, U.-P. Apfel and R. Cao, Angew. Chem., 2021, 60(15), 8472–8476 CrossRef CAS PubMed.
- A. A. Vernekar and G. Mugesh, Chem.–Eur. J., 2012, 18, 15122–15132 CrossRef CAS PubMed.
- K. Li, R. Qin, K. Liu, W. Zhou, N. Liu, Y. Zhang, S. Liu, J. Chen, G. Fu and N. Zheng, ACS Appl. Mater. Interfaces, 2021 DOI:10.1021/acsami.1c11548.
- S. Singh, S. Ghosh, V. K. Pal, M. Munshi, P. Shekar, D. T. Narasimha Murthy, G. Mugesh and A. Singh, EMBO Mol. Med., 2021, 13(5), e13314, DOI:10.15252/emmm.202013314.
- M. V. Irazabal and V. E. Torres, Cells, 2020, 9(6), 1342, DOI:10.3390/cells9061342.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Chem. Soc. Rev., 2019, 48, 3683–3704 RSC.
- D. Bartolini, A. M. Stabile, S. Bastianelli, D. Giustarini, S. Pierucci, C. Busti, C. Vacca, A. Gidari, D. Francisci, R. Castronari, A. Mencacci, M. Di Cristina, R. Focaia, S. Sabbatini, M. Rende, A. Gioiello, G. Cruciani, R. Rossi and F. Galli, Redox Biol., 2021, 45, 102041 CrossRef CAS PubMed.
- M. Chochevska, E. J. Seniceva, S. K. Veličkovska, G. Naumova-Leţia, V. Mirčeski, J. M. F. Rocha and T. Esatbeyoglu, Microorganisms, 2021, 9(9), 1946 CrossRef CAS PubMed.
- Y. Lai, F. Liu, J. Li, Z. Zhang and Y. Liu, J. Electroanal. Chem., 2010, 639, 187–192 CrossRef CAS.
- S. Seyedmahmoudbaraghani, S. Yu, J. Lim and N. V. Myung, Front. Chem., 2020, 8, 785 CrossRef CAS PubMed.
- A. A. Vernekar and G. Mugesh, Chem.–Eur. J., 2012, 18, 15122–15132 CrossRef CAS PubMed.
- https://www.who.int/water_sanitation_health/dwq/chemicals/selenium.pdf.
- S. B. Goldhaber, Regul. Toxicol. Pharmacol., 2003, 38, 232–242 CrossRef CAS PubMed.
- O. Demkiv, N. Stasyuk, R. Serkiz, G. Gayda, M. Nisnevitch and M. Gonchar, Appl. Sci., 2021, 11, 777, DOI:10.3390/app11020777.
- S. Patil, S. Raut, R. Gore and B. Sankapal, New J. Chem., 2015, 39, 9124–9131 RSC.
- W. Brand-Williams, M. E. Cuvelier and C. Berset, LWT--Food Sci. Technol., 1995, 28, 25–30 CrossRef CAS.
- R. Chaisuksant, K. Damwan and A. Poolkasem, Acta Hortic., 2012, 943, 297–302 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |