Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Investigation of acyl transfer auxiliary-assisted glycoconjugation for glycoprotein semi-synthesis†
Kudakwashe
Nyandoro
a,
Charles M. G.
Lamb
a,
Haoran
Yu
b,
Jian
Shi
a and
Derek
Macmillan
 *a
*a
aDepartment of Chemistry, UCL, 20 Gordon Street, London, WC1H 0AJ, UK. E-mail: d.macmillan@ucl.ac.uk
bSchool of Chemical and Biological Engineering, Zhejiang University, Hangzhou, China
First published on 17th October 2022
Abstract
Homogeneous glycoprotein syntheses have become possible in the last decade due to advances in chemical ligation strategies, particularly Native Chemical Ligation (NCL). For native glycoproteins this still requires laborious and technically challenging syntheses of glycopeptide components, combined with multi-segment ligation reactions. Here we explore new reactions between sugar-linked acyl transfer auxiliaries and peptide thioesters. We show that native glycoproteins are difficult to produce using this approach but various related analogues are accessible. The results show that site-specific neoglycoconjugation is a viable route to simply glycosylated proteins, which may be extended using well-documented enzymatic processes.
Introduction
Protein glycosylation remains an unsolved synthetic chemical problem.1 During the biosynthesis of N-linked glycoproteins, specific asparagine (Asn) residues are singled out by the multi-enzyme complex oligosaccharyl transferase (OST) and the carboxamide group of the asparagine sidechain is destabilised to facilitate reaction, as a nucleophile, towards an oligosaccharide acceptor bearing a complex phospholipid leaving group.2 The details concerning recognition and activation of specific Asn residues are beginning to be understood, where conserved OST amino acid residues mediate recognition and twisting of the target Asn amide bond.3The carbohydrate moiety confers desirable properties upon the protein to which it is attached. These include prolonged longevity in the blood, which is particularly relevant to therapeutic proteins, reducing the dosing regimen and consequently the potential for intolerance and error, as well as protecting the protein backbone from proteolysis, and can target the protein to the appropriate site within the body.4 The N-glycans in the Fc domain of antibodies are also crucial for directing effector function.5
Despite over 100 years of carbohydrate chemistry, the biosynthetic process has not been replicated using organic synthesis. Formation of the N-glycopeptide bond typically involves amide bond formation between suitably protected aspartic acid and glycosylamine building blocks in a process introduced by Lansbury.6 Further advances have increased the efficiency of N-glycoside bond formation7 but N-glycoprotein chemical synthesis remains the endeavour of specialized synthetic groups that not only construct the short peptides decorated with complex sugars, but also combine those short peptides into larger bioactive proteins, using chemoselective coupling processes such as native chemical ligation (NCL).8 In an alternative strategy full length proteins of biological origin, and with N-acetylglucosamine (GlcNAc) pre-installed at a specific location as a result of pre-existing cellular glycosylation, are redesigned using endo-glycosidases.1,9 An important challenge is how to attach this first required GlcNAc residue in recombinant proteins, at a site of your choice?
Here we explore a new paradigm for semi-synthetic glycoprotein synthesis by uniting synthetic monosaccharide building blocks with fully recombinant protein, containing the unnatural amino acid “ThioD” (1),10 or “BnE” (2),11 and their corresponding hydrazides (Scheme 1).
Results and discussion
Garner et al. demonstrated that thioester containing amino acid ThioD could be introduced to recombinant proteins,10 and oligosaccharide structures were ligated to green fluorescent protein using cysteine terminated carbohydrates.12 There is only one example, albeit in fully synthetic peptides, reporting NCL-type chemistry, employing “traceless” acyl transfer auxiliaries to unite carbohydrates with sidechain thioester containing peptides.13 In a related study, we examined the ability of an acyl transfer auxiliary to unite synthetic sugar derivatives, linked to an acyl transfer auxiliary, with synthetic peptide thioesters derived from the corresponding acyl hydrazides.14 Our reactions were successful, but only when employing an additional methylene spacer (a C-glycoside) in the carbohydrate building block. Currently, proteins adorned with sidechain hydrazides of aspartic acid are not directly accessible using genetic means, although can be obtained from the corresponding genetically encoded benzyl ester.11,15 We concluded that our failure to form native glycopeptides was perhaps a result of incompatibility between the sugar-linked auxiliary and conditions required for hydrazide pre-activation.This work encouraged us to prepare sugar-linked acyl transfer auxiliaries that may enable site-specific attachment of GlcNAc to fully recombinant proteins with pre-installed thioesters such as ThioD (Scheme 2). To this end, a reductive amination between glycosylamine 2a16 and aldehyde 317 was investigated, but found to produce a mixture of products including imine 4 and over-reduced product 5, with the desired product 6 forming in only 36% yield at best. This was not unexpected, considering the dearth of examples of reductive aminations featuring glycosylamines in the literature. However, once isolated, p-methoxybenzyl (PMB) cleavage, using 3-nitro-2-pyridylsulfenyl chloride (NPysCl) produced the mixed disulfide 7 in 41% yield. In an attempt to overcome the troublesome reductive amination, a transamination process was also investigated, using the fully unprotected glycosyl amine 2b.18 A test transamination with benzylamine proceeded efficiently and, following purification was observed to be exclusively β-configured. Upon extension to benzylic amine 8,19 the reaction was found to proceed similarly and 9 was isolated in 40% yield. Once more the S-PMB protecting group was exchanged to form the mixed disulfide. However 10 proved unstable to chromatography and was used without further purification. For comparison, our methylene extended building block 11 was also prepared. Whilst unnatural, it has precedent as a valuable glycopeptide analogue.20 As expected, the synthesis of 11 was more straightforward. Glycosyl nitrile 1221 was reduced by catalytic hydrogenation in the presence of Boc2O allowing isolation of pure 13 in 82% yield.20a,22 Utilising 13 in place of 2a simplified the reductive amination and subsequent steps, confirming that the hemi-aminal motif present within 2 was the source of synthetic complications. It is noteworthy that, en-route to 7, 10 or 11, the intermediates were not significantly exposed to aqueous conditions. The NPys unsymmetrical disulfide 11, formed from treatment of 14 with NPysCl, existed as a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of desired product and symmetrical disulfide, which complicated analysis. We found that 11 could be reduced with tris-carboxyethylphosphine (TCEP) and the thiol 15 was isolated by preparative HPLC, which, although adding an additional step and compromising yield, afforded a compound that was significantly simpler to characterise. In contrast, 7 or 10 were not stable to the same reducing conditions. Nevertheless, both were used in NCL reactions, assuming that their disulfides would be reduced under ligation conditions.
1 mixture of desired product and symmetrical disulfide, which complicated analysis. We found that 11 could be reduced with tris-carboxyethylphosphine (TCEP) and the thiol 15 was isolated by preparative HPLC, which, although adding an additional step and compromising yield, afforded a compound that was significantly simpler to characterise. In contrast, 7 or 10 were not stable to the same reducing conditions. Nevertheless, both were used in NCL reactions, assuming that their disulfides would be reduced under ligation conditions.
With building blocks in hand, we turned our attention to model ligation reactions using preformed peptide thioesters.
In these experiments (Fig. 1) C-terminal peptide thioester 16, derived from β-interferon residues 76–83 (sequence: H-STGWNETIG-SCH2CH2SO3H) bearing a natural glycosylation site (N80) was initially employed. The C-terminal thioester was first investigated to avoid complicating factors that may arise from additional reagents used for in situ thioester formation at the sidechain, and potentially problematic aspartimide formation. Otherwise we considered it an acceptable model for thioD containing peptides. 16 was incubated with sugar-linked auxiliaries 7, 10 and 15. Reactions progressed to consume 16 but, in our hands, and under various reaction conditions (varying pH, thiol additive, and temperature), only 15 furnished the expected product, with 17 being isolated as the major species. When using 7 or 10, the only peptide-based material that could be identified in the reaction was 18, derived from hydrolysis of the glycosyl amine followed by ligation to the unglycosylated auxiliary. LC-MS also showed two signals, (labelled 7*) corresponding to 7 (by mass) which may have arisen from anomerisation. We reasoned that, since the expected product is a glycosyl amide and relatively stable, hydrolysis of 7, followed by ligation of the free auxiliary was the likely source of the undesirable conjugate. We attempted to combat this by conducting the ligation in organic solvent (DMF, Et3N)23 but no reaction was observed under these conditions. This phenomenon was not unique to the trimethoxybenzyl (Tmb) class of acyl transfer auxiliary. We encountered similar difficulties in the preparation and ligation of “naturally-linked” conjugates to the 2-mercapto-2-phenethyl auxiliary developed by Seitz.24 Again, the introduction of a methylene spacer to afford 19 (Fig. 2a) solved the problem of building block hydrolysis during ligation. 19 was prepared in a similar manner to 15, employing reductive amination between 13 and aldehyde 20.2519 was obtained upon exposure of 21 to triethylsilane and TFA in DCM. To make progress towards protein glycosylation we further examined ligation reactions between 15 or 19 and free ThioD and reactions were complete within 24 h (see ESI†). Whilst encouraging, it proved difficult to isolate the conjugates to free ThioD and so ligation using 19 was also performed against peptide 16 (Fig. 2b). In this case the product 22 was easily isolated and the auxiliary removed to afford the methylene bridged N-glycopeptide analogue 23 in 74% yield. In a final model reaction 19 was ligated to peptide 24 (H-STGWNETIG-OH), exploring reaction at the N80 asparagine (bold and underlined) residue of β-interferon (Fig. 3a). Because the sidechain thioester is not stable to Fmoc-based SPPS, it was produced in situ from the corresponding hydrazide.14 In contrast to the reaction with 16, the ligation product was clearly observed as an inseparable pair of epimers, which returned to a single species, 25, following cleavage of the racemic auxiliary (see ESI†). Confident that 15 and 19 served as reliable building blocks we turned to recombinant protein modification although, due to the ease of synthesis and use of 19 relative to 15, only his building block was progressed.
 | ||
| Fig. 2 (a) Synthesis of sugar-linked auxiliary 19. Reagents and conditions (i) TFA, DCM, 3 h, rt, 96%, then 20, NaB(OAc)3H, DCM, 2% v/v acetic acid, rt, 4 h, 50% over 2 steps. (ii) TFA, Et3SiH, DCM, rt, 2 h, 96%. (b) Ligation reaction between 19 and 16 with accompanying HPLC traces of isolated products. Reagents and conditions (iii) 19 (1.3 equiv.), 0.1 M Na phosphate; pH 7, 6 M guanidine.HCl, 0.2 M MPAA, 60 mM TCEP, 25 °C, 16 h, 34%. (iv) 0.5 M TCEP, 2 M morpholine (pH 8.5), rt, 5 h, 74%.24b | ||
Sperm whale myoglobin, expressed from bacterial cells co-transformed with plasmid pMyo4TAG26 and cognate ThioD tRNA synthetase (ThioDRS)10 was an available model system and so was employed in proof of concept experiments. Using published procedures for protein expression we could isolate the protein containing ThioD (26) in yields of approximately 1 mg L−1 and characterise it by SDS-PAGE and mass spectrometry (Fig. 3b). The presence of the thioester was confirmed by treatment of the protein, typically obtained at concentrations of 18.5 μM, with 2.5% v/v hydrazine hydrate, which showed complete conversion to the hydrazide within 2 h at room temperature (see ESI†). In contrast to reaction with hydrazine, myoglobin reacted only slowly with 19. The reaction progressed to approximately 20% conversion after 6 h as judged by LC-MS despite employing a large excess of 19, and heating to 37 °C. Heating to 37 °C for prolonged periods of time was generally unhelpful resulting in visible sample precipitation, loss of the of the protein's characteristic red/brown colour, and deterioration of the MS Signal. Encouragingly, only initial protein thioester 26, product 27a, and disulfide linked adduct between 27b and 19, were the major species observed in reactions by LC-MS. However, despite significant attempts to improve the reaction by (i) raising or lowering the reaction temperature (ii) conducting the reaction in 3 M guanidine hydrochloride, (iii) addition of MPAA, we were unable to improve the data. Unlike in our model systems there was no evidence of an MPAA thioester intermediate. We considered that the low conversion may be due to either the increased steric bulk of 19 and protein 29 relative to the model systems, or the oxidising nature of the model protein. Whilst 19 was significantly more stable towards oxidation relative to 15, it still contains a benzylic thiol that is susceptible to oxidation. However, the oxidised dimeric form of 19 was not observed to accumulate significantly under the reaction conditions in the first 6 h and conducting reactions in the presence of 10 mM EDTA had a negligible effect on the outcome. Evidence of expected by-products such as thioester hydrolysis or aspartimide formation were only observed after significantly longer reaction times (>6 h). Just as we were keen to investigate whether extending the glycoside by a methylene unit aided ligation of GlcNAc to model peptides, we additionally became interested in whether extending the protein sidechain from Asp to Glu might allow us to overcome the obstacle of low ThioD reactivity towards 19. The corresponding “ThioE” is not available and so we prepared Glu thioesters from the benzyl ester (BnE) using the method recently reported by Xuan and co-workers.11 From an inhouse PylRS mutant (N311A), we were able to recreate the BnE RNA synthetase (BnERS) by simple site directed mutagenesis of this template. We confirmed that the enzyme selectively introduced BnE (Fig. 4a) and once again expressed myoglobin containing the unnatural amino acid. Using an adaptation of the published procedure we could convert the BnE containing protein to the γ-glutamyl hydrazide (Fig. 4b). This procedure required exposing the protein to 30% v/v hydrazine hydrate and heating the sample at 37 °C for 2.5 h. Removal of excess hydrazine was accomplished by snap-freezing and lyopillisation of the reaction mixture. The presence of a viable protein hydrazide 28 was confirmed by reducing the pH of the sample to approximately pH 4 and incubating with 1 M GlcNAc at 40 °C for 4 h. In a reaction analogous to that recently reported at Asp β-hydrazides15 we observed near quantitative conversion to novel neoglycoprotein 29 (Fig. 4c). We did not test the longer-term stability of this linkage and neither confirm the β-stereochemistry at the anomeric linkage nor that the cyclic nature of sugar had been preserved, although preceding studies suggest both features are retained.27
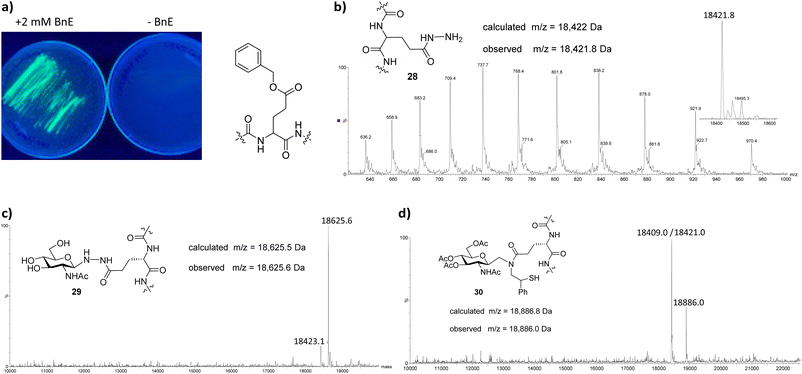 | ||
| Fig. 4 (a) Selectivity of the γ-Benzyl glutamic acid pylRS mutant was confirmed by co-transformation with plasmid pREPpylT for resistance to 50 μg ml−1 chloramphenicol in the presence (left) and absence (right) of 2 mM BnE.26 (b) Purified benzyl ester is converted to the γ-hydrazide by exposure to 30% v/v hydrazine hydrate at 37 °C for 2 h. (c) The presence of the hydrazide was confirmed by GlcNAc bioconjugation. Reagents and conditions: 28 (18.5 μM), 50 mM Na phosphate buffer; pH 4, 1 M GlcNAc, 40 °C, 4 h >80%. (d) Ligation between 28 and 19. Reagents and conditions: 50 mM Na phosphate buffer; pH 4, 1 mM NaNO2, 0 °C, 20 min, then 19, 2.0 M guanidine.HCl 50 mM MPAA, 25 mM TCEP, pH 7, 25 °C, 4 h. | ||
Confident that the hydrazide 28 should serve as a suitable thioester precursor we investigated ligation with 19 using both oxidative28 and pyrazole forming29 methods for hydrazide activation. In preliminary investigations, and in contrast to experiments with model peptide 24, only oxidative activation of the hydrazide with NaNO2 proved successful. However, as with thioD containing protein 26, only partial conversion to the desired neoglycoconjugate was observed after 3 h. A significant quantity of material had also failed to activate and/or hydrolysed during hydrazide activation and ligation, which is a disadvantage of not utilising a preformed thioester. However the results suggested that the nature of the amino acid itself (Asp vs. Glu) was not the only factor affecting the extent of protein modification. Since the reactions between protein samples and 19 gave rise to a mixture of starting material and product, no attempt was made to remove the auxiliary.
Conclusions
In summary we have explored a novel paradigm for glycoprotein synthesis which has the potential to reduce the synthetic burden required to assemble them. If site-specific glycosylation can be directed by the amino acid, and extended enzymatically after conjugation,9,30 then homogeneous semi-synthetic glycoproteins could be accessed without recourse to lengthy peptide synthesis-based processes. By utilizing novel unnatural C-glycosides (15 or 19), glycoside hydrolysis was abolished and ligation to amino acids and model peptides could be easily accomplished in reasonable yields. Whilst reaction of myoglobin thioester 26 with hydrazine hydrate was rapid at room temperature, reaction between 26 and 19 was less efficient. Investigation of ligation employing 19 and γ-glutamyl thioester 28 again showed modest conversion. It is likely that these reactions are compromised for different reasons. In the case of ThioD, the reactivity of the alkyl thioester appears low, whereas the need for pre-activation of 28 may increase hydrolysis and aspartimide formation. Reactions failed to reach completion within a time compatible with the fragile nature of the model protein myoglobin, which additionally displays a wide range of catalytic activities.31Overall, we have demonstrated an important proof of concept, uniting sugar-linked auxiliaries, via an amide linkage, with recombinant protein sidechains. After cleavage of the auxiliary the glycoconjugate is extended by a single methylene unit when compared to a natural N-linked oligosaccharide. Unlike cysteine based “tag and modify” approaches32 the introduction of ThioD or the corresponding benzyl esters of aspartic and glutamic acids don't create the potential for disulfide scrambling and provide access to stable amide linked products. Owing to its more sustainable and less synthetically demanding components, as well as the potential to combine with developed enzymatic GlcNAc extension protocols,9,30 this approach may be developed into a useful glycoprotein and neoglycoprotein synthesis strategy.
Experimental details
General experimental details
Preparative reversed-phase high performance liquid chromatography (RP-HPLC) was performed using a Dionex Ultimate 3000 system equipped with a Phenomenex Jupiter Proteo 90A, C12, 250 × 21.2 mm column. Separations involved a mobile phase of 0.1% TFA (v/v) in water (solvent A)/acetonitrile (solvent B) employing a 5–60% acetonitrile gradient over 60 min, and were monitored at wavelengths 230 nm, 254 nm, and 280 nm. Analytical reversed-phase high performance liquid chromatography (RP-HPLC) was performed using a Dionex Ultimate 3000 equipped with a Phenomenex SphereClone 5μ ODS, C18 250 × 4.6 mm column. Separations used a mobile phase of 0.1% TFA (v/v) in water (solvent A)/acetonitrile (solvent B) employing a 5–95% acetonitrile gradient over 45 min, and were monitored at wavelengths 230 nm, 254 nm, and 280 nm. Analytical LC-MS was carried out on Waters uPLC/SQD-LC mass spectrometer instrument equipped with C18, 2.1 × 50 mm column. Separations were conducted with a linear gradient of 5–95% acetonitrile containing 0.1% formic acid over 10 minutes using a flow rate of 0.2 ml min−1. High resolution mass spectrometry was obtained from a Thermo Q-Exactive Vanquish Orbitrap mass spectrometer equipped with a liquid chromatography for separations. The liquid chromatography was equipped with Thermo Scientific Hypersil Gold C4 50 × 2.1 column and a linear gradient of 5–95% acetonitrile containing 0.1% Formic acid was used over 6 minutes.General method for manual solid phase peptide synthesis
Manual peptide synthesis was carried out using standard Fmoc amino acids on NovaSyn TGT resin pre-loaded with Fmoc-Cys(Trt)-OH for the synthesis of 16 (loading = 0.19 mmol g−1) and Fmoc-Gly-OH (loading = 0.21 mmol g−1) for the synthesis of sidechain peptide hydrazide 24. Syntheses were performed on 0.05 mmol scale. Deprotection of the N-terminal Fmoc protecting groups was achieved with 20% piperidine in DMF. The resin was then washed exhaustively with DMF and CH2Cl2. Fmoc-protected amino acids (5 equiv.) were pre-activated with HBTU/HOBt (5 equiv.) and DIPEA (10 equiv.) in DMF and then added to the reaction vessel for coupling. Each coupling reaction was conducted for 2 to 3 h. Subsequent couplings and deprotections led to the sequential assembly of the model peptide on resin. After chain assembly, deprotection of the peptide from the resin and of the side chain protecting groups was achieved with 4 mL of TFA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) EDT: water (95
EDT: water (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5
2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5) for 4 h. The resin was filtered off and to the filtrate added to diethyl ether (12 mL) which induced peptide precipitation. The mixture was centrifuged at 4000 rpm, 4 °C for 20 min. The ether layer was decanted and fresh Et2O (12 mL) was added followed by further centrifugation. Ether was then decanted, and the peptide precipitate was dissolved in a minimum volume of water and purified by preparative reverse phase (RP) HPLC. Fractions containing product were identified by LC-MS and lyophilised to afford pure peptide.
2.5) for 4 h. The resin was filtered off and to the filtrate added to diethyl ether (12 mL) which induced peptide precipitation. The mixture was centrifuged at 4000 rpm, 4 °C for 20 min. The ether layer was decanted and fresh Et2O (12 mL) was added followed by further centrifugation. Ether was then decanted, and the peptide precipitate was dissolved in a minimum volume of water and purified by preparative reverse phase (RP) HPLC. Fractions containing product were identified by LC-MS and lyophilised to afford pure peptide.
Synthesis of model peptide thioester 16
Peptide H-STGWNETIGC-OH was first constructed using the general method for manual peptide synthesis on Fmoc-Cys(Trt)-NovaSyn-TGT resin. After lyophilisation of the RP-HPLC fractions (tR = 28.5 min), the peptide was isolated as a white fluffy solid (26 mg, 49%). ESI-HRMS: m/z calculated for C44H66N12O17S [M + H]+ 1067.4467, found [M + H] 1067.4458.This peptide (10 mg, 9.4 μmol) was dissolved in 0.1 M sodium phosphate buffer (pH 7.0) to a concentration of 1 mg mL−1. To this solution was added 10% w/v MESNa (1.0 g, 6.1 mmol), 10% w/v hydrazine acetate (1.0 g, 10.9 mmol) and 0.5% w/v TCEP (0.05 g, 0.2 mmol). The reaction was mixed on an Eppendorf thermomixer (600 rpm) at 50 °C for 30 h. The crude mixture was purified by RP-HPLC (tR = 26.0 min) and fractions containing product were lyophilised to afford a pure peptide hydrazide (8.0 mg, 87%) as a white solid. ESI-HRMS: m/z calculated for C41H63N13O15 [M + H]+ 978.4645, found [M + H]+ 978.4634.
Finally, the peptide C-terminal hydrazide (14 mg, 14.3 μmol) was dissolved in 7.0 mL of ligation buffer comprised of 0.1 M sodium phosphate buffer pH 3.0–4.0 and 6.0 M guanidine.HCl. The solution was cooled to −15 °C and NaNO2 (0.7 mL of 0.2 M stock solution dissolved in deionised water) was then added dropwise and the reaction was stirred at −15 °C for 20 min.28 The pH of the reaction was then adjusted to pH 7.0 by dropwise addition of 2 M NaOH. MESNa (2.3 g, 14 mmol) was dissolved in 6M guanidine.HCl (7.0 mL) and added to the reaction mixture followed by TCEP to final concentration of 30 mM. The reaction was stirred at room temperature for 2 h. The crude mixture was purified by RP-HPLC (tR = 27.5 min) and fractions containing 16 were lyophilised to afford pure peptide thioester (9.0 mg, 60%) as a white solid. ESI-HRMS: m/z calculated for C43H66N11O18S2 [M + H]+ 1088.4029, found [M + H]+ 1088.4015.
Ligations between 16 and 7, 10, or 15
Thioester peptide 16 was dissolved in ligation buffer comprised of 0.1 M sodium phosphate buffer; pH 7.0 in 6.0 M guanidine.HCl to a concentration of 2 mg mL−1. 0.5 mL of the solution was added to an Eppendorf tube. 7, 10 or 15 (1.2 eq.) were dissolved in ligation buffer (0.5 mL) and then added, followed by MPAA to a final concentration of 0.2 M, and TCEP to a final concentration of 60 mM. Reactions were shaken in an Eppendorf thermomixer (750 rpm) at room temperature. LC-MS analysis of ligation reactions between 16 and 7 or 10 failed to identify any desired product and the by-products were not isolated. Successful reaction between 16 and 15 was scaled up 10-fold and the reaction was monitored for 72 h. The crude mixture was purified by preparative reverse-phase HPLC (tR = 39 min) and lyophilised to yield 17 (5.7 mg, 40%). ESI-HRMS: m/z calculated for C66H96N13O26S [M + H]+ 1518.6310, found [M + H]+ 1518.6316.Ligation reaction between 16 and 19, and removal of the auxiliary
16 (10 mg, 9.2 μmol) was dissolved in ligation buffer comprised of 0.1 M sodium phosphate buffer pH 7.0 in 6.0 M guanidine.HCl to a concentration of 2 mg mL−1. To this solution (5.0 ml) was added 19 (5.8 mg, 0.01 mmol, 1.2 eq.) dissolved in ligation buffer (5.0 mL) with MPAA (0.5 g, 3.0 mmol), 60 mM TCEP (260 mg, 1.04 mmol) and the reaction was stirred at room temperature. After 12 h, the product was purified from the reaction mixture by semi-preparative reverse-phase HPLC (tR = 39.0 min) and lyophilised to yield 22 (4 mg, 34%). ESI-MS: m/z calculated for C64H91N13O23S [M + H]+ 1442.62, found [M + H]+ 1443.73.22 (1.8 mg, 1.25 μmol) was dissolved in 0.5 mL solution of 2 M morpholine in deionised water. TCEP (62.5 mg, 0.25 mmol) was added to the reaction and the pH was adjusted to pH 8.5 using 2 M NaOH. The mixture was placed in an Eppendorf thermomixer (750 rpm) and shaken at room temperature. After 5 h the reaction was complete and product isolated via preparative HPLC (tR = 31.0 min). Lyophilization resulted in a white solid of 23 (1.2 mg, 74%). ESI-MS: m/z calculated for C56H84N13O23 [M + H]+ 1306.58, found [M + H]+ 1307.0.
Synthesis of model peptide 24
IFN (76–83) with aspartic acid sidechain benzyl ester at position 80 was constructed using the general method for manual peptide synthesis on Fmoc-Gly-NovaSyn-TGT preloaded resin. Commercially available Fmoc-L-aspartic acid 4-benzyl ester was used for the coupling of Asp(OBzl)-OH at position 80. After lyophilisation of the semi-preparative reverse phase HPLC fractions (tR = 35.5 min), peptide ester was isolated as a white fluffy solid (9.6 mg, 19%). The peptide was characterised by mass spectrometry. ESI-MS: m/z calculated for C48H67N10O17 [M + H]+ 1056.48, found [M + H] 1056.39.To form the hydrazide 50–60% v/v hydrazine hydrate (200 μmol) was added to (8 mg, 7.59 μmol) of peptide benzyl ester dissolved in 8 mL of water. The mixture was stirred at room temperature for 1 h then purified by RP-HPLC (tR = 26.0 min) and lyophilised to yield 24 (6.8 mg, 92%). ESI-HRMS: m/z calculated for C41H63N12O16[M + H]+ 979.4485, found [M + H] 979.4484.
Ligation between peptide 24 and 19
Peptide hydrazide was dissolved in ligation buffer comprised of 0.1 M sodium phosphate buffer pH 3.0–4.0 in 6.0 M guanidine.HCl to a concentration of 2 mg ml−1 (5.0 mL). Acetylacetone (125 mg, 1.25 mmol) was added from a 0.25 M stock solution along with 0.2 M MPAA (168 mg, 1.0 mmol) and the mixture was stirred at room temperature. After 2 h, 19 (1.2 eq.) and 60 mM TCEP (75 mg, 0.3 mmol) were added and the pH adjusted to pH 7.0 using 2 M NaOH. The reactions were stirred at room temperature and monitored by LC-MS. After 12 h the crude mixture was purified by preparative reverse phase (RP) HPLC (tR = 40.5 min) and lyophilised to yield the product as a epimeric mixture (4 mg, 34%). ESI-MS: m/z calculated for C64H91N12O24S [M + H]+ 1443.56, found [M + H]+ 1443.69. The product (4 mg, 2.77 μM) was dissolved in a solution of 0.5 M TCEP and 2 M morpholine to a concentration of 4 mg mL−1. The pH was adjusted to pH 8.5 using 2 M NaOH and the mixture was stirred at rt for 16 h. The crude mixture was purified by preparative reverse phase HPLC (tR = 31.0 min) and lyophilised to yield 25 (3 mg, 83%). ESI-MS: m/z calculated for C56H83N12O24 [M + H]+ 1307.56, found [M + H]+ 1307.46.Expression and purification of Sperm whale myoglobin containing ThioD or BnE
E. coli strain DH10B was co-transformed with pBK-ThioDRS (or pBK-BnERS) and pMyo4TAG-pylT.26 Successfully transformed clones were selected on LB-KT agar plates agar plates supplemented with kanamycin (25 μg mL−1) and tetracycline (12.5 μg mL−1). A single colony was picked and inoculated into LB-KT medium (10.0 mL and grown overnight with shaking (250 rpm) at 37 °C. The culture was then diluted 100-fold into LB-KT medium (0.5 L) and grown at 37 °C. When the OD600 = 0.2, solid ThioD (or BnE) was directly added to the culture to a final concentration of 2 mM, and the culture was grown for another 30 min at 37 °C before induction with 0.2% w/v arabinose. The induced cells were grown for 6 h at 37 °C. The cells were pelleted by centrifugation at 8000 rpm for 10 min. The cells were resuspended in His-Bind buffer (50 mM Tris.HCl, 0.5 M NaCl, 5 mM imidazole; pH 8.0) and lysed with a sonic disruptor (10 × 30s cycles) in the presence of protease inhibitor (0.01% PMSF). Insoluble protein and cell debris were removed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The protein was purified by Ni2+ affinity chromatography, eluting from the column in 50 mM Tris.HCl, 0.5 M NaCl, 250 mM imidazole; pH 8.0. Prior to analysis and ligation reactions, samples were buffer-exchanged into 50 mM sodium phosphate buffer (pH 8.0), and concentrated, using an Amicon Ultra-15 centrifugal filter unit (3000 MWCO) according to the manufacturers instructions. Yields were typically 0.9 mg L−1 after 6 h induction. After concentration to 1.0 mL, typical working protein concentrations of 18.5 μM were obtained. Expected mass for MbThioD (26) M = 18554.1 Da, found M = 18553.0 Da. Expected mass for Mb4BnE M+, 18487.2 Da, found M = 18497.5 Da. Note: The isolated protein containing BnE has a mass anomaly of +10 Da, presumably arising from a post translational modification, but all subsequent masses are correct relative to each other).
000 rpm for 15 min. The protein was purified by Ni2+ affinity chromatography, eluting from the column in 50 mM Tris.HCl, 0.5 M NaCl, 250 mM imidazole; pH 8.0. Prior to analysis and ligation reactions, samples were buffer-exchanged into 50 mM sodium phosphate buffer (pH 8.0), and concentrated, using an Amicon Ultra-15 centrifugal filter unit (3000 MWCO) according to the manufacturers instructions. Yields were typically 0.9 mg L−1 after 6 h induction. After concentration to 1.0 mL, typical working protein concentrations of 18.5 μM were obtained. Expected mass for MbThioD (26) M = 18554.1 Da, found M = 18553.0 Da. Expected mass for Mb4BnE M+, 18487.2 Da, found M = 18497.5 Da. Note: The isolated protein containing BnE has a mass anomaly of +10 Da, presumably arising from a post translational modification, but all subsequent masses are correct relative to each other).
Ligation between 26 and 19
In an Eppendorf tube 26 (0.35 mg, 18.9 nmol) dissolved in 50 mM sodium phosphate buffer pH 7.0 (0.5 mL) was added to excess 19 (2.5 mg, 50.3 μmol) and TCEP (0.25 mg, 1.0 μmol). The reaction mixture was shaken in an Eppendorf thermomixer for 18 h at 37 °C. The formation of 27a/27b was monitored by LC-MS. After 18 h, the reaction was stopped as no further changes to the LC-MS spectra were observed. 27 was not isolated. ESI-MS: calculated mass for 27a = 18861.7 Da, observed mass = 18861.4 Da.Hydrazinolysis of Myo4BnE
Hydrazine hydrate 30% v/v was added to concentrated (18.5 μM) samples of Myo4BnE in 50 mM Na phosphate buffer (pH 8.0). The reaction was shaken in an Eppendorf thermomixer (37 °C, 750 rpm), monitored by LC-MS. Conversion was judged to be complete within 3 h. Samples were then snap-frozen in liquid nitrogen and lyophilised to afford crude 28. Expected mass M = 18421.5 Da, found M = 18421.8 Da.Ligation between 28 and N-acetyl D-glucosamine
Lyopilised hydrazide was reconstituted in deionised water to the original concentration (18.5 μM) and the pH was lowered to pH 4 with 2 M HCl. The solution was added to a fresh Eppendorf tube containing solid GlcNAc such that the final concentration of GlcNAc was 1 M. The reaction was shaken in an Eppendorf thermomixer (40 °C, 750 rpm), monitored by LC-MS and conversion was judged to be >80% complete within 4 h. The ligated product was separated from excess GlcNAc by centrifugal filtration (10 kDa MWCO) against 50 mM sodium phosphate buffer; pH 8.0. Expected mass = 18625.0 Da, observed mass = 18625.6 Da.Ligation between hydrazide 28 and 19
Lyophilised hydrazide was reconstituted to its original concentration (18.5 μM) in deionised water and the pH was lowered to pH 4 using 2 M HCl. The protein sample was cooled in an ice/NaCl bath and freshly prepared 0.1 M NaNO2 solution was added to final concentration of 1 mM. After 20 min at 0 °C 19 (2.5 mg, nmol, predissolved in 6 M guanidine hydrochloride such that the final guanidine.HCl concentration was 2.0 M) was added along with MPAA (50 mM final concentration) and TCEP (25 mM final concentration). The pH was then carefully adjusted to pH 7 with 10 M NaOH and the reaction was shaken in an Eppendorf thermomixer (25 °C, 750 rpm). The reaction was monitored by LC-MS and conversion was judged to be approximately 25% complete after 4 h.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge support from EPSRC and UCL.References
- L. Krasnova and C.-H. Wong, J. Am. Chem. Soc., 2019, 141, 3735–3754 CrossRef PubMed.
- L. Lehle, S. Strahl and W. Tanner, Angew. Chem., Int. Ed., 2006, 45, 6802–6818 CrossRef PubMed.
- (a) C. Lizak, S. Gerber, S. Numao, M. Aebi and K. P. Locher, Nature, 2011, 474, 350–355 CrossRef PubMed; (b) L. Bai, T. Wang, G. Zhao, A. Kovach and H. Li, Nature, 2018, 555, 328–333 CrossRef CAS PubMed; (c) R. Wild, J. Kowal, J. Eyring, E. M. Ngwa, M. Aebi and K. P. Locher, Science, 2018, 359, 545–550 CrossRef CAS.
- A. Varki, Glycobiology, 2017, 27, 3–49 CrossRef CAS PubMed.
- J. E. Hudak and C. R. Bertozzi, Chem. Biol., 2014, 21, 16–37 CrossRef CAS PubMed.
- S. T. Anisfeld and P. T. Lansbury, J. Org. Chem., 1990, 55, 5560–5562 CrossRef CAS.
- (a) P. Wang, B. Aussedat, Y. Vohra and S. J. Danishefsky, Angew. Chem., Int. Ed., 2012, 51, 11571–11575 CrossRef; (b) V. Ullmann, M. Rädisch, I. Boos, J. Freund, C. Pöhner, S. Schwarzinger and C. Unverzagt, Angew. Chem., Int. Ed., 2012, 51, 11566–11570 CrossRef.
- (a) C. Unverzagt and Y. Kajihara, Chem. Soc. Rev., 2013, 42, 4408–4420 RSC; (b) Y. Wang, S. H. Yang, M. A. Brimble and P. W. R. Harris, ChemBioChem, 2020, 21, 3301–3312 CrossRef; (c) Y. K. Chong, C. Chandrashekar, D. Zhao, Y. Maki, R. Okamoto and Y. Kajihara, Org. Biomol. Chem., 2022, 20, 1907–1915 RSC; (d) K. Nomura, Y. Maki, R. Okamoto, A. Satoh and Y. Kajihara, J. Am. Chem. Soc., 2021, 143, 10157–10167 CrossRef; (e) M. Murakami, T. Kiuchi, M. Nishihara, K. Tezuka, R. Okamoto, M. Izumi and Y. Kajihara, Sci. Adv., 2016, 2, e1500678 CrossRef PubMed; (f) A. Reif, K. Lam, S. Weidler, M. Lott, I. Boos, J. Lokau, C. Bretscher, M. Mönnich, L. Perkams, M. Schmälzlein, C. Graf, J.-P. Fischer, C. Lechner, K. Hallstein, S. Becker, M. Weyand, C. Steegborn, G. Schultheiss, S. Rose-John, C. Garbers and C. Unverzagt, Angew. Chem., Int. Ed., 2021, 60, 13380–13387 CrossRef PubMed; (g) H. Hessefort, A. Gross, S. Seeleithner, M. Hessefort, T. Kirsch, L. Perkams, K. O. Bundgaard, K. Gottwald, D. Rau, C. G. F. Graf, E. Rozanski, S. Weidler and C. Unverzagt, Angew. Chem., Int. Ed., 2021, 60, 25922–25932 CrossRef.
- (a) F. Tang, L.-X. Wang and W. Huang, Nat. Protoc., 2017, 12, 1702 CrossRef PubMed; (b) A. J. Fairbanks, Curr. Opin. Chem. Biol., 2019, 53, 9–15 CrossRef PubMed; (c) C.-P. Liu, T.-I. Tsai, T. Cheng, V. S. Shivatare, C.-Y. Wu, C.-Y. Wu and C.-H. Wong, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 720–725 CrossRef.
- W. Xuan, D. Collins, M. Koh, S. Shao, A. Yao, H. Xiao, P. Garner and P. G. Schultz, ACS Chem. Biol., 2018, 13, 578–581 CrossRef.
- X. Yang, H. Miao, R. Xiao, L. Wang, Y. Zhao, Q. Wu, Y. Ji, J. Du, H. Qin and W. Xuan, Chem. Sci., 2021, 12, 9778–9785 RSC.
- N. Holloran, D. Collins, U. Rathnayake, B. Zhang, M. Koh, C. Kang and P. Garner, Bioconjugate Chem., 2020, 31, 2362–2366 CrossRef PubMed.
- H. Chai, K. Le
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Mai
Mai![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Hoang, M. D. Vu, K. Pasunooti, C.-F. Liu and X.-W. Liu, Angew. Chem., Int. Ed., 2016, 55, 10363–10367 CrossRef PubMed.
Hoang, M. D. Vu, K. Pasunooti, C.-F. Liu and X.-W. Liu, Angew. Chem., Int. Ed., 2016, 55, 10363–10367 CrossRef PubMed. - N. G. Barnes, K. Nyandoro, H. Jin and D. Macmillan, Chem. Commun., 2021, 57, 1006–1009 RSC.
- Q. Wu, W. Dong, H. Miao, Q. Wang, S. Dong and W. Xuan, Angew. Chem., Int. Ed., 2022, 61, e202116545 Search PubMed.
- B. Premdjee, A. L. Adams and D. Macmillan, Bioorg. Med. Chem. Lett., 2011, 21, 4973–4975 CrossRef CAS.
- F. Burlina, A.-B. M. Abdel-Aal, R. Raz, I. Pinzuti, G. Papageorgiou, J. Li, R. Antrobus, S. R. Martin, S. Kunzelmann, B. Stieglitz and J. Offer, Commun. Chem., 2019, 2, 111 CrossRef PubMed.
- A. V. K. Prasad and J. C. Richards, US Pat, US5668272A, 1997 Search PubMed.
- J. Offer, C. N. C. Boddy and P. E. Dawson, J. Am. Chem. Soc., 2002, 124, 4642–4646 CrossRef CAS.
- (a) L.-X. Wang, M. Tang, T. Suzuki, K. Kitajima, Y. Inoue, S. Inoue, J.-Q. Fan and Y. C. Lee, J. Am. Chem. Soc., 1997, 119, 11137–11146 CrossRef CAS; (b) W. Lai-Xi, F. Jian-Qiang and Y. C. Lee, Tetrahedron Lett., 1996, 37, 1975–1978 CrossRef.
- D. Carrière, S. J. Meunier, F. D. Tropper, S. Cao and R. Roy, J. Mol. Catal. A: Chem., 2000, 154, 9–22 CrossRef.
- D. H. Lenz, G. E. Norris, C. M. Taylor and G. C. Slim, Tetrahedron Lett., 2001, 42, 4589–4591 CrossRef CAS.
- M. Dittmann, J. Sauermann, R. Seidel, W. Zimmermann and M. Engelhard, J. Pept. Sci., 2010, 16, 558–562 CrossRef CAS.
- (a) O. Fuchs, S. Trunschke, H. Hanebrink, M. Reimann and O. Seitz, Angew. Chem., Int. Ed., 2021, 60, 19483–19490 CrossRef CAS; (b) S. F. Loibl, Z. Harpaz and O. Seitz, Angew. Chem., Int. Ed., 2015, 54, 15055–15059 CrossRef CAS PubMed.
- C. M. G. Lamb, J. Shi, J. D. Wilden and D. Macmillan, Org. Biomol. Chem., 2022, 20, 7343–7350 RSC.
- H. Neumann, S. Y. Peak-Chew and J. W. Chin, Nat. Chem. Biol., 2008, 4, 232–234 CrossRef CAS PubMed.
- S. Peluso and B. Imperiali, Tetrahedron Lett., 2001, 42, 2085–2087 CrossRef CAS.
- G.-M. Fang, Y.-M. Li, F. Shen, Y.-C. Huang, J.-B. Li, Y. Lin, H.-K. Cui and L. Liu, Angew. Chem., Int. Ed., 2011, 50, 7645–7649 CrossRef CAS.
- D. T. Flood, J. C. J. Hintzen, M. J. Bird, P. A. Cistrone, J. S. Chen and P. E. Dawson, Angew. Chem., Int. Ed., 2018, 57, 11634–11639 CrossRef PubMed.
- (a) W. Huang, Q. Yang, M. Umekawa, K. Yamamoto and L.-X. Wang, ChemBioChem, 2010, 11, 1350–1355 CrossRef; (b) A. J. Fairbanks, Chem. Soc. Rev., 2017, 46, 5128–5146 RSC.
- (a) V. Tyagi, G. Sreenilayam, P. Bajaj, A. Tinoco and R. Fasan, Angew. Chem., Int. Ed., 2016, 55, 13562–13566 CrossRef; (b) V. Tyagi, R. B. Bonn and R. Fasan, Chem. Sci., 2015, 6, 2488–2494 RSC; (c) J. R. Marshall, J. Mangas-Sanchez and N. J. Turner, Tetrahedron, 2021, 82, 131926 CrossRef.
- S. B. Gunnoo and A. Madder, ChemBioChem, 2016, 17, 529–553 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures for synthesis of 7, 10, 11, 15 and 19. LC-MS data for characterisation of model peptides and spectra of selected compounds. See DOI: https://doi.org/10.1039/d2ob01633h |
| This journal is © The Royal Society of Chemistry 2022 |