Nucleic acid isothermal amplification-based soft nanoarchitectonics as an emerging electrochemical biosensing platform
Jing
Liu
 *a,
Ruke
Wang
a,
Hong
Zhou
*a,
Ruke
Wang
a,
Hong
Zhou
 b,
Motilal
Mathesh
b,
Motilal
Mathesh
 c,
Mukul
Dubey
d,
Wengan
Zhang
a,
Bo
Wang
c,
Mukul
Dubey
d,
Wengan
Zhang
a,
Bo
Wang
 *a and
Wenrong
Yang
*a and
Wenrong
Yang
 *c
*c
aCollege of Chemical and Biological Engineering, Shandong University of Science and Technology, Qingdao, 266590, PR China. E-mail: jingliu@sdust.edu.cn; wb@sdust.edu.cn
bShandong Key Laboratory of Biochemical Analysis; College of Chemistry and Molecular Engineering, Qingdao University of Science and Technology, Qingdao 266042, China
cSchool of Life and Environmental Sciences, Deakin University, Geelong, Victoria 3217, Australia. E-mail: wenrong.yang@deakin.edu.au
dTERI-Deakin Nanobiotechnology Centre, TERI Gram, Gwal Pahari, Gurugram, Haryana, India
First published on 20th June 2022
Abstract
The emergence of nucleic acid isothermal amplification strategies based on soft nanoarchitectonics offers a new dimension to the traditional electrochemical technique, particularly because of its flexibility, high efficiency, and increased sensitivity for analytical applications. Various DNA/RNA isothermal amplification strategies have been developed for the design and fabrication of new electrochemical biosensors for efficient and important biomolecular detection. Herein, we provide an overview of recent efforts in this research field and the strategies for signal-amplified sensing systems, with their biological applications, current challenges and prospects in this promising new area.
1 Introduction
Various electrochemical techniques such as traditional voltammetry, amperometry and electrochemiluminescence have been employed in the development of bioassays with high sensitivity.1–3 The design and development of electrochemical biosensors have attracted great attention from researchers in recent decades owing to the wide applications of biosensors, such as in environmental monitoring, early diagnosis of diseases, healthcare and drug delivery. The key challenges involved in biosensor progress are the inefficient capture of biorecognition signals and the transformation of these signals into electrochemical responses (transduction process) with high sensitivity, short response time, satisfactory reproducibility, and low detection limits.4,5 These challenges can be overcome by using soft “nanoarchitectonics”.Nanoarchitectonics was first proposed in 2000 (using suprainteractions) by Masakazu Aono6,7 and first used in a scientific publication by S. Hecht in 2003.8 Soft nanoarchitectonics is an arrangement of nanostructures or soft molecules in an intended configuration through the harmonization of a series of actions including self-assembly, chemical reactions or molecular recognition. They have been widely used as a key enabling technology to realize the integration of predefined molecular biosensing functionalities into biosensor devices.9 The main consideration for constructing soft nanoarchitectures for electrochemical biosensing requires ordered self-assembly of biomolecules with retained activity (such as protein, peptide and nucleic acid) on the electrode with efficient signal transduction to act as an efficient biosensor. For example, the Niu group developed a peptide-modified gold electrode surface via the N-terminus for trypsin sensing. They used the free carboxyl termini from the tryptic cleavage events for the site-specific labelling of reversible addition–fragmentation chain transfer (RAFT) agents via carboxylate–Zr(IV)–carboxylate chemistry. A considerable amount of Fc redox tags were grafted from the cleaved sites in the presence of an ultralow level of trypsin activity, thus allowing for the signal-on electrochemical interrogation of trypsin activity at ultralow levels.10 Further, protein kinase A (PKA) substrate peptides were phosphorylated through the PKA-catalyzed reaction using ATP as the source of phosphate and the synthesized Pbpy-Ru-TiO2 NP realized the aggregation and amplification of the signal in TiO2.11 Besides, using peptides, nucleic acid-based nanoarchitectonic approaches have attracted great attention owing to their diverse manipulation possibilities for electrochemical biosensing. These affinity probes show high programmability, addressability, mechanical flexibility, and biocompatibility. For instance, EpCAM aptamers tightly bind to tumor cells.12 Following this, poly(A) tails were programmatically synthesized by terminal deoxynucleotidyl transferase (TdT) at the 3′ end of the aptamer using adenosine as a substrate. This allowed the tumor cells to be spontaneously assembled onto the electrode surface due to the strong affinity between the poly(A) tails and gold surface. This method involved multiple signal amplification by thousands of ligands on each tumor cell and negatively charged multiple poly(A) tails, which greatly enhanced the electrochemical signal on the electrode surface.
Among these nucleic acids-assisted amplification methods, the isothermal cascade/exponential amplification method is a powerful and promising tool that can offer high signal amplification of the signal from targets in a robust and efficient way under a mild temperature.13 Currently, a few efficient techniques with isothermal amplification have been proposed, such as rolling circle amplification, strand displacement amplification, nuclease or ribonuclease-assisted amplification and ions-specific DNAzyme.14 Nucleic acid isothermal amplification with electrochemical detection technology has received much attention in the design of high-performance biosensors for various applications.15
Nucleic acid amplification occurs in a homogeneous system, however, the signal generation and receiving occur in (or around) the electrode that can be easily influenced by the reaction, or signal transmittance during the detection. Moreover, the samples are generally from human serum, saliva, and urine, requiring an efficient and cascade/exponential amplification for specific target analytes, which is a big challenge due to the complexity of molecules present in them. In order to obtain profound signal feedback from the probe–target analyte interaction, a new approach was proposed for the design and fabrication of probe molecules in advance to construct smart and multifunctional nanoarchitectures, which could provide a specific interaction and efficient sensing response. Soft nanoarchitectonics based strategy is aimed at designing, fabricating, and achieving optimal spatial organization, and the efficient signal obtained from the target–probe interaction to an analyte and promoting biosensing activity for electrochemical biosensing applications.
Nucleic acid-based soft nanoarchitectonics based interfaces provided flexible design, ordered assembly, and predictable function for functional electrochemical sensing. Meanwhile, the fabrication process and the surface chemistry of these soft nanoarchitectonics interfaces influence the efficiency of signal recognition and amplification, thus providing opportunities for high-efficient target recognition and sensing through well-designed nucleic acid amplification technology.
In this review, we aim to provide the reader with a concise view of new advances in areas ranging from soft nanoarchitectonics on electrodes to strategies for nucleic acid isothermal amplification for electrochemical biosensors. Firstly, we discuss the basic principles of different nucleic acid isothermal amplification strategies, including enzyme-based and enzyme-free exponential amplification techniques. Secondly, the working principle of nucleic acid-assisted nanoarchitectonics for electrochemical biosensing, including new breakthroughs and seminal studies that have been reported in the past three years are discussed. Finally, we will discuss the present challenges and further perspectives of this field for next-generation biosensing mode.
2 Classical electrochemical biosensing
Classical electrochemical (voltammetry and amperometry) biosensors have been widely developed due to their ease of operation, high sensitivity, simplicity, and economical providing wide applications in the detection of chemical and biological targets. The conventional amplification strategies such as the polymerase chain reaction (PCR)16,17 based electrochemical sensors require complicated thermocycling to mediate denaturation, annealing, and subsequent extension. In contrast, nucleic acid isothermal amplification can, however, be performed in one pot and realize exponential signal amplification in a robust, convenient, and efficient way.18 Many isothermal amplification techniques have been applied in the electrochemical sensing methods to build various amperometric or electrochemical impedance sensors, which we will discuss below in detail.2.1 Rolling circle amplification (RCA) for classical electrochemical biosensing
Rolling circle amplification (RCA) is one of the powerful amplification techniques that imitate internal rolling-circle replication by a large number of repetitive sequences. The circular DNA template has a small gap, which in the presence of DNA polymerase, and a primer extends and generates a long ssDNA with thousands of repeated complementary sequences. These repeated sequences could further be tagged with probes for amplified detection of various analytes, including DNA, microRNA, proteins, glutathione, and Pseudomonas aeruginosa.19 For example, to obtain high selectivity, DNA tetrahedron was modified on the gold electrode through the thiols on the three vertices, and a pendant linear structure was designed on the top of the DNA to form a hairpin-like structure with Hg2+ through thymine–Hg2+–thymine interaction. In the presence of glutathione (GSH), the thymine–Hg2+–thymine complex breaks the hairpin structure because of the stronger binding affinity of GSH towards Hg2+.20 This leads to the reformation of the pendant linear strand, which initiates the subsequent RCA reaction with the addition of a circular DNA template and DNA polymerase that could bind to DNA–AgNPs conjugates. The multiple repeating sequences binding with DNA–AgNPs conjugates generated significant silver stripping signals on the electrode surface, thereby, increasing its sensitivity. In a similar approach, DNA tetrahedral nanoprobe with AgNC was immobilized on the electrode surface for the detection of HIV DNA fragments. The process involved the addition of substrate reagents to the target fragment to carry out the RCA reaction, producing several mimic sequences (DNA I and II). This was dropped onto the DNA tetrahedron modified electrode, which replaced DNA–AgNCs via the toehold strand displacement reaction leading to a dramatic decrease in electrochemical signal response resulting in an ultra-low detection limit down to 0.1 fM.21 These two examples of different nanoarchitectonic strategies show very high signal-amplification efficiency. Moreover, RCA could be combined with other efficient amplification technologies, such as clustered regularly interspaced short palindromic repeats (CRISPR)/Cas to construct a “signal-on” immobilization-free electrochemical biosensing platform for micro RNA21.22 Notably, in order to obtain multiple detections of various non-nucleic acid targets, it utilizes competitive binding between aptamer or small molecules to construct logic computation circuits.22 The RCA-based nanoarchitectures also provide an efficient sensing platform for the diagnosis of coronavirus disease 2019 (COVID-19). Different from current nucleic acid testing through quantitative reverse transcription PCR (qRT-PCR) for COVID-19, Lertanantawong’ group designed a multiplex RCA based electrochemical biosensing method for the rapid detection of N and S genes of SARS-CoV-2 from clinical samples (Fig. 1A).23 RNA and cDNA samples were employed as the template for RCA, with the product functionalized with large number of redox-active labels, resulting in differential pulse voltammetry (DPV) signal, which was used to detect low to 1 copy per μL of viral N or S genes in 2 h, and thus could be applied as an on-site, real-time testing methods for COVID-19.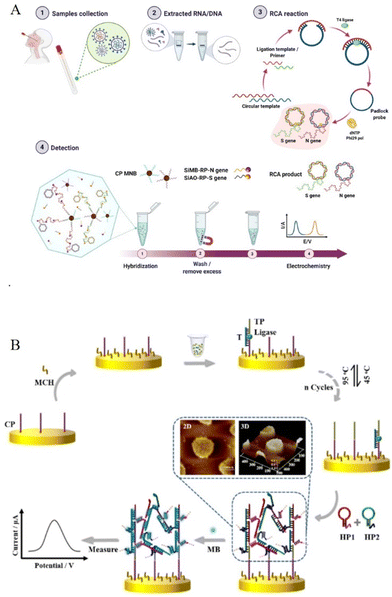 | ||
| Fig. 1 (A) RCA-based classical electrochemical biosensing method for SARS-CoV-2 from clinical samples. Reproduced from ref. 23 with permission from Nat. Commun. Copyright The Author(s) 2021. (B) Schematic illustration of the design and signalling of the HCR-assisted 3D biosensor. Reproduced from ref. 30 with permission from Anal. Chem. Copyright 2021 American Chemical Society. | ||
2.2 DNA strand displacement amplification for classical electrochemical sensing
DNA strand displacement is a powerful tool for the design of ultrasensitive and selective enzyme-free detection systems. DNA is designed with a toehold (a single-stranded domain containing 5–8 bases) where the strand displacement reaction could be initiated through branched-chain migration. The displacement process is fast and predictable and easy to differentiate even single mismatched sequences.24–26 Two prominent examples of DNA strand displacement are the hybridization chain reaction (HCR) and catalyzed hairpin assembly (CHA). In the architecture of HCR, two partially complementary monomer DNAs were employed in a hairpin structure with a long-stem and short-loop. The addition of a target single-stranded DNA (ssDNA) breaks the energy balance in long-stem protected loops, and thus trigger an alternating hybridization reaction, resulting in a long-nicked double-stranded DNA (dsDNA).14 A general architecture method for HCR in electrochemical sensing involves the modification of a hairpin-structured capture probe on the electrode. This is carried out in the presence of the target sequence and sustaining palindromic probes in the solution. The targeted trigger HCR reaction completed on the electrode generates a long double-stranded DNA, which could bind large number of electrochemical tags (such as methylene blue MB labelled DNA strands, RuHex, or hemin/G-quadruplex27) on it. This enables enhanced signal-amplified electrochemical sensing for various targets, including micro RNA21,28 exosomal microRNA,29 and p53DNA30 at fM or even aM level. Different from the traditional architecture of the hairpin capture probe and the generated long linear HCR product on the sensing electrode, some novel architecture strategies have been developed.31 For example, as shown in Fig. 1B, Wu's group constructed an intertwined three-dimensional DNA nanosheet on an electrode surface using two palindromic hairpin probes. A well-aligned ss DNA monolayer was modified on the electrode, and after the reaction of target p53DNA and Taq DNA ligase, a typical sandwich DNA triplex was formed where the nick was located in close proximity to possible cancer-causing point mutations. Then, upon the addition of two palindromic probes, intertwined 3D SDN formed and inserted with methylene blue as an electrochemical indicator, which generated a strong electrochemical signal.30 Miao's recent work designed a three-way junction structure-based dumbbell HCR on the electrode. The dumbbell HCR structure not only keeps the high efficacy of HCR but also shows the tight conformation of the HCR product, which is helpful in shortening the distance between electrochemical species and the electrode interface for enhancement of the electrochemical response, thus obtaining an improved LOD of microRNA to as low as 7.3 aM.32 Furthermore, in order to have better sensitivity to trace targets, some recent electrochemical sensors combine HCR with other nucleic acid amplification technologies.33–35 For example, using target-induced Mg2+-specific DNAzyme amplification in solution, a novel two-dimensional ladder-like HCR reaction was designed, which showed higher catalytic efficiency than those of a traditional HCR reaction with a lower detection limit of 48.5 aM miRNA21.33 In addition, DNA tool enzyme (Nicking endonuclease and polymerase) assisted cascade amplification34 and CRISPR processing technology35 could also be combined in the HCR amplification method for dual signal amplified genetic analysis. Besides the solid electrode, using carbon ink screen printing technology, the architecture for DNA strand displacement amplification could also be constructed on microfluidic cloth-based36 or paper-based analytical devices37 for point-of-care (POC) diagnosis in resource-limited settings, avoiding the cumbersome chip fabrication and high-cost peripheral facilities.Unlike the chain reaction of HCR, CHA catalyzes hairpin DNA assembly through a hybridization reaction. Most CHA reactions were designed in the solution with two toehold DNA hairpins with the target initiating the hybridization of these two hairpins, at the same time, the target sequence is recycled during the CHA process and could be released to initiate another hybridization reaction. The large amount of CHA products generated was captured onto the electrode surface with modification of electrochemical tags to show the enhanced electrochemical response.38 To further achieve higher reaction efficiency, the captured hairpin DNA could also be modified on nanomaterials, such as gold nanomaterials (AuNPs or gold nanorods) and magnetic nanobeads to construct 3D DNA nanomachine-mediated CHA39 or without using nanomaterials, a transduction hairpin (THP) DNA with stem-loop structure is used to form three-leg DNA nanomachine.40 Benefitting from the improved reaction rate and the conversion efficiency, 3D DNA nanomachine-mediated CHA realized a detection limit of 0.27 fM miRNA-155 on gold electrode,40 0.139 fM miRNA-21 on magnetic glassy carbon electrode (MGCE),39 0.17 fM miRNA-21 in enzymatic biofuel cells (EBFCs)41 and 0.32 aM miRNA-21 on magnetic AuNFs/ITO electrodes. Further combining loop-mediated isothermal amplification (LAMP) with CHA, which takes full advantage of the ultrasensitive feature of nucleic acid amplification, an extremely small amount of gene targets (down to 2 copies per μL) was analyzed (Fig. 2A).42
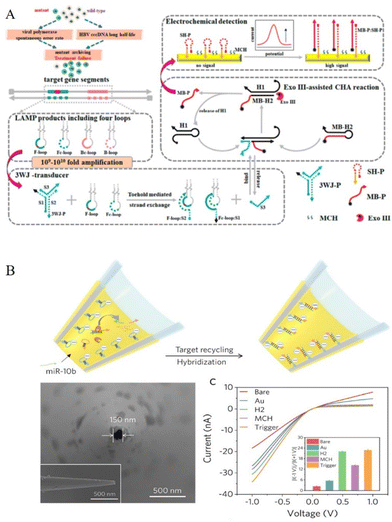 | ||
| Fig. 2 (A) Electrochemical detection of HBV drug-resistant mutants using three-way junction transduction and Exo III-assisted CHA. Reproduced from ref. 42 with permission from Anal. Chem. Copyright 2021 American Chemical Society. (B) Construction of the nanopipette for probing miRNA-10b and stepwise I-V responses of the nanopipette development. Inset: corresponding ICR ratio. Reproduced from ref. 49 with permission from Angew. Chem. Int. Ed. Copyright 2020 Wiley-VCH GmbH. | ||
2.3 Nuclease or ribonuclease-assisted amplification for classical electrochemical sensing
Nuclease mainly includes endonucleases (such as restriction endonucleases and nicking endonucleases, NEase), exonuclease, and duplex-specific nuclease (DSN) with very different cleaving mechanisms. Restriction endonucleases catalyze the cleavage of specific nucleotide sequences (restriction sites) in dsDNA, while nicking endonuclease (NEase) catalyzes the hydrolysis process at specific recognition sites of one strand from dsDNA. An exonuclease (Exos) catalyzes single-stranded or double-stranded DNA with stepwise removal of mononucleotides from the 3′-end or the 5′-end. Recent nuclease-based amplification architectures break through the use of only linear DNA sequences and introduce flexible inorganic nanoparticles, extracellular vesicles, and novel flexible types of DNA structures, including dumbbell or wheel DNA sequences43 to design more smarter electrochemical sensing devices.44 For example, Zr-based metal–organic frameworks-labeled oligonucleotides loaded with methylene blue (MB@Zr-MOFs-cDNA) were designed as signal probes for the fabrication of NEase-assisted sensitive electrochemical sensing of patulin.45 To improve the sensitivity, organic–inorganic hybrid nanoflowers (Cu3(PO4)2 HNFs with high surface area and enhanced catalytic activity were introduced to Exo III-assisted target recycling to build a signal circling amplification detection system for Hg2+.46 Employing aptamer binding small extracellular vesicles complex (aptamer-sEV complex), Zhang's work showed a NEase assistant cycling on the surface of sEV and “AND” logic gate for two tumor-associated proteins, tyrosine kinase-like 7 (PTK7) and prostate-specific membrane antigen (PSMA).47 Miao's recent work combined nuclease-assisted cycling amplification with DNA Dumbbell-Wheel transition and obtained a facile and concise electrochemical method for ctDNA and SARS-CoV-2 RNA detection in clinical samples for exponential amplification dynamics.43Besides endonucleases and exonuclease, duplex-specific nuclease (DSN), or ribonuclease-based amplification methods, the isothermal exponential amplification method of recombinase polymerase amplification (RPA) has also been commonly used in electrochemical sensors. DSN is a type of enzyme that digests DNA in the DNA/RNA duplex, so it has been widely used for the detection microRNAs. The DSN-mediated reaction could be further combined with a DNA walker or DNA machine, initiating cycling hybridization and digestion on the sensing electrode, providing an intense electrochemical response.48 For example, in Xu's recent work, as shown in Fig. 2B, the DSN-assisted target recycling strategy was designed and completed in a nanopipette for stimulating its ionic current rectification (ICR). Benefitting from miniaturized nanodevice, which can extract intracellular fluid under physiological conditions and the efficient amplification effect of DSN-assisted CHA reaction, the work achieved electrochemical measurement of intracellular miR-10b in single live cells.49 Similarly, RNase H specifically degrades the RNA fragment in a DNA–RNA duplex. Actually, they showed similar architecture strategies for the design of cycling amplification and subsequent autonomous movement of a DNA walker or DNA machine.50 Unlike DNA–DNA or DNA–RNA reactions, another method called RPA is a reaction involving DNA and protein. The recombinase combines with the primers to form the protein–DNA complexes, which subsequently scan dsDNA for the homologous sequence, and the subsequent strand-displacement leads to exponential amplification of DNA. In this process, nucleotides could be labelled with electroactive tags or enzymes showing enhanced electrochemical responses.51
3 Electrochemiluminescence (ECL) sensing
Electrochemiluminescence (ECL) is an electrochemical technique that produces chemiluminescence through an electrochemical reaction. It combines integrated advantages from electrochemistry and spectroscopy with improved sensitivity and specificity.2,52–54 As it does not require a light source, it shows low background signals resulting from scattered light and luminescent impurities. Besides, high specificity was obtained because of the adjustable excited states and corresponding co-reactants. After many years of development,55 the technology has rapidly developed and served as an impressive tool for biosensing with high stability, sensitivity, and specificity, especially combining it with nucleic acid amplification strategy.DNA molecules have been widely studied for construction of biosensing interfaces due to their high degree of designability and ease of modification.56 DNA-based ECL sensing signals were affected by the hybridization behavior. The introduction of nucleic acid isothermal amplification reactions could further significantly amplify the ECL signal. These nucleic acids’ isothermal amplification-assisted ECL devices show promising applications that are more sensitive and specific. The basic principle of these biosensors is that when the target DNA binds to the probe DNA, an amplification reaction is initiated to recycle the target DNA, which subsequently signals through conformational or stochiometric changes from the probe DNA. In these ECL systems, ECL luminophores mainly include inorganic, organic, and nanomaterials. Generally, the inorganic systems mainly consist of ruthenium complexes and iridium complexes, while the organic systems include luminol, anthracenes, fluorenes, thienyltriazoles, and their derivatives.4 Since the pioneering work of Bard et al. published in 2002, on the ECL of semiconductor nanocrystals, the ECL behaviors of various nanomaterials have been widely developed and extensively studied using noble metal clusters, carbon nanomaterials, and quantum dots. Various construction methods of ECL devices were designed and developed according to the different characteristics of these luminophores.
3.1 Organic luminophores for amplified ECL biosensing
Luminol is one of the most classical and successful organic luminophores with very broad applications due to its low excitation potential, high quantum yield, and nontoxicity.57,58 Luminol can not only be used by addition in the solution and introduction to the sensing electrode through binding effect with amplification products for ECL emitter,59 but, it can also be modified on various nanomaterials in advance for novel functions.60,61 As shown in Fig. 3A, by doping luminol or diethylamine on polymer dots, a novel ECL array imaging method was constructed by the Ju's group for the high-throughput detection of two microRNAs. After DSN-digestion assisted in-cycling amplification reaction, potential and color-resolved quantitative analysis of miRNA-21 and miRNA-205 was presented with the detection limits of 2.5 and 3.1 pM, respectively.62 Relying on the advantages of luminol, more luminol-related reagents with excellent performance have been explored to increase the luminescence quantum efficiency, such as N-(4-Aminobutyl)-N-(ethylisoluminol) (ABEI). ABEI is a novel derivative of luminol that shows more reactive properties and a stronger ECL signal than luminol.63 Ge's recent work synthesized AuPd NPs as a co-reaction accelerator for the ABEI-H2O2 system and obtained amplified ECL emission of ABEI. Additionally combined with T7 Exo-assisted DNA walker cascade amplification, this work realized multi-strategy signal enhancement for ultra-sensitive detection of miRNA41.64 In luminol or luminol-derivative involved ECL system, H2O2 is a commonly used co-reactant for luminol that exhibits bio-toxicity and instability, thus hampering its application in bioassay. As such, some novel co-reactants were developed in the ECL sensing system.65 For example, a novel H2O2-free ECL system (luminol/dissolved oxygen/Fe@Fe2O3 nanowires) was designed by Yuan's group. Fe@Fe2O3 nanowires could produce H˙ for activating dissolved oxygen into ROS in the detection solution immediately without the application of voltage, which avoided side reactions. Further, with the aid of a highly efficient SDA amplification strategy in the solution, ultrasensitive detection for miRNA was realized with a detection limit down to 52.5 aM.66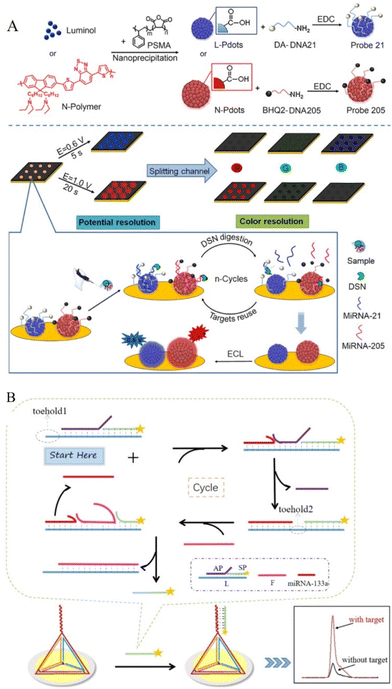 | ||
| Fig. 3 (A) Schematic diagrams of the preparation of Probe 21/205 and ECL imaging array for the detection of miRNA-21/205. Reproduced from ref. 62 with permission from Anal. Chem. Copyright 2021 American Chemical Society. (B) Schematic Illustration of the amplification process and the process of electrode surface modification. Reproduced from ref. 83 with permission from Anal. Chem. Copyright 2021 American Chemical Society. | ||
3.2 Inorganic luminophores for amplified ECL biosensing
In the inorganic luminophores for DNA-assisted amplified ECL biosensing, Ru(phen)32+ is a common luminophore with high ECL emission efficiency and can be intercalated into the grooves of dsDNA. When using Ru(phen)32+ as ECL luminophore, DNA-assisted amplification strategy (such as hyperbranched RCA,67 target induced SDA,68 and HCR69) is well designed to produce a large amount of DNA duplex structures that could load Ru(phen)32+as ECL tags.70 For example, Lin's work combined highly specific Cu+-catalyzed azide–alkyne cycloaddition (CuAAC) with high efficiency of HCR for the detection of pyrophosphatase.71 Besides Ru(phen)32+, tris(2,2′-bipyridine)ruthenium(II) chloride (Ru(bpy)32+) is another most frequently used ECL reporter, which shows ECL emission under ambient conditions in aqueous media.72,73 It is usually combined with a co-reactant, such as a secondary or tertiary amine for an enhanced ECL signal.74 Ru(bpy)32+could be labeled on DNA sequence as signal probes (SP), which could perform satisfactory signal amplified ECL response.75–82 For example, as shown in Fig. 3B, Yang's group constructed a tetrahedral DNA nanostructure on the electrode as capture probes and Ru(bpy)32+ -labeled DNA sequence as signal probes. Relying on the entropy-driven strand displacement reaction, microRNA-133a, which serves as an acute myocardial infarction biomarker was rapidly detected.83 Ru(bpy)32+could also be labelled on nanoparticles as a nanoprobe with a high concentration of Ru(bpy)32+, such as Ru@SiO2 nanoparticles,82,84,85 Ce–Ru-nanostructured coordination polymer,86 Ru(bpy)32+-encapsulated liposomes,87 and Au@Ti3C2@PEI-Ru(dcbpy)32+ nanocomposites88 for the detection of various analysts, including severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).88 During the modification of the sensing interface on electrodes, some anti-fouling coating strategies can also be employed to resist nonspecific adsorption in complex biological environments.89,90 The ECL behavior of Ru(bpy)32+ could be effectively quenched by some nanoparticle compounds and materials, such as carbon dots,91 or nitrogen-doped carbon dots.92 With nuclease-assisted recycling amplification, the quenching effect could be enhanced to achieve higher detection sensitivity.3.3 Nanomaterials as luminophores for amplified ECL biosensing
Since the first report of the ECL phenomenon of silicon QDs,93 many multifarious quantum dots (QDs) have been developed as ECL luminophores, including various QDs, as well as the corresponding alloyed, core–shell structure or doped QDs.4,94–99 In addition to traditional QDs, Ni-doped MoS2 QDs were also synthesized as a novel ECL emitter, which can effectively catalyze co-reactant and generate strong ECL signal.98 Sulfur quantum dots were also synthesized as an ECL emitter that showed excellent aqueous dispersibility, low toxicity, and excellent biological activities in an enzymatic-assisted DNA walker amplified sensing system for the detection of miRNAs.100,101 Besides QDs, other novel nanomaterials have emerged and received considerable attention as effective ECL emitters in recent years, including carbon nanodots, noble metal nanoclusters, and polymer dots.102–107 With the increasing attention to using DNA amplification strategy for the enhanced ECL performance, many of these novel nanomaterials have been studied for DNA-assisted multiple amplification detection for various targets with a co-reaction accelerator.108 For example the construction of Hf-ETTC, metal–organic layer,109 carbon dots,110,111 copper nanoclusters (Cu NCs),112 click chemistry-driven bioanalysis using AuAg nanoclusters,113 graphite phase carbon nitride (g-C3N4),114–117 and AgNCs@MoS2.118 In order to improve the ECL sensing application in terms of good chemical stability and activity, less toxicity or visible light emissions, further improved and efficient nanomaterials were reported such as nitrogen vacancy-rich carbon nitride nanosheets (NV-CN).119 Different from the ECL emission of graphitic carbon nitride at less than 470 nm,120 NV-CN holds stable and highly improved blue-green ECL emissions, avoiding the problem of electrode passivation by trapping the excess electrons in the NV sites, and thus realized as an enhanced ECL aptasensor.121 Other examples involved QDs loaded in mesoporous silica (mSiO2),122 or loaded in MOF,123 all of which have been coupled with DNA-assisted cycling reaction for enhanced performance of ECL sensing.Among the nanomaterial luminophores, it is worth noting that a highly sensitive ECL resonance energy transfer (ECL-RET) method has been reported that greatly improves the performance of ECL bioanalysis. The key thing to designing an efficient ECL-RET system is to construct a donor/acceptor pair that shows a suitable spectral overlap between the ECL spectrum of the donor and the absorption spectrum of the acceptor. One type of ECL-RET system consists of a nanomaterial luminophore and an organic (or inorganic) reagent as a donor/acceptor pair.124,125 As shown in Fig. 4A, Yang's group designed an ECL-RET system based on graphitic carbon nitride nanosheets (GCNNs) and Ru(phen)32+ as a donor/acceptor pairs, in which the signal was regulated by the amount Ru(phen)32+ present. The two optical signals of GCNNs (445 nm) and Ru(phen)32+ (593 nm) showed completely opposite changes through the same one-time DNA-assisted HCR reaction.126 Two different nanomaterials could also be designed as an ECL donor/acceptor pairs for DNA-assisted amplification sensing, such as CdS QDs/Ag NCs pair,127,128 S-doped Lu2O3/Ag2S QDs pair,129 Ag NPs/g-C3N4 pair or g-CNQDs/AgNPs system130 used for RCA, HCR, target binding-triggered DNA walker.
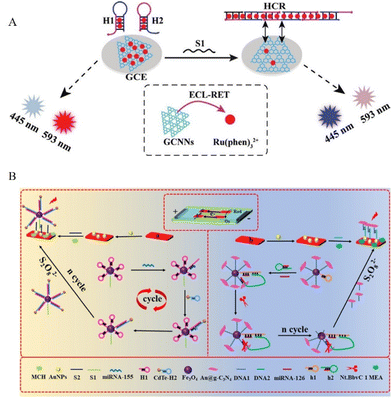 | ||
| Fig. 4 (A) Construction of an immobilization-free ECL-RET biosensor based on GCNNs/Ru(phen)32+ as a donor/acceptor pair. Reproduced from ref. 126 with permission from small. Copyright 2021 Wiley-VCH GmbH. (B) Construction of a paper-based BPE-ECL biosensor bipolar electrode for miRNA-155/126. Reproduced from ref. 136 with permission from Anal. Chem. Copyright 2020 American Chemical Society. | ||
3.4 Ratiometric or dual-modal ECL biosensing
In the ECL-RET system, if the acceptor is also a luminescent material, a dual-wavelength ratiometric assay could be constructed based on the donor's ECL signal and the acceptor's fluorescence signal that shows an opposite trend with the increase of target concentration.5 For example, Au-g-C3N4 (ECL emission at 460 nm) was modified on the electrode surface with assembled DNA tetrahedrons. Upon the targeting of the SARS-CoV-2 RdRp gene, PEI-Ru@Ti3C2 @AuNPs-S7 probes bind with the top of the DNA tetrahedrons, giving a strong ECL emission at 620 nm through the ECL-RET. Coupling with entropy-driven DNA walker cycle amplification strategy, the ratiometric ECL biosensor exhibited sensitive target gene detection to as low as 7.8 aM.131In addition to dual-wavelength ratiometric ECL sensing, other types of ratiometric ECL methods include dual-potential ratiometric, bipolar-electrode (BPE) ratiometric, or internal standard ratiometric biosensors.132–134 For dual-potential biosensors, two ECL emitters are generally required, which show potential-dependent features and the ratio of ECL intensities is differentiable at two excitation potentials. Thus, a pair of anodic/cathodic emitters with independent luminescence is generally employed in this type of biosensor. For example, a CHA-driven DNA walker enhanced biosensor for DNA detection was designed based on the combination of Au@luminol nanoparticles, with Au@CDs nanoflowers as luminophors and CHA-driven bipedal DNA walking nanomachine for DNA detection.135 BPE-ECL ratiometric biosensors typically employ two or more luminophores with distinct emission colours and/or oxidation potentials and are traditionally fabricated on microfluidic devices. For example, CdTe QDs and g-C3N4 NSs were used as dual-potential-resolved ECL signal probes coupled with a 3D DNA machine and obtained enhanced ECL response and improved detection of multiple miRNAs in multiple-channel paper-based sensing microfluidic platform (Fig. 4B).136Fig. 5A shows the construction of the internal standard for a ratiometric analysis,137 the architecture involved in situ assemblies of CsPbBr3 nanocrystals (CPB) into hollow g-C3N4 nanospheres (HCNS) as an ECL emitter. The dual-ECL signal was from an anodic ECL from CPB and a cathodic ECL from HCNS (served as an internal standard for a ratiometric analysis). With the help of DNA assisted-double signal amplification targeting CD44 receptors on the MCF-7 cell surface, the system obtained sensitive detection of MCF-7 cells. From the above examples, these ratiometric strategies could greatly reduce the influences of the instrument or environment, thus providing more precise analysis data than a single signal. This advantage is especially evident at low concentrations of targets in complex biological samples.
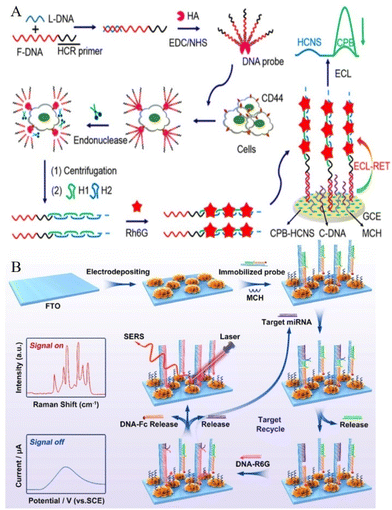 | ||
| Fig. 5 (A) Schematic diagrams of an internal standard assistant ratiometric analysis process. Reproduced from ref. 137 with permission from Anal. Chem. Copyright 2020 American Chemical Society. (B) Schematic diagrams of the proposed strategy for dual-mode enzyme-free target recycling amplification detection of miRNA. Reproduced from ref. 143 with permission from Anal. Chem. Copyright 2021 American Chemical Society. | ||
Another approach based on the dual-modal biosensing method serves as an efficient sensing strategy in the biosensing field where two readouts from two modes are produced and thus avoiding interference from complex sample substrates and obtaining more convincing results. This is advantageous when compared to the above-mentioned ratiometric strategies that also showed two readouts, however, the two readouts are from the same detecting method. For example, electrochemical/ECL dual-mode,138,139 electrochemical/photoelectrochemical (PET) dual-mode,140,141 and ECL/PET dual-mode142 sensing strategies have been reported for improved cross-validating detection. Zhou et al. combined the electrochemistry method and surface-enhanced Raman scattering (SERS) technology to realize a dual-mode assay for miRNA. As shown in Fig. 5B, three-dimensional (3D) popcorn-like gold nanofilms were constructed as novel SERS-electrochemistry active substrates that were further coupled with toehold-mediated SDR amplification reaction to achieve more accurate quantification detection of miRNA. The remarkably low detection limit of 0.12 fM (for the SERS method) and 2.2 fM (for the electrochemical method)143 was reported.
4 Enzyme nanoarchitectonics on 2D nanomaterials
Our group has extensively studied various enzymes and their architectures on 2D carbon nanomaterials. These enzyme-based soft nanoarchitectonics approaches have paved the way to fundamentally understand the process of substrate channeling for cascade reactions and have laid the platform for studying the structure–function relationship of enzymes enhancing its activity and aiding in the development of 2D nanomotors by overcoming key challenges. In this section, the above three will be discussed to showcase how enzyme nanoarchitectonics has proven beneficial for a diverse range of applications.Designing enzyme cascade reactions with perfect substrate channeling has remained a challenging task, requiring the direct transfer of reactants from one active site to another. In general, they involve complex and sophisticated technologies, which are time-consuming and laborious. In order to simplify the process, our group used a simple approach to randomly immobilize the two enzymes participating in cascade reactions onto graphene oxide surfaces and studied their various architectures. It was observed that when the enzymes are immobilized sequentially onto 2D nanomaterials a perfect substrate channeling was possible (Fig. 6A).144 This was due to limiting the diffusion of molecules to two dimensions that enhanced the overall process. Lipases is another enzyme whose architecture has been widely studied by our group. These are enzymes that hydrolyze carboxylic esters in hydrophobic compounds and are used in an array of commercial biocatalysis applications.145 A grand challenge in lipase immobilization to increase the catalytic activity is to open their lids due to interfacial activation in the presence of hydrophobic surfaces.146 We proposed a simple approach for this by immobilizing lipase enzymes onto different tailor-made hydrophobic supports, synthesized by reducing graphene oxide to different time intervals. For the first time using both experimental and theoretical studies, it was concluded that for effective lipase immobilization onto 2D nanomaterials, a balance between surface hydrophobicity/hydrophilicity is required.147
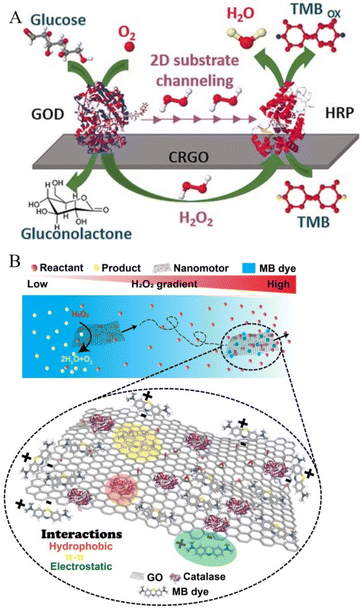 | ||
| Fig. 6 (A) Schematic representations of enzyme nanoarchitectonics on 2D nanomaterials. (A) Perfect substrate channeling between enzymes participating in cascade reactions. Reproduced from ref. 144 with permission from Chem. Eur. J. Copyright 2017 John Wiley and Sons. (B) Enzyme-powered 2D nanomotors with chemotactic behavior for efficient dye removal. Reproduced from ref. 151 with permission from Angew. Chem. Int. Ed. Copyright 2022 John Wiley and Sons. | ||
Nanomotors are a new field of research that has gained increased attention due to their ability to navigate autonomously towards desired targets. This could be exemplified in a variety of applications ranging from biomedical applications148,149 to environmental remediation.150 Major challenges in presently fabricated 2D nanomotors are (i) laborious, time-consuming, and sophisticated technology, (ii) high concentrations of hydrogen peroxide as a fuel source (toxic), (iii) use of the external magnetic field, with (iv) their motion behavior not well understood, thereby impeding its advancement in developing cutting-edge technology. The use of enzyme architectures to power 2D nanostructures is attractive because enzymes can be used as highly efficient motors with high catalytic power and specificity that can operate under ambient conditions. Our group designed the first enzyme-powered 2D nanomotors using soft nanoarchitectonics that could propel at ultra-low fuel concentrations with chemotactic behaviour, and act as an excellent “on-the-move” dye removal capability (Fig. 6B).151
5 Perspectives and conclusions
The flexible design and successful application of DNA-assisted amplification technology-based soft nanoarchitectonics in electrochemical and ECL biosensing have demonstrated significant advantages. In this review, we briefly reviewed recent strategies and controllable assembly of functional components of soft nanoarchitectonics in view of amplified electrochemical/ECL biosensors. These efforts include the fabrication of capture probes, design of DNA-assisted amplification strategies, and construction of a recognition reaction with enhanced electrochemical/ECL response. Benefits from the flexible DNA-assisted amplification strategies and various architectonics methods for the construction of probes provide enhanced and stable readout with ultrasensitive detection of various targets, including DNA, RNA, small molecules, metal ions, pathogens, and cancer cells.Despite the significant progress in the development of these biosensors, further efforts are required to address challenges and expand their applications. Taking COVID-19 virus detection as an example, although the proposed approaches have the advantages of enhanced sensitivity, specificity, and detection speed under gentle isothermal assay conditions, the commercial application of these approaches still faces challenges in the aspect of rapid, low-cost, and robust analysis of large numbers of samples. These challenges also provide new opportunities for further research and development of higher-performance detection strategies, which meet the requirements of commercial applications.
Firstly, even with the wide use of paper or microchips as electrodes, most sensing substrates are still traditional solid electrodes. The development of ECL high-throughput assays based on transistor-based technologies (such as organic electrochemical transistors, OECTs152,153 and field-effect transistors, FETs), lateral flow assay,154 and immobilization-free155 systems are required to meet the criteria of commercial point-of-care (POC) sensing, which is affordable, user-friendly, rapid and robust, and equipment-free. Moreover, to ensure reliable POC sensing, controllable electrode surfaces are required to prevent non-specific protein adsorption in a complex biological sample. Thus, new robust one-step coating strategies for bioactive sensing interfaces show potential prospects.156 Secondly, current DNA-assisted amplification strategies rely on the reaction of DNA/RNA or some specific metal ions, further development of amplification strategies between DNA/RNA and proteins or peptides remains a great challenge. A breakthrough of this limit would greatly expand the application of these amplified sensing strategies to almost all the analytes. Thirdly, a new efficient dual (or multiple)-mode signal response needs to be further explored using novel ECL luminophores or signal response mechanisms to obtain a stable, clear, sensitive, and more convincing response in complex biological samples. With the development of various functional materials, an understanding of the acid nucleic reaction mechanism, together with more systematic nanoarchitectonic approaches are required for improved electrochemical/ECL biosensors development. Finally, autonomous nanoscale systems have recently received tremendous attention due to their vast applications ranging from biomedicine to environmental remediation. It is time to employ a new strategy to develop “truly” two-dimensional (2D) soft nanoarchitectonics-based nanomotors that could enable “chemistry-on-the-fly” for broad applications including biosensing, imaging, and drug delivery.150
Author contributions
The article was written with the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank the funding support from the Shandong Provincial Natural Science Foundation (ZR2021MB090), the National Natural Science Foundation of China with grant no. 21868011, and support by Shandong Key Laboratory of Biochemical Analysis (SKLBA2204). We are also appreciated financial support from the Shandong University of Science and Technology.References
- X. Jia, S. Dong and E. Wang, Biosens. Bioelectron., 2016, 76, 80–90 CrossRef CAS PubMed.
- J. Zhang, L. Zhang, Z. Li, Q. Zhang, Y. Li, Y. Ying and Y. Fu, Small, 2021, 17, 2101665 CrossRef CAS PubMed.
- A. P. F. Turner, I. Karube and G. S. Wilson, Anal. Chim. Acta, 1987, 201, 363–364 CrossRef.
- L. Li, Y. Chen and J.-J. Zhu, Anal. Chem., 2017, 89, 358–371 CrossRef CAS PubMed.
- X.-L. Huo, H.-J. Lu, J.-J. Xu, H. Zhou and H.-Y. Chen, J. Mater. Chem. B, 2019, 7, 6469–6475 RSC | 6469.
- K. Ariga and M. Aono, Jpn. J. Appl. Phys., 2016, 55, 1102A6 CrossRef.
- K. Ariga, Q. Ji, J. P. Hill, Y. Bando and M. Aono, NPG Asia Mater., 2012, 4, e17 CrossRef.
- S. Hecht, Angew. Chem., Int. Ed., 2003, 42, 24–26 CrossRef CAS PubMed.
- J. Liu, H. Zhou, W. Yang and K. Ariga, Acc. Chem. Res., 2020, 53, 644–653 CrossRef CAS PubMed.
- Q. Hu, L. Su, Z. Chen, Y. Huang, D. Qin and L. Niu, Anal. Chem., 2021, 93, 9602–9608 CrossRef PubMed.
- G. Zhang, J. Liu, C. Liu, F. Ding, Y. Li, H. Tang and M. Ma, ACS Sens., 2021, 6, 4451–4460 CrossRef CAS PubMed.
- J. Zheng, H. Shi, M. Wang, C. Duan, Y. Huang, C. Li, Y. Xiang and a. G. Li, Anal. Chem., 2020, 92, 2194–2200 CrossRef CAS PubMed.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491–12545 CrossRef CAS PubMed.
- H. Zhou, J. Liu, J.-J. Xu, S.-S. Zhang and H.-Y. Chen, Chem. Soc. Rev., 2018, 47, 1996–2019 RSC.
- H. Qi, S. Yue, S. Bi, C. Ding and W. Song, Biosens. Bioelectron., 2018, 110, 207–217 CrossRef CAS PubMed.
- F. Dahl, J. Banér, M. Gullberg, M. Mendel-Hartvig, U. Landegren and M. Nilsson, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 4548–4553 CrossRef CAS PubMed.
- Q. Wang, H. Zheng, X. Gao, Z. Lin and G. Chen, Chem. Commun., 2013, 49, 11418–11420 RSC.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Chem. Rev., 2015, 115, 12491 CrossRef CAS PubMed.
- H. Zhou, S. Duan, J. Huang and F. He, Chem. Commun., 2020, 56, 6273–6276 RSC.
- F. Meng, P. Miao, B. Wang, Y. Tang and J. Yin, Anal. Chim. Acta, 2016, 943, 58e63 CrossRef PubMed.
- X. Zhang, Y. Liu, S. Du, Y. Yin, L. Kong, Y. Chang, Y. Chai, Z. Li and R. Yuan, Anal. Chem., 2021, 93, 9568–9574 CrossRef PubMed.
- M. Qing, S. L. Chen, Z. Sun, Y. Fan, H. Luo and N. Li, Anal. Chem., 2021, 93, 7499–7507 CrossRef CAS PubMed.
- T. Chaibun, J. Puenpa, T. Ngamdee, N. Boonapatcharoen, P. Athamanolap, A. P. O’Mullane, S. Vongpunsawad, Y. Poovorawan, S. Lee and B. Lertanantawong, Nat. Commun., 2021, 12, 802 CrossRef CAS PubMed.
- H. Lu, B. Ding, L. Tong, F. Wu, X. Yi and J. Wang, ACS Sens., 2020, 5, 2959–2965 CrossRef CAS PubMed.
- W. Qiu, F. Gao, N. Yano, Y. Kataoka, M. Handa, W. Yang, H. Tanaka and Q. Wang, Anal. Chem., 2020, 92, 11332–11340 CrossRef CAS PubMed.
- L. Zhu, X. Zhang, Y. Chang, S. Xu, R. Yuan and Y. Chai, Chem. Commun., 2021, 57, 5179–5182 RSC.
- Z. Liu, S. Lei, L. Zou, G. Li, L. Xu and B. Ye, Biosens. Bioelectron., 2019, 131, 113–118 CrossRef CAS PubMed.
- Y. Song, M. Wang, Q. Qian, J. Xu, Q. Zhou, S. Lv and P. Miao, ChemElectroChem, 2021, 8, 2778–2782 CrossRef CAS.
- Q. Guo, Y. Yu, H. Zhang, C. Cai and Q. Shen, Anal. Chem., 2020, 92, 5302–5310 CrossRef CAS PubMed.
- W. Wang, Y. Gao, W. Wang, J. Zhang, Q. Li and Z. Wu, Anal. Chem., 2022, 94, 1029–1036 CrossRef CAS PubMed.
- L. Yang, X. Yin, B. An and F. Li, Anal. Chem., 2021, 93, 1709–1716 CrossRef CAS PubMed.
- P. Miao and Y. Tang, Anal. Chem., 2020, 92, 12026–12032 CrossRef CAS PubMed.
- L. Zhu, X. Zhang, R. Yuan and Y. Chai, Anal. Chem., 2022, 94, 1264–1270 CrossRef CAS PubMed.
- Y. Jiang, H. Chai, N. Feng and P. Miao, ACS Sustainable Chem. Eng., 2021, 9, 9257–9263 CrossRef CAS.
- Y. Dai, W. Xu, R. A. Somoza, J. F. Welter, A. I. Caplan and C. C. Liu, Angew. Chem., Int. Ed., 2020, 59, 20545–20551 CrossRef CAS PubMed.
- J. Jiang, H. Wu, Y. Su, Y. Liang, B. Shu and C. Zhang, Anal. Chem., 2020, 92, 7708–7716 CrossRef CAS PubMed.
- X. Liu, X. Gao, L. Yang, Y. Zhao and F. Li, Anal. Chem., 2021, 93, 11792–11799 CrossRef CAS PubMed.
- L. Cui, J. Zhou, X. Yang, J. Dong, X. Wang and C. Zhang, Chem. Commun., 2020, 56, 10191–10194 RSC.
- X.-L. Zhang, Y. Yin, S.-M. Du, L.-Q. Kong, Y.-Q. Chai, Z.-H. Li and R. Yuan, Anal. Chem., 2021, 93, 13952–13959 CrossRef CAS PubMed.
- Y. Xue, Y. Wang, S. Feng, M. Yan, J. Huang and X. Yang, Anal. Chem., 2021, 93, 8962–8970 CrossRef CAS PubMed.
- F. Wang, K. Huang, Y. Hou, X. Tan, X. Wu, X. Yu and X. Zhou, Nanoscale, 2022, 14, 815–822 RSC.
- J. Yu, J. Liu, C. Ma, L. Qi, Y. Du, X. Hu, Y. Jiang, M. Zhou and E. Wang, Anal. Chem., 2022, 94, 600–605 CrossRef CAS PubMed.
- P. Miao, H. Chai and Y. Tang, ACS Nano, 2022, 16, 4726–4733 CrossRef CAS PubMed.
- J. Wang, H. Zhou, J. Liu and W. Yang, J. Electroanal. Chem., 2021, 895, 115419 CrossRef CAS.
- B. He and X. Dong, Chem. Eng. J., 2021, 405, 126642 CrossRef CAS.
- W. He, B. Qiao, F. Li, L. Pan, D. Chen, Y. Cao, J. Tu, X. Wang, C. Lv and Q. Wu, Chem. Commun., 2021, 57, 619–622 RSC.
- Y. Yu, Q. Guo, W. Jiang, H. Zhang and C. Cai, Anal. Chem., 2021, 93, 11298–11304 CrossRef CAS PubMed.
- P. Miao and Y. Tang, Small, 2020, 16, 2004518 CrossRef CAS PubMed.
- H.-Y. Wang, Y.-F. Ruan, L.-B. Zhu, X.-M. Shi, W.-W. Zhao, H.-Y. Chen and J.-J. Xu, Angew. Chem., Int. Ed., 2021, 60, 13244–13250 CrossRef CAS PubMed.
- Y. Wang, N. Hu, C. Liu, C. Nie, M. He, J. Zhang, Q. Yu, C. Zhao, T. Chen and X. Chu, Nanoscale, 2020, 12, 1673–1679 RSC.
- A. Butterworth, P. Pratibha, A. Marx and D. K. Corrigan, ACS Sens., 2021, 6, 3773–3780 CrossRef CAS PubMed.
- W. Miao and A. J. Bard, Chem. Rev., 2008, 108, 2506–2553 CrossRef CAS PubMed.
- P. Wu, X. D. Hou, J.-J. Xu and H.-Y. Chen, Chem. Rev., 2014, 114, 11027–11059 CrossRef CAS PubMed.
- L. Hu and G. Xu, Chem. Soc. Rev., 2010, 39, 3275–3304 RSC.
- D. M. Hercules, Science, 1964, 145, 808–809 CrossRef CAS PubMed.
- J. Wang, H. Zhou, J. Liu, J. He, J. Liu and W. Yang, Microchem. J., 2021, 169, 106581 CrossRef CAS.
- W. Gu, H. Wang, L. Jiao, Y. Wu, Y. Chen, L. Hu, J. Gong, D. Du and C. Zhu, Angew. Chem., Int. Ed., 2020, 59, 3534–3538 CrossRef CAS PubMed.
- F. Chen, D. Wang, J. Chen, J. Ling, H. Yue, L. Gou and H. Tang, Sens. Actuators, B, 2020, 305, 127472 CrossRef CAS.
- Q. Wang, S. Yu, L. Zhang, L. Wang, J. Kong, L. Li and X. Zhang, Chem. Commun., 2022, 58, 1701–1703 RSC.
- S. M. Khoshfetrat, P. Hashemi, A. Afkhami, A. Hajian and H. Bagheri, Sens. Actuators, B, 2021, 348, 130658 CrossRef CAS.
- X. Liu, Q. Wang, J. Chen, X. Chen and W. Yang, Talanta, 2021, 221, 121379 CrossRef CAS PubMed.
- N. Wang, L. Chen, W. Chen and H. Ju, Anal. Chem., 2021, 93, 5327–5333 CrossRef CAS PubMed.
- J. Jiang, P. Zhang, Y. Chai, R. Yuan and K. Peng, Sens. Actuators, B, 2021, 330, 129337 CrossRef CAS.
- Q. Wang, Y. Liu, X. Wang, F. Wang, L. Zhang, S. Ge and J. Yu, ACS Appl. Mater. Interfaces, 2021, 13, 25783–25791 CrossRef CAS PubMed.
- L. Wang, M. Jiang, Y. Chai, R. Yuan and Y. Zhuo, Chem. Commun., 2020, 56, 9000–9003 RSC.
- J. Liu, R. Yang, Y. Chai and R. Yuan, Anal. Chem., 2021, 93, 13334–13341 CrossRef CAS PubMed.
- Y. He, Y. Liu, L. Cheng, Y. Yang, B. Qiu, L. Guo, Y. Wang, Z. Lin and G. Hong, ACS Appl. Mater. Interfaces, 2021, 13, 298–305 CrossRef CAS PubMed.
- H. Xu, S. Zhang, T. Zhang, W. Huang, Y. Dai, R. Zheng and G. Wu, Talanta, 2022, 237, 122967 CrossRef CAS PubMed.
- Y. Xu, Y. Zhang, X. Sui, A. Zhang, X. Liu, Z. Lin and J. Chen, Sens. Actuators, B, 2021, 344, 130226 CrossRef CAS.
- Y. Zhang, G. Xu, G. Lian, F. Luo, Q. Xie, Z. Lin and G. Chen, Biosens. Bioelectron., 2020, 147, 111789 CrossRef CAS PubMed.
- X. Huang, J. Jia, Y. Lin, B. Qiu, Z. Lin and H. Chen, ACS Appl. Mater. Interfaces, 2020, 12, 34716–34722 CrossRef CAS PubMed.
- H. Wei and E. Wang, Luminescence, 2011, 26, 77–85 CrossRef CAS PubMed.
- E. Climent and K. Rurack, Angew. Chem., Int. Ed., 2021, 60, 26287–26297 CrossRef CAS PubMed.
- S. Kirschbaum-Harriman, A. Duerkop and A. J. Baeumner, Analyst, 2017, 142, 2648–2653 RSC.
- X. Ma, Z. Jiang and J. Li, Sens. Actuators, B, 2021, 333, 129562 CrossRef CAS.
- W. Yang, G. Zhang, J. Ni, Q. Wang and Z. Lin, Sens. Actuators, B, 2021, 327, 128872 CrossRef CAS.
- H. Hai, C. Chen, D. Chen, P. Li, Y. Shan and J. Li, Microchim. Acta, 2021, 188, 125 CrossRef CAS PubMed.
- S. Liu, C. Wang, Z. Wang, K. Xiang, Y. Zhang, G. Fan, L. Zhao, H. Han and W. Wang, Biosens. Bioelectron., 2022, 204, 114078 CrossRef CAS PubMed.
- X. Zhang, Y. Zhou, Y. Chai and R. Yuan, Anal. Chem., 2021, 93, 7987–7992 CrossRef CAS PubMed.
- D. Li, Y. Li, F. Luo, B. Qiu and Z. Lin, Anal. Chem., 2020, 92, 12686–12692 CrossRef CAS PubMed.
- Y. Huang, X. Zhu, C. Jin, W. Li, Y. Zhou and R. Yuan, Talanta, 2020, 219, 121274 CrossRef CAS PubMed.
- Z. Ning, Y. Zheng, D. Pan, Y. Zhang and Y. Shen, Biosens. Bioelectron., 2020, 150, 111945 CrossRef CAS PubMed.
- L. Yu, L. Zhu, M. Yan, S. Feng, J. Huang and X. Yang, Anal. Chem., 2021, 93, 11809–11815 CrossRef CAS PubMed.
- X. Fan, S. Wang, Y. Wang, X. Fan and L. Yu, J. Braz. Chem. Soc., 2020, 31, 2620–2627 CAS.
- S. Kesarkar, S. Valente, A. Zanut, F. Palomba, A. Fiorani, M. Marcaccio, E. Rampazzo, G. Valenti, F. Paolucci and L. Prodi, J. Phys. Chem. C, 2019, 123, 5686–5691 CrossRef CAS.
- C. Wang, Q. Han, F. Mo, M. Chen, Z. Xiong and Y. Fu, Anal. Chem., 2020, 92, 12145–12151 CrossRef CAS PubMed.
- G. Liu, J. Ling and J. Li, ACS Sens., 2021, 6, 4185–4192 CrossRef CAS PubMed.
- J. B. Yao, J. Zhang, Z. Fan, Y. Ding, B. Zhou, R. Yang, J. Zhao and K. Zhang, ACS Appl. Mater. Interfaces, 2021, 13, 19816–19824 CrossRef PubMed.
- Q. Hao, Q. Xu, S. Niu, C. Ding and X. Luo, Anal. Chem., 2021, 93, 10679–10687 CrossRef CAS PubMed.
- X. Hu, J. Tian, C. Li, H. Su, R. Qin, Y. Wang, X. Cao and P. Yang, Adv. Mater., 2020, 32, 2000128 CrossRef CAS PubMed.
- J. Yang, Q. Xia, L. Guo, F. Luo, Y. Dong, B. Qiu and Z. Lin, Chem. Commun., 2020, 56, 6692–6695 RSC.
- X. Liu, L. Li, F. Li, W. Zhao, L. Luo, X. Bi, X. Li and T. You, J. Hazard. Mater., 2022, 424, 127480 CrossRef CAS PubMed.
- Z. Ding, B. M. Quinn, S. K. Haram, L. E. Pell, B. A. Korgel and A. J. Bard, Science, 2002, 296, 1293–1297 CrossRef CAS PubMed.
- G. Jie, Y. Qin, Q. Meng and J. Wang, Analyst, 2015, 140, 79–82 RSC.
- X. Lv, X. Pang, Y. Li, T. Yan, W. Cao, B. Du and Q. Wei, ACS Appl. Mater. Interfaces, 2015, 867–872 CrossRef CAS PubMed.
- X. Zhang, B. Zhang, W. Miao and G. Zou, Anal. Chem., 2016, 88, 5482–5488 CrossRef CAS PubMed.
- J. Huan, Q. Liu, A. Fei, J. Qian, X. Dong, B. Qiu, H. Mao and K. Wang, Biosens. Bioelectron., 2015, 73, 221–227 CrossRef CAS PubMed.
- P. Wang, Y. Nie, Y. Tian, Z. Liang, S. Xu and Q. Ma, Chem. Eng. J., 2021, 426, 130732 CrossRef CAS.
- Y. Zhao, L. Tan and G. Jie, Sens. Actuators, B, 2021, 333, 129586 CrossRef CAS.
- L. Liu, Y. Zhang, R. Yuan and H. Wang, Anal. Chem., 2020, 92, 15112–15119 CrossRef CAS PubMed.
- K. Ji, Y. Wang, L. Mao, Y. Wang and X. Zhang, Sens. Actuators, B, 2021, 345, 130405 CrossRef CAS.
- A. Chen, S. Ma, Y. Zhuo, Y. Chai and R. Yuan, Anal. Chem., 2016, 88, 3203–3210 CrossRef CAS PubMed.
- R. Tian, S. Zhang, M. Li, Y. Zhou, B. Lu, D. Yan, M. Wei and D. G. D. Evans, Adv. Funct. Mater., 2015, 25, 5006–5015 CrossRef CAS.
- M. Xiong, Q. Rong, H. M. Meng and X. B. Zhang, Biosens. Bioelectron., 2017, 89, 212–223 CrossRef CAS PubMed.
- Q.-M. Feng, Y.-Z. Shen, M.-X. Li, Z.-L. Zhang, W. Zhao, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2016, 88, 937–944 CrossRef CAS PubMed.
- R. Dai, F. Wu, H. Xu and Y. Chi, ACS Appl. Mater. Interfaces, 2015, 7, 15160–15167 CrossRef CAS PubMed.
