Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescent annulated imidazo[4,5-c]isoquinolines via a GBB-3CR/imidoylation sequence – DNA-interactions in pUC-19 gel electrophoresis mobility shift assay†
M.
Stahlberger
 a,
O.
Steinlein
b,
C. R.
Adam
a,
M.
Rotter
a,
J.
Hohmann
a,
M.
Nieger
c,
B.
Köberle
b and
S.
Bräse
a,
O.
Steinlein
b,
C. R.
Adam
a,
M.
Rotter
a,
J.
Hohmann
a,
M.
Nieger
c,
B.
Köberle
b and
S.
Bräse
 *ad
*ad
aInstitute of Organic Chemistry, Karlsruhe Institute of Technology (KIT), Fritz-Haber-Weg 6, 76131 Karlsruhe, Germany. E-mail: braese@kit.edu
bInstitute of Applied Biosciences, Department of Food Chemistry and Toxicology, Karlsruhe Institute of Technology (KIT), Adenauerring 20, 76131 Karlsruhe, Germany
cDepartment of Chemistry, University of Helsinki, P.O. Box 55 (A. I. Virtasen aukio 1), 00014, Finland
dInstitute of Biological and Chemical Systems – IBCS-FMS, Karlsruhe Institute of Technology (KIT), Herman-von-Helmholtz-Platz 1, 76344 Eggenstein-Leopoldshafen, Germany
First published on 7th April 2022
Abstract
Herein we report the development of a sequential synthesis route towards annulated imidazo[4,5-c]isoquinolines comprising a GBB-3CR, followed by an intramolecular imidoylative cyclisation. X-Ray crystallography revealed a flat 3D structure of the obtained polyheterocycles. Thus, we evaluated their interactions with double-stranded DNA by establishing a pUC-19 plasmid-based gel electrophoresis mobility shift assay, revealing a stabilising effect on ds-DNA against strand-break inducing conditions.
Introduction
The class of multicomponent reactions (MCRs) stands out due to the high degree of diversification accessible through the variation of the individual starting materials.1 Thus, MCRs have emerged as a valuable tool in combinatorial and diversity-oriented synthesis, medicinal chemistry and recently also in polymer chemistry2 and material science.3 Particularly regarding the development of new bioactive structures, MCRs excel due to their unmatched potential in establishing large and diverse compound libraries. However, especially medicinal chemistry demands an even higher level of variability, e.g. for high throughput screenings and lead structure optimisation. To achieve an even higher degree of diversification, post-modification strategies have been developed to convert MCR products into various kinds of heterocyclic scaffolds.4 These transformations are quite elegant since they offer short synthetic pathways to complex structural motifs and help enlarge the accessible chemical space. Due to its widespread applications and high functional group tolerance, most post-MCR strategies have emerged around the Ugi-4CR.5,6 Among these, ring closure via palladium-catalysed cross-coupling reactions are quite prominent.6 Ring expansion can be achieved by inserting C1-synthons like carbon monoxide or isocyanides into the newly coupled bond.7 Chauhan et al. applied this methodology to post-modify Ugi scaffolds and converted these into isoquinolin-1(2H)-ones.8 However, post-cyclisation strategies of the imidazo[1,2-a]pyridine scaffolds accessible via the Groebke–Blackburn–Bienaymé reaction (GBB-3CR)9 remain scarce, despite their tremendous biological relevance.10 Post-modifications of GBB-scaffolds include N-arylations (e.g. Ullman coupling),11,12 Pictet–Spengler reactions,12 Strecker-type cyclisations,13 oxidative ring closures14 or simple intramolecular condensation reactions.15 In most reports, no further studies of these compound classes were conducted to investigate possible bio-applications. Polyheterocycles are promising scaffolds showing various bioactive properties such as anticancer or DNA-intercalation activity.16 Due to their flat structure, extended π-systems, and highly polar nature, interactions with double-strand DNA (ds) are likely. For instance, DNA-intercalating agents can insert between two adjacent base pairs.17 As this induces a structural change of the double-strand, processes involving the unwinding of the DNA like transcription and replication are hindered.18 While this effect is responsible for the cytotoxicity and potential mutagenicity of DNA-intercalators,19 certain chemotherapeutics, the so-called tumour antibiotics like doxorubicin or daunorubicin, also rely on this antiproliferative effect. Aside from this, fluorescent intercalators like ethidium bromide or SYBR Green exhibit enhanced fluorescence upon intercalation into ds-DNA and are therefore commonly employed stains for nucleic acids in biochemical assays, e.g. gel electrophoresis. Thus, we envisioned a modular, sequential synthetic route towards these intriguing scaffolds to access diverse compound libraries and investigate their luminescence properties and interactions with DNA (Scheme 1).Results and discussion
The precursors were synthesised in a GBB-3CR using perchloric acid as the catalyst. In total, a set of five GBB-(4–8) precursors was obtained (Scheme 2). Overall, the 3-(o-bromophenyl)imidazo[1,2-a]heterocycle precursors were obtained in moderate to excellent yields, ranging from 20% to 84%. Considering the mechanism of the GBB-3CR, the yields of the aminopyridine reactions are significantly lower as the electron-withdrawing nature of the additional nitrogen atom decreases the nucleophilicity of the amine, thus hampering the imine formation. Similarly, electron-donating substituents on the aldehyde component also decrease the yields due to the reduced electrophilicity of the carbonyl carbon atom. On the other hand, electron-donating substituents on the amidine component increase the nucleophilicity of the amine, leading to higher yields.Subsequently, these precursors should be intramolecularly cyclised in an imidoylative Hartwig-Buchwald amination reaction to form pyrido[2′,1′:2,3]imidazo[4,5-c]isoquinoline ring systems. Another isocyanide component is inserted between the bromide and the secondary amine in this process. The resulting imine could not be isolated. Presumably, it immediately undergoes an imine–enamine tautomerisation and aromatisation accompanied by the elimination of gaseous iso-butene. Thereby, the π-system is extended to span over the four rings. Hence, tert-butyl isocyanide employed in the initial GBB-3CR can be considered a convertible isocyanide. A mechanistic rationale is depicted in Scheme 3. The constitution of the insertion products could also be verified through X-ray single-crystal analysis for two substances, also confirming their flat 3D structure (9d and 10a, Fig. 1).
 | ||
| Fig. 1 Molecular structure of 9d (left) and 10a (right) (displacement parameters are drawn at 30% (9d) or 50% (10a) probability level). | ||
The reaction conditions were optimised for the insertion of cyclohexyl isocyanide into 4 (Table 1). The desired insertion product was obtained in 83% under imidoylation conditions first reported by Orru et al. to synthesise 4-amino quinazolines.20 Using Pd-Peppsi-iPr as the pre-catalyst, the yield could even be increased to 94%. Under ligand-free conditions, the yield was significantly decreased. In most reactions, potassium acetate was used as a base. Stronger bases were not employed as these would accelerate the amine addition to the intermediate palladium complex, which would compete with the isocyanide insertion into the carbon-palladium bond, leading to a non-imidoylative Hartwig-Buchwald amination process affording the respective five-membered ring.
Using the optimised reaction conditions, the p-bromophenyl imidazo[1,2-a]pyridines and -pyrazines were subjected to imidoylation with various aliphatic isocyanides. The results are depicted in Scheme 4.
 | ||
| Scheme 4 Synthesis of annulated imidazo[4,5-c]isoquinolines via an imidoylative intramolecular Hartwig-Buchwald amination. *Pd(dba)2 (5 mol%) was used as the Pd-source. | ||
The insertion products were obtained in moderate to excellent yields from 20% to 94%. The best results were obtained with precursor 4. The more electron-poor pyrazine analogue 5 was less reactive, resulting in lower yields for 10a and 10b. This trend was also observed for the methoxy-substituted precursors 6 and 7. Among the aliphatic isocyanides evaluated with 4, cyclohexyl isocyanide performed best, followed by pentyl and 1-adamantyl isocyanide, while tert-butyl isocyanide afforded the respective insertion product 9b in only 24% yield. Presumably, this could be due to the increased steric demand of the tert-butyl residue, whereas the 1-adamantyl residue is more rigid. Attempts to incorporate aromatic isocyanides were unsuccessful. A plausible reason for the reduced reactivity of aromatic isocyanides is the formation of more stable palladium complexes. Due to the stronger donating character of aromatic isocyanides, they have a stronger affinity to palladium, leading to the formation of cationic species with multiple isocyanide ligands.21 Additionally, their π-stacking ability adds to this effect. This leads to the deactivation of the catalytically active species, reducing the overall reactivity. On the other side, these multiply coordinated complexes can engage in insertion reactions affording multiple insertion products or isocyanide polymers.22 Another trend in isocyanide reactivity is polarity, resulting in a more difficult purification due to tailing effects during column chromatography. Especially using 2-morpholinoethyl isocyanide, a significant amount of the product could only be obtained in mixed fractions, thus reducing the overall yield of 9e.
The insertion products exhibit a strong blue fluorescence under irradiation with UV light. Thus, the fluorescent properties of the insertion products were investigated. UV/Vis absorption and fluorescence emission spectra were recorded. The normalised spectra are shown in Fig. 2. The selected compounds show similar absorption profiles with two bands at around 250–280 nm and 375–430 nm, consisting of several overlapping bands. The emission spectra all show a maximum emission of around 475 nm. Both absorption and emission profiles are slightly bathochromically shifted for the methoxy substituted substrates.
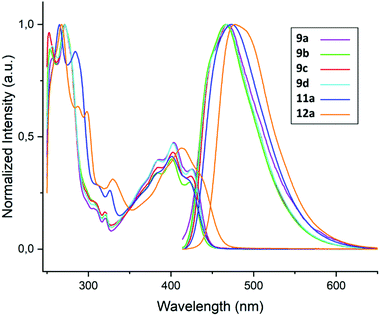 | ||
| Fig. 2 Normalised absorption and emission spectra of selected insertion products (1 mM solution in DMSO, λEx = 390 nm). | ||
pUC19-DNA electrophoretic mobility shift assay
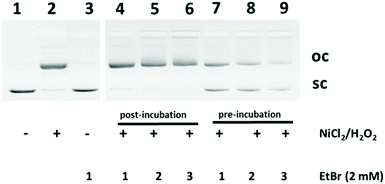
A selection of the synthesised compounds was evaluated in an electrophoretic mobility shift assay (EMSA) to assess their effect on the properties of plasmid DNA. Previously, this assay has been established to investigate the strand-break induction capacity of DNA-damaging substances like transition metal compounds.23 In principle, plasmids of supercoiled DNA are subjected to the substance of interest. Induction of single-strand breaks converts the supercoiled (sc) plasmid DNA into open circular (oc) DNA; double-strand breaks lead to the formation of linear DNA, which can both be separated from sc-DNA by gel electrophoresis.24 Commonly, this assay utilises PM2-DNA, a 10 kb plasmid isolated from the PM2-bacteriophage.25 However, as the isolation of PM2-DNA is a rather challenging process, using different, more easily accessible plasmids would simplify this assay. For example, pUC19 DNA, a small E. coli plasmid of only 2686 bp in length, could provide a more inexpensive and easily obtainable alternative to PM2-DNA. The pUC DNA EMSA was established as an in vitro test system to investigate whether selected annulated imidazo[4,5-c]isoquinolines have the potential to intercalate into DNA. This is shown in Fig. 3 for treatment of pUC19 DNA with NiCl2/H2O2 as DNA strand break inducing agent26 (Fig. 3, lane 2). Ethidium bromide was used as a positive control for a DNA intercalating agent. Ethidium bromide on its own did not affect the migration of pUC19 DNA (Fig. 3, lane 3). However, in combination with NiCl2/H2O2, ethidium bromide affected the formation of oc-DNA. Pre-incubation of pUC19 DNA with ethidium bromide for 1 h followed by treatment with NiCl2/H2O2 for 1 h reduced the amount of oc-DNA dose-dependently (Fig. 3, lanes 7–9), while post-incubation with ethidium bromide had only a small effect on the formation of oc-DNA induced by NiCl2/H2O2 (Fig. 3, lanes 4–6). These data suggest that DNA intercalation prevented the generation of DNA strand breaks, indicating that the pUC assay is a suitable system to assess whether chemicals have the potential to intercalate into DNA.To assess the potential of selected compounds, pUC19 DNA was treated with 2 mM of 4 and 12a, respectively. As observed for treatment with ethidium bromide, pre-incubation with the compounds prevented the generation of NiCl2/H2O2-induced strand breaks and hence resulted in reduced amounts of oc-DNA (Fig. 4, lanes 5, 7), while post-incubation with the compounds showed only a slight effect on the generation of oc-DNA (Fig. 4, lanes 6 and 8).
Applying the annulated imidazo[4,5-c]isoquinolines at a low concentration also strongly reduced the amount of oc-DNA. Pre-treatment of pUC19 DNA with 0.5 mM of the selected compounds for 1 h, followed by post-incubation with NiCl2/H2O2 for 1 h, prevented the generation of strand breaks and, hence, oc DNA formation (Fig. 5, lanes 6–8). In contrast, pre-treatment of pUC19 DNA with cisplatin,27 which was used as a negative control as it has no intercalating potential but damages the DNA by binding to guanine or adenine, showed only a small effect on the formation of oc-DNA and treatment with cisplatin alone did not affect the migration of pUC19 DNA (Fig. 5, lanes 4 and 5).
The formation of oc-DNA by treatment with NiCl2/H2O2 and its inhibition by the selected annulated imidazo[4,5-c]isoquinolines was evaluated by quantification of the sc and oc agarose gel DNA bands (Fig. 6 and 7). Ethidium bromide as positive intercalating control prevented the formation of oc-DNA by 50% while treatment with the negative control cisplatin resulted in only a small reduction of the amount of oc-DNA. Annulated imidazo[4,5-c]isoquinoline mediated inhibition of oc DNA formation was up to 80% for 11a, suggesting that the compounds under investigation are strong DNA intercalating agents.
 | ||
| Fig. 6 Semi-quantification of the agarose gel bands. All compounds were analysed in three independent experiments; the results are shown as mean values ± standard deviation. | ||
Conclusions
A library of twelve annulated imidazo[4,5-c]isoquinolines was synthesised via a novel GBB-3CR/imidoylative Hartwig-Buchwald amination sequence. The absorption and emission spectra of these compounds were recorded. The pUC-19 DNA electrophoresis mobility shift assay was established to investigate the DNA-intercalate properties of the synthesised compounds. We observed a strong inhibitory effect against oxidative strand break induction of the tested compounds, even surpassing ethidium bromide. This stabilising effect indicates DNA-intercalating properties. Due to this DNA-interaction, we further plan to investigate possible antiproliferative and cytostatic activities.Author contributions
The synthesis and characterisation of the compounds were done by MS, CRA and MR. Absorption and emission measurements were performed by JH. The pUC19 relaxation assay was performed by OS. MN provided crystallographic data. Conflicts of interestConflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge DFG support under BR 17530 and the KIT Campus Transfer GmbH for the financial support to MS for her PhD studies. We thank the technical and analytical staff at IOC (KIT) for assistance.Notes and references
- A. Dömling and I. Ugi, Angew. Chem., Int. Ed., 2000, 39, 3168–3210 CrossRef CAS; A. Dömling, W. Wang and K. Wang, Chem. Rev., 2012, 112, 3083–3135 CrossRef PubMed; E. Ruijter and R. V. Orru, Drug Discovery Today: Technol., 2013, 10, e15–e20 CrossRef PubMed; E. Ruijter, R. Scheffelaar and R. V. Orru, Angew. Chem., Int. Ed., 2011, 50, 6234–6246 CrossRef PubMed.
- B. Yang, Y. Zhao, Y. Wei, C. Fu and L. Tao, Polym. Chem., 2015, 6, 8233–8239 RSC.
- M. Stahlberger, N. Schwarz, Z. Hassan, C. Zippel, J. Hohmann, M. Nieger and S. Bräse, Chem. – Eur. J., 2022, 28, e202103511 CAS.
- T. Zarganes-Tzitzikas, A. L. Chandgude and A. Dömling, Chem. Rec., 2015, 15, 981–996 CrossRef CAS PubMed.
- V. Gracias, J. D. Moore and S. W. Djuric, Tetrahedron Lett., 2004, 45, 417–420 CrossRef CAS; C. Kalinski, M. Umkehrer, J. Schmidt, G. Ross, J. Kolb, C. Burdack, W. Hiller and S. D. Hoffmann, Tetrahedron Lett., 2006, 47, 4683–4686 CrossRef; R. Krelaus and B. Westermann, Tetrahedron Lett., 2004, 45, 5987–5990 CrossRef; A. Kumar, D. D. Vachhani, S. G. Modha, S. K. Sharma, V. S. Parmar and E. V. Van der Eycken, Beilstein J. Org. Chem., 2013, 9, 2097–2102 CrossRef PubMed; S. G. Modha, A. Kumar, D. D. Vachhani, J. Jacobs, S. K. Sharma, V. S. Parmar, L. Van Meervelt and E. V. Van der Eycken, Angew. Chem., Int. Ed., 2012, 51, 9572–9575 CrossRef PubMed; S. Pandey, S. Khan, A. Singh, H. M. Gauniyal, B. Kumar and P. M. Chauhan, J. Org. Chem., 2012, 77, 10211–10227 CrossRef PubMed; K. Pérez-Labrada, I. Brouard, I. Méndez, C. S. Pérez, J. A. Gavín and D. G. Rivera, Eur. J. Org. Chem., 2014, 3671–3683 CrossRef; A. A. Peshkov, V. A. Peshkov, O. P. Pereshivko and E. V. Van der Eycken, Tetrahedron, 2015, 71, 3863–3871 CrossRef; A. D. Santos, L. El Kaim, L. Grimaud and C. Ronsseray, Beilstein J. Org. Chem., 2011, 7, 1310–1314 CrossRef PubMed; J. Shi, J. Wu, C. Cui and W. M. Dai, J. Org. Chem., 2016, 81, 10392–10403 CrossRef PubMed; Z. Xu, F. De Moliner, A. P. Cappelli and C. Hulme, Angew. Chem., Int. Ed., 2012, 51, 8037–8040 CrossRef PubMed.
- U. K. Sharma, N. Sharma, D. D. Vachhani and E. V. Van der Eycken, Chem. Soc. Rev., 2015, 44, 1836–1860 RSC.
- J. W. Collet, T. R. Roose, E. Ruijter, B. U. W. Maes and R. V. A. Orru, Angew. Chem., Int. Ed., 2020, 59, 540–558 CrossRef CAS PubMed; J. W. Collet, T. R. Roose, B. Weijers, B. U. W. Maes, E. Ruijter and R. V. A. Orru, Molecules, 2020, 25(21), 4906 CrossRef PubMed; S. Lang, Chem. Soc. Rev., 2013, 42, 4867–4880 RSC; B. Liu, Y. Li, H. Jiang, M. Yin and H. Huang, Adv. Synth. Catal., 2012, 354, 2288–2300 CrossRef; G. Qiu, Q. Ding and J. Wu, Chem. Soc. Rev., 2013, 42, 5257–5269 RSC; S. Sharma and A. Jain, Tetrahedron Lett., 2014, 55, 6051–6054 CrossRef; T. Tang, X. D. Fei, Z. Y. Ge, Z. Chen, Y. M. Zhu and S. J. Ji, J. Org. Chem., 2013, 78, 3170–3175 CrossRef PubMed; T. Vlaar, E. Ruijter, B. U. Maes and R. V. Orru, Angew. Chem., Int. Ed., 2013, 52, 7084–7097 CrossRef PubMed.
- V. Tyagi, S. Khan, A. Giri, H. M. Gauniyal, B. Sridhar and P. M. S. Chauhan, Org. Lett., 2012, 14, 3126–3129 CrossRef CAS PubMed.
- H. Bienaymé and K. Bouzid, Angew. Chem., 1998, 110, 2349–2352 CrossRef CAS; C. Blackburn, B. Guan, P. Fleming, K. Shiosaki and S. Tsai, Tetrahedron Lett., 1998, 39, 3635–3638 CrossRef; K. Groebke, L. Weber and F. Mehlin, Synlett, 1998, 661–663 CrossRef.
- A. Boltjes and A. Dömling, Eur. J. Org. Chem., 2019, 7007–7049 CrossRef CAS PubMed; N. Devi, R. K. Rawal and V. Singh, Tetrahedron, 2015, 71, 183–232 CrossRef; G. Richter, V. W. Y. Liao, P. K. Ahring and M. Chebib, Front. Neurosci., 2020, 14, 599812 CrossRef PubMed; S. Shaaban and B. F. Abdel-Wahab, Mol. Divers., 2016, 20, 233–254 CrossRef PubMed.
- A. El Akkaoui, M.-A. Hiebel, A. Mouaddib, S. Berteina-Raboin and G. Guillaumet, Tetrahedron, 2012, 68, 9131–9138 CrossRef CAS.
- V. Tyagi, S. Khan, V. Bajpai, H. M. Gauniyal, B. Kumar and P. M. S. Chauhan, J. Org. Chem., 2012, 77, 1414–1421 CrossRef CAS PubMed.
- Z. Tber, M. A. Hiebel, H. Allouchi, A. El Hakmaoui, M. Akssira, G. Guillaumet and S. Berteina-Raboin, RSC Adv., 2015, 5, 35201–35210 RSC.
- Y. Li, J. H. Huang, J. L. Wang, G. T. Song, D. Y. Tang, F. Yao, H. K. Lin, W. Yan, H. Y. Li, Z. G. Xu and Z. Z. Chen, J. Org. Chem., 2019, 84, 12632–12638 CrossRef CAS PubMed.
- O. Chavignon, M. Raihane, P. Deplat, J. L. Chabard, A. Gueiffier, Y. Blanche, G. Dauphin and J. Teulad, Heterocycles, 1995, 41, 2019–2027 CrossRef CAS; N. Devi, D. Singh, R. K. Sunkaria, C. C. Malakar, S. Mehra, R. K. Rawal and V. Singh, ChemistrySelect, 2016, 1, 4696–4703 CrossRef.
- A. Favier, M. Blackledge, J.-P. Simorre, S. Crouzy, V. Dabouis, A. Gueiffier, D. Marion and J.-C. Debouzy, Biochemistry, 2001, 40, 8717–8726 CrossRef CAS PubMed.
- L. S. Lerman, J. Mol. Biol., 1961, 3, 18–30 CrossRef CAS PubMed.
- E. C. Long and J. K. Barton, Acc. Chem. Res., 1990, 23, 271–273 CrossRef CAS.
- C.-S. Lee, Y. Hashimoto, K. Shudo and M. Nagao, Heterocycles, 1984, 22, 2249–2253 CrossRef CAS; M. J. Waring, J. Mol. Biol., 1965, 13, 269–282 CrossRef.
- G. Van Baelen, S. Kuijer, L. Rycek, S. Sergeyev, E. Janssen, F. J. de Kanter, B. U. Maes, E. Ruijter and R. V. Orru, Chem. – Eur. J., 2011, 17, 15039–15044 CrossRef CAS PubMed.
- L. A. Perego, P. Fleurat-Lessard, L. El Kaïm, I. Ciofini and L. Grimaud, Chem. – Eur. J., 2016, 22, 15491–15500 CrossRef CAS PubMed.
- M. Suginome and Y. Ito, in Polymer Synthesis, Springer Berlin Heidelberg, Berlin, Heidelberg, 2004, pp. 77–136, DOI:10.1007/b95531.
- A. Kellett, Z. Molphy, C. Slator, V. McKee and N. P. Farrell, Chem. Soc. Rev., 2019, 48, 971–988 RSC.
- T. R. Strick, J.-F. Allemand, D. Bensimon and V. Croquette, Biophys. J., 1998, 74, 2016–2028 CrossRef CAS PubMed.
- M. Asmuss, L. H. F. Mullenders, A. Eker and A. Hartwig, Carcinogenesis, 2000, 21, 2097–2104 CrossRef CAS PubMed; R. H. Männistö, H. M. Kivelä, L. Paulin, D. H. Bamford and J. K. H. Bamford, Virology, 1999, 262, 355–363 CrossRef PubMed.
- M. Cetraz, V. Sen, S. Schoch, K. Streule, V. Golubev, A. Hartwig and B. Köberle, Arch. Toxicol., 2017, 91, 785–797 CrossRef CAS PubMed; S. Kawanishi, S. Inoue and K. Yamamoto, Carcinogenesis, 1989, 10, 2231–2235 CrossRef PubMed; G. Pratviel, J. Bernadou and B. Meunier, Angew. Chem., 1995, 34, 746–769 CrossRef.
- N. J. Wheate, S. Walker, G. E. Craig and R. Oun, Dalton Trans., 2010, 39, 8113–8127 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2152372 and 2152373. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d2ob00372d |
| This journal is © The Royal Society of Chemistry 2022 |