 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A new phosphoramidite enables orthogonal double labelling to form combination oligonucleotide probes†
Chunsen
Bai
 a,
Piotr
Klimkowski
a,
Cheng
Jin
a,
Jagannath
Kuchlyan
a,
Piotr
Klimkowski
a,
Cheng
Jin
a,
Jagannath
Kuchlyan
 a,
Afaf H.
El-Sagheer
*ab and
Tom
Brown
a,
Afaf H.
El-Sagheer
*ab and
Tom
Brown
 *a
*a
aDepartment of Chemistry, University of Oxford, 12 Mansfield Road, Oxford, OX1 3TA, UK. E-mail: tom.brown@chem.ox.ac.uk; afaf.el-sagheer@chem.ox.ac.uk
bChemistry Branch, Department of Science and Mathematics, Faculty of Petroleum and Mining Engineering, Suez University, Suez 43721, Egypt
First published on 27th October 2022
Abstract
Oligonucleotides labelled with thiazole orange intercalator and a reporter dye on the same thymine base have been synthesized. The key phosphoramidite (AP-C3 dT) contains an alkyne and amine, enabling dual orthogonal labelling of the nucleobase. Multiple monomers can be added to produce heavily functionalised oligonucleotides. In their DNA and 2′-OMe RNA formats these combination probes display high duplex stability and fluorescence when bound to complementary DNA and RNA.
Fluorogenic oligonucleotide probes are nucleic acid-based biosensors that respond to targets by a change in fluorescence. In general, such probes consist of a modified synthetic DNA or RNA strand for sequence-specific target recognition and a signal transduction element to report on the interaction with their target.1,2 Various fluorogenic oligonucleotide probe designs have been used to detect and image bioanalytes including mRNA, with excellent specificity and sensitivity.3,4 Molecular Beacons (MBs) and HyBeacon probes are typical examples; MBs are hairpin-shaped single-stranded oligonucleotides with a fluorophore at one end and a quencher at the other.5 Due to the intramolecular quenching of the fluorophore, MBs have weak or negligible fluorescence emission in their hairpin-state. However, upon hybridization with their targets, MBs switch from a hairpin to a rigid linear structure which separates the fluorophore from the quencher, producing strong fluorescence emission. HyBeacon probes are single-stranded oligonucleotides that contain two or more fluorophores attached covalently to internal nucleotides. High fluorescence is observed when these probes are hybridised to complementary DNA targets, in contrast to low fluorescence when they are in their unstructured single-stranded state, due to collisional quenching.6,7 Both have been used in diagnostic PCR applications.
Thiazole orange (TO) is a fluorescent DNA binding molecule that has been used for the detection of nucleic acids.8,9 Its utility is based on an increase in fluorescence upon binding to double-stranded DNA or RNA, caused by restricted rotation about the methine bridge of TO.10 The applications of thiazole orange in fluorescent sensing of biomolecules have recently been reviewed.11 Covalent attachment of thiazole orange to oligonucleotide probes has been well studied12–22 and recently we reported combination probes composed of TO and a reporter dye attached to the same thymidine base for RNA detection.23 The fluorescence of the single-stranded probe is switched off by collisional quenching between TO and the adjacent reporter dye and upon binding to the target strand fluorescence is switched on by two mechanisms; TO intercalation via the major groove of the duplex separates the probe from the reporter fluorophore, and rotation about the methine bridge is restricted due to stacking with the bases in the duplex. As a consequence, the probe emits a strong fluorescent signal due to excitation of TO and fluorescence resonance energy transfer (FRET) to the reporter fluorophore. The combination of TO and a bright reporter dye provides a low single-stranded fluorescence background and strong double-stranded signal. A particular advantage of these combination probes is stabilisation of the probe–target duplex by TO-intercalation. In this work, we develop a much-improved synthesis of combination probes involving chemospecific functionalization with TO and fluorescent dyes such as ROX. To achieve this, we have developed a new AP-C3 dT phosphoramidite (5) containing an alkyne and a trifluoroacetyl (TFA)-protected amine (Fig. 1). Multiple incorporations of the modified nucleotide in combination probes leads to excellent fluorescence on hybridization with the complementary target, while TO reverses the detrimental effect on duplex stability caused by the reporter dye. We also show that combination probes composed of 2′-O-methyl ribosyl nucleotides for use in cellular environments have good duplex stability and high fluorescence when bound to RNA targets.
To site-selectively label oligonucleotides with TO azide and ROX NHS ester, AP-C3 dT phosphoramidite was synthesized according to Fig. 2. The reactions between alkyne and azide groups and NHS-protected carboxylic acids and primary amines are orthogonal and both proceed in aqueous solution with high yield and selectivity. To synthesise the required monomer 3-aminopropan-1-ol was reacted with ethyl trifluoroacetate to protect the amine with TFA in 97% yield,24 and the alcohol of compound 1 was oxidized to aldehyde using Dess–Martin periodinane reagent in 51% yield.25 Then, compound 2 was reacted with dipropargylamine and NaBH(OAc)3 to provide compound 3 with two alkyne groups in 95% yield. Compound 3 was coupled with 5-iodo-5′-DMT-deoxythymidine to give compound 4 (59% yield) which was converted to AP-C3 dT phosphoramidite monomer (compound 5) in 72% yield using a standard phosphitylation protocol.
We assembled oligonucleotides containing single, double and triple incorporations of AP-C3 dT on an ABI 394 DNA synthesizer and deprotected them in concentrated aqueous ammonia at 55 °C for 5 hours (standard conditions). After purification by high-performance liquid chromatography (HPLC), the oligonucleotides were characterized by electrospray mass spectrometry (ES-). The data in Table S1† demonstrates the successful incorporation of AP-C3 dT into DNA. Next, we labelled the AP-C3 dT-modified oligonucleotides with TO azide and separately with ROX NHS ester. For TO-labelling, AP-C3 dT-modified oligonucleotides were coupled with TO azide in triethylammonium acetate (TEAA) buffer with catalysis by CuI:THPTA (tris(3-hydroxypropyltriazolylmethyl)-amine) to produce 1,4-substituted 1,2,3-triazole linkages. For labelling the amino function with ROX (amide formation), AP-C3 dT-modified DNA was reacted with ROX NHS ester in bicarbonate buffer at 37 °C for 3 hours. ROX-labelled oligonucleotides were further incubated with TO azide to produce TO/RX-dual-labelled oligonucleotides (Fig. 3a). Both labelling steps proceeded in near quantitative yield as monitored by HPLC (Fig. S1†). When the reactions were completed, the unreacted dyes were removed by NAP gel-filtration and the labelled oligonucleotides were purified by reversed-phase HPLC. The post-purification analytical HPLC chromatograms and mass spectra of the dye-labelled oligonucleotides indicate high purity as a consequence of efficient site-selective labelling with TO or ROX dyes at the AP-C3 dT sites (Fig. 3b and c). Our new monomer is superior to our previous TO-combination probe monomer which has high polarity,23 making it difficult to purify by chromatography, and requires ultra-mild protective groups on the standard A, G, C monomers in oligonucleotide synthesis due to the sensitivity of thiazole orange to ammonia.
Having confirmed the efficient site-selective labelling of AP-C3 dT-modified oligonucleotides with TO and ROX dyes, we studied the fluorescence response of TO/RX-labelled probes when bound to complementary DNA and RNA oligonucleotides. As described above, TO is a DNA binding molecule that shows bright fluorescence emission after intercalating into duplexes26 due to restricted rotation at its methine bridge. Consistent with this, single TO-labelled DNA (d1-TO/A) shows enhanced fluorescence emission after hybridization with its complementary DNA target (Fig. 4a, black lines). However, no significant change in the fluorescence emission of ROX-labelled probe (d1-Y/RX) was observed upon hybridization with the DNA target (Fig. 4a, blue lines). For the TO/RX combination probes, weak fluorescence emission was observed in the single strand, but upon hybridization with target DNA, the single labelled probe showed a slight increase in fluorescence at the maximum emission wavelength of ROX (red line in Fig. 4a). Comparing d1-TO/RX, d2-TO/RX and d3-TO/RX, triple incorporation of TO/RX (d3-TO/RX) shows far greater fluorescence enhancement after hybridization to complementary DNA with a ratio of >16.7-fold in ds/ss ROX fluorescence. This is an interesting and useful observation, suggesting possible uses in real-time PCR. The d3-TO/RX probe is also effective in detecting a complementary RNA target (Fig. 4d and Fig. S2†). These results demonstrate that DNA combination probes with multiple additions of the labelled AP C3-dT monomer (e.g. probes d2-TO/RX and d3-TO/RX) are highly effective in the fluorescence detection of complementary nucleic acids.
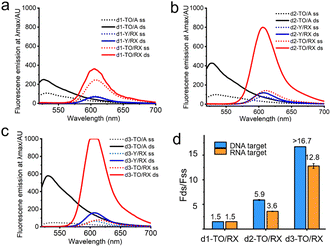 | ||
| Fig. 4 Fluorescence spectra of TO, RX or TO/RX probes bound to DNA target. (a) Fluorescence spectra of 0.25 μM single TO, RX or TO/RX-labelled DNA probes to 0.275 μM DNA target. (b) Fluorescence spectra of 0.25 μM double TO, RX or TO/RX-labelled DNA probes to 0.275 μM DNA target. (c) Fluorescence spectra of 0.25 μM triple TO, RX or TO/RX-labelled DNA probes to 0.275 μM DNA target. Solid red curve shows saturation of detector (Table S4†). (d) Fold-change in fluorescence intensity of TO/RX-labelled DNA probes before and after hybridization with complementary DNA (blue columns) or RNA (orange columns). ss: single strand. ds: double strand. Conditions: 10 mM phosphate buffer, 200 mM NaCl, pH 7.0. Oligonucleotide sequences are shown in Table S1.† | ||
Thermal stability (melting temperature), as measured by the temperature of dissociation of nucleic acid duplexes, is an important factor when designing oligonucleotide probes. We studied the thermal stability of DNA combination probes containing TO, RX and TO/RX modifications against DNA and RNA targets to determine the effects of the labels on duplex stability (Table S8,† left). The duplexes labelled with TO show improved stability which can be attributed to the intercalation of TO. In principle, the greater number of TO moieties, the greater should be the thermal stability of the duplexes, and this is indeed found to be the case, with an increase of around 8.9 °C for three TO-incorporations against its DNA complement. Labelling of ROX decreases the melting temperature of the duplexes, whether or not they contain TO, probably because in the single strand the ROX dye can bury itself in the hydrophobic environment of the coiled DNA strand created by the aromatic nucleobases, whereas in the rigid duplex it is exposed to the aqueous environment, making duplex formation entropically unfavourable. As a result, ROX-only labelled duplexes show low thermal stability compared to TO/RX dual-labelled duplexes (drop of 3.5 °C for three ROX-incorporations). We also investigated the duplex stability of d1-TO/A and d1-TO/RX probes against targets containing a single-nucleotide mismatch. All DNA duplexes and DNA/RNA targets with a G:T mismatch opposite to TO, or with an A:C mismatch five-bases away from TO, gave lower melting temperatures than the fully complementary double strands (Table S9†). For example, the Tm of the duplex between d1-TO/RX and its DNA target is 60.5 °C, compared to 55 °C for its G:T mismatch, and 48.5 °C for its A:C mismatch DNA target. These results indicate that combination probes can discriminate clearly between matched and mismatched targets.
Given that the biomedical applications of fluorogenic oligonucleotides probes require high fluorescence as well as stability against enzymatic degradation in biological fluids, we constructed combination probes containing nucleotides that stabilise oligonucleotides against degradation in vivo. Probes composed of 2′- modifications are known to possess this property, and have been employed in the clinical development of therapeutic oligonucleotides.27,28 We synthesised 2′-OMe RNA probes containing AP-C3 dT (1, 2 and 3 additions), labelled them with TO and ROX and investigated the properties of the resulting 2′-OMe RNA combination probes (Fig. 5). The 2′-OMe RNA probes with single (r1-TO/RX), double (r2-TO/RX) and triple (r3-TO/RX) incorporations of TO and ROX (Table S2†) all displayed good duplex stability when hybridized with an RNA target, 65 °C for r1-TO/RX, 63 °C for r2-TO/RX and 50 °C for r3-TO/RX (Fig. 5a and Table S8,† right). However, in the case of r3-TO/A and r3-TO/RX, the presence of three TO moieties did not improve duplex stability against DNA or RNA compared to two TO moieties (r2-TO/A). This indicates that there should be a minimum separation between the TO intercalators for them to produce the optimum increase in duplex stability and emphasises the extreme duplex destabilisation due to the presence of the ROX dye which is evident from the low melting temperature of r3-Y/RX. The very low stability of the 2′-O-methyl RNA probes against a DNA target is striking, and suggests that careful sequence design could lead to TO/RX probes that are highly selective for RNA over DNA. Both r2-TO/RX and r3-TO/RX showed clear enhancement of fluorescence emission after hybridization with their RNA target (Fig. 5b and Fig. S4†). However, only a 1.5-fold increase of fluorescence intensity of r3-TO/RX after hybridization with its complementary DNA target was observed (Fig. 5b and Fig. S3†). This suggests that in the DNA–RNA hybrid structure, the AP-C3 modifications in the triply modified case are too close, allowing fluorescence quenching to occur.
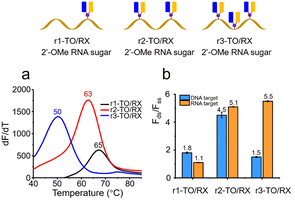 | ||
| Fig. 5 Thermal stability and fluorescence of TO/RX-functionalised 2′-OMe RNA probes when bound to complementary targets. (a) Fluorescence melting temperature of 0.4 μM single, double and triple TO/RX-incorporated 2′-OMe RNA probes bound to 0.44 μM RNA target. (b) Ratios of fluorescence intensity of 0.25 μM TO/RX-incorporated 2′-OMe RNA probes before and after hybridization with 0.275 μM DNA or RNA targets when the excitation wavelength is 510 nm. Conditions: 10 mM phosphate buffer, 200 mM NaCl, pH 7.0. Oligonucleotides sequences are shown in Table S2.† | ||
We have demonstrated that combination probes can be used in real time-PCR-based applications. (Fig. 6). This will enable target analyte concentrations to be determined, and the post-PCR melting temperature of the probe-amplicon duplex to be analysed for point mutation detection.29 The TO-intercalator increases the stability of the probe–target duplex allowing shorter PCR probes to be used with consequently superior mismatch/mutation detection. In support of this we have shown that mismatch discrimination with the new combination probes is excellent (Table S9†).
 | ||
| Fig. 6 RT-PCR (left) of TO/RX probe and fluorescence melting curve (right) of PCR product. A represents 0.15 μM of probe and B represents 0.30 μM of probe. 1–3 represent 1.5 nM, 0.15 nM and 0.015 nM of template. 4 indicates no template (negative control). Oligonucleotide sequence information and details of target DNA are in Table S3.† | ||
Fluorescence lifetime studies support the steady state fluorescence results (Fig. S5 and Table S10†). Lifetimes of all APC3-TO probes with one, two, and three incorporation of TO are bi-exponential with one long (τ2) and one short lifetime (τ1). The short lifetimes are due to relatively unrestricted rotation of TO around the methine bridge and the long lifetimes are attributed to TO in a more constrained environment due to hybridisation with target DNA or RNA. The average lifetimes of the constructs are enhanced in the duplexes because the relative contribution of the short lifetime is diminished. The ratios of lifetimes in the double strands compared to single strands are generally slightly higher for RNA than DNA targets, suggesting that TO is more constrained/planar in an A-like helix than a B-form helix.
In conclusion, this work describes a method to efficiently label single thymidine sites in oligonucleotides with two different fluorophores, one of which is also a duplex-stabiliser. To achieve this, a new AP-C3 dT phosphoramidite monomer containing a terminal alkyne and a TFA-protected primary amine has been developed for the labelling of oligonucleotides with a thiazole orange anchor dye and a ROX reporter dye. The approach takes advantage of the orthogonality of the CuAAC click and amide bond formation reactions which enables multiple TO and ROX labelled oligonucleotides to be produced, and a high density of labels to be introduced into short oligonucleotides. Thermal duplex stability and fluorescence studies demonstrate that TO/RX combination probes can detect nucleic acid targets with highly favourable signal-background-ratios. To enhance resistance against the enzymatic degradation that is encountered in a cellular environment, 2′-OMe RNA combination probes were synthesised and showed good duplex stability and excellent fluorescence properties on binding to complementary RNA targets. Future work will involve optimisation of the length and nature of the linkers between the nucleobase and the fluorescent labels, and attaching thiazole orange to AP-C3 by the quinoline moiety rather than benzthiazole to determine if this offers any advantages in terms of fluorescence or duplex stability. Finally, the use of AP-C3 dT to introduce multiple alkyne and amine moieties into oligonucleotides will be advantageous in other applications where densely-labelled oligonucleotides are required. For such applications many small molecule azides and NHS esters are commercially available, including affinity ligands, as well as alternative intercalators and reporter dyes.
Conflicts of interest
There are no conflicts to declare.References
- P. Kwok, Annu. Rev. Genomics Hum. Genet., 2001, 2, 235–258 CrossRef CAS
.
- R. T. Ranasinghe and T. Brown, Chem. Commun., 2005, 44, 5487–5502 RSC
.
- E. Garcia, L. Lebruska, M. Laurent, R. Shen and D. Barker, Methods Enzymol., 2006, 410, 57–73 Search PubMed
.
- J. Shendure and H. Ji, Nat. Biotechnol., 2008, 26, 1135–1145 CrossRef CAS
.
- S. K. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS PubMed
.
- D. J. French, C. L. Archard, T. Brown and D. G. McDowell, Mol. Cell. Probes, 2001, 15, 363–374 CrossRef CAS PubMed
.
- D. J. French, C. L. Archard, M. T. Andersen and D. G. McDowell, Mol. Cell. Probes, 2002, 16, 319–326 CrossRef CAS
.
- L. G. Lee, C. H. Chen and L. A. Chiu, Cytometry, 1986, 7, 508–517 CrossRef CAS
.
- J. Nygren, N. Svanvik and M. Kubista, Biopolymers, 1998, 46, 39–51 CrossRef CAS
.
- V. Karunakaran, J. L. Pérez Lustres, L. Zhao, N. P. Ernsting and O. Seitz, J. Am. Chem. Soc., 2006, 128, 2954–2962 CrossRef CAS PubMed
.
- O. Suss, L. Motiei and D. Margulies, Molecules, 2021, 26, 2828 CrossRef CAS PubMed
.
- P. Klimkowski, S. De Ornellas, D. Singleton, A. H. El-Sagheer and T. Brown, Org. Biomol. Chem., 2019, 17, 5943–5950 RSC
.
- S. Walsh, A. H. El-Sagheer and T. Brown, Chem. Sci., 2018, 9, 7681–7687 RSC
.
- Q. Lu, Z. Zhou, Y. Mei, W. Wei and S. Liu, Talanta, 2013, 116, 958–963 CrossRef CAS
.
- Y. Kimura, T. Hanami, Y. Tanaka, M. J. L. de Hoon, T. Soma, M. Harbers, A. Lezhava, Y. Hayashizaki and K. Usui, Biochemistry, 2012, 51, 6056–6067 CrossRef CAS
.
- L. Bethge, I. Singh and O. Seitz, Org. Biomol. Chem., 2010, 8, 2439–2448 RSC
.
- S. Berndl and H. A. Wagenknecht, Angew. Chem., Int. Ed., 2009, 48, 2418–2421 CrossRef CAS PubMed
.
- S. Ikeda, T. Kubota, K. Kino and A. Okamoto, Bioconjugate Chem., 2008, 19, 1719–1725 CrossRef CAS
.
- O. Kohler, D. Venkatrao, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69–77 CrossRef
.
- D. V. Jarikote, O. Kohler, E. Socher and O. Seitz, Eur. J. Org. Chem., 2005, 3187–3195 CrossRef CAS
.
- E. Privat, T. Melvin, F. Merola, G. Schweizer, S. Prodhomme, U. Asseline and P. Vigny, Photochem. Photobiol., 2002, 75, 201–210 CrossRef CAS
.
- G. M. Fang, J. Chamiolo, S. Kankowski, F. Hovelmann, D. Friedrich, A. Lower, J. C. Meier and O. Seitz, Chem. Sci., 2018, 9, 4794–4800 RSC
.
- J. Q. Qiu, A. Wilson, A. H. El-Sagheer and T. Brown, Nucleic Acids Res., 2016, 44, e138 CrossRef
.
- A. Milelli, C. Marchetti, M. L. Greco, F. Moraca, G. Costa, E. Turrini, E. Catanzaro, N. Betari, C. Calcabrini, C. Sissi, S. Alcaro, C. Fimognari, V. Tumiatti and A. Minarini, Eur. J. Med. Chem., 2017, 128, 107–122 CrossRef CAS PubMed
.
- R. M. Franzini and E. T. Kool, J. Am. Chem. Soc., 2009, 131, 16021–16023 CrossRef CAS
.
- M. J. Blommers, F. Natt, W. Jahnke and B. Cuenoud, Biochemistry, 1998, 37, 17714–17725 CrossRef CAS
.
- R. Maruyama and T. J. G. Yokota, Methods Mol. Biol., 2020, 2176, 49–56 CrossRef CAS PubMed
.
- B. T. Le, P. Raguraman, T. R. Kosbar, S. Fletcher, S. D. Wilton and R. N. Veedu, Sci. Rep., 2019, 9, 1–10 CrossRef
.
- E. Socher and O. Seitz, Methods Mol. Biol., 2008, 429, 187–198 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ob01899c |
| This journal is © The Royal Society of Chemistry 2022 |



