Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Introducing the penicillamine moiety into a metallopeptide mimicking the NiSOD enzyme: electronic and kinetic effects†
Dóra
Bonczidai-Kelemen
a,
Giuseppe
Sciortino
 b,
Nóra V.
May
c,
Eugenio
Garribba
b,
Nóra V.
May
c,
Eugenio
Garribba
 d,
István
Fábián
d,
István
Fábián
 ae and
Norbert
Lihi
ae and
Norbert
Lihi
 *ae
*ae
aDepartment of Inorganic and Analytical Chemistry, University of Debrecen, H-4032, Debrecen, Hungary. E-mail: lihi.norbert@science.unideb.hu
bInstitute of Chemical Research of Catalonia (ICIQ), The Barcelona Institute of Technology, 43007 Tarragona, Spain
cCentre for Structural Science, Research Centre for Natural Sciences, H-1117, Budapest, Hungary
dDipartimento di Scienze Mediche, Chirurgiche e Sperimentali, Università di Sassari, I-07100 Sassari, Italy
eMTA-DE Redox and Homogeneous Catalytic Reaction Mechanisms Research Group, University of Debrecen, H-4032, Debrecen, Hungary
First published on 1st December 2021
Abstract
Multidisciplinary protein design is reported on a novel metallopeptide mimicking the binding loop of nickel containing SOD enzyme (NiSOD). D-Penicillamine, a natural decomposition product of penicillin, was introduced into the peptide chain yielding the H(Pen)DLPCGLY (wtPen) peptide. The nickel(II) binding ability of wtPen was characterized by thermodynamic (pH-potentiometry), spectroscopic (UV-Vis, CD, MS, NMR) and computational techniques (full DFT and Molecular Dynamics methods). Oxidation of the Ni(II) complex by KO2 yields a square pyramidal Ni(III) species coordinated by the axial His-N in a well-defined α-helix folding state. The structure of the Ni(III) species was analyzed by EPR spectroscopy and theoretical methods confirming that the donor set involved in the metal ion coordination and the folding state are retained after oxidation. The complex exhibits superior SOD activity which was studied by sequential stopped-flow method. Thorough analysis of the data shows that the Ni(III) species rapidly accumulates in the nickel catalyzed decomposition of superoxide anion. Accordingly, the presence of the penicillamine moiety close to the catalytic center increases the life-time of the Ni(III) transient species. In contrast, Ni(III) exists only at relatively low concentration level in the dismutation reaction catalyzed by the native NiSOD enzyme fragment.
Introduction
Protein design via the systematic active site tailoring approach may provide deeper insight into the mechanism of enzyme catalysis. The catalytic reaction can satisfactorily be tuned by exploring the structure–function relationships, the enzyme mechanism in substrate recognition and the kinetic features of the process. Consequently, the controlled design of proteins is a subject of great interest due to their widespread application in industrial biotechnology and chemical synthesis.1–5 Peptides have also been extensively studied in these reactions due to the easier control of the folding processes compared to proteins.6 These compounds are easily accessible via the routinely used solid phase synthetic methods. Peptides and proteins mimicking the active site of oxygenases have been intensively studied because their controlled regio- and stereochemical features have essential role in the oxygenation processes.7–9Superoxide dismutases (SOD) are a dedicate class of enzyme family which are capable of catalyzing the decomposition of superoxide anion radical in the extracellular matrix.10–12 The dismutation reaction yields molecular dioxygen and hydrogen peroxide that degrades through various pathways in further reactions catalyzed by catalase or glutathione peroxidase to yield harmful products.13,14 The absence of these enzymes results in an elevated level of superoxide anion concentration which can cause oxidative stress, the formation of reactive oxygen species (ROS) and extensive cellular damage.15,16 Several studies have also reported that intensive ROS generation may act as a mediator of a number of inflammatory diseases.17–20
Copper/zinc, manganese and iron containing SOD enzymes have been characterized by several methods including structural, equilibrium and computational analysis.10,21 So far, only limited information is available on the peptide-based controlled design for the relatively novel class of Ni-containing SOD enzymes (NiSOD).22–25
The NiSOD is expressed by sodN gene and found in several Streptomyces and cyanobacteria.26,27 The structure of the NiSOD obtained from X-ray crystallography revealed a unique metal coordination environment, where the first six amino acid residues exhibit a loop. This is the so-called NiSOD binding motif. For the reduced form, nickel(II) is accommodated by the binding of the terminal amino group, the first peptide nitrogen and the thiolate of cysteine resulting in a (5,5)-linked chelate system. This coordination mode is supported by macrochelation due to the distant Cys residue. The (2N,2S) coordination environment is featured by square planar structure and diamagnetic behavior. The oxidation of the nickel(II) complex yields a paramagnetic nickel(III) species, its formation is being accompanied by an axial ligation which originates from the binding of the imidazole-N of histidine. Earlier studies have demonstrated that the first six amino acid residues (HCDLPC, in the amino acid one letter code) are the minimal peptide sequence to model both the coordination mode and catalytic cycle of NiSOD enzymes.23,28–31 It was also demonstrated that model complexes are capable of assisting the decomposition of superoxide anion O2− and that the redox cycle also leads to the oxidation of the cysteine residues.
In our previous works, detailed equilibrium, spectroscopic and catalytic activities were reported on nickel complexes formed with NiSOD related peptides.30,32,33 Our results and previous literature studies confirmed the essential role of both cysteine residues in the dismutation of superoxide ion.34,35 The combined application of pH-potentiometry with spectroscopic methods revealed that the cysteine residue in the second position of the peptide chain is crucial to induce spin pairing of nickel(II), while the distant cysteine affects the redox potential of the Ni(II)/Ni(III) couple.32 We have also studied the coordination ability and catalytic features of multiple NiSOD binding loops and oxidation of the cysteine residues was observed similarly to those reported earlier. The results revealed that the binuclear nickel complex exhibits superior SOD activity, the Ni(III) species rapidly accumulates and is removed through a fast degradation process; consequently, the nickel assisted catalysis ceases.33 The oxidative stability of a set of NiSOD related metallopeptides was studied and correlated with their catalytic activities.36 It was confirmed by stopped-flow kinetic method that one of the key factors is the replacement of carboxamide group for secondary amine in the nickel coordination sphere. Such an alternation leads to improved catalytic activity and higher sensitivity for oxidation compared to the model metallopeptides investigated in that study.36 On the basis of these results, it is an intriguing issue how the active form(s) of the catalyst can be stabilized to improve the efficiency in the dismutation of superoxide anion.
In this paper, we report thorough equilibrium, spectroscopic, computational and SOD activity studies on the new metallopeptide containing the NiSOD binding motif (denoted as wtPen throughout the text, Scheme 1). Its sequence is H(Pen)DLPCGLY, where the first cysteine was replaced by penicillamine which features two-electrons donating methyl groups. This study demonstrates that both the steric and electronic features of penicillamine may alter the activity of the catalyst during the decomposition of superoxide anion.
 | ||
| Scheme 1 Structural formulae of the NiSOD binding loop (wtSOD) and the peptide investigated in this study (wtPen). Mutation on the nonapeptide backbone is highlighted by red color. | ||
Results and discussion
Protonation equilibria of wtPen were investigated by using pH-potentiometry and the acid dissociation constants are collected in Table 1. The protonation processes were also followed by UV-vis spectroscopy which offers the possibility to determine the individual spectra of each species involved in the acid–base processes (Fig. S1–S4†). Details on the acid–base character are reported in the ESI.†![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ki) of the peptides and the equilibrium constants (log
Ki) of the peptides and the equilibrium constants (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kpqr) of the nickel(II) complexes formed with wtPen and wtSOD (L)a
Kpqr) of the nickel(II) complexes formed with wtPen and wtSOD (L)a
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ki Ki |
wtPen | wtSOD | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kpqr Kpqr |
wtPen | wtSOD | |
|---|---|---|---|---|---|---|
| Species | Species | |||||
a 3σ standard deviations are indicated in parentheses. I = 0.2 M KCl, T = 298 K.
b Data are taken from ref. 30.
c Equilibrium constant for the deprotonation of amide N. log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K(N−) = log K(N−) = log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β(NiHL) − log β(NiHL) − log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β(NiL). β(NiL).
|
||||||
| [H6L]2+ | 3.78(5) | 3.58 | [NiH3L]+ | 4.87(2) | 4.76 | |
| [H5L]+ | 5.42(7) | 5.48 | [NiHL]− | 5.40(1) | 5.61 | |
| [H4L] | 7.09(6) | 7.20 | [NiL]2− | 6.86(3) | 6.52 | |
| [H3L]− | 8.26(6) | 8.25 | [NiH−1L]3− | 9.89(4) | 9.26 | |
| [H2L]2− | 9.15(6) | 8.92 | [NiH−2L]4− | 11.21(5) | ||
| [HL]3− | 10.07(5) | 9.88 | ||||
∑log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ki Ki |
43.77 | 43.31 | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K(N−)c K(N−)c |
6.86 | 6.52 | |
| Fitted pH range | 2.6–11.6 | Fitted pH range | 3.9–11.3 | |||
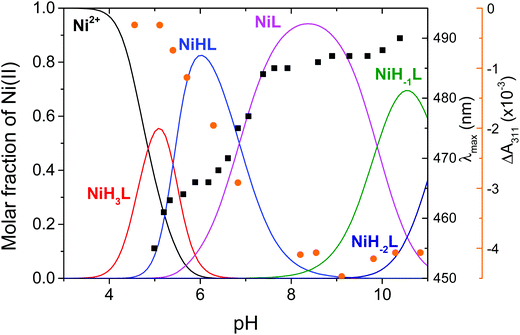
The overall stability constants of the nickel(II)–wtPen complexes were obtained by pH-potentiometric method and are reported in Table 1 and Table S1,† while the corresponding concentration distribution of the complexes as a function pH is shown in Fig. 1.
In general, the complex formation processes of wtPen show similarities to those reported in the nickel(II)/wtSOD system.30 The complex formation reactions between nickel(II) and the peptide start in the slightly acidic pH range with the [NiH3L]+ species, where L indicate wtPen. In this complex, nickel(II) is accommodated by the (NH2,NIm) donor groups (histamine-like coordination) in an octahedral crystal field. This feature is confirmed by the lack of the d–d absorption between 400–500 nm in the UV-vis spectra. The equilibrium constant for the formation of histamine-like coordination mode is calculated, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KHistamine = 5.93, which is close to the log
KHistamine = 5.93, which is close to the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K of histamine-like coordination of [NiH3L]+ complex of wtSOD (log
K of histamine-like coordination of [NiH3L]+ complex of wtSOD (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K = 6.39).30 The agreement of these stability constants strongly supports the same coordination mode and confirms that the carboxylate group of aspartic acid residue – which is already deprotonated in this pH range – does not have any significant contribution to the metal binding.
K = 6.39).30 The agreement of these stability constants strongly supports the same coordination mode and confirms that the carboxylate group of aspartic acid residue – which is already deprotonated in this pH range – does not have any significant contribution to the metal binding.
Upon increasing the pH, a further base consumption process yields the [NiHL]− complex, while the formation of [NiH2L] can be excluded on the basis of pH-potentiometric data, Fig. S1.† The formation of [NiHL]− exhibits cooperative feature and is accompanied by characteristic changes both in the CD and UV-vis spectra (Fig. 2 and Fig. S5†). An intense band was observed in the UV-vis spectra at 465 nm which proves that the complex has a square planar geometry and indicates that the amide nitrogen of the peptide backbone is involved in the metal binding.
 | ||
Fig. 2 CD spectra of the Ni(II)/wtPen 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 system as a function of pH. The spectra were recorded between pH 4.56 and 11.58. Inset: Selected part of the d–d transition. cNi(II) = 2 mM. 1 system as a function of pH. The spectra were recorded between pH 4.56 and 11.58. Inset: Selected part of the d–d transition. cNi(II) = 2 mM. | ||
In parallel, the CD spectra exhibit a new band at 270 nm. In this wavelength range, the Ni(II)–imidazole and Ni(II)–N− charge transfer bands are characteristic. In addition, the CD spectra exhibit a band at 330 nm which confirms the binding of one of the thiolate groups. On the basis of the spectroscopic data, the (NH2,NIm,N−,S−(Cys)) coordination mode is attributed to [NiHL]−, although a weak axial interaction is expected between the Ni(II) and imidazole-N of histidine (vide infra).
This coordination mode features a (5,6)-membered chelate system which is supported by the macrochelation of the distant cysteinyl residue. Additional base consumption process provides the active structure of NiSOD due to the binding of the thiolate group of penicillamine residue. The well resolved CD spectra exhibit a new band at 311 nm with negative Cotton-effect which is a clear evidence for the binding of the penicillamine moiety, e.g.D-penicillamine was used in the synthesis and the introduction of methyl substituents of penicillamine results in higher excitation energies than that of cysteine. Such a feature cannot be observed in the UV-vis spectra, these transitions collapse into a single absorption band with maximum at 340 nm. Therefore, the formation of this complex can selectively be followed by CD spectroscopy at 310 nm and the good correlation between ΔA and the species distribution further confirms the proposed coordination mode (Fig. 1). This [NiL]2− complex has a stoichiometry of NiH−1L(H) where the tyrosine residue remains protonated and is dominant at the physiological pH-range. The UV-vis and CD spectra provide further evidence for the formation of (NH2,N−,S−,S−) coordinated species, although the spectroscopic parameters differ from those observed for the wild-type fragment of NiSOD.37 This is most probably due to the presence of penicillamine moiety in the coordination sphere which significantly affects the electronic features of the complex. The coordination mode was proved by NMR spectroscopy. In Fig. S6,† the 1H–13C HSQC spectra are shown in the presence and absence of nickel(II) at pH 7.8. The addition of nickel(II) causes a large shift of several NMR signals. Drastic effect was observed on the methyl groups of penicillamine moiety which is presumably due to the binding of the thiolate group and amide nitrogen of the peptide backbone. In addition, the His-CH2 signals are affected by the deprotonation and coordination of peptide nitrogen. These results are consistent with the binding of one of the amide nitrogen atoms from the peptide backbone and the thiolate group of penicillamine.
To corroborate the exclusive existence of this complex, ESI-TOF-MS experiments were carried out. The MS spectrum recorded in the Ni(II)/wtPen 0.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 system is shown in Fig. S7.† The major peaks are attributed to the mononuclear nickel(II) complex [NiL]2− (
1 system is shown in Fig. S7.† The major peaks are attributed to the mononuclear nickel(II) complex [NiL]2− (![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) [NiH−1L(H)]2−) with m/z = 543.1789, to the adduct with sodium ([NiH−1L + Na+]2−) with m/z = 554.1699, and to the ligand L− with m/z = 1031.4450. The good agreement between all of the observed and calculated m/z values supports the existence of the postulated species (Table S2 and Fig. S8†). In addition, the MS spectra do not exhibit any indication for the formation of dimeric or bis-ligand species. Further deprotonation processes lead to the formation of [NiH−1L]3− and [NiH−2L]4− complexes. Since these species are relevant in strongly alkaline solutions only, their complex formation equilibria are discussed in the ESI, Fig. S9.†
[NiH−1L(H)]2−) with m/z = 543.1789, to the adduct with sodium ([NiH−1L + Na+]2−) with m/z = 554.1699, and to the ligand L− with m/z = 1031.4450. The good agreement between all of the observed and calculated m/z values supports the existence of the postulated species (Table S2 and Fig. S8†). In addition, the MS spectra do not exhibit any indication for the formation of dimeric or bis-ligand species. Further deprotonation processes lead to the formation of [NiH−1L]3− and [NiH−2L]4− complexes. Since these species are relevant in strongly alkaline solutions only, their complex formation equilibria are discussed in the ESI, Fig. S9.†
The structure, the effect of the metal binding on the peptide conformation and the electronic absorption spectrum of the major complex present under physiological pH in the Ni(II)/wtPen system, [NiL]2− (![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) [NiH−1L(H)]2−), were elucidated with a multistep modelling strategy. The results were compared with those obtained for similar species of wtSOD. The approach consisted in: (i) build a starting model for both the metal free and mononuclear Ni(II)–wtSOD and Ni(II)–wtPen peptide fragments (a.a. 1–9) based on the X-ray structure of Ni(II)–SOD; (ii) carry out a MD simulation in order to find their most stable conformation in solution and elucidate the effect of Ni(II) coordination on the folding; (iii) optimise such conformation at full-DFT theory level; and (iv) predict its electronic vertical transitions comparing the results with the experimental bands.
[NiH−1L(H)]2−), were elucidated with a multistep modelling strategy. The results were compared with those obtained for similar species of wtSOD. The approach consisted in: (i) build a starting model for both the metal free and mononuclear Ni(II)–wtSOD and Ni(II)–wtPen peptide fragments (a.a. 1–9) based on the X-ray structure of Ni(II)–SOD; (ii) carry out a MD simulation in order to find their most stable conformation in solution and elucidate the effect of Ni(II) coordination on the folding; (iii) optimise such conformation at full-DFT theory level; and (iv) predict its electronic vertical transitions comparing the results with the experimental bands.
The metal free peptides display the behaviour of typical intrinsically disordered peptides with continuous transition between folded and unfolded states with a main extended conformation. Upon Ni(II) coordination, the metal induces a significant loss of flexibility. Cluster analysis performed on the full length MD experiments shows one predominant conformation in which the square planar Ni(II) coordination geometry is highly retained and a mainly α-helix conformation is adopted by residues 3–9. Overall, the results highlight that both models are stable during 200 ns of the MD and thus they were selected for subsequent DFT analysis. The representative frames of the most sampled conformations for wtSOD (the native fragment of NiSOD without metal coordination), Ni(II)–wtSOD and Ni(II)–wtPen species are reported in Fig. 3a–c. It is plausible that, after the deprotonation of first peptide-NH and SH of Pen and Cys, the interaction of NIm changes from an equatorial to an axial position.
DFT optimization of the structures confirms the square planar Ni(II) coordination geometry with no binding of NIm of His residue. The final Ni(II)–donor bond lengths are: Ni–NH2 is 2.007 Å (2.008 Å for wtSOD), Ni–N− 1.895 Å (1.901 Å), Ni–SPen2− 2.190 Å (Ni–SCys2− 2.213 Å), Ni–SCys6− 2.250 Å (2.261 Å), while the apical Ni–NHis1 distance is very long, 4.168 Å (4.129 Å). The Cartesian coordinates of the corresponding complexes are collected in Tables S3 and S4.† Noticeably, the introduction of penicillamine moiety results in a shorter bond distance between nickel(II) and the thiolate group in second position than that calculated for wtSOD.
The predicted spectra at TD-DFT theory level are qualitatively in agreement with the experimental results at physiological pH for [NiL]2−. In the case of Ni(II)–wtPen, the principal fitted bands display their maxima between 332 and 502 nm, composed by clear d–d transitions from internal occupied MOs with metal character to the Ni-dx2−y2 LUMO orbital. The two intense transitions around 300 nm are assigned to metal-to-ligand or ligand-to-metal charge transfers (MLCT and LMCT). The analysis and the list of the calculated vertical transitions for the [NiL]2− species formed in the Ni(II)/wtPen and Ni(II)/wtSOD systems are reported in Tables S5 and S6 of the ESI.†
Cyclic voltammetry (CV) and differential pulse voltammetry (DPV) were used to obtain electrochemical data of the Ni(II)-wtPen and Ni(II)-wtSOD complexes at pH 7.6 (Fig. S10†). The cyclic voltammogram shows irreversible feature for these species (the anodic peak occurs at 0.591 V and 0.428 V versus Ag/AgCl for Ni(II)-wtPen and Ni(II)-wtSOD, respectively). The anodic peaks clearly confirm that the two complexes show somewhat different redox properties.
The formation of nickel(III) transient species was followed by EPR spectroscopy. KO2 stabilized in 18-crown-6 was used as an agent to oxidize in situ the corresponding nickel(II) complexes. The EPR spectra are shown in Fig. 4 and the spin-Hamiltonian parameters are collected in Table S7.†
 | ||
| Fig. 4 X-band EPR spectra recorded at 77 K in the Ni(II)/wtPen system after in situ oxidation (black) and the simulated EPR spectra (red). | ||
The EPR spectra unambiguously prove that the oxidation of nickel(II) occurs, and the unpaired electron is located in the dz2 orbital resulting in a gx ∼ gy > gz ∼ ge relationship between the g values.38,39 The interaction between the unpaired electron of nickel(III) and one nitrogen donor with nuclear spin I = 1 yields a three-band superhyperfine splitting centered around g ∼ 2. Although such square pyramidal coordination environment features the oxidized form of wild-type fragment of NiSOD,28,30 the EPR parameters of wtPen differ.
To predict the conformation of the metallopeptides and the spin Hamiltonian EPR parameters of monomeric Ni(III)–wtSOD and Ni(III)–wtPen complexes, a computational protocol similar to that described for the nickel(II) complexes was followed (vide infra). The g and A tensors of 63Ni and 14N nuclei were computed with the functional BP and basis set 6-311 g(d,p) after a validation on a small benchmark consisting of four square planar, square pyramidal and octahedral Ni(III) complexes formed by maleonitriledithiolato, N,N′-disalicylidene-4,5-dichloro-1,2-phenylenediamine and pyridine ligands (see Tables S8–S12†). MD analyses unveil stable square pyramidal Ni(III) coordination geometry and, particularly for Ni(III)–wtPen, more flexible structures from mainly folded to extended conformations of the residues 3–9. The representative frames of the most sampled conformation for Ni(III)–wtSOD and Ni(III)–wtPen are reported in Fig. S11† and their Cartesian coordinates are summarized in Tables S13–S16.†
Considering the flexibility displayed by the oxidized form of the metallated peptides, the fast oxidation times and by analysing the most sampled conformations, the α-helix folding derived from the previous simulations was also selected for DFT analysis.
The energy order of the Ni-based MOs, Ni-dxy ∼ Ni-dxz ∼ Ni-dyz < Ni-dz2 < Ni-dx2−y2, agrees well with the prediction of the ligand field theory.40 Analysis of the MOs composition and spin density shows that the unpaired electron is mainly on the Ni-dz2 orbital, in line with the order of the g values (see Table S7†). This MO is shown in Fig. S12 of the ESI.†
On the optimized structures, the spin Hamiltonian parameters were computed and compared with the EPR experimental results (Table 2). The highest spin density was found on the Ni atom confirming the Ni(III) oxidation state. Moreover, the equatorial nitrogen donors, NH2 and N−, display negligible contribution to the superhyperfine constant ANz, in line with literature results for other Cu(II)41 and Ni(III) complexes.42–46 Therefore, the superhyperfine resonances observed in the EPR spectra (Fig. 4) with wtPen were attributed to the axial Nim of His1 residue. Such an observation for Ni(III) can be explained assuming that the delocalisation of the spin density to the imidazole ring occurs through the MO that describes the coordinative bond formed by Ni-dz2 and sp2 orbital on NIm. This reinforces the finding that for such systems the unpaired electron is located on Ni-dz2 orbital. In contrast, the delocalization on the coordinated equatorial nitrogens is not possible and the spin density on them is almost negligible. This accounts for the small, lower than 3 × 10−4 cm−1 values of AN in these systems. Analysis of the total spin density (α–β) shows that the unpaired electron is localized on Ni and NIm atoms (Fig. 5).
 | ||
| Fig. 5 Total (α–β) spin density (isovalue: 0.007) plot for Ni(III)-wtPen species computed for the DFT energy minimum. | ||
| Species (folding) |
A
N
z
exptl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b,c b,c |
A
N
z
calcd![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b,d b,d |
g exptl x |
g
calcd
x
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e e |
g exptl y |
g
calcd
y
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e e |
g exptl z |
g
calcd
z
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e e |
|---|---|---|---|---|---|---|---|---|
| a Calculations carried out at BP/6-311 g(d,p) theory level with ORCA 4.0 (see ESI† for selection of the method). b Value in 10−4·cm−1. c A N z for the axial His1-N donor. d Percent deviation in parenthesis, calculated as [(ANzcalcd − ANzexptl)/ANzexptl] × 100. e Percent deviation calculated as [(gcalcdi − gexptli)/gexptli] × 100 with i = x, y, z. f Representative structure of the most populated cluster. g α-Helix structure optimized from the Ni(II) MDs. | ||||||||
| Ni(III)–wtSOD (extended)f | 23.7 | 11.6 (−51.1) | 2.289 | 2.148 (−6.2) | 2.220 | 2.124 (−4.3) | 2.012 | 2.020 (0.4) |
| Ni(III)–wtSOD (α-helix)g | 23.9 (0.8) | 2.165 (−5.4) | 2.151 (−3.1) | 2.019 (0.3) | ||||
| Ni(III)–wtPen (extended)f | 22.1 | 12.0 (−45.7) | 2.257 | 2.164 (−4.1) | 2.251 | 2.142 (−4.8) | 2.013 | 2.020 (0.3) |
| Ni(III)–wtPen (α-helix)g | 23.4 (5.9) | 2.164 (−4.1) | 2.150 (−4.5) | 2.020 (0.3) | ||||
The comparison of the experimental and the computed spin Hamiltonian parameters (Table 2) indicates that α-helix folding state is retained in the transient Ni(III) species. In fact, the deviations from the experimental values are huge for the extended conformations (∼50%), suggesting that no fast conformational changes are induced upon the 1e− oxidation process. The coordination environment around Ni(III) is square pyramidal. In contrast with Ni(II), the axial interaction with NIm of His1 residue is strong with bond lengths of 2.075 and 2.073 Å for wtPen and wtSOD. The other distances in the DFT optimized structures are: Ni–NH2 is 2.033 Å (2.034 Å for wtSOD), Ni–N− 1.934 Å (1.934 Å), Ni–SPen2− 2.203 Å (2.220 Å), and Ni–SCys6− 2.281 Å (2.277 Å). The optimized geometries accounting for the experimental EPR parameters are reported in Fig. 6.
 | ||
| Fig. 6 DFT optimized conformations (B97D) accounting for the experimental EPR parameters of Ni(III)–wtSOD (left, orange backbone) and Ni(III)–wtPen (right, blue backbone). | ||
In a dedicated stopped-flow experiment, the Ni(II)-wtPen complex was mixed with KO2 in DMSO to produce the corresponding Ni(III) complex. The progress of the reaction was monitored in a stopped-flow experiment using the PDA detector which offers a possibility to record the electronic absorption spectra of the species. Mixing the Ni(II)-wtPen complex with equimolar KO2 solution rapidly yields the corresponding Ni(III) complex which exhibits characteristic absorption at around 385 nm that is assigned to S/N(π) → Ni(dπ*) charge transfer band according to earlier literature data (Fig. S13†).47 Since only a few studies are reported on the electronic absorption spectra of Ni(III) complexes, the MO's involved in the transitions were analysed by further TD-DFT calculations. The principal fitted band displays a maximum at around 340 nm in good agreement with the experimental observations. It is composed by several LMCT transitions with the most intense involving one-electron promotion from internal occupied MOs mainly localized on His1, Cys6 and Asp3 residues to the Ni-dx2−y2 based LUMO or LUMO+1 orbitals. Other LMCT transitions involve excitations with main LHis1/Asp3/Cys6/Tyr9 → Ni-dx2−y2 character (see Table S17†).
Stability of the metallopeptides
Earlier studies indicated that the addition of KO2, O2 or H2O2 to the reduced form of metallopeptides leads to substantial sulfur-based oxidation processes yielding sulfenate, sulfinate, sulfonate, cysteine-S-monoxide or cysteine-S-dioxide products.36,48 The Ni(II)-wtPen and wtSOD complexes remain intact when they are exposed to air for 12 hours. In contrast, drastic changes were observed in the UV-Vis spectra of these species upon the addition of one H2O2 equivalent (Fig. S14†). However, this degradation process occurs on a much longer time scale than the catalytic dismutation reaction.Catalytic activity
The SOD activity of the nickel complex of wtPen was studied by monitoring the decay of the superoxide anion at 260 nm in water/DMSO 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solvent mixture and the results were compared with those obtained for the nickel complex of wtSOD. Sequential stopped-flow measurements were used to avoid the spectral disturbances. Details of the experimental protocol are reported in our previous paper.33
1 solvent mixture and the results were compared with those obtained for the nickel complex of wtSOD. Sequential stopped-flow measurements were used to avoid the spectral disturbances. Details of the experimental protocol are reported in our previous paper.33
First, the spontaneous decomposition of the superoxide anion was investigated in the absence of catalyst. Moreover, the initial absorbance of the kinetic traces allows us to determine the concentration of superoxide anion after the incubation time (40 s). A simple second-order decay was observed in the spontaneous decomposition of superoxide anion and the second order rate constant, k1 = (3.84 ± 0.03) × 104 M−1 s−1, is in excellent agreement with that reported earlier.49
In the presence of the NiSOD mimic, the absorbance drops very fast from its initial value (ca. 0.5) to below 0.3 in the first measured point (at 2 ms) of the kinetic traces. This proves that a substantial part (40%) of the reaction proceeds within the dead-time of the instrument (ca. 1.5 ms). Since the non-catalyzed decay of superoxide occurs on considerably longer time-scale, these results unambiguously prove that the nickel complex of wtPen exhibits superior SOD activity. Unfortunately, the kinetic traces cannot be fitted with a simple first-order expression assuming steady-state conditions for the NiIII complex. In reality, the absorbance steadily changes in a slow process, that can only be explained by considering that the nickel assisted dismutation of superoxide kinetically couples with the degradation of the complex. The postulated reaction scheme is shown in Scheme S1.† The model incorporates the spontaneous decomposition of the superoxide anion (k1), the dismutation cycle (k2, k3) and the degradation of the NiIII complex (k4). Since the NiII complex is stable in the absence of oxidant, it is reasonable to assume that the degradation occurs in the NiIII complex. In this species, NiIII may be reduced in an intramolecular redox step, and the nickel assisted dismutation ceases yielding an unidentified nickel complex (Ni*). The kinetic model includes reactions (1)–(4) and the corresponding differential equation system is given by eqn (5)–(9).
| 2H+ + 2O2− → H2O2 + O2 k1 | (1) |
| NiIIL + 2H+ + O2− → NiIIIL + H2O2 k2 | (2) |
| NiIIIL + O2− → NiIIL + O2 k3 | (3) |
| NiIIIL → Ni*L k4 | (4) |
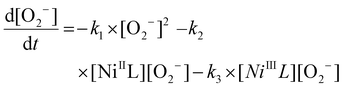 | (5) |
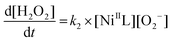 | (6) |
 | (7) |
 | (8) |
 | (9) |
The experimental data were evaluated by fitting the absorbance of each kinetic trace (at 260 and 376 nm) on the basis of this differential equation system by using a non-linear least-squares approach.50 At 260 nm, there are several absorbing species, and the absorbance is expressed by eqn (10):
 | (10) |
The kinetic traces were also evaluated at 376 nm where the spectral contribution of superoxide anion is negligible, and the absorbance is expressed by eqn (11):
| Abs = εNi(II) × [NiIIL] + εNi(III) × [NiIIIL] + εNi* × [Ni*L] | (11) |
Since the intramolecular redox degradation (eqn (4)) significantly affects the dismutation cycle, the process was independently examined at comparable concentrations of the catalyst and superoxide anion at 260 and 376 nm using constant complex concentration. Under such conditions, the pseudo-first order approach is not applicable for reactions (2) and (3). The corresponding kinetic traces are shown in Fig. 7 and Fig. S15.†
Evaluation of the data yields the first-order rate constant for the degradation of the NiIII complex, k4 = 292 ± 2 s−1 and the molar absorptivities of NiIII and Ni* complexes at 260 nm, εNi(III) = (2.58 ± 0.02) × 104 M−1 cm−1, εNi* = (1.57 ± 0.06) × 104 M−1 cm−1, respectively. Noticeably, the estimated first-order rate constants at 260 and 374 nm show excellent agreement supporting the postulated kinetic model. In the final fitting process, k3 was allowed to float, while k2 was fixed at 3 × 108 M−1 s−1 and k1 as well as the molar absorptivities of the NiII/III and Ni* complexes, O2− and H2O2 were involved with fixed values obtained from independent experiments. The strong cross-correlation between k2 and k3 does not allow simultaneous fitting of these parameters. However, the best fit (e.g. considering the smallest standard deviation) was observed when k2 was fixed at 3 × 108 M−1 s−1 or higher. Below this threshold, unrealistic results were obtained. The same experimental and evaluation protocol were used for wtSOD (Fig. S16†), when simultaneous fitting of k2 and k3 was feasible. The results are collected in Table 3 and the corresponding kinetic traces are shown in Fig. 8.
| Parameter | wtPen | wtSOD | Unit |
|---|---|---|---|
| Value | Value | ||
| k 1 | (3.84 ± 0.03) × 104 | (3.84 ± 0.03) × 104 | M−1 s−1 |
| k 2 | >3.0 × 108 | (9.6 ± 0.3) × 107 | M−1 s−1 |
| k 3 | (3.15 ± 0.04) × 107 | (1.72 ± 0.08) × 108 | M−1 s−1 |
| k 4 | 292 ± 2 | 258 ± 2 | s−1 |
| ε Ni(II) | (1.99 ± 0.02) × 104 | (1.48 ± 0.06) × 104 | M−1 cm−1 |
| ε Ni(III) | (2.58 ± 0.02) × 104 | (2.44 ± 0.09) × 104 | M−1 cm−1 |
| ε Ni* | (1.57 ± 0.06) × 104 | (1.8 ± 0.1) × 104 | M−1 cm−1 |
| ε O2− | 2686 | 2686 | M−1 cm−1 |
| ε H2O2 | 38 | 38 | M−1 cm−1 |
It needs to be emphasized that most of the fitted parameters are expected to be pH dependent, therefore the reported values are applicable only at pH 7.6 in water (50 mM HEPES)/DMSO 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solvent mixture. Developing a more appropriate kinetic model for the enzyme activity under biological conditions would require experiments in aqueous solution instead of water/DMSO mixture and the use of a much broader concentration range of the reactants. However, experimental limitations prevent such studies. Most prominently, the preparation of stable aqueous O2− is not feasible. In spite of the noted shortcomings, the results presented here offer an excellent possibility for comparing the SOD activities of the NiSOD models.
1 solvent mixture. Developing a more appropriate kinetic model for the enzyme activity under biological conditions would require experiments in aqueous solution instead of water/DMSO mixture and the use of a much broader concentration range of the reactants. However, experimental limitations prevent such studies. Most prominently, the preparation of stable aqueous O2− is not feasible. In spite of the noted shortcomings, the results presented here offer an excellent possibility for comparing the SOD activities of the NiSOD models.
Fig. 9 demonstrates how the concentration of each species varies during the dismutation process. For wtPen, the results clearly show that the oxidation of nickel(II) (k2) is considerably faster than the reduction of the corresponding nickel(III) complex (k3). Therefore, the nickel(III) complex rapidly accumulates after the initiation of the reaction. During this period of time, a substantial amount of the superoxide anion undergoes dismutation (ca. 80%) but eventually the decay of the O2− concentration becomes considerably slower. In contrast, oxidation of the nickel(II) complex of wtSOD is slower than the reduction process, thus the NiII form is always in excess over the oxidized form. This kinetic feature yields a more effective catalytic activity than that observed for the wtPen. The results unambiguously show that the presence of electron donating methyl substituents stabilizes the nickel(III) species which makes the dismutation less favorable.
The results confirm that the disproportionation of the superoxide anion can be controlled by modifying the active site of the catalyst. Unfortunately, the formation of the nickel(III) complex is followed by intramolecular redox degradation yielding the unidentified Ni* species and, consequently, the active form of the catalyst is removed.
Conclusion
Modification on the peptide sequence of NiSOD binding loop by using the natural decomposition product of the antibiotic penicillin yields a new metallopeptide containing D-penicillamine moiety. The excellent metal binding ability is due to the coordination of (NH2,N−,SPen2−,SCys6−) donor set which corresponds to the coordination environment in the reduced form of NiSOD enzyme. Oxidation of the Ni(II) complex yields a Ni(III) species with a stable square pyramidal coordination mode which is due to the binding of the apical histidine and α-helix folding state. Both the EPR parameters and superhyperfine resonances were analyzed by DFT computations and the results confirmed that the unpaired electron is localized on Ni and NIm atoms.The metallopeptide exhibits superior catalytic activity in the decomposition of superoxide anion. The decomposition reaction was studied both under catalytic and second-order conditions. This kinetic approach made possible to establish a multi-step kinetic model which incorporates the decomposition of the superoxide anion, the dismutation cycle and the degradation of the catalyst. We unequivocally confirmed that the metallopeptide efficiently catalyzes the dismutation process, and that the NiIII form undergoes redox self-degradation. This side-reaction significantly alters the interpretation of dismutation activity of NiSOD related metallopeptides. The concentration profiles as a function of time clearly show that the superoxide anion is quickly dismutated in the presence of the catalysts. Simultaneous intramolecular redox decomposition of the Ni(III) form of the catalyst eventually ceases the dismutation process.
Earlier studies have reported contradictory conclusions on the role of axial ligands and the hydrogen bond system surrounding the catalytically active center of NiSOD models. It has been proposed that the axial coordination of histidine moieties is not required in the metal assisted degradation of superoxide, i.e. an alternative degradation process might exist in the absence of an apical donor group.36 However, it has also been demonstrated that the presence of an axial ligand in the coordination sphere of nickel(III) enhances the rate of the superoxide disproportionation reaction by affecting the redox activity of the NiII/NiIII redox couple.23 Buntkowsky et al. have demonstrated that the hydrogen bond network orients the imidazole ring of histidine and helps the stabilization of the Ni(III) oxidation state.36 Other authors suggested that the hydrogen bonds protect the sulfur atoms of cysteine moieties and enhance the robustness of these groups against oxidative damage of O2 or H2O2.51,52,53 By introducing penicillamine moiety, the formation of hydrogen bond network can be hindered via the bulky methyl substituents. The reaction between the Ni-wtPen complex and superoxide accumulates the NiIII species which is involved in a fast self degradation process. Therefore, we propose an alternative role of hydrogen bond network which can involve the stabilization of the NiIII oxidation state toward redox degradation processes. This explains the observed kinetic features, suggesting that an alternative superoxide degradation process may proceed via a different path in the Ni/wtPen system.
Finally, the presence of the penicillamine moiety (i.e. the electron donating substituents of methyl groups) close to the catalytic center is capable to increase the life-time of the Ni(III) transient species. However, the results clearly show that the following features have to be controlled in an effective dismutation reaction: (i) the concentration of the Ni(III) species needs to be kept at relatively low concentration, (ii) the intramolecular redox degradation process needs to be prevented. Further protein designing may solve these issues.
Experimental and computational section
Equilibrium studies
The protonation constants (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ki) of the ligands and the overall stability constants (log
Ki) of the ligands and the overall stability constants (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) βpqr) were determined by pH-potentiometric titration method. The pH reading was converted to hydrogen ion concentration as described by Irving et al.54 Protonation constants and the overall stability constants, βpqr = [NipHqLr]/[Ni]p[H]q[L]r of the nickel(II) complexes were calculated by using the computational programs, SUPERQUAD55 and PSEQUAD.56
βpqr) were determined by pH-potentiometric titration method. The pH reading was converted to hydrogen ion concentration as described by Irving et al.54 Protonation constants and the overall stability constants, βpqr = [NipHqLr]/[Ni]p[H]q[L]r of the nickel(II) complexes were calculated by using the computational programs, SUPERQUAD55 and PSEQUAD.56
Spectroscopic techniques
UV-visible spectra of the nickel(II) complexes were recorded with an Agilent Technologies Cary 60 UV-VIS spectrophotometer between 200 and 800 nm using the same concentration range as in the pH-potentiometric titrations. The circular dichroism spectra were registered with a Jasco J-810 spectropolarimeter using 1 mm and/or 1 cm cells in the 250–800 nm wavelength range. The individual spectra of the complexes were calculated by solving the overdetermined linear equation system with Matlab on the basis of the estimated stability constants.57 All CW-EPR spectra were recorded with a BRUKER EleXsys E500 spectrometer. The measured spectra were simulated using the EPR program designated by Rockenbauer and Korecz.58 Rhombic g-tensor have been used for the simulation of the components. Orientation dependent linewidths were considered. ESI-TOF-MS measurements were carried out with a Bruker maXis II MicroTOF-Q type Qq-TOF-MS instrument (Bruker Daltonik, Bremen, Germany) in negative mode. 1D 1H and 2D (1H–13C HSQC) NMR spectra were recorded on a Bruker Avance I 400 spectrometer equipped a BB inverse z gradient probe at 298 K.Computational methods
The molecular modelling approach consisted in a multistep strategy including Molecular Dynamics (MD), Density Functional Theory (DFT) and Time-Dependent-Density Functional Theory (TD-DFT) calculations. (i) The starting models for both the metal free and mononuclear Ni(II)/Ni(III)–wtSOD and Ni(II)/Ni(III)–wtPen peptide fragments (a.a. 1–9) were obtained from the X-ray structure of Ni(II)–SOD and Ni(III)–SOD (PDB code 1T6U). (ii) MD simulations in TIP3P periodic water box along 200 ns were carried out with OpenMM66 package in order to find their most stable conformation in solution and elucidate the effect of Ni coordination on the folding. Ni-Bonding force constants and equilibrium parameters were obtained through the Seminario method, using the MCPB.py protocol.59 The trajectories convergence was assessed by RMSI, RMSF, counting clustering method and PCA analysis.60 (iii) The most stable conformation was optimized at full-DFT theory level with Gaussian 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 61 using the functional B97D in continuum model for water. (iv) The UV-vis electronic vertical transitions of the optimized complexes were computed at TD-DFT theory level using the HSE06 functional in continuum model for water according to the method established previously.62 (v) For the Ni(III)–wtSOD and Ni(III)–wtPen peptide fragments the g and A tensors of 63Ni and 14N nuclei were computed through the method implemented into the ORCA package (vers, 4.0),63,64 with the functional BP.65 The method was further calibrated in this work toward a dataset of four Ni(III) complexes (Table S8†). All the predicted spectroscopic parameters were compared with the experimental data, either to confirm the conformation of the peptide fragments or to clarify their electronic structure properties. See the ESI† for further technical details.
61 using the functional B97D in continuum model for water. (iv) The UV-vis electronic vertical transitions of the optimized complexes were computed at TD-DFT theory level using the HSE06 functional in continuum model for water according to the method established previously.62 (v) For the Ni(III)–wtSOD and Ni(III)–wtPen peptide fragments the g and A tensors of 63Ni and 14N nuclei were computed through the method implemented into the ORCA package (vers, 4.0),63,64 with the functional BP.65 The method was further calibrated in this work toward a dataset of four Ni(III) complexes (Table S8†). All the predicted spectroscopic parameters were compared with the experimental data, either to confirm the conformation of the peptide fragments or to clarify their electronic structure properties. See the ESI† for further technical details.
Electrochemistry
Electrochemical measurements (cyclic voltammetry (CV) and differential pulse voltammetry (DPV)) were performed using a BASI Epsilon Eclipse potentiostat (Bioanalytical Systems Inc., West Lafayette, USA) equipped with a three-electrode arrangement that consists of a platinum wire auxiliary electrode (ALS Co. Japan), a glassy carbon electrode (CHI104) and a Ag/AgCl/3 M KCl reference electrode.SOD activity measurements
The catalytic activities of the Ni(II) complexes in the decomposition of O2− and the formation of Ni(III) transient species and further nickel complexes were studied using an Applied Photophysics SX-20 stopped-flow instrument equipped with a photomultiplier tube as the detector.Author contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the text.Conflicts of interest
There are no conflicts to declare.Acknowledgements
N. L. and I. F. are grateful for the financial support of the Hungarian National Research, Development and Innovation Office (NKFIH PD-128326 and K-124983). The research was also financed by the EU and co-financed by the European Regional Development Fund (under the projects GINOP-2.3.2-15-2016-00008). E. G. and G. S. thank Regione Autonoma della Sardegna (grant RASSR79857) and Fondazione di Sardegna (FdS2017Garribba) for financial support. G.S. also thank Spanish MICINN' Juan de la Cierva program, FJC2019-039135-I. This work is also supported by the ÚNKP-21-4-II New National Excellence Program of the Ministry for Innovation and Technology from the source of the National, Research, Development and Innovation Fund and prepared with the professional support of the Doctoral Student Scholarship Program of the Co-operative Doctoral Program of the Ministry of Innovation and Technology financed from the National Research, Development and Innovation Office.References
- I. Khan and M. Waheed Akhtar, Different Approaches For Protein Engineering In Industrial Biotechnology, Nat. Prec., 2011 DOI:10.1038/npre.2011.5601.1.
- D. J. Mikolajczak, J. Scholz and B. Koksch, Tuning the Catalytic Activity and Substrate Specificity of Peptide-Nanoparticle Conjugates, ChemCatChem, 2018, 10, 5665–5668 CrossRef CAS.
- S. P. Godehard, C. P. S. Badenhorst, H. Müller and U. T. Bornscheuer, Protein Engineering for Enhanced Acyltransferase Activity, Substrate Scope, and Selectivity of the Mycobacterium smegmatis Acyltransferase MsAcT, ACS Catal., 2020, 10, 7552–7562 CrossRef CAS.
- R. Fasan, Tuning P450 Enzymes as Oxidation Catalysts, ACS Catal., 2012, 2, 647–666 CrossRef CAS.
- N. Ma, W. Fang, C. Liu, X. Qin, X. Wang, L. Jin, B. Wang and Z. Cong, Switching an Artificial P450 Peroxygenase into Peroxidase via Mechanism-Guided Protein Engineering, ACS Catal., 2021, 11, 8449–8455 CrossRef CAS.
- T. Giraud, S. Bouguet-Bonnet, P. Marchal, G. Pickaert, M.-C. Averlant-Petit and L. Stefan, Improving and fine-tuning the properties of peptide-based hydrogels via incorporation of peptide nucleic acids, Nanoscale, 2020, 12, 19905–19917 RSC.
- P. C. Cirino and F. H. Arnold, Protein engineering of oxygenases for biocatalysis, Curr. Opin. Chem. Biol., 2002, 6, 130–135 CrossRef CAS.
- D. R. Boyd, N. D. Sharma and C. C. R. Allen, Aromatic dioxygenases: molecular biocatalysis and applications, Curr. Opin. Biotechnol., 2001, 12, 564–573 CrossRef CAS.
- Z. Li, J. B. van Beilen, W. A. Duetz, A. Schmid, A. de Raadt, H. Griengl and B. Witholt, Oxidative biotransformations using oxygenases, Curr. Opin. Chem. Biol., 2002, 6, 136–144 CrossRef CAS PubMed.
- I. Fridovich, in Encyclopedia of Biological Chemistry, ed. M. D. Lane, Elsevier, New York, 2004, pp. 135–138 Search PubMed.
- C. R. Kliment, J. M. Tobolewski, M. L. Manni, R. J. Tan, J. Enghild and T. D. Oury, Extracellular superoxide dismutase protects against matrix degradation of heparan sulfate in the lung, Antioxid. Redox Signaling, 2008, 10, 261–268 CrossRef CAS PubMed.
- S. K. Sah, G. Agrahari and T.-Y. Kim, Insights into superoxide dismutase 3 in regulating biological and functional properties of mesenchymal stem cells, Cell Biosci., 2020, 10, 22 CrossRef PubMed.
- I. A. Abreu and D. E. Cabelli, Superoxide dismutases—a review of the metal-associated mechanistic variations, Biochim. Biophys. Acta, 2010, 1804, 263–274 CrossRef CAS PubMed.
- Y. Sheng, I. A. Abreu, D. E. Cabelli, M. J. Maroney, A.-F. Miller, M. Teixeira and J. S. Valentine, Superoxide Dismutases and Superoxide Reductases, Chem. Rev., 2014, 114, 3854–3918 CrossRef CAS PubMed.
- S. W. Ragsdale, Nickel-based Enzyme Systems, J. Biol. Chem., 2009, 284, 18571–18575 CrossRef CAS PubMed.
- D. M. Kurtz, Microbial Detoxification of Superoxide: The Non-Heme Iron Reductive Paradigm for Combating Oxidative Stress, Acc. Chem. Res., 2004, 37, 902–908 CrossRef CAS PubMed.
- H. Wiseman and B. Halliwell, Damage to DNA by reactive oxygen and nitrogen species: role in inflammatory disease and progression to cancer, Biochem. J., 1996, 313, 17–29 CrossRef CAS PubMed.
- T. Nishikawa and E. Araki, Impact of Mitochondrial ROS Production in the Pathogenesis of Diabetes Mellitus and Its Complications, Antioxid. Redox Signaling, 2007, 9, 343–353 CrossRef CAS PubMed.
- P. Newsholme, V. F. Cruzat, K. N. Keane, R. Carlessi and P. I. H. de Bittencourt Jr., Molecular mechanisms of ROS production and oxidative stress in diabetes, Biochem. J., 2016, 473, 4527–4550 CrossRef CAS PubMed.
- A. Umeno, V. Biju and Y. Yoshida, In vivo ROS production and use of oxidative stress-derived biomarkers to detect the onset of diseases such as Alzheimer's disease, Parkinson's disease, and diabetes, Free Radical Res., 2017, 51, 413–427 CrossRef CAS.
- A.-F. Miller, Superoxide dismutases: active sites that save, but a protein that kills, Curr. Opin. Chem. Biol., 2004, 8, 162–168 CrossRef CAS PubMed.
- H. D. Youn, E. J. Kim, J. H. Roe, Y. C. Hah and S. O. Kang, A novel nickel-containing superoxide dismutase from Streptomyces spp, Biochem. J., 1996, 318, 889–896 CrossRef CAS PubMed.
- K. P. Neupane, K. Gearty, A. Francis and J. Shearer, Probing Variable Axial Ligation in Nickel Superoxide Dismutase Utilizing Metallopeptide-Based Models: Insight into the Superoxide Disproportionation Mechanism, J. Am. Chem. Soc., 2007, 129, 14605–14618 CrossRef CAS PubMed.
- M. Schmidt, S. Zahn, M. Carella, O. Ohlenschläger, M. Görlach, E. Kothe and J. Weston, Solution Structure of a Functional Biomimetic and Mechanistic Implications for Nickel Superoxide Dismutases, ChemBioChem, 2008, 9, 2135–2146 CrossRef CAS.
- J. Domergue, P. Guinard, M. Douillard, J. Pécaut, O. Proux, C. Lebrun, A. Le Goff, P. Maldivi, P. Delangle and C. Duboc, A Bioinspired NiII Superoxide Dismutase Catalyst Designed on an ATCUN-like Binding Motif, Inorg. Chem., 2021, 60, 12772–12780 CrossRef CAS.
- J. Wuerges, J.-W. Lee, Y.-I. Yim, H.-S. Yim, S.-O. Kang and K. D. Carugo, Crystal structure of nickel-containing superoxide dismutase reveals another type of active site, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8569–8574 CrossRef CAS.
- B. Palenik, B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb and J. Waterbury, The genome of a motile marine Synechococcus, Nature, 2003, 424, 1037–1042 CrossRef CAS PubMed.
- J. Shearer and L. M. Long, A Nickel Superoxide Dismutase Maquette That Reproduces the Spectroscopic and Functional Properties of the Metalloenzyme, Inorg. Chem., 2006, 45, 2358–2360 CrossRef CAS PubMed.
- J. Shearer, Use of a Metallopeptide-Based Mimic Provides Evidence for a Proton-Coupled Electron-Transfer Mechanism for Superoxide Reduction by Nickel-Containing Superoxide Dismutase, Angew. Chem., Int. Ed., 2013, 52, 2569–2572 CrossRef CAS PubMed.
- N. Lihi, G. Csire, B. Szakács, N. V. May, K. Várnagy, I. Sóvágó and I. Fábián, Stabilization of the Nickel Binding Loop in NiSOD and Related Model Complexes: Thermodynamic and Structural Features, Inorg. Chem., 2019, 58, 1414–1424 CrossRef CAS PubMed.
- G. Csire, A. Kolozsi, T. Gajda, G. Pappalardo, K. Várnagy, I. Sóvágó, I. Fábián and N. Lihi, The ability of the NiSOD binding loop to chelate zinc(II): the role of the terminal amino group in the enzymatic functions, Dalton Trans., 2019, 48, 6217–6227 RSC.
- N. Lihi, D. Kelemen, N. V. May and I. Fábián, The Role of the Cysteine Fragments of the Nickel Binding Loop in the Activity of the Ni(II)-Containing SOD Enzyme, Inorg. Chem., 2020, 59, 4772–4780 CrossRef CAS PubMed.
- D. Kelemen, N. V. May, M. Andrási, A. Gáspár, I. Fábián and N. Lihi, High Enzyme Activity of a Binuclear Nickel Complex Formed with the Binding Loops of the NiSOD Enzyme**, Chem. – Eur. J., 2020, 26, 16767–16773 CrossRef CAS.
- V. Pelmenschikov and P. E. M. Siegbahn, Nickel Superoxide Dismutase Reaction Mechanism Studied by Hybrid Density Functional Methods, J. Am. Chem. Soc., 2006, 128, 7466–7475 CrossRef CAS PubMed.
- J. Shearer, Insight into the Structure and Mechanism of Nickel-Containing Superoxide Dismutase Derived from Peptide-Based Mimics, Acc. Chem. Res., 2014, 47, 2332–2341 CrossRef CAS PubMed.
- D. Tietze, J. Sartorius, B. Koley Seth, K. Herr, P. Heimer, D. Imhof, D. Mollenhauer and G. Buntkowsky, New insights into the mechanism of nickel superoxide degradation from studies of model peptides, Sci. Rep., 2017, 7, 17194 CrossRef PubMed.
- A. T. Fiedler, P. A. Bryngelson, M. J. Maroney and T. C. Brunold, Spectroscopic and Computational Studies of Ni Superoxide Dismutase: Electronic Structure Contributions to Enzymatic Function, J. Am. Chem. Soc., 2005, 127, 5449–5462 CrossRef CAS PubMed.
- W.-Z. Lee, C.-W. Chiang, T.-H. Lin and T.-S. Kuo, A Discrete Five-Coordinate NiIII Complex Resembling the Active Site of the Oxidized Form of Nickel Superoxide Dismutase, Chem. – Eur. J., 2012, 18, 50–53 CrossRef CAS PubMed.
- E. Garribba and G. Micera, The Determination of the Geometry of Cu(II) Complexes: An EPR Spectroscopy Experiment, J. Chem. Educ., 2006, 83, 1229–1232 CrossRef CAS.
- Y. Jean, Molecular Orbitals of Transition Metal Complexes, Oxford University Press, Oxford, 2005 Search PubMed.
- B. R. McGarvey, Theory of the Spin Hamiltonian Parameters for Low Spin Cobalt(II) Complexes, Can. J. Chem., 1975, 53, 2498–2511 CrossRef CAS.
- E. S. Gore and D. H. Busch, Stable octahedral, low-spin nickel(III) complexes of a tetradentate macrocyclic ligand having saturated nitrogen donors, Inorg. Chem., 1973, 12, 1–3 CrossRef CAS.
- J. M. Bemtgen, H. R. Gimpert and A. Von Zelensky, Nickel(III) complexes of 3,9-dimethyl-4,8-diazaundeca-3,8-diene-2,10-dione dioxime with two axial halide ligands. ESR spectra, electronic spectra, and ligand-exchange kinetics, Inorg. Chem., 1983, 22, 3576–3580 CrossRef CAS.
- A. Desideri and J. B. Raynor, Electron spin resonance spectroscopy of some dianiono(1,4,8,11-tetra-azacyclotetradecane)-iron(III) and -nickel(III) salts, J. Chem. Soc., Dalton Trans., 1977, 2051–2054 RSC.
- C. Freire and B. de Castro, Spectroscopic characterisation of electrogenerated nickel(III) species. Complexes with N2O2 Schiff-base ligands derived from salicylaldehyde, J. Chem. Soc., Dalton Trans., 1998, 1491–1498 RSC.
- B. De Castro and C. Freire, EPR and electrochemical study of nickel(III) complexes of bis(3,5-dichlorosalicylaldehyde) o-phenylenediimine. Evidence for adduct formation with pyridines, Inorg. Chem., 1990, 29, 5113–5119 CrossRef CAS.
- P. A. Stenson, A. Board, A. Marin-Becerra, A. J. Blake, E. S. Davies, C. Wilson, J. McMaster and M. Schröder, Molecular and Electronic Structures of One-Electron Oxidized NiII–(Dithiosalicylidenediamine) Complexes: NiIII–Thiolate versus NiII–Thiyl Radical States, Chem. – Eur. J., 2008, 14, 2564–2576 CrossRef CAS PubMed.
- J. Shearer, Dioxygen and superoxide stability of metallopeptide based mimics of nickel containing superoxide dismutase: The influence of amine/amidate vs. bis-amidate ligation, J. Inorg. Biochem., 2013, 129, 145–149 CrossRef CAS PubMed.
- B. J. Bolann, H. Henriksen and R. J. Ulvik, Decay kinetics of O2˙− studied by direct spectrophotometry. Interaction with catalytic and non-catalytic substances, Biochim. Biophys. Acta, Gen. Subj., 1992, 1156, 27–33 CrossRef CAS.
- Scientist, version 2.0; Micromath Software, Salt Lake City, 1995 Search PubMed.
- J. Shearer, K. L. Peck, J. C. Schmitt and K. P. Neupane, Cysteinate Protonation and Water Hydrogen Bonding at the Active-Site of a Nickel Superoxide Dismutase Metallopeptide-Based Mimic: Implications for the Mechanism of Superoxide Reduction, J. Am. Chem. Soc., 2014, 136, 16009–16022 CrossRef CAS PubMed.
- E. M. Gale, B. S. Narendrapurapu, A. C. Simmonett, H. F. Schaefer and T. C. Harrop, Exploring the Effects of H-Bonding in Synthetic Analogues of Nickel Superoxide Dismutase (Ni-SOD): Experimental and Theoretical Implications for Protection of the Ni−SCys Bond, Inorg. Chem., 2010, 49, 7080–7096 CrossRef CAS.
- C. S. Mullins, C. A. Grapperhaus and P. M. Kozlowski, Density functional theory investigations of NiN2S2 reactivity as a function of nitrogen donor type and N–H⋯S hydrogen bonding inspired by nickel-containing superoxide dismutase, JBIC, J. Biol. Inorg. Chem., 2006, 11, 617–625 CrossRef CAS PubMed.
- H. M. Irving, M. G. Miles and L. D. Pettit, A study of some problems in determining the stoicheiometric proton dissociation constants of complexes by potentiometric titrations using a glass electrode, Anal. Chim. Acta, 1967, 38, 475–488 CrossRef CAS.
- P. Gans, A. Sabatini and A. Vacca, SUPERQUAD: an improved general program for computation of formation constants from potentiometric data, J. Chem. Soc., Dalton Trans., 1985, 1195–1200 RSC.
- L. Zékány and I. Nagypál, Computational Methods for the Determination of Formation Constants, Plenum Press, New York, NY, USA, 1985 Search PubMed.
- T. M. MATLAB and, Statistics Toolbox Release 2012b, Inc, Natick, Massachusetts, United States.
- A. Rockenbauer and L. Korecz, Automatic computer simulations of ESR spectra, Appl. Magn. Reson., 1996, 10, 29–43 CrossRef CAS.
- P. Li and K. M. Merz Jr., MCPB. py: A Python Based Metal Center Parameter Builder, J. Chem. Inf. Model., 2016, 56, 599–604 CrossRef CAS PubMed.
- G. Sciortino, J.-E. Sánchez-Aparicio, J. Rodríguez-Guerra Pedregal, E. Garribba and J.-D. Maréchal, Computational insight into the interaction of oxaliplatin with insulin, Metallomics, 2019, 11, 765–773 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr., J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, Ö. Farkas, J. B. Foresman, J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 16, revision B.01, Gaussian, Inc., Wallingford, CT, 2010 Search PubMed.
- G. Sciortino, N. Lihi, T. Czine, J.-D. Maréchal, A. Lledós and E. Garribba, Accurate prediction of vertical electronic transitions of Ni(II) coordination compounds via time dependent density functional theory, Int. J. Quantum Chem., 2018, 118, e25655 CrossRef.
- F. Neese, ORCA – An Ab Initio, DFT and Semiempirical Program Package, Version 4.0, Max-Planck-Institute for Chemical Energy Conversion, Mülheim a. d. Ruhr, 2017 Search PubMed.
- F. Neese, Software update: the ORCA program system, version 4.0, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2017, 8, e1327 Search PubMed.
- F. Neese, Metal and ligand hyperfine couplings in transition metal complexes: The effect of spin–orbit coupling as studied by coupled perturbed Kohn–Sham theory, J. Chem. Phys., 2003, 118, 3939–3948 CrossRef CAS.
- P. Eastman, J. Swails, J. D. Chodera, R. T. McGibbon, Y. Zhao, K. A. Beauchamp, L.-P. Wang, A. C. Simmonett, C. D. Harrigan, C. D. Stern, R. P. Wiewiora, B. R. Brooks and V. S. Pande, OpenMM 7: Rapid Development of High Performance Algorithms for Molecular Dynamics, PLoS Comput. Biol., 2017, 13, e1005659 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental and computational data. See DOI: 10.1039/d1qi01025e |
| This journal is © the Partner Organisations 2022 |