Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of pyrrothiamine, a novel thiamine analogue, and evaluation of derivatives as potent and selective inhibitors of pyruvate dehydrogenase†
Alex H. Y.
Chan
,
Terence C. S.
Ho
,
Kwasi
Agyei-Owusu
and
Finian J.
Leeper
 *
*
Yusuf Hamied Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK. E-mail: fjl1@cam.ac.uk
First published on 31st October 2022
Abstract
Inhibition of thiamine pyrophosphate (TPP)-dependent enzymes with thiamine/TPP analogues that have the central thiazolium ring replaced with other rings is well established, but a limited number of central rings have been reported. We report a novel analogue, pyrrothiamine, with a central pyrrole ring. We further develop pyrrothiamine derivatives as potent and selective inhibitors of pyruvate dehydrogenase, which might have anti-cancer potential.
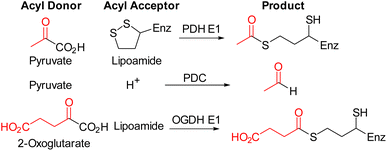
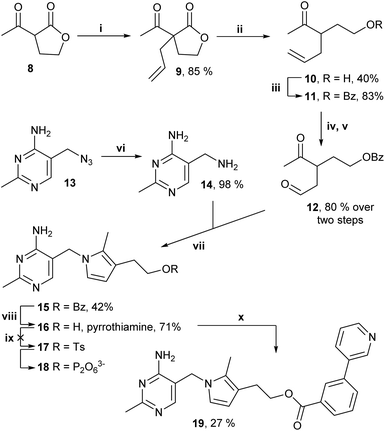
Thiamine pyrophosphate (TPP)-dependent enzymes have diverse activities and include pyruvate dehydrogenase E1-subunit (PDH E1), pyruvate decarboxylase (PDC), oxoglutarate dehydrogenase E1-subunit (OGDH E1) (Fig. 1).1,2 Individual TPP-dependent enzymes differ in substrate binding and chemistry, but they all need the coenzyme TPP 2 for activity and possess a highly similar TPP-binding pocket (key interactions summarized in Fig. 2a).1–7 The use of small molecule inhibitors to inhibit these enzymes for cellular studies and for medicinal or agrochemical purposes has long been of interest, and a common strategy lies in the use of thiamine/TPP analogues having a neutral central ring, in place of the thiazolium ring, to capture the strong stabilising interactions between the enzyme and the catalytically active high-energy TPP ylide 3 (Fig. 2a).8–25 Many derivatives with a central triazole ring 4 and modifications on the hydroxyethyl tail have been described in the literature (Fig. 2b).18–25 Although other central rings have also been reported - thiophene (deazathiamine) 6,8 furan 7,13 benzene8 and pyrazole16 - they have received much less attention than triazole 4 probably because of their longer synthetic routes. Comparison between TPP analogues clearly indicates that the choice of central rings directly impacts potency (Fig. 2c). In this study, we develop a novel thiamine analogue, pyrrothiamine 16 with a central pyrrole ring.
 | ||
| Fig. 2 (a) Binding mode of the catalytically active TPP ylide 3 in the TPP pocket and reaction mechanism for PDC. (b) Structures of thiamine/TPP analogues. (c) Inhibitory data of some TPP analogues. | ||
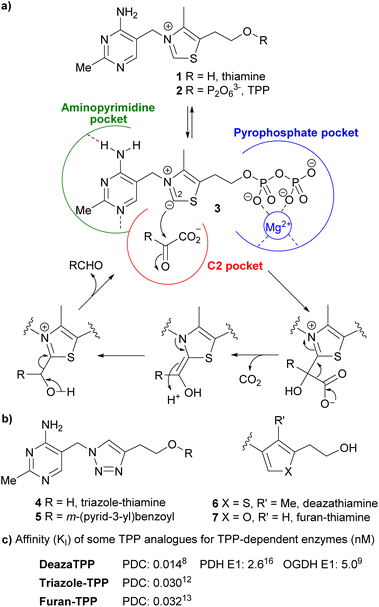
Pyrrothiamine 16 was synthesised via a Paal–Knorr reaction between primary amine 14 and 1,4-dicarbonyl 12 (Scheme 1a). The 1,4-dicarbonyl 12 was prepared from 2-acetylbutyrolactone 8 by alkylation with allyl bromide to give racemic lactone 9, then hydrolysis and decarboxylation to alcohol 10. An equilibrium between acyclic alcohol 10 and its cyclic hemiacetal complicated the purification and analysis of the NMR spectra, so 10 was benzoylated to ester 11. Oxidative cleavage of the alkene was implemented in two-steps via a 1,2-diol by Sharpless dihydroxylation followed by sodium periodate cleavage, giving aldehyde 12 in 80% yield over two steps. The amine 14 was prepared by hydrogenating azide 13, in turn obtained in a single-step reaction13 from thiamine hydrochloride. Aldehyde 12 was then coupled with amine 14 to yield the protected pyrrole 15, which was then hydrolysed to give pyrrothiamine 16.
Pyrrothiamine 16 was not expected to be a potent inhibitor per se, as it lacks any group to occupy the pyrophosphate pocket or interact with the Mg2+. We attempted to make its pyrophosphate 18 in the same way as we had done for other thiamine analogues.8,13 However the first step, formation of the tosylate 17, failed. We suspect that 17 may have formed but was unstable, due to the electron-rich pyrrole ring assisting SN1 displacement of tosylate by nucleophiles. Although pyrroTPP 18 would be expected to be very potent inhibitor of TPP-dependent enzymes, it would not be useful in cellular studies and/or medicinal applications as the polyanionic pyrophosphate would make it membrane-impermeable.17,26 To obtain a potentially membrane-permeable inhibitor, we coupled the alcohol of pyrrothiamine 16 to m-(pyrid-3-yl)benzoic acid to generate ester 19 (Scheme 1b). This is because our recent study27 of dozens of triazole-thiamine esters identified 5 as the most potent PDH E1 inhibitor (Table 1). Recent evidence28–30 has shown that selective PDH E1 inhibitors have anti-cancer potential.
| Compounds | PDH E1 | PDC | OGDH E1 | ||
|---|---|---|---|---|---|
| IC50 (μM ± SEM)a,b | × TPPc | K I (nM ± SEM)d | Inhibition (%)a,e | Inhibition (%)a,f | |
| a Data are the means of measurements in three technical replicates. b IC50 values determined at [TPP] = 10 μM. c Affinity of the inhibitor versus that of TPP, i.e. [TPP]/IC50. d K I is based on the previously reported27KD of TPP = 50 nM using [TPP]/IC50 = KD(TPP)/KI. e Percentage inhibition determined for compounds at 200 μM with [TPP] = 200 μM. f Percentage inhibition determined for compounds at 40 μM with [TPP] = 40 μM. g Ref. 27 | |||||
| 16 | 19.7 ± 1.9 | 0.51 | 98.5 ± 9.5 | 22 ± 6 | 30 ± 5 |
| 19 | 1.2 ± 0.1 | 8.42 | 6.0 ± 0.5 | <10 | <10 |
| 5 | 3.0 ± 0.4 | 3.33 | 15.0 ± 2.0 | Not determined | Not determined |
The potency of 16 and 19 against PDH E1 (Table 1) was evaluated in enzyme assays using porcine PDH E1, which is nearly identical (>95%) to human PDH E1 and is a widely used model for it.3–7,18–24,27,31 Ester 19 is a much better inhibitor (>15-fold) than unesterified 16 and 2.5-fold better than the equivalent triazole analogue 5. Their TPP-competitive nature was confirmed as the observed potency decreased with increasing [TPP] (Table S1†). The affinity of 19 for PDH E1 was found to be 8.4 times greater than that of TPP and the KI is in the low-nanomolar range.
Pyrrothiamine ester 15, an intermediate in the synthesis of 16, was also tested as an inhibitor under conditions where [inhibitor] = [TPP]. The percentage inhibition on PDH E1 was found to be 32%, 41% and 78% for compounds 16, 15 and 19 respectively (Table S1†) so the affinity of 15 is intermediate between the affinities of 16 and 19. No further enzymatic evaluation was conducted on 15 but the result of this assay implies that both the benzoyl ester and the terminal m-pyrid-3-yl ring independently contribute to binding.
To test their selectivity, compounds 16 and 19 were tested on PDC and OGDH E1 because these enzymes process either the same donor or acceptor substrate as PDH E1 (Fig. 1). The assays showed that 16 binds to all three enzymes with similar (low) affinity but ester 19 was almost inactive towards PDC and OGDH E1 (Table 1).
Insights into the binding mode were pursued through in silico studies: docked using the program GOLD into the active site of human PDH E1, 19 overlays well with TPP (Fig. 3b), thus supporting its TPP-competitive relationship. While the pyrophosphate of TPP interacts with the Mg2+, the bi-aryl tail of 19 has a pi-cation interaction with the Mg2+ and occupies a relatively hydrophobic extension of the pyrophosphate pocket, with the nitrogen atom of the pyridine hydrogen-bonded to an arginine residue (Fig. 3d). The binding interactions are shown in Fig. 3e.
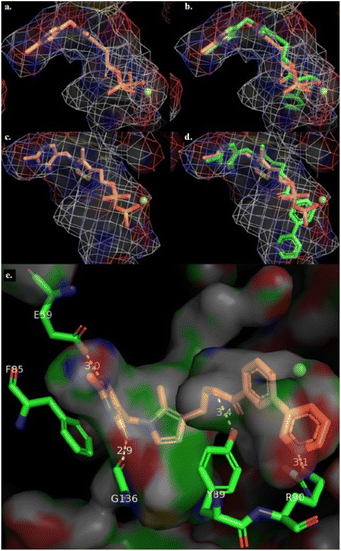 | ||
| Fig. 3 (a) Binding mode of TPP in human PDH E1 (PDB: 6CFO) showing the V-shaped conformation between the aminopyrimidine and the thiazolium ring. (b) Predicted binding mode of 19 (green atoms) overlayed with TPP as in view (a). (c) Binding mode of TPP showing the coordination between the pyrophosphate moiety and Mg2+. (d) Predicted binding mode of 19 overlayed with TPP as in view (c); (e) interactions between ester 19 and PDH E1 binding pocket shown as surface representation with interactions and distances shown as dashed lines; Mg2+ is represented as a yellowish-green sphere. | ||
When 19 was docked into the active site of OGDH E1, no reasonable docking was found due to substantial intermolecular steric clashes. This suggests that the smaller pyrophosphate pocket of OGDH, relative to PDH, cannot accommodate the bulky bi-aryl ester tail, consistent with earlier findings.27 In short, the introduction of the bi-aryl ester group of 19 onto the terminus of pyrrothiamine 16 enhanced both binding affinity for PDH E1 (KI![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 vs. 98 nM) and selectivity over other TPP-dependent enzymes (Table 1), it seems due to steric reasons.
6 vs. 98 nM) and selectivity over other TPP-dependent enzymes (Table 1), it seems due to steric reasons.
Unlike deazaTPP (pyrophosphate of 6), which has nanomolar (or subnanomolar) affinities for multiple TPP-dependent enzymes (Fig. 1c), ester 19 is selective towards PDH E1 with comparable potency. Furthermore, without the pyrophosphate moiety, ester 19 is likely to be hydrophobic enough to diffuse through membranes in cellular studies. Pyrrole 19 binds 2.5 times tighter to PDH E1 than the equivalent triazole 5 (Table 1) and in addition, pyrrole 13 has a free C2-position (next to the pyrrolic nitrogen) that will allow introduction of substituents at that position. C2-substituents should project into the C2-pocket10,15,27 where the substrate binding and catalytic groups are located (Fig. 1a).1–7 It has been verified that exploring the distinctive electronic/steric elements of the C2-pocket can lead to selective inhibition.3–15,27 Thus, the current pyrrolic scaffold has an additional possibility for functionalisation to enhance its selectivity that triazole 5 does not.
In conclusion, we report herein a novel thiamine analogue – pyrrothiamine 16, which expands the range of thiamine/TPP analogues. A derivative of 16, ester 19, was biochemically and computationally established as a potent and selective PDH E1 inhibitor, which may have anti-cancer potential towards cancers over-expressing PDH.28–30 The synthesis of 19 is relatively short and could have been two steps shorter if 10 had been protected with the eventual m-(pyrid-3-yl)benzoyl group instead of a simple benzoyl group. Our group is hoping to combine C2-substitution and tail modification to prepare compounds selective to transketolase for cancer treatment.32 Notably, this electron-rich pyrrolic scaffold makes it exceptionally suitable for C2-functionalisation via Friedel–Crafts acylation reaction as pyrroles are among the most reactive aromatic rings in electrophilic substitution reactions. We anticipate that this novel thiamine analogue will encourage researchers to shift from the triazole scaffold 4 to pyrrole 16, as well as thiophene 6 and furan 7. Not only are these scaffolds (6, 7 and 16) more potent inhibitors than their triazole equivalents, but also their free C2-position allows the introduction of C2-substituents for selectivity (and affinity) improvement.27
Author contributions
A. H. Y. C. synthesised all the compounds; T. C. S. H. performed most of the enzyme assays and the molecular docking; K. A.-O. pioneered the synthetic route to 16; F. J. L. supervised the project. A. H. Y. C., T. C. S. H. and F. J. L. all contributed to writing the paper.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
A. H. Y. C. and T. C. S. H were supported by K. M. Medhealth. K. A.-O. was supported by a studentship from EPSRC.References
- R. A. W. Frank, F. J. Leeper and B. F. Luisi, Cell. Mol. Life Sci., 2007, 64, 892–905 CrossRef CAS PubMed.
- O. H. Wieland, Rev. Physiol., Biochem. Pharmacol., 1983, 94, 123–170 Search PubMed.
- J. M. Smith, R. J. Vierling and C. Freel Meyers, MedChemComm, 2012, 3, 65–67 RSC.
- S. Sanders, R. J. Vierling, D. Bartee, A. A. DeColli, M. J. Harrison, J. L. Aklinski, A. T. Koppisch and C. L. Freel Meyers, ACS Infect. Dis., 2017, 3, 467–478 CrossRef CAS PubMed.
- D. Bartee and C. L. Freel Meyers, Biochemistry, 2018, 57, 4349–4356 CrossRef CAS PubMed.
- D. Bartee and C. L. Freel Meyers, Acc. Chem. Res., 2018, 51, 2546–2555 CrossRef CAS PubMed.
- D. Bartee, S. Sanders, P. D. Phillips, M. J. Harrison, A. T. Koppisch and C. L. Freel Meyers, ACS Infect. Dis., 2019, 5, 406–417 CrossRef CAS PubMed.
- S. Mann, C. Perez Melero, D. Hawksley and F. J. Leeper, Org. Biomol. Chem., 2004, 2, 1732–1741 RSC.
- F. J. Leeper, D. Hawksley, S. Mann, C. Perez Melero and M. D. H. Wood, Biochem. Soc. Trans., 2005, 33, 772–775 CrossRef CAS PubMed.
- T. Masini, B. Lacy, L. Monjas, D. Hawksley, A. R. de Voogd, B. Illarionov, A. Iqbal, F. J. Leeper, M. Fischer, M. Kontoyianni and A. K. H. Hirsch, Org. Biomol. Chem., 2015, 13, 11263–11277 RSC.
- L. Monjas, L. J. Y. M. Swier, A. R. de Voogd, R. C. Oudshoorn, A. K. H. Hirsch and D. J. Slotboom, MedChemComm, 2016, 7, 966–971 RSC.
- L. J. Y. M. Swier, L. Monjas, A. Guskov, A. R. de Voogd, G. B. Erkens, D. J. Slotboom and A. K. H. Hirsch, ChemBioChem, 2015, 16, 819–826 CrossRef CAS PubMed.
- K. M. Erixon, C. L. Dabalos and F. J. Leeper, Org. Biomol. Chem., 2008, 6, 3561–3572 RSC.
- A. Iqbal, E.-H. Sahraoui and F. J. Leeper, Beilstein J. Org. Chem., 2014, 10, 2580–2585 CrossRef PubMed.
- S. Lüdtke, P. Neumann, K. M. Erixon, F. Leeper, R. Kluger, R. Ficner and K. Tittmann, Nat. Chem., 2013, 5, 762–767 CrossRef PubMed.
- A. A. Thomas, J. De Meese, Y. Le Huerou, S. A. Boyd, T. T. Romoff and S. S. Gonzales, et al. , Bioorg. Med. Chem. Lett., 2008, 18, 509–512 CrossRef CAS PubMed.
- E. Grabowska, M. Czerniecka, U. Czyżewska, A. Zambrzycka, Z. Łotowski and A. Tylicki, J. Enzyme Inhib. Med. Chem., 2021, 36, 122–129 CrossRef CAS PubMed.
- A. H. Y. Chan, I. Fathoni, T. Ho, K. J. Saliba and F. J. Leeper, RSC Med. Chem., 2022, 13, 817–821 RSC.
- A. H. Y. Chan, T. C. S. Ho, I. Fathoni, R. Pope, K. J. Saliba and F. J. Leeper, submitted.
- J. Feng, H. He, Y. Zhou, M. Cai, H. Peng, H. Liu, L. Liu, L. Feng and H. He, Bioorg. Med. Chem., 2019, 27, 115159 CrossRef CAS PubMed.
- Y. Zhou, J. Feng, L. Feng, D. Xie, H. Peng, M. Cai and H. He, J. Agric. Food Chem., 2019, 67, 12538–12546 CrossRef CAS PubMed.
- Y. Zhou, M. Cai, H. Zhou, L. Hou, H. Peng and H. He, Pestic. Biochem. Physiol., 2021, 177, 104894 CrossRef CAS PubMed.
- Y. Zhou, S. Zhang, M. Cai, K. Wang, J. Feng, D. Xie, L. Feng, H. Peng and H. He, J. Agric. Food Chem., 2021, 69, 5804–5817 CrossRef CAS PubMed.
- Y. Zhou, Y. Qin, H. Zhou, T. Zhang, J. Feng, D. Xie, L. Feng, H. Peng, H. He and M. Cai, Pestic. Biochem. Physiol., 2022, 105098 CrossRef CAS PubMed.
- H. He, H. He, L. Feng, H. Peng and X. Tan, Chinese Patent, CN106588887A, 2017 Search PubMed.
- E. S. Rudge, A. H. Y. Chan and F. J. Leeper, RSC Med. Chem., 2022, 13, 375–391 RSC.
- F. J. Leeper, A. H. Y. Chan, T. C. S. Ho and D. Parle, ChemRxiv, 2022, DOI:10.26434/chemrxiv-2022-scv2t. This content is a preprint and has not been peer-reviewed.
- S. M. Davidson, T. Papagiannakopoulos, B. A. Olenchock, J. E. Heyman, M. A. Keibler and A. Luengo, et al. , Cell Metab., 2016, 23, 517–528 CrossRef CAS PubMed.
- C. T. Hensley, B. Faubert, Q. Yuan, N. Lev-Cohain, E. Jin, J. Kim, L. Jiang and B. Ko, et al. , Cell, 2016, 164, 681–694 CrossRef CAS PubMed.
- J. Chen, I. Guccini, D. Di Mitri, D. Brina, A. Revandkar, M. Sarti and E. Pasquini, et al. , Nat. Genet., 2018, 50, 219–228 CrossRef CAS PubMed.
- D. A. Walsh, R. H. Cooper, R. M. Denton, B. J. Bridges and P. J. Randle, Biochem. J., 1976, 157, 41–67 CrossRef CAS PubMed.
- M. Li, X. Zhao, H. Yong, J. Xu, P. Qu, S. Qiao, P. Hou, Z. Li, S. Chu, J. Zheng and J. Bai, Cell Death Dis., 2022, 13, 99 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Methods and results of the molecular docking and enzyme assays; synthesis methods, analytical data and NMR spectra. See DOI: https://doi.org/10.1039/d2ob01819e |
| This journal is © The Royal Society of Chemistry 2022 |