- N. Zhang, Z. Y. Zhao, H. Gao, Y. Yu, J. Pan, H.-Y. Chen and J.-J. Xu, J. Electroanal. Chem., 2021, 900, 115743 CrossRef CAS.
- K. Zhao, L. Wang, P. Liu, X. Hang, H. Wang, S. Ye, Z. Liu and G. Liang, Sens. Actuators, B, 2021, 346, 130485 CrossRef CAS.
- Y. Yang, G. Hu, W. Liang, L. Yao, W. Huang, Y. Zhang, J. Zhang, J. Wang, R. Yuan and D. Xiao, Nanoscale, 2020, 12, 5932–5941 RSC.
- L. Wang, K. Zhao, Z. Liu, Y. Zhang, P. Liu, S. Ye, Y. Zhang and G. Liang, Sens. Actuators, B, 2021, 348, 130660 CrossRef CAS.
- S. Ye, C. Pan, Y. Dai and G. Liang, Microchem. J., 2021, 165, 106123 CrossRef CAS.
- M. Pan, Y. Lei, Y. Chai, R. Yuan and Y. Zhuo, Anal. Chem., 2020, 92, 13581–13587 CrossRef CAS PubMed.
- F. Yang, X. Jiang, W. Liang, Y. Chai, R. Yuan and Y. Zhuo, Anal. Chem., 2020, 92, 2566–2572 CrossRef CAS PubMed.
- Q. Zhang, Y. Tian, Z. Liang, Z. Wang, S. Xu and Q. Ma, Anal. Chem., 2021, 93, 3308–3314 CrossRef CAS PubMed.
- Y. Tang, J. Li, Q. Guo and G. Nie, Sens. Actuators, B, 2019, 282, 824–830 CrossRef CAS.
- X. Zhou, W. Zhang, Z. Wang, J. Han, G. Xie and S. Chen, Biosens. Bioelectron., 2020, 148, 111795 CrossRef CAS PubMed.
- H. Lv, A. Chen, W. Cheng, L. Kong, M. Zhao, S. Ding, H. Ju and W. Cheng, Anal. Chem., 2020, 92, 15624–15631 CrossRef CAS PubMed.
- F. Li, M. Wang, Y. Zhou, H. Yin and S. Ai, Microchim. Acta, 2021, 188, 68 CrossRef CAS PubMed.
- R. Zou, Y. Lin and C. Lu, Anal. Chem., 2021, 93, 2678–2686 CrossRef CAS PubMed.
- Z. Feng, R. Zhao, Z. Lu, L. Jia, R. Ma, W. Zhang, L. Shang, Q. Xue and H. Wang, Chem. Commun., 2020, 56, 11074–11077 RSC.
- S. A. Kitte, F. A. Bushira, C. Xu, Y. Wang, H. Li and Y. Jin, Anal. Chem., 2022, 94, 1406–1414 CrossRef CAS PubMed.
- H. Zhu and S. Ding, Biosens. Bioelectron., 2019, 134, 109–116 CrossRef CAS PubMed.
- G. Mo, D. Qin, X. Jiang, X. Zheng, W. Mo and B. Deng, Sens. Actuators, B, 2020, 310, 127852 CrossRef CAS.
- S. Xiao, X. Wang, C. Yang, Y. Jiang, S. Zhen and C. Huang, Anal. Chem., 2022, 94, 1178–1186 CrossRef CAS PubMed.
- J. Guo, M. Xie, P. Du, Y. Liu and X. Lu, Anal. Chem., 2021, 93, 10619–10626 CrossRef CAS PubMed.
- L. Zhu, L. Yu, T. Meng, Y. Peng and X. Yang, Small, 2021, 17, 2102881 CrossRef CAS PubMed.
- S. Liu, H. Yang, P. Wang and Q. Feng, Anal. Chim. Acta, 2021, 1144, 68e75 Search PubMed.
- J. Ge, C. Li, Y. Zhao, X. Yu and G. Jie, Chem. Commun., 2019, 55, 7350–7353 RSC.
- H. Gao, J. Zhang, Y. Liu, W. Tu, T. Wei and Z. Dai, Anal. Chem., 2019, 91, 12038–12045 CrossRef CAS PubMed.
- Z. Liu, J. Wang, Y. Lu, C. Cui, L. Zheng and L. Hu, J. Electroanal. Chem., 2021, 899, 115697 CrossRef CAS.
- Z. Fan, B. Yao, Y. Ding, D. Xu, J. Zhao and K. Zhang, Chem. Eng. J., 2022, 427, 131686 CrossRef CAS PubMed.
- Y. Xu, Z. Wang, C. Ding and X. Luo, Sens. Actuators, B, 2020, 322, 128613 CrossRef CAS.
- Y. He, F. Hu, J. Zhao, G. Yang, Y. Zhang, S. Chen and R. Yuan, Anal. Chem., 2021, 93, 8783–8790 CrossRef CAS PubMed.
- J. Zhao, Y. He, K. Tan, J. Yang, S. Chen and R. Yuan, Anal. Chem., 2021, 93, 12400–12408 CrossRef CAS PubMed.
- L. Wang, P. Liu, Z. Liu, H. Cao, S. Ye, K. Zhao, G. Liang and J. Zhu, Sens. Actuators, B, 2021, 327, 128890 CrossRef CAS.
- F. Wang, Y. Liu, C. Fu, N. Li, M. Du, L. Zhang, S. Ge and J. Yu, Anal. Chem., 2021, 93, 1702–1708 CrossRef CAS PubMed.
- Y. Cao, W. Zhu, H. Wei, C. Ma, Y. Lin and J. Zhu, Anal. Chem., 2020, 92, 4123–4130 CrossRef CAS PubMed.
- H. Ge, X. Wang, J. Xu, H. Lin, H. Zhou, T. Hao, Y. Wu and Z. Guo, Anal. Chem., 2021, 93, 14885–14891 CrossRef CAS PubMed.
- L. Zhu, M. Zhang, J. Ye, M. Yan, Q. Zhu, J. Huang and X. Yang, Anal. Chem., 2020, 92, 8614–8622 CrossRef CAS PubMed.
- Q. Zhang, Y. Fu, K. Xiao, C. Du, X. Zhang and J. Chen, Anal. Chem., 2021, 93, 6801–6807 CrossRef CAS PubMed.
- M. Liu, G. Chen, Y. Qin, J. Li, L. Hu, W. Gu and C. Zhu, Anal. Chem., 2021, 93, 9897–9903 CrossRef CAS PubMed.
- Q. Cai, D. Wu, H. Li, G. Jie and H. Zhou, Biosens. Bioelectron., 2021, 191, 113455 CrossRef CAS PubMed.
- H. Zhou, J. Zhang, B. Li, J. Liu, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2021, 93, 6120–6127 CrossRef CAS PubMed.
- M. Mathesh, J. Liu, C. J. Barrow and W. Yang, Chem. – Eur. J., 2017, 23, 304–311 CrossRef CAS PubMed.
- F. Hasan, A. A. Shah and A. Hameed, Enzyme Microb. Technol., 2006, 39, 235–251 CrossRef CAS.
- C. Bernal, A. Illanes and L. Wilson, Langmuir, 2014, 30, 3557–3566 CrossRef CAS PubMed.
- M. Mathesh, B. Luan, T. O. Akanbi, J. K. Weber, J. Liu, C. J. Barrow, R. Zhou and W. Yang, ACS Catal., 2016, 6, 4760–4768 CrossRef CAS.
- M. Mathesh, J. Sun, F. van der Sandt and D. A. Wilson, Nanoscale, 2020, 12, 22495–22501 RSC.
- M. Mathesh, J. Sun and D. A. Wilson, J. Mater. Chem. B, 2020, 8, 7319–7334 RSC.
- E. Karshalev, B. Esteban-Fernández de Ávila and J. Wang, J. Am. Chem. Soc., 2018, 140, 3810–3820 CrossRef CAS PubMed.
- M. Mathesh, E. Bhattarai and W. Yang, Angew. Chem., Int. Ed., 2022, 134, e202113801 Search PubMed.
- P. Romele, P. Gkoupidenis, D. A. Koutsouras, K. Lieberth, Z. M. Kovács-Vajna, P. W. M. Blom and F. Torricelli, Nat. Commun., 2020, 11, 3743 CrossRef CAS PubMed.
- A. Nawaz, Q. Liu, W. L. Leong, K. E. Fairfull-Smith and P. Sonar, Adv. Mater., 2021, 33, 2101874 CrossRef CAS PubMed.
- C. Srisomwat, A. Yakoh, N. Chuaypen, P. Tangkijvanich, T. Vilaivan and O. Chailapakul, Anal. Chem., 2021, 93, 2879–2887 CrossRef CAS PubMed.
- X. Xie, Z. Wang, M. Zhou, Y. Xing, Y. Chen, J. Huang, K. Cai and J. Zhang, Small Methods, 2021, 5, 2101072 CrossRef CAS PubMed.
- Z. F. Wu and P. Yang, Adv. Mater. Interfaces, 2015, 2, 1400401 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |






