Open Access Article
Open Access ArticleRole of detergents and nuclease inhibitors in the extraction of RNA from eukaryotic cells in complex matrices†
Cian
Holohan‡
 a,
Nathan
Feely‡
a,
Peng
Li
b,
Gerard
Curran
b and
Gil U.
Lee
a,
Nathan
Feely‡
a,
Peng
Li
b,
Gerard
Curran
b and
Gil U.
Lee
 *a
*a
aConway Institute for Biomedical Research and School of Chemistry, University College Dublin, Ireland. E-mail: gilulee@gmail.com
bMagnostics Ltd, 2 Clifton Lane, Monkstown, Co Dublin, Ireland
First published on 9th August 2022
Abstract
The potential for liquid biopsy samples to be used in place of more invasive tissue biopsies has become increasingly revalent as it has been found that nucleic acids (NAs) present in the blood of cancer patients originate from tumors. Nanomagnetic extraction has proven to be a highly effective means to rapidly prepare NA from clinical samples for molecular diagnostics. In this article, the lysis reaction used to extract RNA from the human epithelial melanoma cells have been optimized using silica coated superparamagnetic nanoparticles (SPM NP). The lysis buffer (LB) is composed of several agents that denature cells, i.e., surfactant and guanidinium isothiocyanate (GITC), and agents that inhibit the degradation of circulated nucleic acids (cfNAs). The surfactant Triton X-100 has been widely used in LB but has been placed on the European Union REACH list. We have compared the qRT-PCR sensitivity resulting from LBs composed of Triton X-100 to several sustainable surfactants, i.e., Tergitol 15-S-7, Tergitol 15-S-9 and Tween-20. Surprisingly, the inclusion of these surfactants in the LB was not found to significantly improve cell lysis, and subsequently the sensitivity of qRT-PCR. The role of the sample matrix was also examined by performing extractions from solutions containing up to 30 mg mL−1 serum albumin. The qRT-PCR sensitivity was found to decrease as the concentration of this protein was increased; however, this was linked to an increased RNase activity and not the concentration of the protein itself. These results lead us to recommend a reformulation of LB for clinical samples, and to conclude that sensitive qRT-PCR RNA analysis can be performed in serum with the timely addition of an RNase inhibitor.
Introduction
Tissue biopsies are widely used to analyze cancer tumors. However, extracting tumor tissue samples from patients can be an invasive process and may have large associated risks. To circumvent these issues, a recent shift towards the use of circulating tumor cells (CTCs) extracted from liquid biopsies, such as blood samples, has occurred. CTCs are tumor cells that are shed into the circulatory system by detaching from tumors, and provide an alternative sample type to tissue biopsies.1 Importantly, the RNA extracted from CTCs can be used as a biomarker to map the dynamic changes to the transcriptional landscape of a tumor over the course of treatment, therefore monitoring the impact of the treatment on the tumor.2 Circulating tumor RNA (ctRNA) are short strands of RNA originating from tumor cells that are present independently of cells in bodily fluids and can be used in a similar manner to CTCs.3 Although exceptionally useful, the relative abundance of CTCs and cell free NAs (cfNAs) in liquid biopsies can vary greatly between the tumor type and stage.4,5 Therefore, the RNA extraction method employed must be high yielding. Furthermore, the purified NA must be free of impurities as this can negatively impact downstream analysis.6 The first step to extract RNA from liquid biopsy samples is to lyse the CTCs, thus releasing the cellular RNA into bulk solution alongside the ctRNA for further purification.The LB is a key component of NA extraction kits. This buffer lyses bio-membranes in order to release NA into the bulk solution for downstream purification. However, LBs perform many tasks beyond membrane lysis. The LB must serve to inactivate endogenous proteins and RNases present in the sample itself or in the sample transport media. Failure to remove RNases from liquid biopsies can result in RNA degradation which negatively impacts RNA detection via molecular techniques.7 RNase inactivation is achieved by the presence of EDTA, a chelator of divalent cations, important cofactors for several types of RNase and chaotropic agents, such as GITC, which disrupt the hydrogen bonds holding together the secondary and tertiary structure of proteins.8 Additionally, this inactivates highly infectious samples to accommodate easier handling by lab staff. Ethanol is frequently used in the LB as it reduces the solubility of NAs in solution and facilitates their “crashing out” onto the surface of SPM adsorbents.9 Lastly, surfactants are included in LB formulations in order to solubilize bio-membranes resulting in cell lysis. This is achieved through the incorporation of phospholipids and transmembrane proteins into surfactant micelles due to the reduction in the surface free energy of water.10
Triton X-100 is an example of a non-ionic surfactant that is widely used as a component in LB to lyse cell membranes and solubilize proteins. Despite its routine use in laboratories around the world, questions surrounding its environmental impact have been raised. Like all surfactants, Triton X-100 has two key structural elements, hydrophobic and hydrophilic domains. The hydrophilic domain is a linear polymer chain containing 10 to 12 ethylene glycol monomers and the hydrophobic domain is based on octyl phenol (Fig. 1A). Following environmental degradation, Triton X-100 breaks into short chain ethylene glycols and octyl phenol derivatives, the latter raising significant environmental concerns due to hormone mimetic activity in fish, birds and mammals (Fig. 1B).11 Therefore, Triton X-100 has been added to the European Union (EU) REACH authorization list with a view to phasing out commercial use of this surfactant within the EU.12,13 Although the sustainable applications of Triton X-100 have been investigated,14 research has mainly focused on the replacement of Triton X-100 with alternative surfactants.15,16 “Designed to degrade” principles have been employed when creating new types of surfactant, such as Tergitol and Tween. These surfactants maintain ethylene glycol as the hydrophilic domain; however, the hydrophobic domain has been replaced with an aliphatic chain (Fig. 1A). Additionally, Tergitol and Tween type surfactants are not known to degrade into products exhibiting hormone mimetic activity and are therefore more environmentally sustainable than Triton X-100.
In considering alternatives to Triton X-100, we have selected surfactants with a critical micelle concentration (CMC) lower than that of Triton X-100 (Fig. 1A). The CMC reflects the power of the surfactant to solubilize lipid membranes and proteins, whereby surfactants with lower CMC values require a lower concentration to effectively solubilize hydrophobic components. To our knowledge there has not been a comprehensive comparison of multiple sustainable surfactants for the purpose of RNA extraction. We aimed to identify important parameters to consider for sustainable LB formulations by comparing the efficiency of RNA extractions performed on A375 human epithelial melanoma cells using LBs containing Triton X-100, Tween-20, Tergitol 15-S-7 and Tergitol 15-S-9.
Once the cells are lysed, the NA must then be isolated from the bulk solution. Developments made in the field of SPM nanomaterials have improved the workflow of this process to a point where it has become capable of processing a large quantity of samples. Key to the application of SPM nanomaterials is the design and manufacture of nanostructured SPM NPs with core–shell morphologies, in particular the coating of a SPM core with silica, a material previously used as the solid phase of more antiquated NA extraction columns. Fig. 1C illustrates the application of silica coated SPM NPs to extract human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA from A375 human epithelial melanoma cells as done in this study. The SPM NPs used were synthesized via templated self-assembly, resulting in the formation of high iron content magnetite NPs with sizes ranging from 200–1000 nm achieving saturation magnetization values on the order of 60–80 emu g−1 as reported previously.17 Despite yielding a lower surface area to volume ratio, the large magnetization values result in fast and high efficiency separation, key to enabling highly reproducible and scalable NA extraction and purification. The application of nanostructured silica SPM NPs synthesized using a modified Stöber process enables us to determine the precise role of surfactants in NA extraction systems and to quantitatively assess the performance of the more sustainable alternatives.18
Experimental
Materials
All reagents used in this study were molecular biology grade and were used as received without further alterations unless otherwise specified. A375 epithelial human melanoma cells were acquired from ATCC (ATCC® CRL-1619™). RNase-free filter pipette tips (STARLAB, UK), 96 deep well plates (Sarstedt, Germany), sealing mats (Fisher Scientific, USA) and microtubes (Eppendorf, Germany) were used through-out. Plasticware that was not certified RNase free was treated using RNaseZap® (Sigma-Aldrich, USA) immediately prior to use. The surfactants Triton X-100 (Fisher Scientific, USA), Tergitol 15-S-7 (DOW, USA), Tergitol 15-S-9 (DOW, USA) and Tween-20 (Sigma-Aldrich, USA) were suplemented into the LB. Sample preparation and handling were carried out in an aseptic manner in a sterile, RNase-free, BSL-2 hood unless otherwise stated. The model serum was 30 mg mL−1 of protease free bovine serum albumin (BSA) (Sigma-Aldrich, USA) in phosphate buffer saline (PBS). The fluorometric RNaseAlert® kit (Thermofisher, USA) was employed to measure RNase activity in all prepared buffers. All qRT-PCR reactions were carried out using the SuperScript™ III Platinum™ one-step qRT-PCR kit (Invitrogen, USA) in conjunction with GAPDH endogenous control gene primers/probes (Applied Biosystems, USA).Nucleic acid extraction using SPM NPs
High magnetization SPM NPs were provided by Magnostics Ltd (Silica Aurosphere 800 nm and 200 nm beads). These beads were prepared using the templated self-assembly process described by O'Mahony et al.19 The SPM bead cores were silica coated by a variant of the Stöber process.18 The basal LB was composed of 6 M GITC, 25 mM EDTA and was supplemented with 1, 2, or 3% (w/v) of each surfactant. Wash buffer (WB) 1 consisted of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) pure ethanol and basal LB, and WB 2 consisted of 80% (v/v) ethanol in nuclease-free water.
1 (v/v) pure ethanol and basal LB, and WB 2 consisted of 80% (v/v) ethanol in nuclease-free water.
The RNA extraction was performed using a four step process illustrated in Fig. 1B as described by Holohan, et al. using deep well 96-well plates and the M96D-400 magnetic separator (Magnostics Ltd).20 Prior to sample introduction, 1,4-dithiotheritol (2% (w/v) DTT) and polyadenylic acid carrier RNA (poly(A), 2 mg mL−1) were added to the LBs. Samples containing 0, 5, 50, 500, 5000 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A375 cells were treated with 250 μL of LB at r.t. for 10 min. The full contents of each tube were transferred to a 96-deep well plate containing 50 μL silica coated SPM NPs and 250 μL ethanol in each well and was shaken at 1000 rpm for 10 min. The SPM NPs were isolated using the magnetic separator and washed twice with WB1, followed by two washes with WB 2 to remove impurities. The purified RNA was eluted from the SPM NPs into 50 μL nuclease-free water and stored at −80 °C prior to qRT-PCR analysis. All extractions were performed in triplicate.
000 A375 cells were treated with 250 μL of LB at r.t. for 10 min. The full contents of each tube were transferred to a 96-deep well plate containing 50 μL silica coated SPM NPs and 250 μL ethanol in each well and was shaken at 1000 rpm for 10 min. The SPM NPs were isolated using the magnetic separator and washed twice with WB1, followed by two washes with WB 2 to remove impurities. The purified RNA was eluted from the SPM NPs into 50 μL nuclease-free water and stored at −80 °C prior to qRT-PCR analysis. All extractions were performed in triplicate.
A375 cell sample preparation
The sample was composed of A375 cells that were cultured in T-75 flasks in RPMI-1640 medium supplemented with 5% (v/v) fetal bovine serum (FBS, qualified & heat-inactivated, Thermo), L-glutamine and 1% (v/v) penicillin/streptomycin at 37 °C in a 5% CO2 atmosphere. The cells were sub-cultured every 2–3 days and tested negative for mycoplasma contamination. Cells were detached from the flask using accutase and adjusted to a concentration of 1 × 106 mL−1 in PBS. Serial dilutions of this stock were prepared and used immediately for fresh cell samples or stored at −80 °C to be used for frozen cell samples. The 0.03, 0.3 and 3% (w/v) protein-containing samples were created by adding 1 mL of 1 × 106 mL−1 A375 cells to 4 mL PBS containing 0.0375, 0.375 and 3.75% (w/v) BSA. In some cases, the samples were prepared with BSA solutions that had been pre-treated to inactivate RNases. This matrix was prepared by adding basal LB, and basal LB supplemented with 1, 2, 3, or 6% (w/v) Triton X-100 in a 1.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to PBS containing 3.75% (w/v) BSA. These were then incubated for 1 h at 37 °C. 50
1 ratio to PBS containing 3.75% (w/v) BSA. These were then incubated for 1 h at 37 °C. 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A375 cells were subsequently added, so that the final volume was 500 μL. The samples were incubated for 10 min at r.t.
000 A375 cells were subsequently added, so that the final volume was 500 μL. The samples were incubated for 10 min at r.t.
Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
A master-mix was created using the SuperScript™ III Platinum™ one-step qRT-PCR kit in conjunction with GAPDH endogenous control gene primers/probes with the following composition: SuperScript™ III RT/Platinum™ Taq mix (24.8 μL), 2× reaction mix (620 μL), ROX™ reference dye (2.5 μL), GAPDH primers/probes (62 μL, final concentration: 10 nM primers and 17 nM VIC labelled probes) and nuclease-free water (220 μL). 15 μL aliquots of the master-mix were used per 5 μL of the extracted samples (corresponding to 0, 5, 50, 500 and 5000 cells) for the qRT-PCR analysis. 5 μL containing 5000 A375 cells, extracted using LB containing 3% (w/v) Triton X-100, which previously amplified successfully, acted as the qRT-PCR positive control. The samples were kept on ice immediately prior to qRT-PCR analysis using a QuantStudio™ 7 Flex real-time PCR (Applied Biosystems®, USA). The run parameters followed a standard PCR cycle beginning with reverse transcription at 50 °C for 15 min, denaturation at 95 °C for 2 min, followed by 50 amplification cycles consisting of a denaturation step at 95 °C for 15 s and annealing and extension at 60 °C for 1 min with a temperature ramp of 1.6 °C s−1 and fluorescence acquisition in the annealing/extension phase. QuantStudio Software v1.3 was used to analyze the data. The cycle threshold was set at 0.05 and baseline set to automatic. Two-tailed t-tests were performed using GraphPad software to make comparisons between samples. These comparisons were regarded as statistically significant for p < 0.05.Drop-shape analysis to determine surfactant CMC
The critical micelle concentration (CMC) of each surfactant was characterized using the droplet shape contact angle analysis technique. Basal LB containing increasing concentrations of surfactants were prepared and made up with PBS and ethanol in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio to mimic the surfactant concentration under sample lysis conditions. Contact angles of these solutions were measured by pipetting a 10 μL droplet of each solution onto a polydimethylsiloxane substrate (prepared using a two-part silicone elastomer, DowsilSygard™ 184, as per manufacturer instructions) using a Kruss DSA10 Mk2 system. Images of the droplets were captured and analyzed using the Kruss drop shape analysis software (DSA v1.80.0.2), using automatic baseline and droplet contour detection to extract the contact angle. All samples were measured in triplicate.
1 ratio to mimic the surfactant concentration under sample lysis conditions. Contact angles of these solutions were measured by pipetting a 10 μL droplet of each solution onto a polydimethylsiloxane substrate (prepared using a two-part silicone elastomer, DowsilSygard™ 184, as per manufacturer instructions) using a Kruss DSA10 Mk2 system. Images of the droplets were captured and analyzed using the Kruss drop shape analysis software (DSA v1.80.0.2), using automatic baseline and droplet contour detection to extract the contact angle. All samples were measured in triplicate.
Results and discussion
Characterization of silica coated SPM NPs
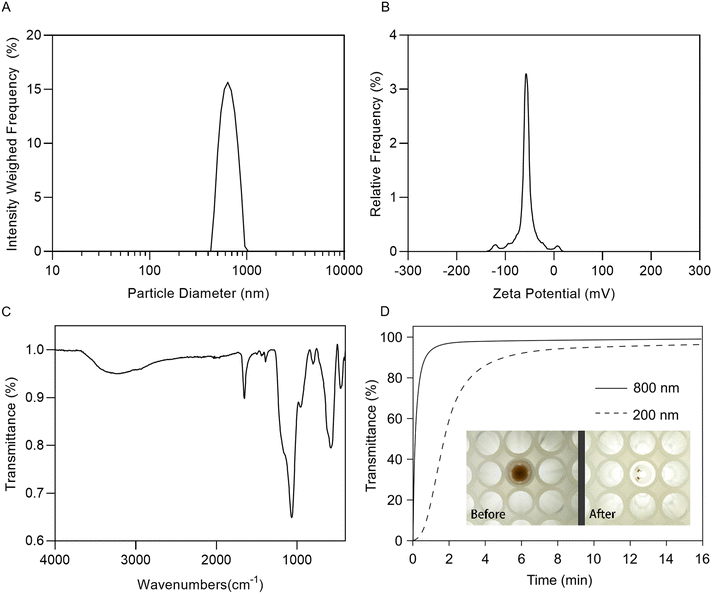
The purification of RNA from biological samples is performed using silica separation matrices in many clinical laboratories due to the speed and reliability of the process. The mechanism of RNA-silica binding has been attributed to hydrogen bonding interactions between surface silanol groups and the phospho-ribose backbone of RNA, although there is evidence that attractive hydrophobic interactions between the RNA base pairs and hydrophobic pockets in the silica structure may also play a role in the adsorption process.21 We have previously described the preparation of SPM NPs using an oil/water emulsion templated assembly of Fe3O4 NPs.19,20 These SPM cores were coated with a thin layer of silica using the Stöber process to enable RNA extraction.18ESI Fig. 1† presents transmission electron micrographs of nominal 800 nm SPM NPs in which the dense iron oxide metal cores coated with a silica shell, which appears to be 26 ± 3 nm thick. Dynamic light scattering indicated that the NPs have a hydrodynamic diameter of 770 nm and a zeta potential of −57 mV in pH 7.0 deionized water (Fig. 2A and B). The surface composition of the NPs was confirmed using IR spectroscopy (Fig. 2C). The bands at 460, 590, and 630 cm−1 are characteristic of Fe–O vibration modes of the SPM core, while the bands at 470, 800, 960, 1080 cm−1 are consistent with Si–O vibration modes of a silica coating with a sharp peak at 1650 cm−1 representing the H–O–H stretching modes of adsorbed water.
To maximize our RNA extraction yields and minimize the time required for RNA extraction the separation performance of the silica coated SPM NPs was assessed. Fig. 2D presents the separation of the SPM NPs in a 96-well plate by measuring the change in transmitted light. We observed that 95% of the 800 nm silica coated SPM NPs were captured after 90 s, this contrasted with a separation time of more than 10 min for 200 nm silica coated SPM NPs which were prepared in a similar manner. This observation is due to the increased surface area: volume ratio of the 200 nm SPM MPs causing a decrease in the magnetic force acting to pull the particles towards the magnetic separator, therefore we applied the 800 nm MPs with a 5 min separation time in our extractions to ensure maximum RNA yields.
Influence of surfactant composition on RNA extraction efficiency
The A375 melanoma cell line was used as a model system to characterize the influence of surfactant on RNA extraction efficiency. Fig. S2, ESI† depicts a representative qRT-PCR amplification plot of extracted GAPDH mRNA and presents the fluorescence intensity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ΔRn) plotted as a function of thermal cycle number to detect the target sequences. Three samples are plotted: positive controls comprised of RNA extracted from samples containing 50
ΔRn) plotted as a function of thermal cycle number to detect the target sequences. Three samples are plotted: positive controls comprised of RNA extracted from samples containing 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (green) and 50 (blue) A375 cells using LB supplemented with 3% (w/v) Triton X-100 and a sample representative of a single failed triplicate amplification which contained RNA extracted from 50
000 (green) and 50 (blue) A375 cells using LB supplemented with 3% (w/v) Triton X-100 and a sample representative of a single failed triplicate amplification which contained RNA extracted from 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A375 cells using basal LB containing no surfactant (red). The Rn values were obtained by dividing the fluorescence of the VIC reporter dye (GAPDH assays) by the fluorescence of a passive reference dye (ROX™). The ΔRn values were determined by subtracting the Rn value of the baseline signal from that of the experimental reaction. After multiple thermal cycles the target sequence is amplified to a point whereby the emitted fluorescence signal crosses a threshold for detection. The cycle number at which the threshold is crossed is defined as the cycle threshold (Ct) value. In this study the threshold was set at a ΔRn value of 0.05. A lower Ct value indicates the presence of a larger quantity of the target RNA sequence in the original sample as fewer thermal cycles are required to cross the threshold and vice versa. Therefore, the quantity of RNA present in solution i.e., the ability of surfactant to lyse cells can be inferred by comparing the Ct values.
000 A375 cells using basal LB containing no surfactant (red). The Rn values were obtained by dividing the fluorescence of the VIC reporter dye (GAPDH assays) by the fluorescence of a passive reference dye (ROX™). The ΔRn values were determined by subtracting the Rn value of the baseline signal from that of the experimental reaction. After multiple thermal cycles the target sequence is amplified to a point whereby the emitted fluorescence signal crosses a threshold for detection. The cycle number at which the threshold is crossed is defined as the cycle threshold (Ct) value. In this study the threshold was set at a ΔRn value of 0.05. A lower Ct value indicates the presence of a larger quantity of the target RNA sequence in the original sample as fewer thermal cycles are required to cross the threshold and vice versa. Therefore, the quantity of RNA present in solution i.e., the ability of surfactant to lyse cells can be inferred by comparing the Ct values.
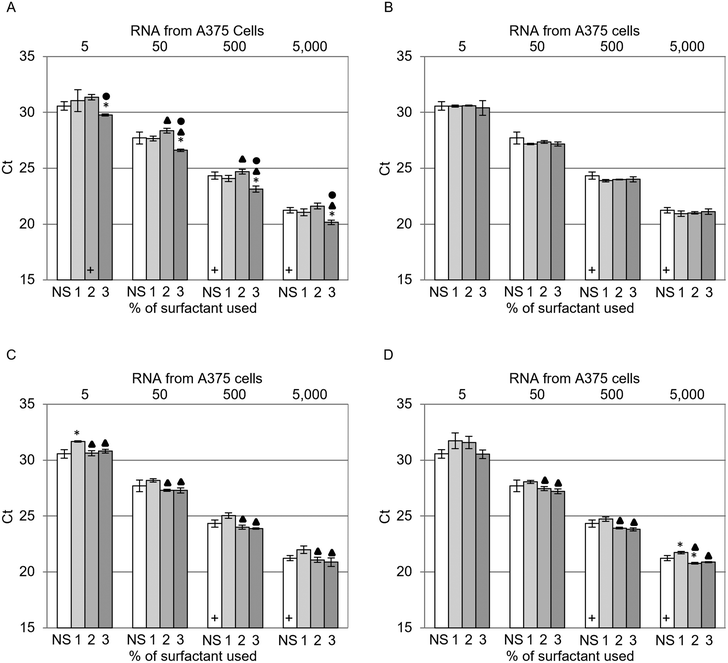
The impact of the sustainable surfactants on cell lysis was examined by comparing the Ct values achieved with RNA extracted from cells using LBs supplemented with different surfactants. Fig. 3 presents the qRT-PCR results for each surfactant tested, i.e., Triton X-100 (T100) (Fig. 3A), Tergitol 15-S-7 (T7) (Fig. 3B), Tergitol 15-S-9 (T9) (Fig. 3C) and Tween-20 (T20) (Fig. 3D). These surfactants were examined at concentrations of 1, 2 and 3% (w/v) and were used to extract RNA from samples containing 0, 50, 500, 5000 and 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A375 cells. Frozen A375 cells were used throughout this study as freeze–thaw induced cell lysis was not observed to significantly influence RNA extraction (Fig. S3, ESI†). The Ct values decrease with increasing cell concentration, with a log-linear behavior that is consistent with normal qRT-PCR analysis. The results for samples containing no cells were omitted from Fig. 3 as they failed to amplify. Statistical analysis was performed using two-tailed t-tests (p < 0.05) to compare the increasing surfactant concentrations to each other and to their NS counterparts. The inclusion of surfactant in LB did not significantly improve the Cts, with the exception of 2% (w/v) T20 5000 cells and all 3% (w/v) T100 samples. However, changing the significance value to p < 0.01 rendered all samples insignificant compared to their NS counterparts. Statistically significant improvements (p < 0.05) in Cts were observed on moving from 2 to 3% (w/v) of T100 (Fig. 3A) and 1 to 2% (w/v) for both T9 and T20 (Fig. 3C and D). No statistically significant improvements in Cts were observed for any T7 concentration tested (Fig. 3B). Despite the minimal impact observed when including surfactant in LBs in comparison to NS extractions, the statistically significant improvements in Cts within each surfactant concentration series suggested that the surfactants may play a minor role in the mechanism of RNA extraction.
000 A375 cells. Frozen A375 cells were used throughout this study as freeze–thaw induced cell lysis was not observed to significantly influence RNA extraction (Fig. S3, ESI†). The Ct values decrease with increasing cell concentration, with a log-linear behavior that is consistent with normal qRT-PCR analysis. The results for samples containing no cells were omitted from Fig. 3 as they failed to amplify. Statistical analysis was performed using two-tailed t-tests (p < 0.05) to compare the increasing surfactant concentrations to each other and to their NS counterparts. The inclusion of surfactant in LB did not significantly improve the Cts, with the exception of 2% (w/v) T20 5000 cells and all 3% (w/v) T100 samples. However, changing the significance value to p < 0.01 rendered all samples insignificant compared to their NS counterparts. Statistically significant improvements (p < 0.05) in Cts were observed on moving from 2 to 3% (w/v) of T100 (Fig. 3A) and 1 to 2% (w/v) for both T9 and T20 (Fig. 3C and D). No statistically significant improvements in Cts were observed for any T7 concentration tested (Fig. 3B). Despite the minimal impact observed when including surfactant in LBs in comparison to NS extractions, the statistically significant improvements in Cts within each surfactant concentration series suggested that the surfactants may play a minor role in the mechanism of RNA extraction.
Characterization of surfactant CMC in the LB
The CMC of each surfactant in water and LB was characterized using the drop shape contact angle technique.22Fig. 4A and B present the contact angle data plotted as a function of surfactant concentration in water and under sample lysis conditions. As the concentration of surfactant increases, the interfacial tension and contact angle of the droplet decrease until the CMC is reached. Further increases in surfactant concentration do not reduce the contact angle further, rather micelles are formed inside the drop. The CMC can be determined as the point of intersection between the lines of best fit for the rapidly decreasing and plateau regions of the plot. Fig. 4C presents the CMCs calculated from these plots. All four surfactants demonstrated CMC increases of at least one order of magnitude when in LB compared to water. T100 presented the highest CMC in LB at a concentration of approximately 3.4% (w/v) (Fig. 4C). This correlates well with the corresponding qRT-PCR results (Fig. 3A), where statistically significant improvements in Cts were observed as the T100 concentration increased from 2 to 3% (w/v). There was no correlation between statistically significant improvements in Cts for LBs that contained T7, T9 or T20 (Fig. 3B–D) with each surfactant having a CMC value less than 1% (w/v) (Fig. 4C). The poor correlations observed between the CMC data and RNA extraction efficiency suggested that the surfactants did not play a significant role in membrane solubilization. Indeed, the chaotropic destabilization of surfactant micelles by GITC and ethanol alone appear to effectively lyse membranes in the absence of surfactants. | ||
| Fig. 4 Characterization of the surfactant CMC in A. water, and B. under sample lysis conditions using the drop shape analysis technique. C. Summary of CMC data obtained from plots A and B. | ||
RNA extraction from serum models
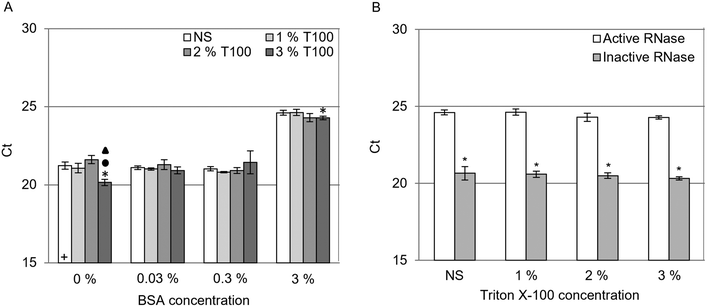
To assess the effect of surfactants in RNA extractions from serum, BSA was supplemented into samples prior to RNA extraction. Fig. 5 presents the qRT-PCR results for samples supplemented with 0.03, 0.3 and 3% (w/v) BSA extracted using NS and 1, 2 and 3% (w/v) T100. Two-tailed t-tests (p < 0.05) were used to make comparisons between samples. The presence of BSA did not significantly impact Cts until 3% (w/v). The inclusion of T100 in the LB did not significantly affect RNA extraction from samples containing protein in comparison to samples extracted using NS. The extraction performed in 3% (w/v) BSA with 3% (w/v) T100 had a statistically significant, lower Ct compared to its NS counterpart, although changing the significance value to p < 0.01 rendered this comparison insignificant. This suggests that the impact of protein on RNA extraction was concentration dependent, with surfactant having a negligible effect on RNA extraction from samples containing protein.The impact of serum on RNA extractions was examined by measuring the RNase activity present in BSA. An RNaseAlert® QC V2 kit was used to measure the RNase activity of the extraction buffers (Fig. S4, ESI†) and samples containing BSA (Fig. S5, ESI†). The samples were prepared by adding 3% (w/v) BSA in PBS to deionized water, basal LB and basal LB containing 2% (w/v) DTT in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio and monitoring the RNase activity in duplicate. The extraction buffer components and each sample buffer prior to the addition of BSA tested negative for RNase activity (Fig. S4 and S5 ESI†). 3% (w/v) BSA prepared in deionized water tested positive for RNase activity, whereas 3% (w/v) BSA prepared in basal LB tested negative for RNase activity, irrespective of DTT content. This data indicates that RNase activity was responsible for the increase in Cts observed in Fig. 5A rather than the protein concentration in the extraction.
1 ratio and monitoring the RNase activity in duplicate. The extraction buffer components and each sample buffer prior to the addition of BSA tested negative for RNase activity (Fig. S4 and S5 ESI†). 3% (w/v) BSA prepared in deionized water tested positive for RNase activity, whereas 3% (w/v) BSA prepared in basal LB tested negative for RNase activity, irrespective of DTT content. This data indicates that RNase activity was responsible for the increase in Cts observed in Fig. 5A rather than the protein concentration in the extraction.
The influence of the RNases in samples prepared with 3% (w/v) BSA was further investigated by inactivating endogenous RNases before cells were added to the sample. Fig. 5B presents a comparison of Cts between samples prepared with standard BSA (active RNase) and pre-inactivated BSA (inactive RNase). Pre-inactivation was achieved by adding BSA to NS and 1, 2, 3 and 6% (w/v) T100 LBs and incubating these for 1 h at 37 °C before adding 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 A375 cells. Two-tailed t-tests were used to make comparisons between active and inactive RNase samples. Inactive RNase samples demonstrated statistically significant lower Cts compared to their active RNase counterparts and were consistent with their 0% (w/v) BSA counterpart presented in Fig. 5A. Increasing the T100 concentration to 6% (w/v) did not significantly improve RNA extraction in the inactive RNase sample (data not shown). These results indicated that while the presence of BSA had no detectable influence on extraction, the endogenous RNases present in 3% (w/v) BSA negatively impacted RNA extraction.
000 A375 cells. Two-tailed t-tests were used to make comparisons between active and inactive RNase samples. Inactive RNase samples demonstrated statistically significant lower Cts compared to their active RNase counterparts and were consistent with their 0% (w/v) BSA counterpart presented in Fig. 5A. Increasing the T100 concentration to 6% (w/v) did not significantly improve RNA extraction in the inactive RNase sample (data not shown). These results indicated that while the presence of BSA had no detectable influence on extraction, the endogenous RNases present in 3% (w/v) BSA negatively impacted RNA extraction.
Conclusions
We have identified improvements that can be made to LB formulations by examining the impact of the composition of LBs on qRT-PCR sensitivity. Although the chaotropic nature of the LB destabilized the micelles, making the formation of such structures less thermodynamically favourable,23,24 the surfactants concentration exceeded, or came close to, their CMCs in the LB. Thus, the inclusion of surfactants in LB had a negligible impact on RNA extraction. Instead, the chaotropic agents in the LB formulation provided a mechanism through which the lipid bilayers of cellular membranes were effectively lysed in the absence of surfactants, causing the release of NAs into the bulk solution.25–27 This is an important observation as it is widely accepted that surfactants are required in LB formulations for cell lysis to occur. Throughout the current SARS-CoV-2 pandemic >750 million NAAT tests have been performed in USA alone. Given that 7.5 mg of surfactant is used per sample in the 3% (w/v) LB, the removal of surfactant from LB formulations could eliminate 1000 of kg of surfactant worldwide. This is especially relevant due to the rate of testing during the current SARS-CoV-2 pandemic and the resulting wide scale implementation of infectious disease molecular diagnostic testing, suggesting a modification in the LB could significantly decrease the environmental impact of Triton X-100 and cost of molecular diagnostic testing.28The results presented in this article demonstrate timely inactivation of RNases was more critical for high efficiency RNA extraction than the addition of surfactants. The inactivation of RNases resulted in 8× increase in the efficiency of extraction of mRNAs from the model serum samples due to the rapid and highly efficient SPM NPs used in this study. The LB successfully inactivated endogenous RNases, with pre-inactivated samples presenting with no detrimental effect to the Ct. However, failing to pre-inactivate the RNases present in protein-containing samples was detrimental to the Ct. Therefore, endogenous RNases degrade sample RNA when RNase inactivation and cell lysis occur in the same step. We recommend that nuclease inhibitors, e.g., GITC, EDTA and DTT, be added immediately post sample acquisition to reduce the influence of endogenous nucleases. This would help to inactivate endogenous RNases prior to cell lysis. Furthermore, the concentration of carrier RNA (poly-(A)) in the LB should be increased so as to provide an alternative target for any potentially still active RNases. Adhering to these recommendations would ensure minimal sample RNA degradation and near quantitative RNA capture.
Author contributions
CH and NF methodology, validation, analysis, investigation, visualization, and writing. PL and GC resources. GL concept, writing, supervision.Conflicts of interest
The authors do not have a conflict of interest to declare.Acknowledgements
This work was supported by the Science Foundation of Ireland (15/IA/3127 and 20COV0054) and the Higher Education Authority of Ireland (covid-19 emergency grant).Notes and references
- F. Cheng, L. Su and C. Qian, Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer, Oncotarget, 2016, 7, 48832–48841 CrossRef PubMed.
- S. Hassan, T. Blick, E. D. Williams and E. W. Thompson, Applications of RNA characterisation in circulating tumor cells, Front. Biosci.-Landmark, 2020, 25, 874–892 CrossRef CAS PubMed.
- M. H. Larson, et al., A comprehensive characterization of the cell-free transcriptome reveals tissue- and subtype-specific biomarkers for cancer detection, Nat. Commun., 2021, 12, 2357 CrossRef CAS PubMed.
- A. J. Rushton, G. Nteliopoulos, J. A. Shaw and R. C. Coombes, A review of circulating tumor cell enrichment technologies, Cancers, 2021, 13, 1–33 CrossRef PubMed.
- C. Bettegowda, et al., Detection of circulating tumor DNA in early- and late-stage human malignancies, Sci. Transl. Med., 2014, 6, 1–12 CrossRef PubMed.
- B. Yalçinkaya, K. Coşkun, M. Akgoz and S. Pence, A simple silica-based DNA isolation method for cell-free DNA analysis from liquid biopsy, Turkish J. Biochem., 2020, 45, 701–705 CrossRef.
- Y. Nishibata, et al., RNase in the saliva can affect the detection of severe acute respiratory syndrome coronavirus 2 by real-time one-step polymerase chain reaction using saliva samples, Pathol., Res. Pract., 2021, 220, 153381 CrossRef CAS PubMed.
- G. Salvi, P. De Los Rios and M. Vendruscolo, Effective interactions between chaotropic agents and proteins, Proteins: Struct., Funct., Genet., 2005, 61, 492–499 CrossRef CAS PubMed.
- S. E. Walker and J. Lorsch, RNA purification - Precipitation methods. Methods in Enzymology, Elsevier Inc., 2013, vol. 530 Search PubMed.
- D. Lichtenberg, Characterization of the solubilization of lipid bilayers by surfactants, Biochim. Biophys. Acta, Biomembr., 1985, 821, 470–478 CrossRef CAS.
- E. H. Jho, S. H. Yun, P. Thapa and J. W. Nam, Changes in the aquatic ecotoxicological effects of Triton X-100 after UV photodegradation, Environ. Sci. Pollut. Res., 2021, 28, 11224–11232 CrossRef CAS PubMed.
- European Chemicals Agency, SVHC SupportDocument: 4-(1,1,3,3-Tetramethylbutyl)phenol, ethoxylated, as substances of very high concern, 2012 Search PubMed.
- European Union, REGULATION (EC) No 1907/2006 OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL of 18 December 2006 concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH), establishing a European Chemicals Agency, amending Directive 1999/4, Official Journal of the European Union 7–13, 2006, DOI:10.4324/9781315270326-156.
- L. Abu-Ghunmi, M. Badawi and M. Fayyad, Fate of Triton X-100 Applications on Water and Soil Environments: A Review, J. Surfactants Deterg., 2014, 17, 833–838 CrossRef CAS.
- J.-B. Farcet, J. Kindermann, M. Karbiener and T. R. Kreil, Development of a Triton X–100 replacement for effective virus inactivation in biotechnology processes, Eng. Rep., 2019, 1(5), e12078 CAS.
- J. Farcet, et al., Synthesis of “Nereid,” a new phenol–free detergent to replace Triton X–100 in virus inactivation, J. Med. Virol., 2021, 93, 3880–3889 CrossRef CAS PubMed.
- C. Fields, P. Li, J. J. O'Mahony and G. U. Lee, Advances in affinity ligand-functionalized nanomaterials for biomagnetic separation, Biotechnol. Bioeng., 2016, 113, 11–25 CrossRef CAS PubMed.
- W. Stober and A. Fink, Controlled Growth of Monodisperse Silica Spheres in the Micron Size Range, J. Colloid Interface Sci., 1968, 26, 62–69 CrossRef.
- J. J. O'Mahony, M. Platt, D. Kilinc and G. Lee, Synthesis of superparamagnetic particles with tunable morphologies: The role of nanoparticle-nanoparticle interactions, Langmuir, 2013, 29, 2546–2553 CrossRef PubMed.
- C. Holohan, et al., Influence of viral transport media and freeze-thaw cycling on the sensitivity of qRT-PCR detection of SARS-CoV-2 nucleic acids, Nanoscale, 2021, 13, 15659–15667 RSC.
- B. Shi, Y. K. Shin, A. A. Hassanali and S. J. Singer, DNA Binding to the Silica Surface, J. Phys. Chem. B, 2015, 119, 11030–11040 CrossRef CAS PubMed.
- M. Y. Alkawareek, et al., Simple Experiment to Determine Surfactant Critical Micelle Concentrations Using Contact-Angle Measurements, J. Chem. Educ., 2018, 95, 2227–2232 CrossRef CAS.
- K. I. Assaf and W. M. Nau, The Chaotropic Effect as an Assembly Motif in Chemistry, Angew. Chem., Int. Ed., 2018, 57, 13968–13981 CrossRef CAS PubMed.
- R. J. Midura and M. Yanagishita, Chaotropic Solvents Increase the Critical Micellar Concentrations of Detergents, Anal. Biochem., 1995, 228, 318–322 CrossRef CAS PubMed.
- M. S. Islam, A. Aryasomayajula and P. R. Selvaganapathy, A review on macroscale and microscale cell lysis methods, Micromachines, 2017, 8(3), 83 CrossRef.
- V. Prachayasittikul, C. Isarankura-Na-Ayudhya, T. Tantimongcolwat, C. Nantasenamat and H. J. Galla, EDTA-induced membrane fluidization and destabilization: Biophysical studies on artificial lipid membranes, Acta Biochim. Biophys. Sin., 2007, 39, 901–913 CrossRef PubMed.
- L. O. Ingram, Mechanism of lysis of Escherichia coli by ethanol and other chaotropic agents, J. Bacteriol., 1981, 146, 331–336 CrossRef CAS PubMed.
- J. Hasell, E. Mathieu and D. Beltekian, et al., A cross-country database of COVID-19 testing, Sci. Data, 2020, 7, 345 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr02850f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |