Open Access Article
Open Access ArticleRapid measurement of waterborne bacterial viability based on difunctional gold nanoprobe†
Junlin Wen a,
Jianbo Liua,
Jialin Wua and
Daigui He*b
a,
Jianbo Liua,
Jialin Wua and
Daigui He*b
aGuangdong Key Laboratory of Environmental Catalysis and Health Risk Control, School of Environmental Science and Engineering, Institute of Environmental Health and Pollution Control, Guangdong University of Technology, Guangzhou 510006, P. R. China
bCollege of Artificial Intelligence, Guangdong Mechanical & Electrical Polytechnic, Guangzhou 510550, P. R. China. E-mail: hedaigui@gdmec.edu.cn; Tel: +86-20-36552429; Fax: +86-20-36552429
First published on 11th January 2022
Abstract
Rapid measurement of waterborne bacterial viability is crucial for ensuring the safety of public health. Herein, we proposed a colorimetric assay for rapid measurement of waterborne bacterial viability based on a difunctional gold nanoprobe (dGNP). This versatile dGNP is composed of bacteria recognizing parts and signal indicating parts, and can generate color signals while recognizing bacterial suspensions of different viabilities. This dGNP-based colorimetric assay has a fast response and can be accomplished within 10 min. Moreover, the proposed colorimetric method is able to measure bacterial viability between 0% and 100%. The method can also measure the viability of other bacteria including Staphylococcus aureus, Shewanella oneidensis, and Escherichia coli O157H7. Furthermore, the proposed method has acceptable recovery (95.5–104.5%) in measuring bacteria-spiked real samples. This study offers a simple and effective method for the rapid measurement of bacterial viability and therefore should have application potential in medical diagnosis, food safety, and environmental monitoring.
1. Introduction
Waterborne bacterial infections are conditions caused by bacteria transmitted in water,1,2 and can lead to acute or chronic syndromes including cholera, typhoid fever and diarrhea.3,4 These waterborne diseases give rise to millions of deaths a year according to reports of the World Health Organization, and pose severe threats to public health.5,6 Living bacteria such as Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) commonly cause these waterborne diseases. Rapid measurement of waterborne bacterial viability is critical to ensure public health security.Enormous efforts have been made to detect waterborne bacterial viability. Plate culturing methods are considered the “gold standard” and are frequently used to measure bacterial viability but suffer from time-consuming operational procedures, requiring 24 to 48 hours to obtain results.7–9 Molecular methods, despite requiring less detection time, depend heavily on laborious sample preparations.10,11 Fluorescent methods employ various organic dyes to differentiate living and dead bacterial cells.12–15 Although rapid and easy to operate, these fluorescent methods rely on costly fluorescent dyes and require complex instruments such as laser scanning confocal microscopy16 or flow cytometry,17 which impede their widespread applications in resource-limited regions. Therefore, there is an urgent need to develop a rapid, simple, and low-cost method for measuring bacterial viability.
Colorimetric assays have received widespread attention due to their simpleness, fast response, and visual detection.18–20 Developing a high-performance sensing probe is the core guarantee to construct an effective colorimetric assay. Various enzyme-based probes have been used for colorimetric assays, in which different enzymes are conjugated to organic or inorganic materials and specifically generate color signals by catalyzing the oxidation of chromogenic substrates.21–23 These enzyme-based colorimetric assays commonly involve multistep incubations and washings, which are time-consuming and readily generate false positive results. Gold nanoprobes (GNPs) are gold particles with diameters between 1 and 100 nm. GNPs have unique optical properties and can change their color along with the dispersion state by means of a surface plasma resonance mechanism.24,25 Moreover, GNPs can be easily synthesized at low cost and have excellent colloidal stability.26 Consequently, it is desirable to explore a GNP-based colorimetric assay for measuring bacterial viability.
In this study, we proposed a novel colorimetric method for the rapid measurement of waterborne bacterial viability by using a difunctional GNP (dGNP). dGNPs can differentiate bacterial suspensions of different viabilities and generate color signals through surface plasma resonance. The measurement ranges of this dGNP-based method cover 0% to 100% bacterial viability. This proposed method possesses a fast response and can be accomplished within 10 min. Furthermore, the developed method has acceptable recovery when challenged with bacteria-spiked real water samples. This study advances a novel strategy for rapid measurement of waterborne bacterial viability, and is expected to have applications in the field of medical, food, and environmental science.
2. Results and discussion
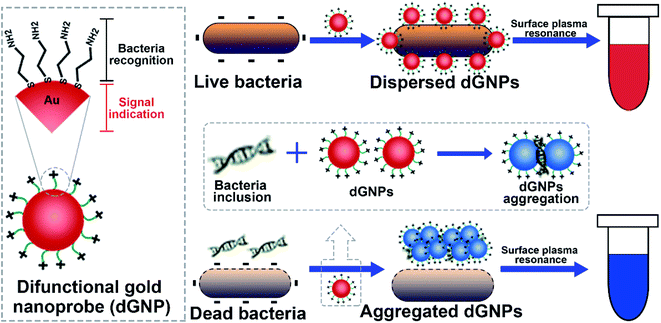
2.1. Principle of the proposed colorimetric method
Principle of the proposed colorimetric method is illustrated in Scheme 1. As shown, the dGNP is composited of bacteria recognizing part and the signal indicating part. This versatile dGNP can differentiate bacterial suspensions of different viabilities through the molecule cysteamine and generate various levels of color change via surface plasmon resonance. Living bacterial suspension with intact cells, when incubated with dGNPs, displays characteristic wine red color due to the dispersed state of dGNPs. In contrast, dead bacterial suspensions containing dGNPs develops significant color change because of the reduced interparticles distance of dGNPs induced by the released bacterial inclusions, such as nucleic acids. The intensity of the color development is positively correlated with bacterial viability, which allows quantitative measurement of bacterial viability.2.2. Characterization of the synthesized dGNP
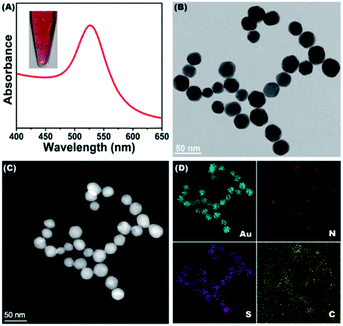
The dGNPs were prepared by the reduction of chloroauric acid with sodium borohydride in the presence of cysteamine. As shown in Fig. 1A, the synthesized dGNP is a wine-red colloid (inserted picture), which has a concentration of 1.62 nM and a strong absorbance peak at a wavelength of 528 nm. As demonstrated in the TEM image (Fig. 1B), the dGNPs are monodispersed and have an average particle size of 29.5 nm in diameter (Fig. S1†). For further characterization, EDS-coupled scanning TEM (STEM-EDS) mapping was carried out. Fig. 1C is a HAADF image of the dGNPs, where elemental mapping analysis was conducted. Fig. 1D shows the mapping of Au, N, S and C. Elemental mapping of N and S also reveals that the dGNPs are coated with cysteamine. These results demonstrated the successful synthesis of dGNPs.2.3. Method feasibility evaluation
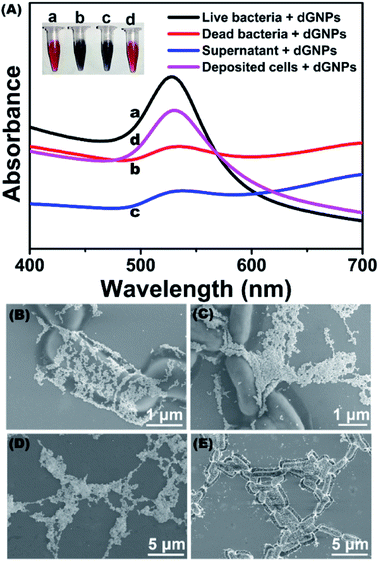
To evaluate the feasibility of the proposed method, dGNPs were incubated with E. coli K12 to perform color development. As shown in Fig. 2A, the dGNP that incubated with living bacteria remained characteristic wine red (inserted photo, tube a), while the dGNP that reacted with dead bacteria developed a significant color change (inserted photo, tube b). The dGNP/bacteria mixture was characterized with UV-vis spectroscopy. As displayed in the spectra (Fig. 2A, line a), living bacteria/dGNPs have strong absorption at 528 nm, which can be attributed to the surface plasmon resonance (SPR) absorption of dispersed dGNPs.27 GNPs have been reported to possess antibacterial properties and may cause bacterial death.28,29 However, this effect was not observed in the present study, which could be attributed to the short incubation of dGNPs with bacteria. dGNPs employed in this study were incubating with bacteria for only 3 min, comparing with 120–240 min in the literatures, which have minor harmful effect on the bacterial cells. Compared with the living bacteria, the dead bacteria exhibited a significant decrease in absorbance at 528 nm (Fig. 2A, line b), which is in accordance with previous reports.30,31Since bacterial cell and lysates have been reported to be able to develop color changes,32–34 to investigate their effect in this study, the dead bacterial suspension was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, after which the supernatant and settled cells (suspended in ultrapure water) were collected for color-developing reactions. As suggested in the inserted photo, the supernatant (tube c) had a color change, while the settled cells (tube d) remained wine-red. The difference was also observed in the UV-vis spectra (Fig. 2A, lines c and d).
000 rpm, after which the supernatant and settled cells (suspended in ultrapure water) were collected for color-developing reactions. As suggested in the inserted photo, the supernatant (tube c) had a color change, while the settled cells (tube d) remained wine-red. The difference was also observed in the UV-vis spectra (Fig. 2A, lines c and d).
The above result was further confirmed by SEM assay. The dGNPs were dispersed in the presence of living bacterial suspension (Fig. 2B) or in settled dead bacterial cells (Fig. 2E), whereas the dGNPs reacted with dead bacterial suspension (Fig. 2C) or dead bacterial supernatant (Fig. 2D) were significantly aggregated. This result indicated that color development may be mostly caused by released intracellular components. To demonstrate this, UV-vis spectroscopy was carried out. The supernatant presents a strong absorption at 260 nm (Fig. S2†), which could be attributed to the absorbance of nucleic acids.35 Agarose gel electrophoresis further confirms the presence of large number of nucleic acids in the supernatant. Nucleic acids have been reported to induce the aggregation of GNPs to develop a color-changing reaction.36–38 These results demonstrated the feasibility of the proposed method in measuring bacterial viability.
2.4. Optimization of working conditions
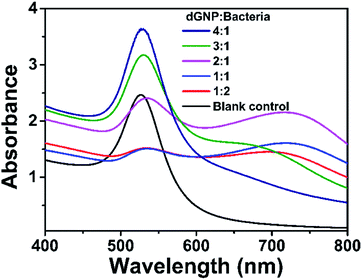
The proposed colorimetric method is based on the color development between dGNPs and bacterial suspensions. The performance of this method could be affected by the concentration of bacteria/dGNPs and color development time. Therefore, these parameters were optimized by using 1.62 nM dGNP and 1.0 × 109 CFU mL−1 dead E. coli K12.To evaluate the influence of bacteria/dGNP concentration. The ratios of bacteria/dGNP (v/v), including 1/4, 1/3, 1/2, 1/1, and 2/1, were employed in the assay. As shown in Fig. 3, the absorbance value at 528 nm decreased as the volume ratio increased from 1/4 to 1/1; thereafter, the value remained stable. The minimum absorbance value is presented at 1/1. As compared with blank control (dGNP/water, 1/1), the dGNP/bacteria at ratio 1/1 also has significant color change. Therefore, 1/1 was set as the optimized volume ratio of bacteria/dGNPs in the following study, where the final concentrations of dGNPs and bacteria were 0.81 nM and 5.0 × 108 CFU mL−1, respectively.
To investigate the effect of color development time, 5.0 × 108 CFU mL−1 dead E. coli K12 suspension and 0.81 nM dGNP were mixed and subjected to 30 s, 60 s, 90 s, 120 s, 150 s, 180 s, 210 s and 240 s of color development. As shown in Fig. S4,† the color signal decreases gradually as the color-developing time is prolonged from 30 s to 180 s, after which the signal remains almost unchanged, indicating that the color development reaches an equilibrium state. Therefore, 180 s was considered the optimal reaction time for further experiments.
2.5. Assay performance of the proposed method
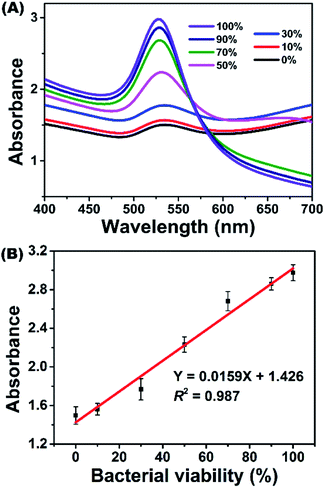
Under optimized parameters, E. coli K12 suspensions with different viabilities, which were prepared by using living and dead bacterial suspensions, were measured with the proposed colorimetric method. Fig. 4 depicts the typical response of the method challenged with E. coli K12 suspensions of 0% to 100% viability. In the UV-vis spectra, the absorbance value at 528 nm (A528) decreased gradually as bacterial viability decreased. By plotting A528 against bacterial viability, a linear relationship was observed between 0% and 100% viability (Fig. 4B). It could be described by the equation Y = 0.0159X + 1.426 with a correlation coefficient (R2) of 0.987, where Y is A528 and X represents the bacterial viability.This performance is comparable to those of colorimetric bienzyme systems39 and fluorescence spectroscopy40 but better than those of bioimaging,13,41 continuous-wave terahertz transmission imaging42 and metabolic monitoring.43 Moreover, the proposed method possesses a fast response, which could be accomplished within 10 min. This method only requires a simple apparatus and low-cost dGNP without enzymes, antibodies, or DNA/RNA probes. Beyond these advantages, this colorimetric method is homogeneous and room-temperature, which make it possible to automate by standard microplate manipulation and to implement a high-throughput assay. Furthermore, this method can also form the basis for an instrument-free quantitative assay by means of smart devices or cloud computing techniques. These results demonstrated the excellent performance of the proposed dGNP based colorimetric method toward bacterial viability measurement.
2.6. Method generality evaluation
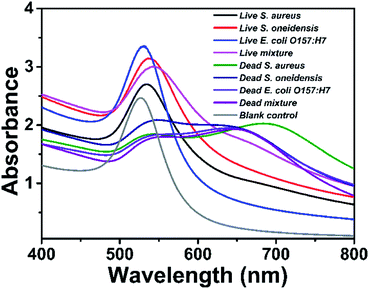
The proposed method was challenged with the other three bacteria to assess its generality. Bacterial strains such as S. aureus, S. oneidensis, and E. coli O157:H7 (1.0 × 109 CFU mL−1) were tested. As shown in Fig. 5, the absorption peaks of the tested samples are shifting between 530 nm and 542 nm dependent on the bacterial species, and all the tested bacteria observed color signal differences between the dead and living samples. Compared with blank control, the dead bacterial samples also show significant difference in color signal. Since multiple bacterial strains are commonly found in nature, mixed bacterial samples containing S. aureus, S. oneidensis, and E. coli O157:H7 were tested. The bacterial mixtures were of 0% and 100% viability. Similar to those of single bacterial species, mixed bacterial samples also showed obvious signal differences between the live and dead bacterial samples. These results demonstrates that the proposed method can be generally applied to measure the viability of other bacterial species.2.7. Real sample detection
The proposed method was investigated with bacteria-spiked water samples to test its practical application in measuring real samples. Unfiltered drinking water and tap water samples were diluted 10 times with ultrapure water and spiked with E. coli K12 to prepare bacterial suspensions with 0%, 50% and 100% viability. River water samples were treated with membrane filtration and diluted 10 times with ultrapure water before spiking with E. coli K12. The prepared bacterial suspensions were mixed with dGNP and then subjected to spectrophotometer measurement. Table S1† shows that the recoveries are between 95.5% and 104.5%, and the relative standard deviations of all water samples are less than 5%. This result demonstrated the excellent anti-referring ability of the proposed method and its potential application in measuring real water samples.3. Experimental
3.1. Reagents and materials
Chloroauric acid was purchased from Sangon Biotech Co., Ltd. (Shanghai, China). Cysteamine and sodium borohydride were provided by Macklin Biochemical Co., Ltd. (Shanghai, China). Tetramethyl-benzidine (TMB) solution was obtained from Sigma-Aldrich (St. Louis, MO). Other chemical reagents were of analytical reagent grade. All solutions were prepared using ultrapure water with a resistivity of 18.2 MΩ cm.3.2. Bacterial strain and culture
The bacterial strain E. coli K12 was purchased from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures. The bacterial glycerol stock was added to Luria-Bertani (LB) broth containing 10 g L−1 tryptone, 5 g L−1 yeast extract, and 10 g L−1 NaCl before shaking at 180 rpm in an incubator.44 After incubating overnight at 37 °C, the bacterial cultures were harvested by centrifugation at 5000g for 10 min. The deposited bacterial cells were washed three times with sterilized ultrapure water. The obtained bacterial cells were suspended in sterilized ultrapure water and diluted to an optical density at 600 nm (OD600) equal to 1.0, which contained approximately 1.0 × 109 colony forming units per milliliter (CFU mL−1). Dead bacterial suspensions were prepared by incubating the living bacterial suspensions at 100 °C for 60 min. The living and dead bacteria suspensions were all stored at 4 °C prior to measurement.3.3. Synthesis of dGNP
dGNPs were prepared according to previous reports with modifications.45,46 Briefly, an aqueous solution of cysteamine (400 μL, 213 mM) was mixed with 40 mL of 1.42 mM chloroauric acid solution in a 100 mL glass bottle. After stirring for 20 min at room temperature, sodium borohydride solution (10 mM, 10 μL) was added to the mixture, which was vigorously stirred for 30 min in the dark. The obtained wine-red colloid was stored at 4 °C prior to use.3.4. Transmission electron microscopy (TEM) assay
TEM images of the synthesized dGNPs were obtained by using an FEI Talos F200S field emission transmission electron microscope operated at 200 kV.47,48 The mapping images were acquired with energy-dispersive X-ray spectroscopy (EDS) coupled to TEM. To prepare samples for TEM imaging, the synthesized dGNPs were dropped onto a carbon-coated copper grid before drying at room temperature.3.5. Scanning electron microscopy (SEM) assay
SEM images were taken with a Hitach SU8220 field-emission scanning electron microscope at an applied voltage of 3 kV.49,50 Prior to imaging, the SEM samples were prepared by mixing the dGNPs with the bacterial samples, which were then dropped onto a conductive Si substrate and dried at room temperature.3.6. Colorimetric detection of bacterial viability
In a typical assay, 500 μL of dGNPs was mixed with 500 μL of bacterial suspension (OD600 = 1) in a 1.5 mL centrifugal tube. The mixtures containing dGNP and bacteria were left to develop color changes by incubating 3 min at room temperature. The color change of the bacteria/dGNP mixture was determined using a UV-2600 ultraviolet-visible (UV/vis) spectrometer (Shimadzu, Japan).4. Conclusions
In summary, we have proposed a novel and practical colorimetric method for rapid measurement of bacterial viability by using a dGNP. The proposed method only uses label-free dGNP, takes less than 10 min, requires a simple apparatus, is able to measure the viability of bacteria other than E. coli K12, and has favorable anti-interfering ability in measuring spiked real water samples. The method has additional benefit beyond its fast response and simplicity. The colorimetric method is homogeneous and room temperature, which makes it possible to automate by standard microplate manipulation and to implement high-throughput screening. Furthermore, we should point out that this proposed method can also form the basis for an instrument-free quantitative assay by means of smart devices and cloud computing techniques. Overall, this study advances an ingenious strategy for waterborne bacterial viability detection and should have application potential in medical diagnosis, food safety, and environmental monitoring.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2021A1515010173) and the One-Hundred Young Talents of the Guangdong University of Technology (Grant No. 1143–220413696).References
- P. K. Pandey, P. H. Kass, M. L. Soupir, S. Biswas and V. P. Singh, Contamination of water resources by pathogenic bacteria, AMB Express, 2014, 4, 51 CrossRef PubMed.
- W. Pons, I. Young, J. Truong, A. Jones-Bitton, S. McEwen, K. Pintar and A. Papadopoulos, A Systematic Review of Waterborne Disease Outbreaks Associated with Small Non-Community Drinking Water Systems in Canada and the United States, PLoS One, 2015, 10(10), e0141646 CrossRef PubMed.
- F. Y. Ramirez-Castillo, A. Loera-Muro, M. Jacques, P. Garneau, F. J. Avelar-Gonzalez, J. Harel and A. L. Guerrero-Barrera, Waterborne Pathogens: Detection Methods and Challenges, Pathogens, 2015, 4(2), 307–334 CrossRef PubMed.
- A. B. Hoyer, S. G. Schladow and F. J. Rueda, A hydrodynamics-based approach to evaluating the risk of waterborne pathogens entering drinking water intakes in a large, stratified lake, Water Res., 2015, 83, 227–236 CrossRef CAS PubMed.
- M. M. Osiemo, G. M. Ogendi and C. M'Erimba, Microbial Quality of Drinking Water and Prevalence of Water-Related Diseases in Marigat Urban Centre, Kenya, Environ. Health Insights, 2019, 13, 1178630219836988 Search PubMed.
- M. D. Kirk, S. M. Pires, R. E. Black, M. Caipo, J. A. Crump, B. Devleesschauwer, D. Dopfer, A. Fazil, C. L. Fischer-Walker, T. Hald, A. J. Hall, K. H. Keddy, R. J. Lake, C. F. Lanata, P. R. Torgerson, A. H. Havelaar and F. J. Angulo, World Health Organization Estimates of the Global and Regional Disease Burden of 22 Foodborne Bacterial, Protozoal, and Viral Diseases, 2010: A Data Synthesis, PLoS Med., 2015, 12(12), e1001921 CrossRef PubMed.
- H. X. Liu, F. F. Zhan, F. Liu, M. J. Zhu, X. M. Zhou and D. Xing, Visual and sensitive detection of viable pathogenic bacteria by sensing of RNA markers in gold nanoparticles based paper platform, Biosens. Bioelectron., 2014, 62, 38–46 CrossRef CAS PubMed.
- M. Eryilmaz, E. A. Soykut, D. Cetin, I. H. Boyaci, Z. Suludere and U. Tamer, SERS-based rapid assay for sensitive detection of Group A Streptococcus by evaluation of the swab sampling technique, Analyst, 2019, 144(11), 3573–3580 RSC.
- N. Parraga-Nino, S. Quero, A. Ventos-Alfonso, N. Uria, O. Castillo-Fernandez, J. J. Ezenarro, F. X. Munoz, M. Garcia-Nunez and M. Sabria, New system for the detection of Legionella pneumophila in water samples, Talanta, 2018, 189, 324–331 CrossRef CAS PubMed.
- J. T. Keer and L. Birch, Molecular methods for the assessment of bacterial viability, J. Microbiol. Methods, 2003, 53(2), 175–183 CrossRef CAS PubMed.
- L. Birch, C. E. Dawson, J. H. Cornett and J. T. Keer, A comparison of nucleic acid amplification techniques for the assessment of bacterial viability, Lett. Appl. Microbiol., 2001, 33(4), 296–301 CrossRef CAS PubMed.
- L. Boulos, M. Prevost, B. Barbeau, J. Coallier and R. Desjardins, Live/Dead (R) BacLight (TM): application of a new rapid staining method for direct enumeration of viable and total bacteria in drinking water, J. Microbiol. Methods, 1999, 37(1), 77–86 CrossRef CAS PubMed.
- Y. Si, C. Grazon, G. Clavier, J. Rieger, Y. Y. Tian, J. F. Audibert, B. Sclavi and R. Meallet-Renault, Fluorescent Copolymers for Bacterial Bioimaging and Viability Detection, ACS Sens., 2020, 5(9), 2843–2851 CrossRef CAS PubMed.
- J. D. Franke, A. L. Braverman, A. M. Cunningham, E. E. Eberhard and G. A. Perry, Erythrosin B: a versatile colorimetric and fluorescent vital dye for bacteria, Biotechniques, 2020, 68(1), 7–13 CrossRef CAS PubMed.
- L. Zhang, L. L. Guo, X. Shan, X. H. Lin, T. T. Gu, J. K. Zhang, J. L. Ge, W. D. Li, H. X. Ge, Q. Jiang and X. H. Ning, An elegant nitroreductase responsive fluorescent probe for selective detection of pathogenic Listeria in vitro and in vivo, Talanta, 2019, 198, 472–479 CrossRef CAS PubMed.
- X. Y. Jing, X. Liu, C. S. Deng, S. S. Chen and S. G. Zhou, Chemical signals stimulate Geobacter soli biofilm formation and electroactivity, Biosens. Bioelectron., 2019, 127, 1–9 CrossRef CAS PubMed.
- A. D. Cabral, N. Rafiei, E. D. de Araujo, T. B. Radu, K. Toutah, D. Nino, B. I. Murcar-Evans, J. N. Milstein, D. Kraskouskaya and P. T. Gunning, Sensitive Detection of Broad-Spectrum Bacteria with Small-Molecule Fluorescent Excimer Chemosensors, ACS Sens., 2020, 5(9), 2753–2762 CrossRef CAS PubMed.
- H. N. Kim, W. X. Ren, J. S. Kim and J. Yoon, Fluorescent and colorimetric sensors for detection of lead, cadmium, and mercury ions, Chem. Soc. Rev., 2012, 41(8), 3210–3244 RSC.
- B. Pejcic, P. Eadington and A. Ross, Environmental monitoring of hydrocarbons: A chemical sensor perspective, Environ. Sci. Technol., 2007, 41(18), 6333–6342 CrossRef CAS PubMed.
- K. Shrivas, R. Shankar and K. Dewangan, Gold nanoparticles as a localized surface plasmon resonance based chemical sensor for on-site colorimetric detection of arsenic in water samples, Sens. Actuators, B, 2015, 220, 1376–1383 CrossRef CAS.
- D. R. Chang, S. Zakaria, S. E. Samani, Y. Y. Chang, C. D. M. Filipe, L. Soleymani, J. D. Brennan, M. Liu and Y. F. Li, Functional Nucleic Acids for Pathogenic Bacteria Detection, Acc. Chem. Res., 2021, 54(18), 3540–3549 CrossRef CAS PubMed.
- C. Bayrac, F. Eyidogan and H. A. Oktem, DNA aptamer-based colorimetric detection platform for Salmonella Enteritidis, Biosens. Bioelectron., 2017, 98, 22–28 CrossRef CAS PubMed.
- O. R. Miranda, X. N. Li, L. Garcia-Gonzalez, Z. J. Zhu, B. Yan, U. H. F. Bunz and V. M. Rotello, Colorimetric Bacteria Sensing Using a Supramolecular Enzyme-Nanoparticle Biosensor, J. Am. Chem. Soc., 2011, 133(25), 9650–9653 CrossRef CAS PubMed.
- D. A. Giljohann, D. S. Seferos, W. L. Daniel, M. D. Massich, P. C. Patel and C. A. Mirkin, Gold Nanoparticles for Biology and Medicine, Angew. Chem., Int. Ed., 2010, 49(19), 3280–3294 CrossRef CAS PubMed.
- S. Hu, P. J. Huang, J. Wang and J. Liu, Dissecting the Effect of Salt for More Sensitive Label-Free Colorimetric Detection of DNA Using Gold Nanoparticles, Anal. Chem., 2020, 92(19), 13354–13360 CrossRef CAS PubMed.
- R. A. Sperling, P. Rivera gil, F. Zhang, M. Zanella and W. J. Parak, Biological applications of gold nanoparticles, Chem. Soc. Rev., 2008, 37(9), 1896–1908 RSC.
- I. H. El-Sayed, X. H. Huang and M. A. El-Sayed, Surface plasmon resonance scattering and absorption of anti-EGFR antibody conjugated gold nanoparticles in cancer diagnostics: Applications in oral cancer, Nano Lett., 2005, 5(5), 829–834 CrossRef CAS PubMed.
- Y. Cui, Y. Zhao, Y. Tian, W. Zhang, X. Lu and X. Jiang, The molecular mechanism of action of bactericidal gold nanoparticles on Escherichia coli, Biomaterials, 2012, 33(7), 2327–2333 CrossRef CAS PubMed.
- B. Lee and D. G. Lee, Synergistic antibacterial activity of gold nanoparticles caused by apoptosis-like death, J. Appl. Microbiol., 2019, 127(3), 701–712 CrossRef CAS PubMed.
- Y. Choi, N. H. Ho and C. H. Tung, Sensing phosphatase activity by using gold nanoparticles, Angew. Chem., Int. Ed., 2007, 46(5), 707–709 CrossRef CAS PubMed.
- H. B. Dong, F. Zou, X. J. Hu, H. Zhu, K. Koh and H. X. Chen, Analyte induced AuNPs aggregation enhanced surface plasmon resonance for sensitive detection of paraquat, Biosens. Bioelectron., 2018, 117, 605–612 CrossRef CAS PubMed.
- M. S. Verma, J. L. Rogowski, L. Jones and F. X. Gu, Colorimetric biosensing of pathogens using gold nanoparticles, Biotechnol. Adv., 2015, 33(6), 666–680 CrossRef CAS PubMed.
- M. S. Verma, P. Z. Chen, L. Jones and F. X. Gu, Branching and size of CTAB-coated gold nanostars control the colorimetric detection of bacteria, RSC Adv., 2014, 4(21), 10660–10668 RSC.
- Y. Mao, T. T. Fan, R. Gysbers, Y. Tan, F. Liu, S. Lin and Y. Y. Jiang, A simple and sensitive aptasensor for colorimetric detection of adenosine triphosphate based on unmodified gold nanoparticles, Talanta, 2017, 168, 279–285 CrossRef CAS PubMed.
- Y. G. Ke, S. Lindsay, Y. Chang, Y. Liu and H. Yan, Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays, Science, 2008, 319(5860), 180–183 CrossRef CAS PubMed.
- P. Miao and Y. G. Tang, Gold Nanoparticles-Based Multipedal DNA Walker for Ratiometric Detection of Circulating Tumor Cell, Anal. Chem., 2019, 91(23), 15187–15192 CrossRef CAS PubMed.
- F. Zhang, S. Y. Wang and J. W. Liu, Gold Nanoparticles Adsorb DNA and Aptamer Probes Too Strongly and a Comparison with Graphene Oxide for Biosensing, Anal. Chem., 2019, 91(22), 14743–14750 CrossRef CAS PubMed.
- J. L. Wen, J. H. Chen, L. Zhuang and S. G. Zhou, Designed diblock hairpin probes for the nonenzymatic and label-free detection of nucleic acid, Biosens. Bioelectron., 2016, 79, 656–660 CrossRef CAS PubMed.
- J. H. Sun, J. Huang, A. R. Warden and X. T. Ding, Real-time detection of foodborne bacterial viability using a colorimetric bienzyme system in food and drinking water, Food Chem., 2020, 320, 126581 CrossRef CAS PubMed.
- F. Ou, C. McGoverin, S. Swift and F. Vanholsbeeck, Rapid and cost-effective evaluation of bacterial viability using fluorescence spectroscopy, Anal. Bioanal. Chem., 2019, 411(16), 3653–3663 CrossRef CAS PubMed.
- Y. X. Song, H. Li, F. Lu, H. B. Wang, M. L. Zhang, J. J. Yang, J. Huang, H. Huang, Y. Liu and Z. H. Kang, Fluorescent carbon dots with highly negative charges as a sensitive probe for real-time monitoring of bacterial viability, J. Mater. Chem. B, 2017, 5(30), 6008–6015 RSC.
- X. Yang, J. Shi, Y. Y. Wang, K. Yang, X. Zhao, G. Y. Wang, D. G. Xu, Y. X. Wang, J. Q. Yao and W. L. Fu, Label-free bacterial colony detection and viability assessment by continuous-wave terahertz transmission imaging, J. Biophotonics, 2018, 11(8), e201700386 Search PubMed.
- S. Ayyash, W. I. Wu and P. R. Selvaganapathy, Fast and Inexpensive Detection of Bacterial Viability and Drug Effectiveness through Metabolic Monitoring, Sensors, 2016, 16(11), 22–25 CrossRef PubMed.
- D. D. Smith, D. Girodat, H. J. Wieden and L. B. Selinger, Streamlined purification of fluorescently labeled Escherichia coli phosphate-binding protein (PhoS) suitable for rapid-kinetics applications, Anal. Biochem., 2017, 537, 106–113 CrossRef CAS PubMed.
- X. M. Miao, Z. Y. Cheng, H. Y. Ma, Z. B. Li, N. Xue and P. Wang, Label-Free Platform for MicroRNA Detection Based on the Fluorescence Quenching of Positively Charged Gold Nanoparticles to Silver Nanoclusters, Anal. Chem., 2018, 90(2), 1098–1103 CrossRef CAS PubMed.
- Y. Jv, B. X. Li and R. Cao, Positively-charged gold nanoparticles as peroxidiase mimic and their application in hydrogen peroxide and glucose detection, Chem. Commun., 2010, 46(42), 8017–8019 RSC.
- Y. Li, M. Yang, M. Pentrak, H. He and Y. Arai, Carbonate-Enhanced Transformation of Ferrihydrite to Hematite, Environ. Sci. Technol., 2020, 54(21), 13701–13708 CrossRef CAS PubMed.
- F. Xie, L. Ma, M. Y. Gan, H. M. He, L. Q. Hu, M. H. Jiang and H. H. Zhang, One-pot construction of the carbon spheres embellished by layered double hydroxide with abundant hydroxyl groups for Pt-based catalyst support in methanol electrooxidation, J. Power Sources, 2019, 420, 73–81 CrossRef CAS.
- R. Hou, C. Luo, S. F. Zhou, Y. Wang, Y. Yuan and S. G. Zhou, Anode potential-dependent protection of electroactive biofilms against metal ion shock via regulating extracellular polymeric substances, Water Res., 2020, 178, 115845 CrossRef CAS PubMed.
- X. R. Zhang, Y. Q. Wang, Y. H. Du, M. Qing, F. Yu, Z. Q. Tian and P. K. Shen, Highly active N,S co-doped hierarchical porous carbon nanospheres from green and template-free method for super capacitors and oxygen reduction reaction, Electrochim. Acta, 2019, 318, 272–280 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra07287k |
| This journal is © The Royal Society of Chemistry 2022 |