Open Access Article
Open Access ArticleSynthesizing the biochemical and semiconductor worlds: the future of nucleic acid nanotechnology
Jacob M.
Majikes†
 and
J. Alexander
Liddle†
and
J. Alexander
Liddle†
 *
*
Physical Measurement Laboratory, National Institute Standards and Technology, 100 Bureau drive, Gaithersburg, MD 20878, USA. E-mail: Liddle@nist.gov
First published on 13th October 2022
Abstract
Since its inception nearly 40 years ago [Kallenbach, et al., Nature, 1983, 305, 829; N. C. Seeman, J. Theoretical Biology, 1982, 99, 237], Nucleic Acid Nanotechnology (NAN) has matured and is beginning to find commercial applications. For the last 20 years, it has been suggested that NAN might be an effective replacement for parts of the semiconductor lithography or protein engineering workflows. However, in that time, these incumbent technologies have made significant advances, and our understanding of NAN's strengths and weaknesses has progressed, suggesting that the greatest opportunities in fact lie elsewhere. Given the commitment of resources necessary to bring new technologies to the market and the desire to use those resources as wisely as possible, we conduct a critical examination of where NAN may benefit from, and provide benefit to, adjacent technologies and compete least with market incumbents. While the accuracy of our conclusions may be limited by our ability to extrapolate from the current state of NAN to its future commercial success, we conclude that the next promising direction is to create a bridge between biology and semiconductor technology. We also hope to stimulate a robust conversation around this technology's capabilities with the goal of building consensus on those research and development efforts that would advance NAN to the greatest effect in real-world applications.
Significance statement
Nucleic acid nanotechnology has the potential to act as a bridge between the worlds of biology and semiconductor technology. This potential will only be realized if new groups of researchers with a foot in each world, and an eye on the revolutionary applications enabled by NAN's ability to connect the two, enter the field. The context and analysis we provide here is designed to attract these new groups and provide signposts to exciting new avenues of research.Introduction
The phrase “fools rush in where angels fear to tread” is particularly apt in the case of technological soothsaying. We do not relish the possibility of making a foolish prediction, especially since this possibility may be high in the case of Nucleic Acid Nanotechnology (NAN) because of its unique mix of top-down design with bottom-up assembly. However, for technologies to develop fruitfully, a robust dialogue is needed, provoked by the competition between bold, if imperfect, visions. Our only recourse is to think carefully, step into the breach, present a vision, and hope to be on the side of the angels. To do so honestly is our intent, so we hope that subjecting NAN to an intentionally critical view serves to counterbalance the more optimistic claims regarding NAN's capabilities and will help our community to invest its resources effectively.We are inspired to offer the following assessment by efforts half a decade ago to define a useful intersection between semiconductor technology and synthetic biology, or SemiSynBio.1,2 These efforts were in turn inspired by the obvious scalability and energy efficiency of computation and information storage in biological organisms combined with the successful application of DNA functionalized nanoparticles3 and development of DNA nanostructures.4,5 Since then, RNA nanotechnology has flourished,6 the falling cost of phosphoramidite synthesis has plateaued, and nucleic acid nanotechnology has advanced significantly.5,7,8 Based on these developments, we believe it is timely to revisit the future of the field and its growth outside of academe.
We predict that NAN will revolutionize nanofabrication, but not by replacing any current workflows in either top-down semiconductor fabrication or protein engineering. Instead, it will create a new class of composite nanostructures by integrating a variety of optical, electronic, and biochemical capabilities, while providing rudimentary edge computation in the chemical domain.
This prediction has major implications for NAN. For example, pursuit of integrated circuit fabrication via DNA self-assembly will be shown to be economically fruitless. Other high-interest topics, such as DNA data storage, may reside on the edge of commercial viability; plausibly able to capture niche markets. In common with previous surveys of the field,7,9,10 we believe that products such as super-resolution rulers11,12 and in situ biosensors,13,14 will continue to satisfy small markets of researchers, while complex nucleic acid therapeutics15,16 will soon address larger, as-yet unmet needs.
In the future, NAN will find industrial applications through its singular ability to integrate numerous, heterogenous functional elements and to perform chemical logic, as enabled by DNA computation research.17 Thus, NAN will connect semiconductor devices and biomacromolecular systems. If this is the case, any call-to-action to implement NAN must necessarily include the following:
| Design | To handle multi-level conceptual hierarchies |
| Purification | To mitigate thermodynamic limits on yield |
| Yield verification/metrology | To optimize pre-production and manufacturing quality control |
We will first provide context for this prediction, describe how NAN has built momentum, elucidate its principal strengths and weaknesses, highlight initial market entry points, and present our prediction in depth before concluding.
Technological soothsaying
It's tough to make predictions, especially about the future -Yogi BerraSome technology predictions are successful, such as Moore's Law which correctly augured the trajectory of the semiconductor industry: Moore observed a link between manufacturing cost and computational performance that allowed him to extrapolate four data points onto a trend that held true for five decades.18 He was aided in this by the preexisting paradigm of a market for information processing.
Other predictions have been less successful, e.g., single carbon nanotube transistors in integrated circuits have yet to materialize commercially.19 However, industrial applications of carbon nanotubes have slowly come into focus, ranging from engineering composites to perfect light absorbers, with transparent conductive films and sensors coming to market.20
These examples emphasize that while technical challenges may stimulate the imagination, commercial considerations are crucial and must be included when predicting real-world impact. Many forecasts for carbon nanotube electronics underestimated the investment required to integrate a new component into an already extremely complex, optimized manufacturing process. Even worse, they underestimated these costs at a time when the integrated circuit industry had access to lower hanging fruit for investment. No technology exists in isolation – both competing and complementary technologies merit consideration.
History is littered with the graves of distinct, and even superior, technologies which tried and failed to unseat market incumbents.21 This is because we live in a world where time and money are finite resources: nascent technologies must be able to compete in the foreseeable future, not just at the limit of time approaching infinity. More lucrative markets can justify greater R&D investments, but the bar becomes higher when nascent technologies are entering an established and already-crowded field. For NAN, these incumbents include click chemistry, protein engineering, and semiconductor fabrication. Despite this formidable array of competitors, NAN has built considerable momentum and is delivering new capabilities which promise to create new markets.
Virtuous cycles
You don't have to swing hard to hit a home run. If you got the timing, it'll go. – Yogi BerraLike any promising new technology, NAN is poised to be an overnight success after decades of hard work and incremental improvements. Developments internal to NAN, have been aided by advances in DNA synthesis, sequencing, and modification driven by the demands of the biomedical industry. Recent events, such as the mass production of mRNA for COVID vaccines will further de-risk biomanufacturing of nucleic acid nanostructures that can be implemented via post-transcriptional folding in RNA.22 While we refer the reader to several exhaustive reviews of progress in NAN both broadly7,8,23–25 and for specific applications,7,24,26,27 we believe it is sufficient for our prediction to say that the virtuous cycles driving technical improvements and field specific capabilities, depicted in Fig. 1, have reached a critical threshold for commercial viability.
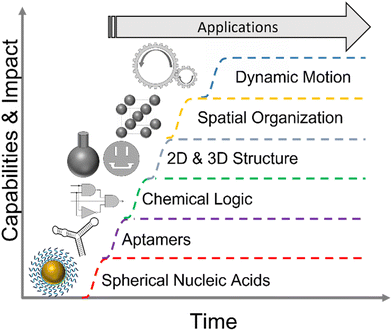 | ||
| Fig. 1 The growth of the nucleic acid nanotechnology toolbox. One such advance, DNA origami, merits explicit discussion.28 | ||
Origami is modular, simple to design,29–31 does not require expensive oligomer purification, and has cooperative energetics leading to relatively high yields of correctly assembled structures. The same origami designs can be reused repeatedly for a wide variety of experiments. Origami allows researchers to fully leverage the inherent sequence modularity of DNA. Modularity enables abstraction, i.e., NAN structural motifs. Abstraction, in turn, enables conceptual and physical hierarchies, e.g., directed origami tiling. Finally, these hierarchies enable the complexity and flexibility that are hallmarks of NAN.
However, we must caution that, while origami is a useful tool in the lab, it may not necessarily be the right method for high-volume manufacturing. Origami fulfils the same purpose as an electronics breadboard at the molecular scale, making it an excellent tool for prototyping. However, like a breadboard, origami is inherently inefficient in the amount of material used relative to each component assembled. This limitation, together with the dearth of efficient macromolecular purification techniques,32 can render applications requiring significant mass of origami, e.g., smart therapeutics, challenging.
Nevertheless, origami has put NAN into the hands of a diverse and ambitious group of researchers, allowing them to rapidly explore its potential, accelerating the virtuous cycle, while identifying applications and maturing its capabilities.
Nucleic acid nanofabrication
If you can't imitate him, don't copy him -Yogi BerraUnfortunately, it takes time for novel technological capabilities to be digested by the engineering community. Until then, any new technology appears, like the laser, to be a “solution looking for a problem”,33 and it is tempting to apply this new “hammer” to a collection of existing “nails” with unsatisfactory results.
This temptation has proven particularly strong in the context of semiconductor manufacturing because NAN's top-down design and molecular patterning characteristics suggest that it can usefully replace parts of the semiconductor workflow. NAN integration with these workflows is hobbled in two critical ways. First, the cations necessary for nucleic acid self-assembly would, if present during fabrication, destroy the electrical properties of most semiconductor devices.34 Second, NAN assembly occurs at, or within a few kT of, thermodynamic equilibrium, under which conditions the defect percentage is high,35–37 greater than one in 102, and scales poorly with size.38,39 The intrinsic defectivity of NAN makes it a poor choice for leading-edge nanoelectronic fabrication, in which defect levels are, and must be, routinely less than one in 1012. In biological organisms, separate complex cellular machineries repair damaged DNA and dispose of misfolded proteins, which comprise as much as 30% of those synthesized.40 These machineries are not interchangeable, and neither can handle misfolded nucleic acid nanostructures. Semiconductor-compatible defect levels will remain out of reach without equivalent machineries, the development of which is a major, unaddressed challenge.
The difficulties of integrating NAN into protein engineering workflows for therapeutics are less severe, but not negligible. In biomanufacturing, functional optimization of aptamers16,41 and proteins is achieved through evolutionary selection: individual random mutations in sequence result in changes in structure that can yield large changes in function. However, because NAN structure depends not on a single sequence, but on a large number of paired sequences, a single mutation on a single strand will not significantly change a structure. Optimization of NAN function requires the mutation of the entire design, which requires simultaneous large changes in sequence coherently amongst all the constituent oligomers. To our knowledge no scheme capable of NAN structural mutations has been advanced. Additionally, there is no NAN equivalent to ATP-powered work that would enable mimicry of advanced biological functionality.
In summary, while NAN provides access to a size regime relevant for both semiconductor and biochemical communities, we emphasize that it has the systems integration capabilities of neither, nor is it compatible with their workflows. This is a significant hurdle to competition with either of these technologies, as NAN must then compete with the whole workflow, rather than one step in it. However, this does not mean that NAN cannot be integrated successfully with the products of those workflows,42–47 and new markets made accessible through its unique capabilities, which we outline below.
First, NAN has an unusually straightforward design process for a bottom-up self-assembly technique.29,48 While this is unlikely to generate applications on its own, it is a critical enabler.
Second, NAN can direct multiple, diverse nanoscale components, through sequence addressability to predetermined locations on a structure.30,49 The directing sequences are often modular, allowing efficient re-engineering. For a small number of functional components, connection by synthetic chemistry, e.g., click chemistry, is often more practical as bifunctional polymer linkers are commercially available with the same moieties used for covalent binding to nucleic acids.50 However, when connecting more than 3 or 4 functional elements at a desired spacing, NAN is uniquely capable.
Third, NAN inherently produces multiple competing conformational states because the energetics of a single base pair or fold are on the order of kT. Because of this, assembly into a single state can never be guaranteed. By contrast, in semiconductor systems defect energies are sufficiently high that there is no equilibrium population, and they can be engineered out.18 NAN structures’ sampling of conformational states enables stimulus response through design of the free-energy landscape. Methods for doing this have been thoroughly explored by the DNA computation community. Of these, strand displacement is by far the most mature, and can generate signaling cascades, oscillators, and basic logic gates.17 DNA computing has the potential to be massively parallel, albeit with much slower individual computations,51,52 and is currently contending with leak reactions and spurious binding which become increasingly burdensome as the number of strands increases.53 More structural NAN features such as physicochemical properties, e.g., stiffness,7,54 small molecule/aptamer recognition,41,55 and pH sensing13,56–58 have also been used to create dynamic responses.
The power of NAN's addressability and control over the free-energy landscape, combined with its straightforward design, yields exceptional flexibility and allows higher levels of conceptual abstraction. These characteristics have enabled early NAN applications.
Initial market entry points
You've got to be very careful if you don't know where you are going, because you might not get there. – Yogi BerraNAN's strengths can be seen clearly in nucleic acid-only smart therapeutic applications currently approaching the market, e.g., reversible anticoagulants.59–61 In this example, anchor aptamers which bind tightly to thrombin (a key protein in the coagulation cascade) but do not inhibit its activity, are precisely spaced with aptamers that bind weakly, but inhibit coagulation. This significantly enhances anticoagulation activity and can be ‘turned off’ via strand displacement logic. The ability to turn coagulation on and off via external signals would be invaluable for surgical applications and cannot be achieved readily without NAN. Other standalone NAN applications in development include targeted drug delivery to cancer cells,15 delivery of siRNA,62in vivo sensors for difficult-to-measure ions,13 or toehold switch riboregulators for viral detection.63 These products are first-to-market because their functional elements are nucleic acid modifications or aptamers, significantly reducing the barriers associated with purification and systems engineering. Progress for NAN therapeutics using heterogeneous functional elements will enter the market later as these barriers are surmounted. All of these therapeutics integrate multiple functional elements in conjunction with logic provided by free energy landscape engineering. We predict this will be a hallmark of successful NAN products.
The future of NAN
The future ain't what it used to be – Yogi BerraGeneral predictions
In the decade prior to the 2016 SemiSynBio workshop and the resulting 2018 roadmap,1,2 the cost of working with DNA decreased dramatically: by an order of magnitude for DNA oligomers [<200 bases], two orders of magnitude for DNA genes [>1000 bases], and more than five orders of magnitude for DNA sequencing.64,65 These breakthroughs defined the context for the SemiSynBio perspective on the future. However, much has changed in the nucleic acid and semiconductor industries since then ref. 66.In the half decade since the SemiSynBio roadmap, the rate of cost reduction for DNA sequencing has diminished67 while those for DNA oligomer synthesis have stayed nearly constant. However, in this same period, gene synthesis technology has continued to improve68,69 and there have been major advances in biomanufacturing scale-up of DNA origami.70 As a result, the environment for NAN is favorable, and the prognosis for production is positive. However, we do not yet know how to transition from breadboard prototypes, e.g., origami, to integrated and optimized manufacturable devices, except in cases with nucleic acid-only functional elements where co-transcriptional folding may be implemented.22
The last half decade has also generated tectonic shifts in the semiconductor industry. Cost and performance improvement can no longer be maintained by scaling transistor feature size, and the path forward via changes in transistor geometry is already defined. The unending demand for better performance is driving innovations in circuit design, heterogeneous integration, advanced packaging, and 3D fabrication. The latter is particularly relevant, since self-assembly is often surmised to have an advantage in 3D patterning. The current commercial state of the art sets an exceedingly high bar – a leading-edge microprocessor may have over 15 levels of metal,71 while NAND flash memory devices currently have 128 device levels.72
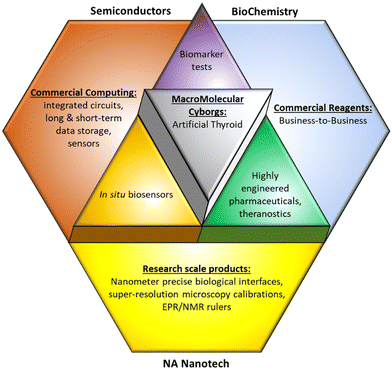
In light of this altered landscape, a revised definition of SemiSynBio may illuminate a more profitable path to the future, illustrated in Fig. 2. Under this new definition, NAN integrates the products of semiconductor and protein engineering workflows: this synthesis will lead to new functionalities which are unachievable by either independently.
 | ||
| Fig. 2 Our revised definition for SemiSynBio as a commercially relevant application of NAN. Tetrahedra and Protein image reproduced from ref. 73 and 74 respectively. | ||
We take a broad view of “Semi” that covers the full range of devices and systems that can be fabricated using the semiconductor manufacturing toolset of deposition, lithography, and etch. In addition to electronic circuits, we include MEMS, microfluidics, photonics, etc. Similarly, “Bio” should be taken to include aptamers, enzymes, DNAzymes, RNAzymes, lipids, carbohydrates, etc.
Semi enables us to build complicated, hierarchical systems to generate and manage signals and information, but it is currently unable to achieve the sensitivity and especially the specificity of biomacromolecular interactions. Conversely, outside the cell, Bio can still provide exceptional specificity, but hierarchy and signal feedback are limited. Until we can engineer biological systems de novo, replicating their functionality with only a few de-integrated, or disjecta membra, components is infeasible. In the meantime, using NAN to synthesize Bio and Semi capabilities will allow practicable recapitulation and engineering of a useful subset of biological functions,75 particularly those reliant on spatial organization of macromolecules.76,77
At the highest level, our prediction for NAN markets comprises SemiSynBio products with the smart therapeutics already discussed, as illustrated in Fig. 3. More detailed predictions follow.
Detailed predictions
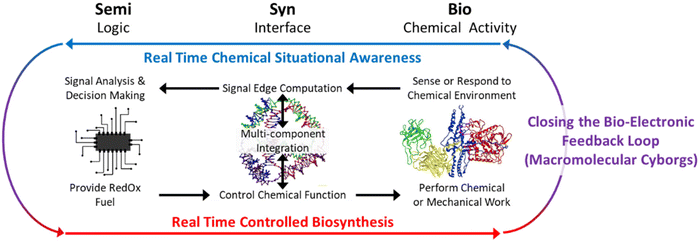
One early step is to augment electronic sensor systems with NAN structures and biomacromolecules for binding targets of interest. Signals linked to changes in NAN structure conformations, as a function of competition between multiple binding events of interest will provide context-rich information. Sensor arrays consisting of such structures, tuned for different concentrations and binding targets could mimic the hierarchy of the olfactory system, providing real time detection of multiple molecules of interest in cell culture or aqueous reaction vessels. This, coupled with AI, could be applied to unknown samples to act as an ‘artificial bloodhound’; the combination of true chemical situational awareness and electronics would be invaluable for monitoring environmental and health systems in real time.At an even higher level of complexity, NAN could create true feedback between electronic and biochemical system components. For this, structures must perform nanoscale chemical or mechanical work78 that is gated to either the biochemical or electronic components, a goal which is in line with ambitions of the molecular machine community78,79. While electric fields have been used to generate controlled nanoscale motion,80 we anticipate difficulty in using them to drive useful work in a complex system where they may interfere with other functions. An alternative to optical, magnetic, or electronic field-driven work would be the use of chemical fuels. This would require the additional, admittedly herculean, task of generating fuel molecules, e.g., Adenosine TriPhosphate (ATP), under electronic control. Bioelectrocatalysis, where electronics drive redox proteins,81–83 is a natural fit for this SemiSynBio paradigm, but while redox proteins in the electron transport chain do drive ATP synthase, integrating all of the components for ATP generation on a chip would be non-trivial. Faster, albeit less elegant, approaches might combine chromatophores, i.e., whole ATP generating vesicles, with microfluidics.84,85 Ultimately, both of these concepts are consistent with early visions of enhanced enzymatic function,86 in which the necessary compartmentalization87–89 is aided by top-down nanofabrication. The resulting system capable of reading in chemical context and acting on it through gated fuel generation could provide dynamic control of biochemical environments that is currently unachievable. We imagine that the most obvious, and most difficult, example of this would be an ‘artificial thyroid’ capable of monitoring and correcting imbalances in hormone and enzyme populations dynamically.
We term these systems ‘macromolecular cyborgs’ as their strengths and weaknesses are similar to those of fictional cyborgs. In the popular imagination, macro-scale cyborgs combine the efficiency and strength of mechanical systems with the flexibility and responsiveness of human, biological ones. Conversely, at the nanoscale, macromolecular cyborgs would combine the efficiency and specificity of biochemical systems with the flexibility and responsiveness of programmed electronic systems. Both fictional macro-scale and potential nanoscale cyborgs cannot simply regenerate components and must be built and repaired intentionally, rather than grown.
This limitation will restrict macromolecular cyborgs to high-value applications in which regular replacement of highly engineered electronic surfaces is tolerable. This same limitation means that applications such as light harvesting will not be economically viable beyond laboratory research. Because of the robustness and efficiency90 of both the semiconductor and non-biological organic material systems employed in photovoltaic devices there is no case for adding a biological component. In contrast, the artificial thyroid concept meets these criteria and could be envisioned as a product similar to the artificial pancreas, capable of dynamically monitoring and maintaining hormones with more finesse than the combination of lab tests and injections.
In the context of specific predictions, while we are not optimistic about it, we would be remiss not to mention DNA-based data storage. It has had lab-scale proof of concept,91,92 funding93 and significant media attention. Its appeal stems from a nearly indefinite chemical lifetime, a theoretically low readout energy, immunity to storage medium obsolescence, and potential synergy with DNA based computation.51 Application of these attributes to the growing market for ‘cold’ archival data makes a convincing argument for many.94
DNA's chemical lifetime minimizes the need for periodic maintenance, i.e., reading and re-writing data, which become the dominant energy and financial costs for storage times longer than 50 to 100 years. However, the lifetime is only indefinite when DNA is dry or frozen, and is much shorter when it is in solution, where reading and writing occur. Currently, read/write speeds are bounded by hybridization kinetics, though this could be addressed via massively parallel nanopore sequencing.94 As such, the physical limit for DNA read/write speeds is still under debate. However, we believe it is uncontroversial to say that it is physically impossible to use DNA in the context of data centers, which can service tens of gigabytes per second. Reducing the 1% of global energy consumption associated with ‘hot’ storage in data centers is a laudable goal but is a poor motivational argument for investment in DNA, which competes with more efficient archival storage.95 This is especially true as data centers reduce their energy footprint.96 Unfortunately, conflation of these data ‘temperature’ categories is common in arguments for DNA as a storage medium.
Our lack of marketplace optimism for archival applications, unsurprisingly, comes from the presence of market incumbents.97 Magnetic tape, which has a 10-to-20-year lifespan, also boasts low-energy consumption. Absent magnetic tape, it would be reasonable to predict that DNA data storage would capture the ultra-long term storage market and use that market to invest in improving read/write times. However, as proponents of DNA data storage pursue this path, the magnetic tape industry will be incentivized to invest in their own technology. In short, every argument for investment in DNA is also a valid for investment in magnetic tape.
Unfortunately for DNA, archival markets are inherently risk averse. Consumers looking for peace of mind in disaster scenarios are unlikely to find the uncertainties of adopting nascent technology palatable. Even worse, the cost-benefit advantage of DNA storage is in maintenance-over-time, with break-even compared to magnetic tape occurring in tens of years.94 DNA storage firms will have to choose between absorbing the upfront costs and offering a service plan whose benefits accrue on a longer timescale than the decision-maker's promotion cycle. We believe this will be a significant impediment to implementation.
Our pessimism addressed, we concede that one need not be victorious to be valuable. If DNA data storage captures the archival market, it will feed a virtuous cycle of development and enable other possibilities. DNA is well suited to storing short pieces of information in a highly encrypt-able format with extraordinarily high copy number.98 This could be valuable in watermarking genetically engineered products, as well as in eventual commercial application of synthetic biology.
However, technological soothsaying is an inherently uncertain endeavor. We would be unsurprised if much of our prediction failed to come true, except DNA based nanoelectronics which we consider to be fundamentally economically unrealizable. Ultimately, realistic projections of how market incumbents will improve over time provides vital context in targeting NAN research. However, even research efforts that prove to be wide of the mark will drive virtuous technical cycles and hasten NAN's growth into a mature technology, if not as effectively as more well-considered efforts.
Further afield
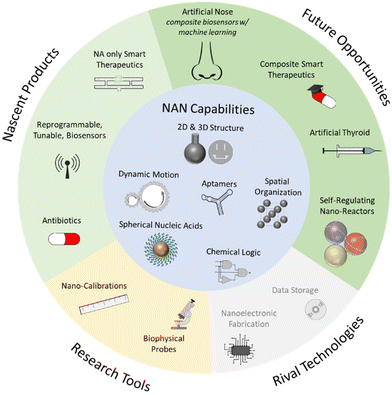
“That's how I kept myself positive—by not getting all negative.” – Yogi BerraWhile we emphasize the nearer, and more foreseeable future, the broader impact of NAN on the technology landscape will dramatically change how we interact with the nanoscale by increasing the scope, complexity, and our ability to engineer molecular recognition systems.
By providing a unique bridge between the micro- and nano- scales, NAN will enable countless advances in biophysical chemistry – beyond the emerging products we have already discussed: for example, the ability to spatially organize antibacterial agents has potential to improve antibiotics.99,100 The vast toolbox of colloidal self-assembly has already benefitted from NAN101 – further integration of these technologies could enable programmable, self-organizing, and self-regulating vesical micro-reactors.
By bridging the programmability and sensitivity of semiconductor systems with the specificity of biological ones, NAN will create a new class of flexible context-rich biosensors – this will culminate in macromolecular cyborg systems capable of sensing the chemical environment, performing internal logic, and then acting on that environment.
By adding chemical logic to synthetic biology, NAN will enable additional layers of complexity and responsivity for a field already rich in both.
These applications, and those closer to fruition discussed earlier, are depicted in Fig. 4, where we have grouped them by their proximity to market, and by their role in the market as either a rival to an incumbent, existing technology, or as a unique set of products.
Critically, in all the cases for which we are optimistic, NAN is an enabling technology that marries other existing capabilities. Capitalizing on these opportunities will require larger collaborations with an emphasis on system engineering.
Conclusions & takeaways
When you come to a fork in the road, take it – Yogi BerraIn both our review, and perspective thereof, we hope to have made a compelling case as to why and how NAN will be successfully commercialized. In addition, we have attempted to illustrate how forcing NAN to fit into either the semiconductor nanofabrication or protein engineering manufacturing paradigms hinders the effective exploitation of its unique features.
The strengths of NAN are its straightforward design (which speeds prototyping), modular addressability (which allows arrangement of large numbers of functional elements), and free energy landscape engineering (which allows computation in the chemical domain and dynamic modification of functional element interactions). NAN's weaknesses are the difficultly of incorporating it into existing manufacturing workflows and its intrinsically high, thermodynamically unavoidable, defect levels. By leveraging its strengths and avoiding its weaknesses, nucleic acid-only therapeutics are already on their way to market.
While there are no fundamental barriers to scale up and manufacturing the weaknesses elucidated above must be addressed to enable the next generation of products – SemiSynBio devices and biocomposite smart therapeutics. Focused investment is required in three areas: first, improvements in high-throughput metrology for yield verification that allow optimization during development and quality control during manufacturing; second, purification methods that mitigate NAN's intrinsic defectivity; third, robust design tools that manage multiple levels of conceptual hierarchy, supporting system-level design.
These calls to action will not be satisfied by the current NAN community alone but can be met with expertise drawn from the larger biomanufacturing and semiconductor communities, which will expand the knowledge base and state of the practice. As NAN products appear in the marketplace, we anticipate that commercial Semi and Bio will begin to develop dedicated tools, accelerating development and deployment.
We believe there is great reason to be optimistic for NAN. Our optimism for applications varies from high (smart therapeutics and macromolecular cyborgs), to moderate (biomarking/watermarking/encryption), to questionable (DNA data storage), to dismal (DNA nanoelectronics). Regardless of whether our optimism is well-placed, the diversity of this portfolio is large enough to guarantee the commercial viability of nucleic acid nanotechnology.
Conflicts of interest
No competing interests.Acknowledgements
We would like to acknowledge Will Hughes for critical feedback.References
- V. Zhirnov, Semiconductor Synthetic Biology, 2018 Search PubMed.
- M. Bathe, et al., Roadmap on biological pathways for electronic nanofabrication and materials, Nano Futures, 2019, 3, 12001 CrossRef CAS.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, Spherical nucleic acids, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef CAS PubMed.
- Y. Hu and C. M. Niemeyer, From DNA Nanotechnology to Material Systems Engineering, Adv. Mater., 2019, 31, 1806294 CrossRef PubMed.
- H. Ramezani and H. Dietz, Building machines with DNA molecules, Nat. Rev. Genet., 2019, 12, 5–26 Search PubMed.
- G. C. Shukla, et al., A boost for the emerging field of RNA nanotechnology report on the First International Conference on RNA nanotechnology, ACS Nano, 2011, 5, 3405–3418 CrossRef CAS PubMed.
- M. Deluca, Z. Shi, C. E. Castro and G. Arya, Dynamic DNA nanotechnology: Toward functional nanoscale devices, Nanoscale Horiz., 2020, 5, 182–201 RSC.
- Y. Hu and C. M. Niemeyer, From DNA Nanotechnology to Material Systems Engineering, Adv. Mater., 2019, 31, 1806294–1806294 CrossRef PubMed.
- K. Sanderson, What to make with DNA origami, Nature, 2010, 464, 158–159 CrossRef CAS PubMed.
- F. Hong, F. Zhang, Y. Liu and H. Yan, DNA Origami: Scaffolds for Creating Higher Order Structures, Chem. Rev., 2017, 117, 12584–12640 CrossRef CAS PubMed.
- J. Schnitzbauer, M. T. Strauss, T. Schlichthaerle, F. Schueder and R. Jungmann, Super-resolution microscopy with DNA-PAINT, Nat. Protoc., 2017, 12, 1198–1228 CrossRef CAS PubMed.
- J. J. Schmied, et al., DNA origami-based standards for quantitative fluorescence microscopy, Nat. Protoc., 2014, 9, 1367–1391 CrossRef CAS PubMed.
- N. Narayanaswamy, et al., A pH-correctable, DNA-based fluorescent reporter for organellar calcium, Nat. Methods, 2019, 16, 95–102 CrossRef CAS PubMed.
- A. Saminathan, et al., A DNA-based voltmeter for organelles, Nat. Nanotechnol., 2021, 16, 96–103 CrossRef CAS PubMed.
- S. M. Douglas, I. Bachelet and G. M. Church, A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads, Science, 2012, 335, 831–834 CrossRef CAS PubMed.
- J. A. Kulkarni, et al., The current landscape of nucleic acid therapeutics, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- F. C. Simmel, B. Yurke and H. R. Singh, Principles and Applications of Nucleic Acid Strand Displacement Reactions, Chem. Rev., 2019, 119, 6326–6369 CrossRef CAS PubMed.
- G. M. Moore, Cramming more components onto integrated circuits, Electronics, 1965, 38, 114 Search PubMed.
- M. Davenport, Much Ado About Small Things, Chem. Eng. News Arch., 2015, 93, 10–15 Search PubMed.
- R. Rao, et al., Carbon Nanotubes and Related Nanomaterials: Critical Advances and Challenges for Synthesis toward Mainstream Commercial Applications, ACS Nano, 2018, 12, 11756–11784 CrossRef CAS PubMed.
- C. Christensen, The Innovator's Dilemma, Harvard Business Review Press, 1997 Search PubMed.
- C. Geary, P. W. K. Rothemund and E. S. Andersen, A single-stranded architecture for cotranscriptional folding of RNA nanostructures, Science, 2014, 345, 799–804 CrossRef CAS PubMed.
- A. R. Chandrasekaran, et al., DNA nanotechnology approaches for microRNA detection and diagnosis, Nucleic Acids Res., 2019, 47, 10489–10505 CrossRef CAS PubMed.
- S. Surana, A. R. Shenoy and Y. Krishnan, Designing DNA nanodevices for compatibility with the immune system of higher organisms, Nat. Nanotechnol., 2015, 10, 741–747 CrossRef CAS PubMed.
- A. Jahanban-Esfahlan, et al., Dynamic DNA nanostructures in biomedicine: Beauty, utility and limits, J. Controlled Release, 2019, 315, 166–185 CrossRef CAS PubMed.
- Y. Ke, C. Castro and J. H. Choi, Structural DNA Nanotechnology: Artificial Nanostructures for Biomedical Research, Annu. Rev., 2018, 375–403 CAS.
- N. Stephanopoulos, Hybrid Nanostructures from the Self-Assembly of Proteins and DNA, Chem, 2020, 6, 364–405 CAS.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- J. M. Majikes and J. A. Liddle, DNA Origami Design: A How-To Tutorial, J. Res. Natl. Inst. Stand. Technol., 2021, 126, 1–33 CrossRef.
- K. F. Wagenbauer, et al., How We Make DNA Origami, ChemBioChem, 2017, 18, 1873–1885 CrossRef CAS PubMed.
- S. Dey, et al., DNA origami, Nat. Rev. Methods Primers, 2021, 1, 13 CrossRef CAS.
- D. Mathur and I. L. Medintz, Analyzing DNA Nanotechnology: A Call to Arms For The Analytical Chemistry Community, Anal. Chem., 2017, 89(5), 2646–2663 CrossRef CAS PubMed.
- A. Chodos, Physics History: May 16, 1960: Maiman Builds First Working Laser, APS News, 2010, vol. 19 Search PubMed.
- R. L. Hance, K. Erington and M. A. Chonko, Mobile ion contamination in CMOS circuits: a clear and present danger, 2002. DOI:10.1109/irws.1994.515819.
- M. T. Strauss, F. Schueder, D. Haas, P. C. Nickels and R. Jungmann, Quantifying absolute addressability in DNA origami with molecular resolution, Nat. Commun., 2018, 9, 1–7 CrossRef CAS PubMed.
- J. M. Majikes, P. N. Patrone, A. J. Kearsley, M. Zwolak and J. A. Liddle, Failure Mechanisms in DNA Self-Assembly: Barriers to Single-Fold Yield, ACS Nano, 2021, 15, 3284–3294 CrossRef CAS PubMed.
- Y. Ke, G. Bellot, N. V. Voigt, E. Fradkov and W. M. Shih, Two design strategies for enhancement of multilayer–DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning, Chem. Sci., 2012, 3, 2587 RSC.
- A. N. Marchi, I. Saaem, B. N. Vogen, S. Brown and T. H. LaBean, Towards larger DNA origami, Nano. Lett., 2014, 14(10), 5740–5747 CrossRef CAS PubMed.
- L. L. Ong, et al., Programmable self-assembly of three-dimensional nanostructures from 10,000 unique components, Nature, 2017, 552, 72–77 CrossRef CAS PubMed.
- U. Schubert, et al., Rapid degradation of a large fraction of newly synthesized proteins by proteasomes, Nature, 2000, 404, 770–774 CrossRef CAS PubMed.
- A. Krissanaprasit, C. M. Key, S. Pontula and T. H. LaBean, Self-Assembling Nucleic Acid Nanostructures Functionalized with Aptamers, Chem. Rev., 2021, 121(22), 13797–13868 CrossRef CAS PubMed.
- A. Gopinath, E. Miyazono, A. Faraon and P. W. K. Rothemund, Engineering and mapping nanocavity emission via precision placement of DNA origami, Nature, 2016, 535, 401–405 CrossRef CAS PubMed.
- X. Ouyang, et al., Docking of Antibodies into the Cavities of DNA Origami Structures, Angew. Chem., Int. Ed., 2017, 56, 14423–14427 CrossRef CAS PubMed.
- T. Kurokawa, et al., DNA Origami Scaffolds as Templates for Functional Tetrameric Kir3 K+ Channels, Angew. Chem., 2018, 8510, 2586–2591 CrossRef PubMed.
- L. Mallik, et al., Electron Microscopic Visualization of Protein Assemblies on Flattened DNA Origami, ACS Nano, 2015, 9, 7133–7141 CrossRef CAS PubMed.
- B. J. H. M. Rosier, et al., Proximity-induced caspase-9 activation on a DNA origami-based synthetic apoptosome, Nat. Catal., 2020, 3, 295–306 CrossRef CAS PubMed.
- A. Gopinath, et al., Absolute and arbitrary orientation of single-molecule shapes, Science, 2021, 371, eabd6179 CrossRef CAS PubMed.
- N. C. Seeman, Structural DNA Nanotechnology, Cambridge University Press, 2016, DOI:10.1017/CBO9781139015516.
- P. Wang, T. A. Meyer, V. Pan, P. K. Dutta and Y. Ke, The Beauty and Utility of DNA Origami, Chem, 2017, 2, 359–382 CAS.
- M. Madsen and K. V. Gothelf, Chemistries for DNA Nanotechnology, Chem. Rev., 2019, 119(10), 6384–6458 CrossRef CAS PubMed.
- X. Song and J. Reif, Nucleic Acid Databases and Molecular-Scale Computing, ACS Nano, 2019, 13, 6256–6268 CrossRef CAS PubMed.
- K. Matange, J. M. Tuck and A. J. Keung, DNA stability: a central design consideration for DNA data storage systems, Nat. Commun., 2021, 12, 1358 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Scaling Up Digital Circuit Computation with DNA strand Displacement Cascades, Science, 2011, 99, 1196–1202 CrossRef PubMed.
- C. E. Castro, H.-J. Su, A. E. Marras, L. Zhou and J. Johnson, Mechanical design of DNA nanostructures, Nanoscale, 2015, 7, 5913–5921 RSC.
- A. Porchetta, A. Vallée-Bélisle, K. W. Plaxco and F. Ricci, Allosterically tunable, DNA-based switches triggered by heavy metals, J. Am. Chem. Soc., 2013, 135, 13238–13241 CrossRef CAS PubMed.
- T. Zhou, et al., pH-responsive size-tunable self-assembled DNA dendrimers, Angew. Chem., Int. Ed., 2012, 51, 11271–11274 CrossRef CAS PubMed.
- D. Liu, et al., A reversible pH-driven DNA nanoswitch array, J. Am. Chem. Soc., 2006, 128, 2067–2071 CrossRef CAS PubMed.
- J. M. Majikes, L. C. C. Ferraz and T. H. LaBean, pH-Driven Actuation of DNA Origami via Parallel I-Motif Sequences in Solution and on Surfaces, Bioconjugate Chem., 2017, 28, 1821–1825 CrossRef CAS PubMed.
- S. Zhao, et al., A DNA origami-based aptamer nanoarray for potent and reversible anticoagulation in hemodialysis, Nat. Commun., 2021, 12, 358 CrossRef CAS PubMed.
- A. Krissanaprasit, et al., Genetically Encoded, Functional Single-Strand RNA Origami: Anticoagulant, Adv. Mater., 2019, 31, 1808262 CrossRef PubMed.
- A. Rangnekar, J. A. Nash, B. Goodfred, Y. G. Yingling and T. H. LaBean, Design of Potent and Controllable Anticoagulants Using DNA Aptamers and Nanostructures, Molecules, 2016, 21(22), 202 CrossRef PubMed.
- P. Zakrevsky, et al., Truncated tetrahedral RNA nanostructures exhibit enhanced features for delivery of RNAi substrates, Nanoscale, 2020, 12, 2555–2568 RSC.
- B. Boettner, Harvard Wyss Institute's nasal swab and toehold switch technologies licensed to Agile Biodetection to facilitate SARS-CoV-2 diagnostic efforts, 2020 Search PubMed.
- R. Carlson, The changing economics of DNA synthesis, Nat. Biotechnol., 2009, 27, 1091–1094 CrossRef CAS PubMed.
- R. Carlson, Time for New DNA Synthesis and Sequencing Cost Curves, 2014 Search PubMed.
- A. Walker, Samsung's 3D V-NAND Flash Product – The Spires of El Dorado? 3D incites, 2014 Search PubMed.
- K. A. Wetterstrand, DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP) Search PubMed.
- A. M. Thayer, Synthetic RNA, DNA providers tackle the oligos market, Chem. Eng. News Arch., 2017, 95, 17 Search PubMed.
- M. Eisenstein, Enzymatic DNA synthesis enters new phase, Nat. Biotechnol., 2020, 38, 1113–1115 CrossRef CAS PubMed.
- B. Kick, F. Praetorius, H. Dietz and D. Weuster-Botz, Efficient Production of Single-Stranded Phage DNA as Scaffolds for DNA Origami, Nano Lett., 2015, 15, 4672–4676 CrossRef CAS PubMed.
- W. Gayde, How CPUs are Designed, Part 3: Building the Chip, Tech Spot, 2019 Search PubMed.
- “3D NAND overview”, 2021.
- R. P. Goodman, I. A. T. Schaap, C. F. Tardin, C. M. Erben, R. M. Berry, C. F. Schmidt and A. J. Turberfield, et al., Rapid Chiral Assembly of Rigid DNA Building Blocks for Molecular Nanofabrication, Science, 2005, 310, 1661–1665 CrossRef CAS PubMed.
- D. B. Lacy, W. Tepp, A. C. Cohen, B. R. DasGupta and R. C. Stevens, Crystal structure of botulinum neurotoxin type A and implications for toxicity, Nat. Struct. Biol., 1998, 5, 898–902 CrossRef CAS PubMed.
- G. Saper and H. Hess, Synthetic Systems Powered by Biological Molecular Motors, Chem. Rev., 2020, 120, 288–309 CrossRef CAS PubMed.
- I. V. Surovtsev and C. Jacobs-Wagner, Subcellular Organization: A Critical Feature of Bacterial Cell Replication, Cell, 2018, 172, 1271–1293 CrossRef CAS PubMed.
- N. Perrimon, C. Pitsouli and B. Z. Shilo, Signaling mechanisms controlling cell fate and embryonic patterning, Cold Spring Harbor Perspect. Biol., 2012, 4, a005975 Search PubMed.
- A. Coskun, M. Banaszak, R. D. Astumian, J. F. Stoddart and B. A. Grzybowski, Great expectations: Can artificial molecular machines deliver on their promise?, Chem. Soc. Rev., 2012, 41, 19–30 RSC.
- I. Aprahamian, The Future of Molecular Machines, ACS Cent. Sci., 2020, 6, 347–358 CrossRef CAS PubMed.
- A. K. Pumm, W. Engelen and E. Kopperger, et al. A DNA origami rotary ratchet motor, Nature, 2022, 607, 492–298 CrossRef CAS PubMed.
- A. Ruff, F. Conzuelo and W. Schuhmann, Bioelectrocatalysis as the basis for the design of enzyme-based biofuel cells and semi-artificial biophotoelectrodes, Nat. Catal., 2020, 3, 214–224 CrossRef CAS.
- H. Chen, F. Dong and S. D. Minteer, The progress and outlook of bioelectrocatalysis for the production of chemicals, fuels and materials, Nat. Catal., 2020, 3, 225–244 CrossRef.
- N. Karimian, P. Hashemi, A. Khanmohammadi, A. Afkhami and H. Bagheri, The Principles and Recent Applications of Bioelectrocatalysis, Anal. Bioanal. Chem. Res., 2020, 7, 281–301 CAS.
- S. K. Ravi and S. C. Tan, Progress and perspectives in exploiting photosynthetic biomolecules for solar energy harnessing, Energy Environ. Sci., 2015, 8, 2551–2573 RSC.
- Y. Kim, S. A. Shin, J. Lee, K. D. Yang and K. T. Nam, Hybrid system of semiconductor and photosynthetic protein, Nanotechnology, 2014, 25(34), 342001 CrossRef PubMed.
- J. Fu, M. Liu, Y. Liu, N. W. Woodbury and H. Yan, Interenzyme Substrate Diffusion for an Enzyme Cascade Organized on Spatially Addressable DNA Nanostructures, J. Am. Chem. Soc., 2012, 134, 5516–5519 CrossRef CAS PubMed.
- S. Tsitkov and H. Hess, Design Principles for a Compartmentalized Enzyme Cascade Reaction, ACS Catal., 2019, 9, 2432–2439 CrossRef CAS.
- O. Idan and H. Hess, Origins of activity enhancement in enzyme cascades on scaffolds, ACS Nano, 2013, 7, 8658–8665 CrossRef CAS PubMed.
- K. S. Rabe, J. Müller, M. Skoupi and C. M. Niemeyer, Cascades in Compartments: En Route to Machine-Assisted Biotechnology, Angew. Chem., Int. Ed., 2017, 56, 13574–13589 CrossRef CAS PubMed.
- R. E. Blankenship, et al., Comparing photosynthetic and photovoltaic efficiencies and recognizing the potential for improvement, Science, 2011, 332, 805–809 CrossRef CAS PubMed.
- S. L. Shipman, J. Nivala, J. D. Macklis and G. M. Church, CRISPR–Cas encoding of a digital movie into the genomes of a population of living bacteria, Nature, 2017, 547, 345–349 CrossRef CAS PubMed.
- W. H. Press, J. A. Hawkins, S. K. Jones, J. M. Schaub and I. J. Finkelstein, HEDGES error-correcting code for DNA storage corrects indels and allows sequence constraints, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18489–18496 CrossRef CAS PubMed.
- Semiconductor Synthetic Biology for Information Storage and Retrieval (SemiSynBio-II), 2021.
- D. D. S. Alliance,Preserving Our Digital Legacy: An Introduction to DNA Data Storage, 2021 Search PubMed.
- B. Johns, Improving Information Technology Sustainability with Modern Tape Storage, 2021 Search PubMed.
- E. Masanet, A. Shehabi, N. Lei, S. Smith and J. Koomey, Recalibrating global data center energy-use estimates, Science, 2020, 367, 984–986 CrossRef CAS PubMed.
- L. Dormehl, Are old-school magnetic tapes the data storage medium of the future?, Digital Trends, 2020 Search PubMed https://www.digitaltrends.com/computing/ibm-magnetic-storage-record/ .
- G. D. Dickinson, et al., An alternative approach to nucleic acid memory, Nat. Commun., 2021, 12, 1–10 CrossRef PubMed.
- I. Mela, et al., DNA nanostructures as a tool for targeted antimicrobial delivery, Angew. Chem., Int. Ed., 2020, 1–6 Search PubMed.
- S. Obuobi and N. Škalko-Basnet, Nucleic acid hybrids as advanced antibacterial nanocarriers, Pharmaceutics, 2020, 12, 1–25 CrossRef PubMed.
- M. Yah, et al., Self-assembled three-dimensional chiral colloidal architecture, Science, 2017, 636, 633–636 Search PubMed.
Footnote |
| † Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |