DOI:
10.1039/D1RA07793G
(Paper)
RSC Adv., 2022,
12, 1871-1884
Oxidative DNA cleavage mediated by a new unexpected [Pd(BAPP)][PdCl4] complex (BAPP = 1,4-bis(3-aminopropyl)piperazine): crystal structure, DNA binding and cytotoxic behavior†
Received
21st October 2021
, Accepted 17th December 2021
First published on 12th January 2022
Abstract
A novel Pd(II) double complex, [Pd(BAPP)][PdCl4], containing the 1,4-bis(3-aminopropyl)piperazine (BAPP) ligand is investigated. X-ray crystallography of a single crystal confirmed the structure of the [Pd(BAPP)][PdCl4] complex. The spectroscopic behavior was also elucidated using elemental analysis, nuclear magnetic resonance and Fourier-transform infrared spectroscopy, and mass spectrometry. The antimicrobial susceptibility of the [Pd(BAPP)][PdCl4] complex against all tested microbial strains was lower than that of the BAPP ligand except for C. albicans. The cytotoxic impacts of the BAPP ligand and its [Pd(BAPP)][PdCl4] complex were evaluated in vitro for HepG2, CaCo-2 and MCF7 cell lines as well as the WI-38 normal cell line. The anticancer activity was markedly improved by the complexation. The [Pd(BAPP)][PdCl4] complex could selectively inhibit the tested cancer cells in a safe way to the non-tumorigenic cell (WI-38). From the DNA binding studies with ultraviolet-visible spectrophotometry, the [Pd(BAPP)][PdCl4] complex interacts more efficiently with the calf thymus DNA than its BAPP ligand through the intercalative binding mode. In the absence of an external reductant, the [Pd(BAPP)][PdCl4] complex cleaved the intact supercoiled pBR322 DNA under physiological conditions in a concentration-dependent manner. Additionally, electrophoretic experiments were performed in the presence of different radical scavengers, namely DMSO, NaN3 and KI, and ruled out the hydrolytic mechanistic pathway of the reaction and suggested that the oxidative mechanism is the preferred one. The results of the binding affinity of the [Pd(BAPP)][PdCl4] complex to human DNA were modeled using a molecular docking study showing that the complex interacts more strongly with human DNA than the ligand. Finally, an in vitro pharmacokinetic study was assessed through in silico ADME predictions.
1. Introduction
Cancer is one of the major causes of the increase in morbidity and mortality rates, second to cardiovascular disease.1 As reported, 9.6 million cancer-related deaths have been enumerated in 2018.2 It is forecasted to increase up to 22.7 million in 2030 worldwide.3 Medicinal inorganic chemistry is a prosperous area, in which most scientists focused their efforts in the design of metal-based drugs which could be considered as a potential chemotherapeutic agent for cancer treatment.4,5 Cisplatin, carboplatin, and oxaliplatin succeeded to play the role of Pt-based anticancer agents, but their unwelcome interaction with non-cancerous cells leads to some unwanted toxic effects and hinder their clinical application6 such as, neurotoxicity, nephrotoxicity, ototoxicity, myelosuppression7 and limited usage for different cancer cells8 in addition to the acquired cellular resistance towards platinum-based drugs.9 Thus, the development of non-Pt(II)-based drugs including the Pd(II) complexes have been devoted special attention owing to the structural similarities between Pt(II) and Pd(II) with the higher kinetic lability of Pd(II) versus Pt(II) analogues.10 It is worth mentioning that judicious choice of the inert chelating ligands can relatively decrease these kinetic labilities to the extent to allow the complex for reaching to target, DNA, without dissociation in passing through the cytoplasm and accordingly enhance their cytotoxic activity.11,12 Hence, the desire to explore more potent and less hazardous Pd(II)-based anticancer agent become a thriving area for drug discoverers. Piperazine is a crucial building block owing to the structural resemblance to glucose and therefore to the cyclodextrins. So, metal-based macrocyclic drugs including nitrogen-containing heterocyclic piperazine derivatives exhibit many biological features such as the antitumor activity, in which a series of clinical anticancer drugs such as entrectinib, palbociclib and olmutinib are convinced to be used in cancer treatment clinically.13 There are some previously reported copper complexes based on piperazine derived ligand 2-((2-piperazin-1-yl-ethylimino)-methyl)-phenol (H2L) namely [Cu(L)(H2O)2(NO3)](NO3), [Cu(L)(ClO4)(N3)] and [Cu2(μ-1,3-NCS)2(L)](ClO4)2·2H2O have been evaluated for anti-cancerous effect against human breast cancer cell line by Pait et al.14 Some reported complexes exhibited better in vitro antiproliferative activity against the tested cancer cell lines than cisplatin.13 Moreover, they may act as inhibitors for HIV-1 replication,15 anti-inflammatory effect16 and antimicrobial activity against drug-resistant strains like pathogenic Staphylococcus.17 Additionally, a wide range of medicinal applications of compounds bearing piperazine scaffold such as cardio-protective, antidiabetic, relieving pain agent, anti-tuberculosis and antimalarial activities.18,19 In addition, generally the presence of a pair of cis-primary amines situated in the precursor ligand is the primary requirement for successful synthesis of the complex. Thus, structural studies and the evaluation of the antimicrobial activity of copper(II), cobalt(II) and nickel(II) metal complexes of a hexadentate ligand 1,4-bis((2-hydroxybenzaldehyde)propyl)piperazine were also reported.20 The super-helical DNA is the primary cellular target for most clinically important antitumoral and antimicrobial drugs. So, in the scope of rationally understanding the cytotoxic behavior of various drugs at the molecular level, DNA binding/cleavage propensities are probed. As reported before, under physiological conditions, the metal complexes possessing efficient DNA binding and cleavage propensities are mostly considered as drugs for cancer treatment and genomic research.21–23
In the light of these facts and in continuation of our previous work on palladium amine complexes,24–29 novel Pd(II) complex, [Pd(BAPP)][PdCl4], based on 1,4-bis(3-aminopropyl)piperazine ligand (BAPP) has been synthesized. The unexpected geometrical features were successfully identified by single-crystal X-ray structure analysis and different spectroscopic tools. In this article, we present the antimicrobial activity of the [Pd(BAPP)][PdCl4] complex compared to its free BAPP ligand. In vitro cytotoxic activity of the complex compared to its parental ligand, BAPP, was evaluated against three human tumorigenic cell lines, breast carcinoma (MCF7), hepatocellular carcinoma (HepG2) and epithelial colorectal adenocarcinoma cells (CaCo-2) in addition to the human fetal lung fibroblast normal cell (WI-38). The binding studies with calf thymus DNA (CT-DNA) were performed using UV-vis absorption. DNA cleavage propensities of [Pd(BAPP)][PdCl4] complex and BAPP ligand were evaluated. A molecular docking study was carried out to assess the potential binding mode of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex to the double-stranded DNA. In addition, in silico ADME expectations were assessed to follow their drug-likeness parameters.
2. Experimental section
2.1. Instruments and programs
Elemental analysis of the complex was realized through Vario EL III (CHN) analyzer at the micro-analytical center, Cairo University. JASCO spectrometer FT/IR-460 plus was used to accomplish the Fourier transform infrared spectra of the complex. The electron impact mass spectrum of the complex was recorded at 70 eV with the aid of a SHIMADZU QP-2010 plus mass spectrometer. The absorption titration measurements of CT-DNA were determined with a Shimadzu UV 1800 spectrophotometer. A Shimadzu DTG-60H apparatus synchronized with DTG/TG was used to analyze the thermal behavior of the complex under nitrogen atmosphere (20 mL min−1) using a platinum crucible with a heating rate of 10 °C min−1. The X-ray diffraction data of [Pd(BAPP)][PdCl4] complex was collected on a Bruker Axs Kappa Apex diffractometer using Mo Kα radiation. The Denzo–Scalepack30 software package was used for cell refinement and data reduction. A multi-scan absorption correction (Sortav31) was applied to the intensities before structure solution. The structure was solved by the intrinsic phasing (SHELXT32) method. Structural refinement was carried out using SHELXL32 software with SHELXLE33 graphical user interface. The NH hydrogen atom was located from the difference Fourier map and refined isotropically. All other hydrogen atoms were positioned geometrically and constrained to ride on their parent atoms, with C–H = 0.97 Å, N–H = 0.89 Å and Uiso = 1.2Ueq (parent atom). The crystallographic details are summarized in Table 1. The molecular modeling studies through the Density Functional Theory (DFT) calculations have been accomplished to investigate the equilibrium geometry of BAPP at the B3LYP/6-311G++(d,p) level of theory and that of the [Pd(BAPP)][PdCl4] complex at the B3LYP/GENCEP level of theory,34 in which C, H, N and Cl atoms at 6-311G++(d,p) and Pd atom at (LANL2DZ),35 respectively, using Gaussian 09 program.36
Table 1 Crystal data and structure refinement for [Pd(BAPP)][PdCl4]
| R1 = ∑‖Fo| − |Fc‖/∑|Fo|. wR2 = [∑[w(Fo2 − Fc2)2]/∑[w(Fo2)2]]1/2. |
| CCDC |
2078974 |
V (Å3) |
914.64(6) |
| Empirical formula |
C10H24Cl4N4Pd2 |
Z |
2 |
| FW |
554.93 |
μ(Mo Kα) (mm−1) |
2.545 |
| Temp. (K) |
298(2) |
No. reflns |
8524 |
| λ (Å) |
0.7107 |
Unique reflns |
4148 |
| Cryst. syst. |
Triclinic |
Completeness to θ = 25.242° |
99.6% |
| Space group |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
GOOF (F2) |
1.030 |
| a (Å) |
8.5750(2) |
Rint |
0.050 |
| b (Å) |
9.0820(4) |
R1a (I ≥ 2σ) |
0.0358 |
| c (Å) |
12.9300(4) |
wR2b (I ≥ 2σ) |
0.0762 |
| α (deg) |
76.030(2) |
|
|
| β (deg) |
83.160(2) |
|
|
| γ (deg) |
69.500(2) |
|
|
2.2. Synthesis of [Pd(BAPP)][PdCl4] complex
Mixture of PdCl2 (177 mg; 1.0 mmol) and KCl (149 mg; 2.0 mmol) in the least amount of H2O was heated to about 50 °C for 2 h with stirring till reaching a clear solution. Then, the aqueous solution of 1,4-bis(3-aminopropyl) piperazine (BAPP) (100 mg; 0.5 mmol) was added dropwise to the clear solution under hot condition with continuous stirring then the solution was left aside for crystallization at room temperature. After a few days, brown needle-like crystals, suitable for X-ray crystallographic study, were obtained. The analytical data indicate that the composition of the crystals corresponds to the compound having the formula [Pd(BAPP)][PdCl4]. Yield: 74% (205 mg, 0.369 mmol). Elemental analysis calculated (%) for C10H24Cl4N4Pd2 (FW = 554.97): C 21.64, H 4.36, N 10.10. Found: C 21.00, H 4.32, N 10.06. IR (KBr, ν in cm−1): 3448 νa(NH2), 2924 νa(CH2), 2870 νs(CH2); νa(C–N) 1458; νs(C–N) 1165. Molar cond. (DMF, 10−3 M, μS cm−1): 9.5. UV-visible (DMSO, 10−4 M, nm): 236, 300, 421. FAB-MS (m/z, %): (M − 2H) 552. 1H NMR (DMSO, 300 MHz, ppm): 1.4–1.5 (m, 12H, (CH2)Aliphatic), 2.2–2.3 (m, 8H, (CH2–CH2)Piperazine), 2.5 (s, 4H, NH2). 13C NMR (DMSO, 300 MHz, ppm) 30.4 (C2, C5), 40.1 (C1, C4), 53.0 (C7, C8, C9, C10), 55.9 (C3, C6).
2.3. Bioactivity assay
2.3.1. In vitro antimicrobial activity. The antimicrobial activity of the compounds was tested using the agar well diffusion method.37 The antimicrobial activity of the [Pd(BAPP)][PdCl4] complex and the BAPP ligand was assessed and repeated three times according to the procedure recommended through the National Committee for Clinical Laboratory Standards.38 The bacterial strains of Staphylococcus aureus ATCC 13565 (Gram+), Bacillus subtilis ATCC 6051 (Gram+), Escherichia coli ATCC 10536 (Gram−), Pseudomonas aeruginosa ATCC 27853 (Gram−) in addition to two fungal strains of Candida albicans ATCC 10231 and Aspergillus flavus ATCC 16872 were evaluated.
2.3.2. In vitro anticancer activity. The [Pd(BAPP)][PdCl4] complex and the BAPP ligand were tested for the hepatocellular carcinoma (HepG2), breast cancer (MCF7), and human epithelial colorectal adenocarcinoma (CaCo-2) cell lines, in addition to human fetal lung fibroblast normal cell (WI-38) to evaluate their cytotoxic behavior through using MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium] standard assay.39,40 Human cell lines were obtained from VACSERA (Giza, Egypt) and maintained in DMEM (Gibco, Life Technologies Inc., UK) supplemented with 10% FBS (Gibco) and 1% penicillin/streptomycin (Gibco) in a humidified incubator containing 5% CO2 at 37 °C. According to the MTT system, the measurement of the level of mitochondrial dehydrogenases reflects well with the cellular metabolic activity.41 The cells (1 × 104 cells per well) were seeded on the 96-multiwell plate for 1 day before the treatment with the tested compounds to allow the attachment of the cells. The untreated cells played the role of the negative control. The cells were treated by [Pd(BAPP)][PdCl4] complex or its BAPP ligand at different concentrations (5, 10, 25, 50 μM) and incubated for 2 days. Each concentration treatment was performed in triplicate, repeated 3 times and the mean values were recorded. Upon the addition of MTT (100 μL, 0.5 mg mL−1), water-soluble yellow dye on the living cell, the tetrazolium solution was reduced by the mitochondrial dehydrogenases, and then the insoluble reduced formazan was dissolved by DMSO to yield the purple solution. The optical density was measured spectrophotometrically by ELISA reader at 490 nm. The relation between the relative cell viability versus concentration was plotted, and IC50 values were evaluated.
2.4. DNA-interaction assays
2.4.1. DNA binding affinity. Calf thymus DNA (CT-DNA) was used in the DNA binding experiments. The CT-DNA was dissolved in Tris–NaCl (5 mM Tris–HCl/50 mM NaCl (pH 6.9–7.1)) buffer. The DNA purity was confirmed by measuring the UV absorbance ratio value of (A260/A280) and it was found to be more than 1.8.42 The concentration of DNA per nucleotide was measured from the absorbance value at A260 (ε = 6600 M−1 cm−1).43,44 In the DNA binding experiments, the changes in the electronic absorption spectra of the [Pd(BAPP)][PdCl4] complex (90 μM) or its BAPP ligand (450 μM) (in 5% DMSO solution) upon the titration with increasing amounts of the buffered CT-DNA over a range of 10–100 μM were monitored at room temperature and each experiment was repeated twice and the mean values were recorded. After each titration, the reaction mixture had been agitated for 2 min before measuring the absorption spectrum which in turn reaching equilibrium. The calculation of the DNA-binding constants (Kb) was performed by applying Wolfe–Shimer eqn (1) where [DNA] is the concentration of DNA, εA = Aobsd/[compound], εf = the extinction coefficient for the free compound and εb = the extinction coefficient for the fully bound form compound.45| | |
[DNA]/(εa − εf) = [DNA]/(εb − εf) + 1/[Kb(εb − εf)]
| (1) |
2.4.2. DNA cleavage ability. DNA cleavage ability was monitored using agarose gel electrophoresis assay. The plasmid pBR322 (∼0.4 μg) in 5 mM Tris buffer (pH = 7.1) and 37 °C were subjected to the treatment with the BAPP ligand or the [Pd(BAPP)][PdCl4] complex (100 μM), then incubated for 2.5 h. The incubation time was followed by electrophoresis for 2 h at 70 V on 1% agarose gel using a Tris–boric acid–EDTA buffer. After electrophoresis, the bands were visualized using UV light and photographed. ImageJ software was utilized to identify the % of the cleavage products.46,47 The nuclease activity of the tested compounds was evaluated through the degree of transformation of the supercoiled form (SC) to the nicked (N) and linear (L) form of pBR322 DNA plasmid. The cleavage mechanism was studied in the presence of different scavengers as DMSO, NaN3, KI or the chelating agent EDTA.
2.4.3. Molecular docking study. The binding energies concerning the DNA–ligand/complex interactions were computed theoretically MOE2019 software. The structures of the BAPP ligand and the corresponding [Pd(BAPP)][PdCl4] complex were converted into the PDB format file from the output of Gaussian 09 software. The crystal structure of B-DNA (1BNA: 5′-D(*CP*GP*CP*GP*AP*TP*TP*CP*GP*CP*G)-3′) was downloaded from Protein Data Bank (http://www.rcsb.org/pdb).
3. Results and discussion
3.1. Synthesis and structural characterization of the [Pd(BAPP)][PdCl4] complex
[Pd(BAPP)][PdCl4] complex was afforded through the reaction of BAPP ligand with the aqueous solution of K2PdCl4 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio) at 50 °C (Scheme 1). The coordination behavior of BAPP ligand towards Pd(II) ion was investigated via the X-ray crystallography, molar conductivity, IR, mass spectrometry, 1H NMR, 13C NMR and thermal studies. The complex behaves as a non-conductor species in DMF solution,48 as the molar conductance value is 9.5 μS cm−1 at 25 °C, which presumably owing to ion pair formation. Similar behavior was previously reported in [Pd(PNHP)Cl][CuCl2]49 in which (PNHP) = aminophosphine-bis[2-(diphenylphosphino)ethyl]amine. The UV-Vis spectra of [Pd(BAPP)][PdCl4] complex and its free parental ligand were recorded in DMSO. The stability of the [Pd(BAPP)][PdCl4] complex was investigated via the observation of the change of the electronic absorption spectra over time (48 h). Slight or no change (Fig. S7†) was monitored. Both BAPP ligand and [Pd(BAPP)][PdCl4] complex exhibited a sharp peak at 236 nm and 300 nm which could be assignable to the transitions of an electron from σ (bonding orbital)/σ* (antibonding) orbital and the nonbonding orbital/σ* of the amine in the BAPP ligand.50 The similarity between the absorption bands of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex reflects that the spectrum of the complex was predominated mainly by ligand transitions. Besides, only the complex displayed one broadband as a shoulder in the region of 389–489 nm due to a combination of nitrogen–Pd(II) charge transfer (L–M) and Pd(II) d–d bands51 as expected for the square planar geometry through the involvement of the BAPP ligand in the coordination to the Pd(II) ion.52 In the mass spectrum (Fig. S1†), the detected molecular ion peak at m/z 552 is convenient with the molecular formula C10H24Cl4N4Pd2 of the complex. IR spectrum of the BAPP ligand (Fig. S2†) shows a broad medium band at 3356 cm−1 belonging to the asymmetric stretching vibration mode of terminal amino groups of the diamine BAPP ligand.53 An asymmetric and symmetric stretching vibration mode of methylene group was noted at 2823 cm−1 and 2939 cm−1 of medium intensity, respectively.54,55 The bands noted at 1465 and 1157 cm−1 assigned for (C–N) asymmetric and symmetric stretching vibrations, respectively.56 Upon complexation, the band assigned for the amino group is shifted to a higher wavenumber 3449 cm−1 ascribing the coordination through the lone pair of the nitrogen atom.57 Similarly, the shift of the (C–N) asymmetric and symmetric vibrations to 1458 and 1165 cm−1, respectively, under incorporation of nitrogen atoms in the formation of [Pd(BAPP)]2+ complex cation. The proton NMR spectrum (Fig. S3†) of the investigated complex recorded in DMSO, showed a multiplet signal in the range 1.4–1.5 ppm is attributable to aliphatic –CH2.58 The signal at 2.5 ppm is particularly for the protons of –NH2. The 13C NMR showed four strong important signals which are consistent with the expected conformational symmetry of the complex. The thermogravimetric profile of the [Pd(BAPP)][PdCl4] was recorded under atmospheric condition and the obtained TG curve was shown in Fig. S8.† The TGA data imply that the complex starts the decomposition at 224 °C confirming that neither water nor solvent molecules were present in the complex.59 The thermal degradation proceeds via two stages, the first one in the temperature range 30–292 °C, derived from the mass loss of 36.70% (calcd 36.04%) corresponds to C10H24N4 moiety of the BAPP ligand. The second one in the temperature range 292–705 °C, derived from the mass loss of 24.31% (calcd 25.55%) corresponds to the loss of two Cl2 leaving two Pd metals as a thermally stable final decomposition residue.
2 molar ratio) at 50 °C (Scheme 1). The coordination behavior of BAPP ligand towards Pd(II) ion was investigated via the X-ray crystallography, molar conductivity, IR, mass spectrometry, 1H NMR, 13C NMR and thermal studies. The complex behaves as a non-conductor species in DMF solution,48 as the molar conductance value is 9.5 μS cm−1 at 25 °C, which presumably owing to ion pair formation. Similar behavior was previously reported in [Pd(PNHP)Cl][CuCl2]49 in which (PNHP) = aminophosphine-bis[2-(diphenylphosphino)ethyl]amine. The UV-Vis spectra of [Pd(BAPP)][PdCl4] complex and its free parental ligand were recorded in DMSO. The stability of the [Pd(BAPP)][PdCl4] complex was investigated via the observation of the change of the electronic absorption spectra over time (48 h). Slight or no change (Fig. S7†) was monitored. Both BAPP ligand and [Pd(BAPP)][PdCl4] complex exhibited a sharp peak at 236 nm and 300 nm which could be assignable to the transitions of an electron from σ (bonding orbital)/σ* (antibonding) orbital and the nonbonding orbital/σ* of the amine in the BAPP ligand.50 The similarity between the absorption bands of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex reflects that the spectrum of the complex was predominated mainly by ligand transitions. Besides, only the complex displayed one broadband as a shoulder in the region of 389–489 nm due to a combination of nitrogen–Pd(II) charge transfer (L–M) and Pd(II) d–d bands51 as expected for the square planar geometry through the involvement of the BAPP ligand in the coordination to the Pd(II) ion.52 In the mass spectrum (Fig. S1†), the detected molecular ion peak at m/z 552 is convenient with the molecular formula C10H24Cl4N4Pd2 of the complex. IR spectrum of the BAPP ligand (Fig. S2†) shows a broad medium band at 3356 cm−1 belonging to the asymmetric stretching vibration mode of terminal amino groups of the diamine BAPP ligand.53 An asymmetric and symmetric stretching vibration mode of methylene group was noted at 2823 cm−1 and 2939 cm−1 of medium intensity, respectively.54,55 The bands noted at 1465 and 1157 cm−1 assigned for (C–N) asymmetric and symmetric stretching vibrations, respectively.56 Upon complexation, the band assigned for the amino group is shifted to a higher wavenumber 3449 cm−1 ascribing the coordination through the lone pair of the nitrogen atom.57 Similarly, the shift of the (C–N) asymmetric and symmetric vibrations to 1458 and 1165 cm−1, respectively, under incorporation of nitrogen atoms in the formation of [Pd(BAPP)]2+ complex cation. The proton NMR spectrum (Fig. S3†) of the investigated complex recorded in DMSO, showed a multiplet signal in the range 1.4–1.5 ppm is attributable to aliphatic –CH2.58 The signal at 2.5 ppm is particularly for the protons of –NH2. The 13C NMR showed four strong important signals which are consistent with the expected conformational symmetry of the complex. The thermogravimetric profile of the [Pd(BAPP)][PdCl4] was recorded under atmospheric condition and the obtained TG curve was shown in Fig. S8.† The TGA data imply that the complex starts the decomposition at 224 °C confirming that neither water nor solvent molecules were present in the complex.59 The thermal degradation proceeds via two stages, the first one in the temperature range 30–292 °C, derived from the mass loss of 36.70% (calcd 36.04%) corresponds to C10H24N4 moiety of the BAPP ligand. The second one in the temperature range 292–705 °C, derived from the mass loss of 24.31% (calcd 25.55%) corresponds to the loss of two Cl2 leaving two Pd metals as a thermally stable final decomposition residue.
 |
| | Scheme 1 Synthesis of [Pd(BAPP)][PdCl4] complex. | |
3.2. Description of the crystal structure
The crystal structure of the [Pd(BAPP)][PdCl4] complex was presented by the ORTEP diagram in Fig. 1.
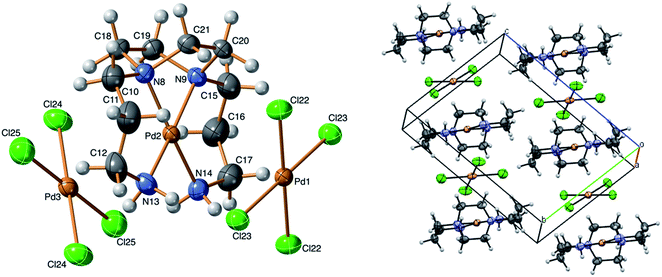 |
| | Fig. 1 ORTEP view for [Pd(BAPP)][PdCl4] complex showing 50% probability ellipsoids and the atom-numbering scheme. | |
Table 1 demonstrates the structural refinement details and the corresponding crystallographic data. The structural data have been deposited at the Cambridge Crystallographic Data Centre: CCDC No. 2078974.† The crystal structure data of the double salt of [Pd(BAPP)][PdCl4] reveals that the asymmetric unit incorporates one [Pd(BAPP)]2+ complex cation and two independent halves of [PdCl4]2− (acting as a counter ion or a complex anion). The anionic parts [PdCl4]2− reside on in the middle of the faces of the unit cell, so that half of each belongs to a different unit cell, Fig. 1. The cationic complex carrying the +2 charges requires two negative charges for electroneutrality. The title complex was crystallized in the triclinic lattice with space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and the crystal packing with Z = 2. In the cationic complex [Pd(BAPP)]2+, the palladium atom was situated in a slightly distorted square planar geometry, surrounded by four nitrogen atoms of the tetradentate ligand (BAPP). This distortion is indicated through the deviation from the linearity of the subtended angles around the palladium center (N1–Pd2–N4 = 171.4(3)° and N2–Pd2–N3 = 170.5(3)°). The sum of bite angle around the Pd2 atom is 360.3°. As reported recently by Houser and co-workers, the geometry of four coordinate complexes could be ascertained quantitatively through the new parameter, τ4 = [360° − (α + β)]/141° where (α = ∠N1–Pd2–N2 = 171.4(3)° and β = ∠N2–Pd2–N3 = 170.5(3)°). The value of τ4 = 0.13 confirms the distortion of square planar geometry of the complex cation.60 Despite the distortion, the environment around the palladium center is planar. The large bending in the bond angle of N1–Pd2–N2 was forced to be 73.4(2)° by the restraint of the five-membered ring. The bond lengths of the four Pd–N are Pd2–N1 = 2.052(6), Pd2–N2 = 2.054(6), Pd2–N3 = 2.053(7) and Pd2–N4 = 2.058(7), which compare well with the previously reported [Pd(BAPP)](ClO4)2 61 analogue and lies in the range of many other reported palladium amine complexes.34,62,63
and the crystal packing with Z = 2. In the cationic complex [Pd(BAPP)]2+, the palladium atom was situated in a slightly distorted square planar geometry, surrounded by four nitrogen atoms of the tetradentate ligand (BAPP). This distortion is indicated through the deviation from the linearity of the subtended angles around the palladium center (N1–Pd2–N4 = 171.4(3)° and N2–Pd2–N3 = 170.5(3)°). The sum of bite angle around the Pd2 atom is 360.3°. As reported recently by Houser and co-workers, the geometry of four coordinate complexes could be ascertained quantitatively through the new parameter, τ4 = [360° − (α + β)]/141° where (α = ∠N1–Pd2–N2 = 171.4(3)° and β = ∠N2–Pd2–N3 = 170.5(3)°). The value of τ4 = 0.13 confirms the distortion of square planar geometry of the complex cation.60 Despite the distortion, the environment around the palladium center is planar. The large bending in the bond angle of N1–Pd2–N2 was forced to be 73.4(2)° by the restraint of the five-membered ring. The bond lengths of the four Pd–N are Pd2–N1 = 2.052(6), Pd2–N2 = 2.054(6), Pd2–N3 = 2.053(7) and Pd2–N4 = 2.058(7), which compare well with the previously reported [Pd(BAPP)](ClO4)2 61 analogue and lies in the range of many other reported palladium amine complexes.34,62,63
3.3. Molecular DFT calculation of BAPP ligand and [Pd(BAPP)][PdCl4] complex
Fig. 2 and 3 show the optimized structures of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex, respectively, in the form of the lowest energy configurations. The natural charges resulted from Natural Bond Orbital Analysis (NBO) were also shown. The two palladium atoms are four-coordinate in square planar geometry. The atoms N1, N2, N4 and N3 are almost in one plane deviated by −7.236°. Also, the atoms Cl1, Cl2, Cl3 and Cl4 are almost in one plane deviated by 0.365°. The bite angles in the distorted square planar geometry are ranging from 72.48° (for N1–Pd2–N2 bite angle) to 94.69° for N3–Pd2–N4 bite angle as shown in Table 2. The optimization implies the conformational changes of the piperazine ring from chair conformation to boat one upon complexation. This was evidenced by the calculated distance between N1–N2 of the piperazine ring to be 2.885 Å in the BAPP ligand decreased by 0.385 Å to 2.500 Å in the complex. Also, the N3–N4 bond length was drastically decreased from 11.413 Å to 3.085 Å upon complex formation as given in Table S2.† The natural charges computed from the NBO-analysis for the BAPP ligand and the [Pd(BAPP)][PdCl4] complex were given in Fig. 2 and 3. The more negative natural charges on the nitrogen atoms coordination centers in the case of uncoordinated BAPP are N1 (−0.564), N2 (−0.567), N3 (−0.945), N4 (−0.944). These charges are decreased upon complexation as shown in Fig. 2 and 3. Moreover, the complex shows more positive natural charges of Pd2 (+0.559) coordinated to nitrogen atoms relative to Pd1 (+0.260) coordinated to chloride atoms implying the Lewis acidity–basicity variation or back donation from the two different coordinating atoms.
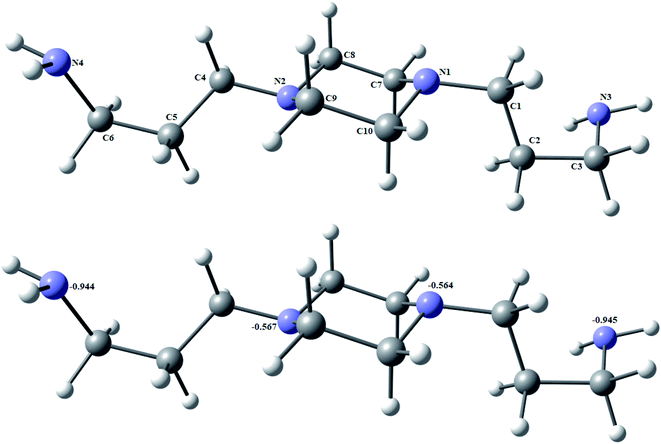 |
| | Fig. 2 The optimized structure of BAPP and the natural charges on atoms of BAPP by density function B3LYP/6-311G++(dp). | |
 |
| | Fig. 3 The optimized structure, the vector of the dipole moment, and the natural charges on active centers of [Pd(BAPP)][PdCl4] complex. | |
Table 2 Comparison of the important X-ray and DFT optimized bond lengths (Å) and bond angles (°) of [Pd(BAPP)][PdCl4] complex
| Type of bond |
Bond length (Å) |
Type of Angle |
Angle (°) |
| X-ray |
DFT |
X-ray |
DFT |
| Dihedral angle. |
| Pd2–N1 |
2.047 |
2.102 |
N1–Pd2–N2 |
73.44 |
72.48 |
| Pd2–N2 |
2.054 |
2.126 |
N1–Pd2–N3 |
98.60 |
93.18 |
| Pd2–N3 |
2.053 |
2.095 |
N2–Pd2–N4 |
98.97 |
99.06 |
| Pd2–N4 |
2.058 |
2.100 |
N3–Pd2–N4 |
89.30 |
94.69 |
| Pd1–Cl1 |
2.313 |
2.473 |
N1–Pd2–N4 |
171.5 |
169.8 |
| Pd1–Cl2 |
2.301 |
2.473 |
N2–Pd2–N3 |
170.6 |
164.9 |
| Pd1–Cl3 |
2.313 |
2.421 |
Cl1–Pd1–Cl2 |
89.73 |
91.49 |
| Pd1–Cl4 |
2.301 |
2.426 |
Cl1–Pd1–Cl4 |
90.27 |
88.24 |
| |
|
|
Cl2–Pd1–Cl3 |
90.27 |
89.73 |
| |
|
|
Cl3–Pd1–Cl4 |
89.73 |
90.07 |
| |
|
|
Cl1–Pd1–Cl3 |
180.0 |
175.1 |
| |
|
|
Cl2–Pd1–Cl4 |
180.0 |
174.3 |
| |
|
|
N1–N2–N4–N3 |
−6.520a |
−7.236a |
| |
|
|
Cl1–Cl2–Cl3–Cl4 |
0.000a |
0.365a |
The calculated total energies, energies of highest occupied molecular orbital (HOMO), energies of lowest unoccupied molecular orbital (LUMO) and dipole moments are reported in Table 3. The more negative value of total energy of the [Pd(BAPP)][PdCl4] complex relative to that of BAPP ligand refers to higher stability. Also, the energy gap (Eg) = ELUMO − EHOMO, is also reported in Table 3 and Fig. 4. Fig. 5 shows the electrostatic potential maps (MEP) which were generated through a new cubic contour to allocate the electron cloud distribution over the molecule functional groups. The electrophilic (blue zone) and the nucleophilic (red zone) areas were clarified. The BAPP ligand map shows red spots in the contour of the amine group and nitrogen atoms coordinating sites which signify their nucleophilic tendency towards the Pd2+ metal ion. For the complex [Pd(BAPP)][PdCl4], the map is divided into two zones, the red zone is around the contour of the anionic complex [PdCl4]2−, while the blue one is around the contour of the cationic complex [Pd(BAPP)]2+.
Table 3 Calculated energies of BAPP and [Pd (BAPP)][PdCl4]complex at B3LYP/6-311G(dp) for all atoms except Pd at B3LYP/LANL2DZ
| |
Ea |
HOMOb |
LUMOc |
Egd |
Dipole momente |
| The total energy (a.u.). Highest occupied molecular orbital (eV). Lowest unoccupied molecular orbital (eV). Eg = ELUMO − EHOMO (eV). Dipole moment (debye). |
| BAPP |
−614.382 |
−4.9307 |
2.5813 |
7.5120 |
1.6566 |
| [Pd(BAPP)][PdCl4] |
−2709.059 |
−5.8461 |
−3.0585 |
2.7876 |
23.9199 |
 |
| | Fig. 4 HOMO and LUMO charge density maps of BAPP and its [Pd(BAPP)][PdCl4] complex. | |
 |
| | Fig. 5 Molecular electrostatic potential (MEP) surface of BAPP (left) and [Pd(BAPP)][PdCl4] complex (right). | |
3.4. Antimicrobial activity
The antimicrobial activity of the BAPP ligand and its [Pd(BAPP)][PdCl4] complex has been tested against different microorganisms such as bacteria and fungi. The antimicrobial activity values for the [Pd(BAPP)][PdCl4] complex are lower than those found for the BAPP ligand for all the tested species except for C. albicans is more susceptible to the [Pd(BAPP)][PdCl4] complex than the BAPP ligand. Generally, it seems to be the coordination of Pd(II) ion to the tetra-dentate BAPP ligand reduced the antimicrobial activities of the corresponding BAPP ligand. Whereby, the activity could not be attributed to the presence of the metal center only,64 as the free ligand exhibited a superior activity than the corresponding cationic complex of [Pd[BAPP]]2+. As reported before, in a large number of palladium complexes bearing N,N-chelate ligands, the free ligand shows an antimicrobial activity while their corresponding metal complexes were devoid to have an antibacterial and antifungal properties such as [Pd(L1)2] and [Pd(L2)2] complexes (where Ll = 3-(1-(phenylamino)ethylidene)-chroman-2,4-dione and L2 = 3-(1-(o-toluidino)ethylidene)-chroman-2,4-dione), L1 and L2 had higher activity towards Staphylococcus aureus and Salmonella enterica, respectively. In addition to other palladium(II) complexes based on quinolinylaminophosphonates ligands such as diethyl [α-anilino-(quinolin-3-ylmethyl)]phosphonate (L3) and dibutyl [α-(quinolin-3-ylamino)-N-benzyl]phosphonates (L4), only L3 and L4 act as bacteriostatic agents against B. thuringiensis var. kurstaki (for L3), B. megaterium and S. epidermidis (for L4).65–68 This indicates that the activity is not depending solely on the existence of metal ions but rather a synergistic effect of many factors.69
3.5. Cytotoxic activity
In the cytotoxicity analysis, in comparison with the BAPP ligand, the [Pd(BAPP)][PdCl4] complex shows more killing potential towards the tested cancer cell lines of MCF7 (IC50 = 49 ± 3.7 μM), HepG2 (IC50 = 14.6 ± 1.1 μM) and CaCo-2 (IC50 = 14.9 ± 1.5 μM) than the BAPP ligand which shows no cytotoxic effect towards the tested carcinogenic cell lines as presented in Fig. 6. On the other hand, the complex and the BAPP ligand induce less damage to the human fetal lung fibroblast normal cell (WI-38). To the best of our knowledge few palladium complexes containing [PdCl2] as a counter ion were encountered and their cytotoxic behavior towards different cell line did not previously discussed.70 However, in light of these results, in vitro studies are necessary to validate the in vivo results in future trends.
 |
| | Fig. 6 Cellular growth inhibition of (A) BAPP ligand (B) [Pd(BAPP)][PdCl4] complex against normal cell (W138), breast carcinoma (MCF7), hepatocellular carcinoma (HepG2) and epithelial colorectal adenocarcinoma cells (CaCo-2). The cell viability was measured by MTT assay after incubation for 48 h. | |
3.6. DNA-interaction assays
3.6.1. CT-DNA binding affinity. In cancer therapy, the mechanism of antitumor activity of the metal-based anticancer therapeutics could be initially considered through the evaluation of their DNA binding affinities.71,72 A wide varieties of different compounds could interact covalently or non-covalently with DNA. The covalent mode of interaction includes the formation of a covalent bond between the compound and the nucleic acid. While in the non-covalent mode, the compounds bind reversibly with the DNA helix through different modes including the electrostatic interaction with the external negatively charged sugar-phosphate backbone of the DNA-helix via Coulomb forces,45,73 groove-binding interaction which might be either major and/or minor, the intercalation binding mode of interaction between the stacked base pairs of the native DNA74 and three-way junction.75,76 The UV-visible absorption titration spectra were monitored to determine the binding affinity of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex towards the CT-DNA. The absorption spectra of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex in the absence and presence of an increasing concentration of CT-DNA were shown in Fig. 7 and 8, respectively. By increasing the concentration of CT-DNA, the absorption band around 236 nm was assigned for n–π* transition in the ligand and its complex was affected, resulting in hyperchromic effect for the BAPP ligand and hypochromic effect for the [Pd(BAPP)][PdCl4] complex. The hyperchromic or the hypochromic spectral changes are reflecting a conformational or structural change in the CT-DNA after the binding to a certain compound.77 By plotting the [DNA]/(εa − εf) versus [DNA], the intrinsic binding constants Kb were deduced from the ratio of the slope to the intercept. The calculated values of Kb are 0.62 ± 0.06 × 104 M−1 for the BAPP ligand and 1.65 ± 0.08 × 105 M−1 for the [Pd(BAPP)][PdCl4] complex. The intrinsic binding constant of the [Pd(BAPP)][PdCl4] complex is higher than many previously reported Pd(II) complex of ligand bearing piperazine moiety such as [Pd(Phenpip)(OH2)2] where (Phenpip = 1-phenylpiperazine) (Kb = 4.14 × 103 M−1),78 [Pd(Pip)(H2O)2]2+ where (Pip = piperazine) (Kb = 4.68 × 103 M−1),79 suggesting an electrostatic and/or groove binding mode for the interaction between the complexes and CT-DNA. Our results imply a greater interaction between the [Pd(BAPP)][PdCl4] complex and DNA than the BAPP ligand. By inspecting the absorption band at 236 nm of the [Pd(BAPP)][PdCl4] complex, the decrease in the molar absorptivity with the increment of DNA concentration suggesting that the complex may act as an intercalative moiety sandwiched between the CT-DNA base pairs.80,81 In addition, there is a slight blue shift in higher concentration levels of DNA. Besides, an isosbestic point was noticed around 244 nm. The negative value of the standard Gibbs free energy change (ΔG = −7.118 kcal mol−1) for the [Pd(BAPP)][PdCl4] complex-DNA binding affinity derived from the following equation ΔG = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb confirms that the association between the complex-DNA to form an adduct is spontaneous.
Kb confirms that the association between the complex-DNA to form an adduct is spontaneous.
 |
| | Fig. 7 Absorption spectrophotometric titration of BAPP ligand (4.5 × 10−4 M) against increasing concentrations of CT-DNA. [DNA] = 0–100 μM, step 20 μM. Arrow (↑) refers to the hyperchromic effect. The inset shows the linear fit of [DNA]/(εa − εf) vs. [DNA]. | |
 |
| | Fig. 8 Absorption spectrophotometric titration of the double complex [Pd(BAPP)][PdCl4] (9.0 × 10−5 M) against increasing concentrations of CT-DNA. [DNA] = 0–100 μM, step 20 μM. Arrow (↓) refers to the hypochromic effect. The inset shows the linear fit of [DNA]/(εa − εf) vs. [DNA]. | |
3.6.2. Plasmid DNA cleaving ability. Metallonucleases are an alternative to the bio-nucleases, which can cleave the phosphodiester bonds of the DNA nucleotides.82 The main advantages of chemical nucleases are their relatively smaller size and their higher stability towards the temperature and over wide pH range.83 Some clinically reported artificial nucleases were approved to treat cancer,84,85 in which the cell death happens by DNA cleavage. The nuclease activity of the [Pd(BAPP)][PdCl4] complex and its BAPP ligand was studied under the physiological pH and temperature. The DNA cleavage propensity was measured through inspecting the gradual degradation of the intact supercoiled form (SC (form I)) to nicked circular (NC (form II)) and linear (L (form III)) forms.86 Upon electrophoresis of the pBR322 plasmid DNA, the supercoiled form will immigrate with the highest speed in the gel. If any scission occurs on one strand of the double-stranded plasmid DNA, the supercoiled form will be relaxed to the slowest moving nicked circular form. Further scission in the plasmid leading to the linear form situated between the intact supercoiled and the nicked circular forms in the gel. The nuclease activity of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex were tested at 100 μM concentration without using any external cofactor as shown in Fig. 9. The result confirmed the better cleavage propensity of the complex relative to the ligand owing to the complete disappearance of all forms in the case of the complex.87 Moreover, the concentration dependence study for the [Pd(BAPP)][PdCl4] complex was performed at various concentrations (10 μM, 50 μM and 100 μM) in which it was concluded that the [Pd(BAPP)][PdCl4] complex can cleave the intact supercoiled form in a concentration-dependent manner as presented in Fig. 9B. To know further about the mechanism of the DNA cleavage process by the complex (hydrolytic vs. oxidative), the effect of different ROS (reactive oxygen species) scavengers has been investigated, which may include DMSO acting as a hydroxyl radical (OH˙) scavenger,88 KI which behaves as a peroxide (O22−) scavenger89,90 and NaN3 which behaves as a singlet (1O2) quencher.91 It is worth noting that the addition of DMSO to the [Pd(BAPP)][PdCl4] complex has no significant effect on DNA degradation, excluding the role of (OH˙) in the mechanism of DNA cleavage. On the other hand, the cleavage efficiency of the [Pd(BAPP)][PdCl4] complex was significantly inhibited by adding NaN3, KI or EDTA as demonstrated in Fig. 10. These facts dictate the involvement of (O22−) and (1O2) as reactive oxygen species in the cleavage process. In addition, the EDTA chelating agent reduces DNA cleavage indicating that Pd(II) complex plays a key role in the DNA breakage. From these results we can deduce that, the [Pd(BAPP)][PdCl4] complex is able of exerting DNA cleavage activity through an oxidative DNA damage pathway.
 |
| | Fig. 9 Gel electrophoresis patterns showing the cleavage of pBR322 plasmid DNA (∼0.4 μg) by (A) the [Pd(BAPP)][PdCl4] complex compared to BAPP ligand and (B) the [Pd(BAPP)][PdCl4] complex at different concentrations (10, 50, 100 μM) in Tris–HCl/NaCl buffer (pH = 7.2) at 37 °C for 1.5 h. | |
 |
| | Fig. 10 Cleavage of pBR322 plasmid DNA (∼0.4 μg) by the ROS scavengers (0.2 M) (DMSO, KI, NaN3) in the presence and absence of the complex (100 μM) in Tris–HCl/NaCl buffer (pH = 7.2) at 37 °C for 1.5 h. | |
3.6.3. Molecular docking of compounds with human DNA. An insight into the interaction of the titled ligand and its corresponding complex with macromolecules could be provided through a docking study. Moreover, the relative binding free energy and the different possible binding modes of the studied compounds to the B-DNA (PDB ID: 1BNA) active site could be well estimated. In the present work, the binding free energy of the BAPP ligand and [Pd(BAPP)][PdCl4] complex to the human DNA receptor were found to be −1.5 and −20.0 kcal mol−1, respectively, Table 4, Fig. 11 and 12. The more negative binding energy of the complex indicates the stronger DNA interaction. The molecular docking data are well correlated with the DNA binding/cleavage studies.
Table 4 The docking interaction data calculations of (A) BAPP and (B) [Pd(BAPP)][PdCl4] with the active sites of the receptor of human DNA (PDB ID: 1BNA)
| |
Receptor |
Interaction |
Distancea (Å) |
E (kcal mol−1) |
| The lengths of H-bonds are in brackets. |
| BAPP |
| N⋯2 |
N2⋯DG⋯16 (B) |
H-Acceptor |
3.44 (2.54) |
−1.0 |
| C⋯9 |
6-Ring DC⋯11 (A) |
H-pi |
4.70 |
−0.5 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| [Pd(BAPP)][PdCl4] |
| PD⋯1 |
OP2⋯DG⋯22 (B) |
Metal |
2.56 |
−1.8 |
| N⋯5 |
OP2⋯DC⋯21 (B) |
Ionic |
3.41 |
−2.3 |
| N⋯5 |
OP2⋯DG⋯22 (B) |
Ionic |
3.15 |
−3.6 |
| N⋯CL |
OP2⋯DG⋯22 (B) |
Ionic |
3.00 |
−4.5 |
| CL⋯41 |
OP1⋯DG⋯22 (B) |
Ionic |
3.16 |
−3.5 |
| CL⋯41 |
OP2⋯DG⋯22 (B) |
Ionic |
3.02 |
−4.3 |
 |
| | Fig. 11 2D plot of the interaction between (A) BAPP ligand and (B) [Pd(BAPP)][PdCl4] complex with the active site of the receptor of human DNA (PDB ID: 1BNA). The interactions with DNA nucleotides are shown with dotted curves. | |
 |
| | Fig. 12 Molecular docking simulation studies of the interaction between BAPP (A) and [Pd(BAPP)][PdCl4] (B) with the active site of the receptor of human DNA (PDB ID: 1BNA). | |
3.7. In silico pharmacokinetic indexes
In silico ADMET (Absorption, Distribution, Metabolism, Elimination, Toxicity) assay is an approach to give an insight if a compound has acceptable pharmacokinetics and pharmacodynamics properties before administration to the body or not.92 The penetrate brain-barrier (BBB), partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w), gastrointestinal absorption (GI), topological polar surface area (TPSA), solubility and skin permeation (log
Po/w), gastrointestinal absorption (GI), topological polar surface area (TPSA), solubility and skin permeation (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp) of the BAPP ligand as well as the [Pd(BAPP)][PdCl4] complex were evaluated (Table S4†) by the aid of SwissADME server.93 The results indicate that the [Pd(BAPP)][PdCl4] complex has high human gastrointestinal intestinal absorption probability (GI) synchronized with no blood–brain barrier permeation which is claimed as an ideal drug candidate. The synthetic accessibility score exists in the range of (1–10) which implies the moderate synthetic accessibility of the [Pd(BAPP)][PdCl4] complex.
Kp) of the BAPP ligand as well as the [Pd(BAPP)][PdCl4] complex were evaluated (Table S4†) by the aid of SwissADME server.93 The results indicate that the [Pd(BAPP)][PdCl4] complex has high human gastrointestinal intestinal absorption probability (GI) synchronized with no blood–brain barrier permeation which is claimed as an ideal drug candidate. The synthetic accessibility score exists in the range of (1–10) which implies the moderate synthetic accessibility of the [Pd(BAPP)][PdCl4] complex.
4. Conclusion
In conclusion, a novel palladium(II) complex bearing 1,4-bis(3-aminopropyl)piperazine ligand has been isolated and fully characterized using different spectroscopic tools. X-ray single-crystal diffraction study points out that the palladium center in the [Pd(BAPP)]2+ cationic complex is four coordinated with slight distortion in the square planar geometry (τ4 value = 0.13). The thermal stability of the [Pd(BAPP)][PdCl4] complex demonstrates the absence of water molecules either inside or outside the coordination sphere. The [Pd(BAPP)][PdCl4] complex showed lower antimicrobial activity than those found for the BAPP ligand for all the tested species except for C. albicans. The in vitro cell viability test by the MTT assay indicates that the [Pd(BAPP)][PdCl4] complex has anticancer activities against MCF7, HepG2 and CaCo-2 without any hazardous effect to the healthy cell (WI-38). The DNA binding study has been investigated using the electronic absorption spectrophotometric measurements. The results suggest that the interaction of the complex with the DNA occurred via an intercalative mode with Kb value equal to 1.65 ± 0.08 × 105 M−1. In addition, the [Pd(BAPP)][PdCl4] complex can cleave the pBR322 plasmid DNA efficiently without any external reducing agent in a concentration-dependent activity. To elucidate the mechanism of action by using different ROS scavengers such as DMSO, KI and NaN3 in the presence and absence of the [Pd(BAPP)][PdCl4] complex, the results suggest the oxidative mechanistic pathway. From DNA cleaving properties, it can be concluded that the new [Pd(BAPP)][PdCl4] complex may inhibit the growth of cancer cells through cleaving of the genome. Accordingly, the [Pd(BAPP)][PdCl4] complex seems to be an adequate choice for further in vitro and in vivo biological studies.
Conflicts of interest
There are no conflicts to declare.
Abbreviation list
| BAPP | 1,4-Bis(3-aminopropyl) piperazine |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium |
| HepG2 | Human hepatocellular carcinoma cell line |
| CaCo-2 | Cancer Coli-2 (human colorectal adenocarcinoma cells) |
| MCF7 | Michigan Cancer Foundation-7 (human breast cancer cells) |
| WI-38 | Wistar Institute-38 (non-malignant human foetal lung fibroblast) |
| ADME | Absorption, Distribution, Metabolism, Elimination, Toxicity |
| m (in NMR) | Multiplet |
| s (in NMR) | Singlet |
| CT | Calf-thymus |
| DMSO | Dimethylsulfoxide |
| DMF | Dimethylformamide |
| ELISA | Enzyme linked immunosorbent assay |
| EDTA | Ethylenediaminetetraacetic acid |
| TGA | Thermogravimetric analysis |
| PDB | Protein data bank |
| HOMO | Highest occupied molecular orbital |
| LUMO | Lowest unoccupied molecular orbital |
References
- R. A. Khan, M. R. Khan, M. Usman, F. Sayeed, H. A. Alghamdi, S. Alrumman, W. Alharbi, N. N. Farshori, M. M. Al-Oqail and M. R. Siddiqui, Saudi J. Biol. Sci., 2020, 27, 2164–2173 CrossRef CAS PubMed.
- Y. O. Ayipo, W. A. Osunniran and M. N. Mordi, Coord. Chem. Rev., 2021, 432, 213746 CrossRef CAS.
- F. Bray, A. Jemal, N. Grey, J. Ferlay and D. Forman, Lancet Oncol., 2012, 13, 790–801 CrossRef PubMed.
- N. V. Loginova, H. I. Harbatsevich, N. P. Osipovich, G. A. Ksendzova, T. V. Koval'chuk and G. I. Polozov, Curr. Med. Chem., 2020, 27, 5213–5249 CrossRef CAS PubMed.
- T. W. Hambley, Science, 2007, 318, 1392–1393 CrossRef CAS PubMed.
- X. Wang and Z. Guo, Chem. Soc. Rev., 2013, 42, 202–224 RSC.
- R. P. Miller, R. K. Tadagavadi, G. Ramesh and W. B. Reeves, Toxins, 2010, 2, 2490–2518 CrossRef CAS PubMed.
- B. Chabner, Nat. Rev. Cancer, 2005, 5, 65–72 CrossRef CAS PubMed.
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- Ž. D. Bugarčić, J. Bogojeski and R. van Eldik, Coord. Chem. Rev., 2015, 292, 91–106 CrossRef.
- N. P. Barry and P. J. Sadler, Chem. Commun., 2013, 49, 5106–5131 RSC.
- Y. Li, Z. Gu, C. Zhang, S. Li, L. Zhang, G. Zhou, S. Wang and J. Zhang, Eur. J. Med. Chem., 2018, 144, 662–671 CrossRef CAS PubMed.
- H. Wang, J. Wei, H. Jiang, Y. Zhang, C. Jiang and X. Ma, Molecules, 2021, 26, 1453 CrossRef CAS PubMed.
- M. Pait, B. Kundu, S. C. Kundu and D. Ray, Inorg. Chim. Acta, 2014, 418, 30–41 CrossRef CAS.
- J. R. Tagat, R. W. Steensma, S. W. McCombie, D. V. Nazareno, S.-I. Lin, B. R. Neustadt, K. Cox, S. Xu, L. Wojcik and M. G. Murray, J. Med. Chem., 2001, 44, 3343–3346 CrossRef CAS PubMed.
- C. Papadopoulou, A. Geronikaki and D. Hadjipavlou-Litina, Il Farmaco, 2005, 60, 969–973 CrossRef CAS PubMed.
- P. Chaudhary, S. Nimesh, V. Yadav, A. K. Verma and R. Kumar, Eur. J. Med. Chem., 2007, 42, 471–476 CrossRef CAS PubMed.
- A. K. Rathi, R. Syed, H.-S. Shin and R. V. Patel, Expert Opin. Ther. Pat., 2016, 26, 777–797 CrossRef CAS PubMed.
- R. Kant and S. Maji, Dalton Trans., 2021, 50, 785–800 RSC.
- A. A. El-Sherif, M. R. Shehata, M. M. Shoukry and M. H. Barakat, Spectrochim. Acta, Part A, 2012, 96, 889–897 CrossRef CAS PubMed.
- D. S. Raja, N. S. Bhuvanesh and K. Natarajan, Dalton Trans., 2012, 41, 4365–4377 RSC.
- P. J. Bednarski, F. S. Mackay and P. J. Sadler, Anti-Cancer Agents Med. Chem., 2007, 7, 75–93 CrossRef CAS PubMed.
- X. Qiao, Z.-Y. Ma, C.-Z. Xie, F. Xue, Y.-W. Zhang, J.-Y. Xu, Z.-Y. Qiang, J.-S. Lou, G.-J. Chen and S.-P. Yan, J. Inorg. Biochem., 2011, 105, 728–737 CrossRef CAS PubMed.
- M. R. Shehata, M. M. Shoukry, M. A. Mabrouk, A. Kozakiewicz and R. van Eldik, J. Coord. Chem., 2019, 72, 2035–2049 CrossRef CAS.
- M. R. Shehata, M. M. Shoukry, M. S. Ragab and R. van Eldik, Eur. J. Inorg. Chem., 2017, 2017, 1877–1887 CrossRef CAS.
- M. R. Shehata, M. M. Shoukry and M. S. Ragab, J. Mol. Struct., 2018, 1159, 216–225 CrossRef CAS.
- M. R. Shehata, M. M. Shoukry, M. A. Mabrouk and R. Van Eldik, J. Coord. Chem., 2016, 69, 522–540 CrossRef CAS.
- M. R. Shehata, Arabian J. Chem., 2019, 12, 1395–1405 CrossRef CAS.
- M. Shehate, M. Shoukry and M. Ragab, Open Chem., 2012, 10, 1253–1261 CAS.
- Z. Otwinowski and W. Minor, Methods Enzymol., 1997, 276, 307–326 CAS.
- R. H. Blessing, Acta Crystallogr., Sect. A: Found. Crystallogr., 1995, 51, 33–38 CrossRef PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Adv., 2015, 71, 3–8 CrossRef PubMed.
- C. B. Hübschle, G. M. Sheldrick and B. Dittrich, J. Appl. Crystallogr., 2011, 44, 1281–1284 CrossRef PubMed.
- M. R. Shehata, M. M. Shoukry and M. S. Ragab, Spectrochim. Acta, Part A, 2012, 96, 809–814 CrossRef CAS PubMed.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 270–283 CrossRef CAS.
- M. Frisch, G. Trucks, H. Schlegel, G. Scuseria, M. Robb, J. Cheeseman, G. Scalmani, V. Barone, B. Mennucci and G. Petersson, Int. J. Quantum Chem., 2013, 113, 2019–2039 CrossRef.
- A. Scott, in Mackie and McCartney practical medical microbiology, ed. J. Colle, J. Duguid, A. Fraser and B. Marmion, Churchill Livingstone, Edinburgh, 13 edn, 1989, vol. 2, pp. 161–181 Search PubMed.
- National Committee for Clinical Laboratory Standards, 2002, M100-S112.
- J. Carmichael, W. G. DeGraff, A. F. Gazdar, J. D. Minna and J. B. Mitchell, Cancer Res., 1987, 47(4), 936–942 CAS.
- J. M. Edmondson, L. S. Armstrong and A. O. Martinez, J. Tissue Cult. Methods, 1988, 11, 15–17 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- J. Marmur, J. Mol. Biol., 1961, 3, 208–218 CrossRef CAS.
- P. Sathyadevi, P. Krishnamoorthy, N. S. Bhuvanesh, P. Kalaiselvi, V. V. Padma and N. Dharmaraj, Eur. J. Med. Chem., 2012, 55, 420–431 CrossRef CAS PubMed.
- M. Reichmann, S. Rice, C. Thomas and P. Doty, J. Am. Chem. Soc., 1954, 76, 3047–3053 CrossRef CAS.
- A. Zianna, G. D. Geromichalos, A. G. Hatzidimitriou, E. Coutouli-Argyropoulou, M. Lalia-Kantouri and G. Psomas, J. Inorg. Biochem., 2019, 194, 85–96 CrossRef CAS PubMed.
- P. Zhao, J. Li, L.-J. Yang, J.-Z. Lu, H.-M. Guo, L.-N. Ma and B.-H. Ou, J. Coord. Chem., 2013, 66, 4220–4236 CrossRef CAS.
- M. A. Ragheb, M. A. Eldesouki and M. S. Mohamed, Spectrochim. Acta, Part A, 2015, 138, 585–595 CrossRef CAS PubMed.
- W. J. Geary, Coord. Chem. Rev., 1971, 7, 81–122 CrossRef CAS.
- M. I. García-Seijo, A. Habtemariam, S. Parsons, R. Gould and M. E. García-Fernández, New J. Chem., 2002, 26, 636–644 RSC.
- A. Pan, I. Mitra, S. Mukherjee, S. Ghosh, U. Chatterji and S. C. Moi, ACS Appl. Bio Mater., 2021, 4, 853–868 CrossRef CAS.
- B. Neelam, N. Fehmida, B. Alok, B. Sudha and A. Amir, Eur. J. Med. Chem., 2000, 35, 481–486 CrossRef CAS PubMed.
- A. B. P. Lever, in Inorganic electronic spectroscopy, Elsevier, Amsterdam, 2nd edn, 1984, vol. 33 Search PubMed.
- S. Chattopadhyay, P. Chakraborty, M. G. Drew and A. Ghosh, Inorg. Chim. Acta, 2009, 362, 502–508 CrossRef CAS.
- Z. Kantarci, T. Raci Sertbakan and E. Kasap, Spectrosc. Lett., 2005, 38, 583–594 CrossRef CAS.
- N. Nishat, M. Haq, T. Ahamad and V. Kumar, J. Coord. Chem., 2007, 60, 85–96 CrossRef CAS.
- B. Qin, J. Lin, Z. Lin, H. Ren and Y. Xue, Spectrochim. Acta, Part A, 2005, 61, 717–720 CrossRef PubMed.
- R. Walton, Spectrochim. Acta, Part A, 1965, 21, 1795–1801 CrossRef CAS.
- B. Biswal and B. Bag, Org. Biomol. Chem., 2013, 11, 4975–4992 RSC.
- L. A. Saghatforoush, F. Chalabian, A. Aminkhani, G. Karimnezhad and S. Ershad, Eur. J. Med. Chem., 2009, 44, 4490–4495 CrossRef CAS PubMed.
- L. Yang, D. R. Powell and R. P. Houser, Dalton Trans., 2007, 955–964 RSC.
- G. A. Lawrance, M. Maeder, M. Napitupulu, A. L. Nolan, M. Rossignoli, V. Tiwow and P. Turner, Inorg. Chim. Acta, 2005, 358, 3227–3235 CrossRef CAS.
- M. R. Shehata, M. M. Shoukry, M. S. Ragab and R. van Eldik, Eur. J. Inorg. Chem., 2017, 2017, 1877–1887 CrossRef CAS.
- G. A. Lawrance, M. Maeder, M. Napitupulu, A. L. Nolan, M. Rossignoli, V. Tiwow and P. Turner, Inorg. Chim. Acta, 2005, 358, 3227–3235 CrossRef CAS.
- L. Oehninger, R. Rubbiani and I. Ott, Dalton Trans., 2013, 42, 3269–3284 RSC.
- Z. Abd El-Wahab, M. M. Mashaly, A. Salman, B. El-Shetary and A. Faheim, Spectrochim. Acta, Part A, 2004, 60, 2861–2873 CrossRef PubMed.
- G. Watanabe, H. Sekiya, E. Tamai, R. Saijo, H. Uno, S. Mori, T. Tanaka, J. Maki and M. Kawase, Chem. Pharm. Bull., 2018, 66, 732–740 CrossRef CAS PubMed.
- M. Juribašić, K. Molčanov, B. Kojić-Prodić, L. Bellotto, M. Kralj, F. Zani and L. Tušek-Božić, J. Inorg. Biochem., 2011, 105, 867–879 CrossRef PubMed.
- E. H. Avdović, D. L. Stojković, V. V. Jevtić, D. Milenković, Z. S. Marković, N. Vuković, I. Potočňák, I. D. Radojević, L. R. Čomić and S. R. Trifunović, Inorg. Chim. Acta, 2019, 484, 52–59 CrossRef.
- C. Vidya Rani, M. P. Kesavan, G. G. Vinoth Kumar, M. J. D. Jeyaraj, J. Rajesh and G. Rajagopal, Appl. Organomet. Chem., 2018, 32, e4538 CrossRef.
- V. Zaitsev, A. Trofimchuk and V. Skopenko, Theor. Exp. Chem., 1984, 19, 583–587 CrossRef.
- S. Kumar, M. K. Ahmad, M. Waseem and A. K. Pandey, Med. Chem., 2015, 5, 2161 Search PubMed.
- A. R. Kapdi and I. J. Fairlamb, Chem. Soc. Rev., 2014, 43, 4751–4777 RSC.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565–4579 RSC.
- L. P. Wakelin, G. J. Atwell, G. W. Rewcastle and W. A. Denny, J. Med. Chem., 1987, 30, 855–861 CrossRef CAS PubMed.
- A. Oleksi, A. G. Blanco, R. Boer, I. Usón, J. Aymamí, A. Rodger, M. J. Hannon and M. Coll, Angew. Chem., 2006, 118, 1249–1253 CrossRef.
- R. Eshkourfu, B. Čobeljić, M. Vujčić, I. Turel, A. Pevec, K. Sepčić, M. Zec, S. Radulović, T. Srdić-Radić and D. Mitić, J. Inorg. Biochem., 2011, 105, 1196–1203 CrossRef CAS PubMed.
- E. A. Lafayette, V. de Almeida, S. Mônica, M. G. Da Rocha Pitta, E. I. Carneiro Beltrão, T. Gonçalves da Silva, R. Olímpio de Moura, I. Da Rocha Pitta, L. B. De Carvalho and M. Do Carmo Alves de Lima, Molecules, 2013, 18, 15035–15050 CrossRef CAS PubMed.
- A. A. Shoukry and M. S. Mohamed, Spectrochim. Acta, Part A, 2012, 96, 586–593 CrossRef CAS PubMed.
- H. H. Elgedany and A. A. Shoukry, Int. J. Basic Appl. Sci., 2018, 18, 1–12 Search PubMed.
- S. Bhattacharya, G. Mandal and T. Ganguly, J. Photochem. Photobiol., B, 2010, 101, 89–96 CrossRef CAS PubMed.
- S. Abedanzadeh, K. Karami, M. Rahimi, M. Edalati, M. Abedanzadeh, A. mohammad Tamaddon, M. D. Jahromi, Z. Amirghofran, J. Lipkowski and K. Lyczko, Dalton Trans., 2020, 49, 14891–14907 RSC.
- D. S. Sigman, A. Mazumder and D. M. Perrin, Chem. Rev., 1993, 93, 2295–2316 CrossRef CAS.
- K. Velugula, A. Kumar and J. P. Chinta, Inorg. Chim. Acta, 2020, 507, 119596 CrossRef CAS.
- R. Loganathan, S. Ramakrishnan, E. Suresh, A. Riyasdeen, M. A. Akbarsha and M. Palaniandavar, Inorg. Chem., 2012, 51, 5512–5532 CrossRef CAS PubMed.
- M. Howe-Grant, K. C. Wu, W. R. Bauer and S. J. Lippard, Biochemistry, 1976, 15, 4339–4346 CrossRef CAS PubMed.
- V. G. Vaidyanathan and B. U. Nair, J. Inorg. Biochem., 2002, 91, 405–412 CrossRef CAS PubMed.
- A. M. Mansour, N. T. Abdel-Ghani and M. S. Ragab, Appl. Organomet. Chem., 2020, 34, e5995 CAS.
- J. E. Repine, O. W. Pfenninger, D. W. Talmage, E. M. Berger and D. E. Pettijohn, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 1001–1003 CrossRef CAS PubMed.
- L. E. Marshall, D. R. Graham, K. A. Reich and D. S. Sigman, Biochemistry, 1981, 20, 244–250 CrossRef CAS PubMed.
- S. S. Massoud, R. S. Perkins, F. R. Louka, W. Xu, A. Le Roux, Q. Dutercq, R. C. Fischer, F. A. Mautner, M. Handa and Y. Hiraoka, Dalton Trans., 2014, 43, 10086–10103 RSC.
- N. Hasty, P. B. Merkel, P. Radlick and D. R. Kearns, Tetrahedron Lett., 1972, 13, 49–52 CrossRef.
- N. Guechtouli, S. Zaater, N. Kichou, S. Bouaziz-Terrachet and H. Meghezzi, J. Mol. Struct., 2019, 1188, 23–30 CrossRef CAS.
- A. Daina, O. Michielin and V. Zoete, Sci. Rep., 2017, 7, 1–13 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2078974. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1ra07793g |
|
| This journal is © The Royal Society of Chemistry 2022 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Mohamed R. Shehata
*a,
Mohamed R. Shehata a,
Mohamed M. Shoukrya,
Matti Haukka
a,
Mohamed M. Shoukrya,
Matti Haukka b and
Mohamed A. Ragheb
b and
Mohamed A. Ragheb c
c
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio) at 50 °C (Scheme 1). The coordination behavior of BAPP ligand towards Pd(II) ion was investigated via the X-ray crystallography, molar conductivity, IR, mass spectrometry, 1H NMR, 13C NMR and thermal studies. The complex behaves as a non-conductor species in DMF solution,48 as the molar conductance value is 9.5 μS cm−1 at 25 °C, which presumably owing to ion pair formation. Similar behavior was previously reported in [Pd(PNHP)Cl][CuCl2]49 in which (PNHP) = aminophosphine-bis[2-(diphenylphosphino)ethyl]amine. The UV-Vis spectra of [Pd(BAPP)][PdCl4] complex and its free parental ligand were recorded in DMSO. The stability of the [Pd(BAPP)][PdCl4] complex was investigated via the observation of the change of the electronic absorption spectra over time (48 h). Slight or no change (Fig. S7†) was monitored. Both BAPP ligand and [Pd(BAPP)][PdCl4] complex exhibited a sharp peak at 236 nm and 300 nm which could be assignable to the transitions of an electron from σ (bonding orbital)/σ* (antibonding) orbital and the nonbonding orbital/σ* of the amine in the BAPP ligand.50 The similarity between the absorption bands of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex reflects that the spectrum of the complex was predominated mainly by ligand transitions. Besides, only the complex displayed one broadband as a shoulder in the region of 389–489 nm due to a combination of nitrogen–Pd(II) charge transfer (L–M) and Pd(II) d–d bands51 as expected for the square planar geometry through the involvement of the BAPP ligand in the coordination to the Pd(II) ion.52 In the mass spectrum (Fig. S1†), the detected molecular ion peak at m/z 552 is convenient with the molecular formula C10H24Cl4N4Pd2 of the complex. IR spectrum of the BAPP ligand (Fig. S2†) shows a broad medium band at 3356 cm−1 belonging to the asymmetric stretching vibration mode of terminal amino groups of the diamine BAPP ligand.53 An asymmetric and symmetric stretching vibration mode of methylene group was noted at 2823 cm−1 and 2939 cm−1 of medium intensity, respectively.54,55 The bands noted at 1465 and 1157 cm−1 assigned for (C–N) asymmetric and symmetric stretching vibrations, respectively.56 Upon complexation, the band assigned for the amino group is shifted to a higher wavenumber 3449 cm−1 ascribing the coordination through the lone pair of the nitrogen atom.57 Similarly, the shift of the (C–N) asymmetric and symmetric vibrations to 1458 and 1165 cm−1, respectively, under incorporation of nitrogen atoms in the formation of [Pd(BAPP)]2+ complex cation. The proton NMR spectrum (Fig. S3†) of the investigated complex recorded in DMSO, showed a multiplet signal in the range 1.4–1.5 ppm is attributable to aliphatic –CH2.58 The signal at 2.5 ppm is particularly for the protons of –NH2. The 13C NMR showed four strong important signals which are consistent with the expected conformational symmetry of the complex. The thermogravimetric profile of the [Pd(BAPP)][PdCl4] was recorded under atmospheric condition and the obtained TG curve was shown in Fig. S8.† The TGA data imply that the complex starts the decomposition at 224 °C confirming that neither water nor solvent molecules were present in the complex.59 The thermal degradation proceeds via two stages, the first one in the temperature range 30–292 °C, derived from the mass loss of 36.70% (calcd 36.04%) corresponds to C10H24N4 moiety of the BAPP ligand. The second one in the temperature range 292–705 °C, derived from the mass loss of 24.31% (calcd 25.55%) corresponds to the loss of two Cl2 leaving two Pd metals as a thermally stable final decomposition residue.
2 molar ratio) at 50 °C (Scheme 1). The coordination behavior of BAPP ligand towards Pd(II) ion was investigated via the X-ray crystallography, molar conductivity, IR, mass spectrometry, 1H NMR, 13C NMR and thermal studies. The complex behaves as a non-conductor species in DMF solution,48 as the molar conductance value is 9.5 μS cm−1 at 25 °C, which presumably owing to ion pair formation. Similar behavior was previously reported in [Pd(PNHP)Cl][CuCl2]49 in which (PNHP) = aminophosphine-bis[2-(diphenylphosphino)ethyl]amine. The UV-Vis spectra of [Pd(BAPP)][PdCl4] complex and its free parental ligand were recorded in DMSO. The stability of the [Pd(BAPP)][PdCl4] complex was investigated via the observation of the change of the electronic absorption spectra over time (48 h). Slight or no change (Fig. S7†) was monitored. Both BAPP ligand and [Pd(BAPP)][PdCl4] complex exhibited a sharp peak at 236 nm and 300 nm which could be assignable to the transitions of an electron from σ (bonding orbital)/σ* (antibonding) orbital and the nonbonding orbital/σ* of the amine in the BAPP ligand.50 The similarity between the absorption bands of the BAPP ligand and the [Pd(BAPP)][PdCl4] complex reflects that the spectrum of the complex was predominated mainly by ligand transitions. Besides, only the complex displayed one broadband as a shoulder in the region of 389–489 nm due to a combination of nitrogen–Pd(II) charge transfer (L–M) and Pd(II) d–d bands51 as expected for the square planar geometry through the involvement of the BAPP ligand in the coordination to the Pd(II) ion.52 In the mass spectrum (Fig. S1†), the detected molecular ion peak at m/z 552 is convenient with the molecular formula C10H24Cl4N4Pd2 of the complex. IR spectrum of the BAPP ligand (Fig. S2†) shows a broad medium band at 3356 cm−1 belonging to the asymmetric stretching vibration mode of terminal amino groups of the diamine BAPP ligand.53 An asymmetric and symmetric stretching vibration mode of methylene group was noted at 2823 cm−1 and 2939 cm−1 of medium intensity, respectively.54,55 The bands noted at 1465 and 1157 cm−1 assigned for (C–N) asymmetric and symmetric stretching vibrations, respectively.56 Upon complexation, the band assigned for the amino group is shifted to a higher wavenumber 3449 cm−1 ascribing the coordination through the lone pair of the nitrogen atom.57 Similarly, the shift of the (C–N) asymmetric and symmetric vibrations to 1458 and 1165 cm−1, respectively, under incorporation of nitrogen atoms in the formation of [Pd(BAPP)]2+ complex cation. The proton NMR spectrum (Fig. S3†) of the investigated complex recorded in DMSO, showed a multiplet signal in the range 1.4–1.5 ppm is attributable to aliphatic –CH2.58 The signal at 2.5 ppm is particularly for the protons of –NH2. The 13C NMR showed four strong important signals which are consistent with the expected conformational symmetry of the complex. The thermogravimetric profile of the [Pd(BAPP)][PdCl4] was recorded under atmospheric condition and the obtained TG curve was shown in Fig. S8.† The TGA data imply that the complex starts the decomposition at 224 °C confirming that neither water nor solvent molecules were present in the complex.59 The thermal degradation proceeds via two stages, the first one in the temperature range 30–292 °C, derived from the mass loss of 36.70% (calcd 36.04%) corresponds to C10H24N4 moiety of the BAPP ligand. The second one in the temperature range 292–705 °C, derived from the mass loss of 24.31% (calcd 25.55%) corresponds to the loss of two Cl2 leaving two Pd metals as a thermally stable final decomposition residue.

![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and the crystal packing with Z = 2. In the cationic complex [Pd(BAPP)]2+, the palladium atom was situated in a slightly distorted square planar geometry, surrounded by four nitrogen atoms of the tetradentate ligand (BAPP). This distortion is indicated through the deviation from the linearity of the subtended angles around the palladium center (N1–Pd2–N4 = 171.4(3)° and N2–Pd2–N3 = 170.5(3)°). The sum of bite angle around the Pd2 atom is 360.3°. As reported recently by Houser and co-workers, the geometry of four coordinate complexes could be ascertained quantitatively through the new parameter, τ4 = [360° − (α + β)]/141° where (α = ∠N1–Pd2–N2 = 171.4(3)° and β = ∠N2–Pd2–N3 = 170.5(3)°). The value of τ4 = 0.13 confirms the distortion of square planar geometry of the complex cation.60 Despite the distortion, the environment around the palladium center is planar. The large bending in the bond angle of N1–Pd2–N2 was forced to be 73.4(2)° by the restraint of the five-membered ring. The bond lengths of the four Pd–N are Pd2–N1 = 2.052(6), Pd2–N2 = 2.054(6), Pd2–N3 = 2.053(7) and Pd2–N4 = 2.058(7), which compare well with the previously reported [Pd(BAPP)](ClO4)2 61 analogue and lies in the range of many other reported palladium amine complexes.34,62,63
and the crystal packing with Z = 2. In the cationic complex [Pd(BAPP)]2+, the palladium atom was situated in a slightly distorted square planar geometry, surrounded by four nitrogen atoms of the tetradentate ligand (BAPP). This distortion is indicated through the deviation from the linearity of the subtended angles around the palladium center (N1–Pd2–N4 = 171.4(3)° and N2–Pd2–N3 = 170.5(3)°). The sum of bite angle around the Pd2 atom is 360.3°. As reported recently by Houser and co-workers, the geometry of four coordinate complexes could be ascertained quantitatively through the new parameter, τ4 = [360° − (α + β)]/141° where (α = ∠N1–Pd2–N2 = 171.4(3)° and β = ∠N2–Pd2–N3 = 170.5(3)°). The value of τ4 = 0.13 confirms the distortion of square planar geometry of the complex cation.60 Despite the distortion, the environment around the palladium center is planar. The large bending in the bond angle of N1–Pd2–N2 was forced to be 73.4(2)° by the restraint of the five-membered ring. The bond lengths of the four Pd–N are Pd2–N1 = 2.052(6), Pd2–N2 = 2.054(6), Pd2–N3 = 2.053(7) and Pd2–N4 = 2.058(7), which compare well with the previously reported [Pd(BAPP)](ClO4)2 61 analogue and lies in the range of many other reported palladium amine complexes.34,62,63


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb confirms that the association between the complex-DNA to form an adduct is spontaneous.
Kb confirms that the association between the complex-DNA to form an adduct is spontaneous.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)


![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Po/w), gastrointestinal absorption (GI), topological polar surface area (TPSA), solubility and skin permeation (log
Po/w), gastrointestinal absorption (GI), topological polar surface area (TPSA), solubility and skin permeation (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp) of the BAPP ligand as well as the [Pd(BAPP)][PdCl4] complex were evaluated (Table S4†) by the aid of SwissADME server.93 The results indicate that the [Pd(BAPP)][PdCl4] complex has high human gastrointestinal intestinal absorption probability (GI) synchronized with no blood–brain barrier permeation which is claimed as an ideal drug candidate. The synthetic accessibility score exists in the range of (1–10) which implies the moderate synthetic accessibility of the [Pd(BAPP)][PdCl4] complex.
Kp) of the BAPP ligand as well as the [Pd(BAPP)][PdCl4] complex were evaluated (Table S4†) by the aid of SwissADME server.93 The results indicate that the [Pd(BAPP)][PdCl4] complex has high human gastrointestinal intestinal absorption probability (GI) synchronized with no blood–brain barrier permeation which is claimed as an ideal drug candidate. The synthetic accessibility score exists in the range of (1–10) which implies the moderate synthetic accessibility of the [Pd(BAPP)][PdCl4] complex.







