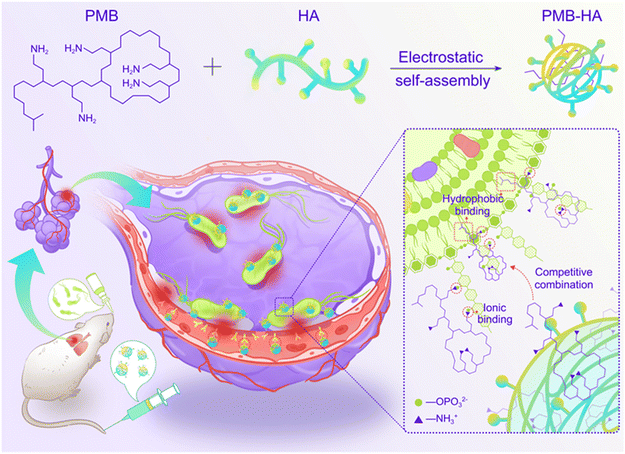
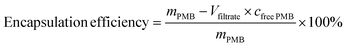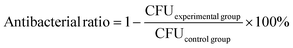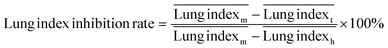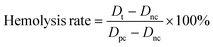An inflammation-targeted nanoparticle with bacteria forced release of polymyxin B for pneumonia therapy†
Peisen
Zhang‡
a,
Qiuhong
Ouyang‡
a,
Tianshu
Zhai‡
b,
Jing
Sun
c,
Jun
Wu
a,
Feng
Qin
d,
Ni
Zhang
d,
Saisai
Yue
a,
Xinchen
Yang
 a,
Hanyi
Zhang
a,
Yi
Hou
a,
Hanyi
Zhang
a,
Yi
Hou
 a,
Li
Deng
a,
Fang
Wang
a,
Qingyuan
Zhan
*b,
Qingsong
Yu
a,
Li
Deng
a,
Fang
Wang
a,
Qingyuan
Zhan
*b,
Qingsong
Yu
 *a,
Meng
Qin
*a,
Meng
Qin
 *a and
Zhihua
Gan
a
*a and
Zhihua
Gan
a
aBeijing Advanced Innovation Centre for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing 100029, PR China. E-mail: yuqs@mail.buct.edu.cn; qinmeng212@mail.buct.edu.cn
bDepartment of Pulmonary and Critical Care Medicine, Center of Respiratory Medicine, China-Japan Friendship Hospital, Beijing 100029, PR China. E-mail: drzhanqy@163.com
cInstitute of Chinese Materia Medica, China Academy of Chinese Medical Sciences, Chaoyang District, Beijing 100029, PR China
dNational Chengdu Center for Safety Evaluation of Drugs, State Key Laboratory of Biotherapy/Collaborative Innovation Center for Biotherapy, Department of Psychiatry, West China Hospital of Sichuan University, Chengdu, Sichuan 610041, PR China
First published on 3rd August 2022
Abstract
The epidemic of multidrug-resistant Gram-negative bacteria is an ever-growing global concern. Polymyxin B (PMB), a kind of “old fashioned” antibiotic, has been revived in clinical practice and mainly used as last-line antibiotics for otherwise untreatable serious infections because the incidence of the resistance to PMB is currently relatively low in comparison with other antibiotics in vivo owing to the unique bactericidal mechanism of PMB. However, serious adverse side effects, including nephrotoxicity and neurotoxicity, hamper its clinical application. Herein, we describe the development of a nanoparticle that can target sites of inflammation and forcedly release PMB specifically in the area of Gram-negative bacteria. This particle was constructed through the electrostatic self-assembly of hyaluronic acid (HA) and PMB molecules in order to realize the safe and effective treatment of pneumonia. After systemic administration, PMB-HA nanoparticles were found to actively accumulate in the lungs, precisely target the CD44 receptors over-expressed on the membrane of activated endothelial cells in inflammatory sites, and then come into contact with the bacteria resident in the damaged alveolar-capillary membrane. Due to the electrostatic and hydrophobic interactions between PMB and the lipopolysaccharide (LPS) in the outer membranes of bacteria, the PMB molecules in the PMB-HA nanoparticles are expected to escape from the nanoparticles to insert into the bacteria via competitive binding with LPS. Through shielding the cationic nature of PMB, PMB-HA nanoparticles also possess outstanding biosafety performance in comparison to free PMB. It is thus believed that this smart delivery system may pave a new way for the resurrection of PMB in the future clinical treatment of bacterial inflammatory diseases.
1 Introduction
The epidemic of multidrug-resistant Gram-negative bacteria is an ever-growing global concern. For example, the acquiring of an infection with such a pathogen, especially carbapenem-resistant strains, has led to increased risks of mortality to patients of Intensive Care Units and other hospital facilities.1–3 One strategy to change this situation is the revival of “old fashioned” antibiotics. Polymyxin B (PMB), one member of the polymyxin family, acts as an antibiotic via its strong interactions with the lipid A moiety of lipopolysaccharide (LPS) which induces permeabilization of the outer membranes of Gram-negative bacteria. PMB was formerly widely used for pneumonia, bacteremia, and tissue infections.4 Unfortunately, intravenous polymyxins were limited in clinical practice in the 1960s due to their toxic side effects, such as allergic reactions, nephrotoxicity, and neurotoxicity.5,6 Hence, modification of PMB to minimize its toxicity is essential if it is to be widely applied to clinical applications.The amino group on the α,γ-diaminobutyric acid residues of cationic PMB is a double-edged sword. On one hand, it binds to and disrupts the negatively charged LPS in the outer membrane of Gram-negative bacteria. On the other hand, it also protonates under physiological conditions and interacts with phosphate anions in the cell membranes of healthy cells, especially in the kidney, where PMB accumulates.7 That is the most critical mechanism of PMB-induced nephrotoxicity: PMB increases the permeability of tubular epithelial cell membranes, leading to an influx of cations, anions and water.8 In the last two decades of the twentieth century, several approaches have been used to encapsulate PMB through electrostatic interactions, shielding the cationic portion or reducing the number of cationic residues.9–13 However, although these strategies can greatly reduce the toxicity of PMB, the antibacterial activity is sacrificed due to the shielding of the LPS-binding positive charge.
In recent years, nano-based smart drug delivery systems with outstanding targeting abilities and excellent controlled drug release performances have demonstrated an exciting potential for the precise treatment of various diseases with minimized side effects.14–16 In our previous publications, we have described the development of a nano system to precisely deliver hydrophobic dihydroxytestosterone molecules through the blood–brain barrier to the areas of intracranial stroke, which may enhance the therapeutic efficacy of dihydroxytestosterone.17 In addition, we constructed a pH-triggered smart UCNP@GA-Fe(III) nanoprobe based on rare earth nanoparticles, which can responsively release Fe3+ in the tumor microenvironment to induce the apoptosis of tumor cells through ROS accumulation.18
Very recently, Jin and coworkers prepared CS-DA/PMB nanocomplexes, which reduce the toxicity of the drug by shielding the positive charge through the electrostatic interaction of positively charged PMB and negatively charged 2,3-dimethylmaleic anhydride (DA) grafted chitosan (CS). Through locoregional administration, the nanoparticles can realize acid-triggered PMB release, which significantly reduces the side effects of this highly toxic antibiotic without reducing its intrinsic antibacterial activity.19 Unfortunately, the acidic feature is not specific to the inflammatory site in the organism. Lysosomes, especially those of various lung-resident phagocytic cells, may also trigger the release of the PMB from acid-responsive nano system, which hampers the anti-inflammation activity of PMB.20 In addition, the locoregional administration route may limit the application of nanoparticles for deep lesion treatments due to poor accessibility. Therefore, a new PMB-based formulation that can not only sensitively release PMB to bacteria, but also precisely target sites of inflammation upon systemic administration with limited adverse side effects is very much needed to ensure both the high anti-inflammatory efficacy and high biosafety of PMB.
Hyaluronan (HA) is a natural polysaccharide that, like CS, has good biocompatibility and biodegradability.21–23 More importantly, HA is the principal ligand of CD44, a cell-adhesion molecule that is ubiquitously expressed on leukocytes, endothelial and tumor cells.24–26 It has been suggested that CD44 is up-regulated and over-expressed on various cells under inflammatory conditions, and the interaction of CD44 and its HA ligand is very active in inflammatory diseases.27–32 Therefore, we considered HA to be a candidate drug carrier for PMB improve the targeting of the drug to the site of infection.
In this work, an inflammation-targeted smart nanoparticle with bacteria-triggered PMB forced release ability was successfully constructed through the electrostatic self-assembly of HA and PMB molecules for realizing both safe and effective treatment of pneumonia (Scheme 1). Through shielding the cationic nature of PMB, PMB-HA nanoparticles are expected to possess outstanding biosafety performance in comparison to free PMB. After intravenous injection, PMB-HA nanoparticles are expected to accumulate in the lungs, precisely target the CD44 receptors over-expressed on the membranes of activated endothelial cells in inflammatory sites, and come into contact with the bacteria resident in the damaged alveolar-capillary membrane. As is known, the amino groups in PMB can bind with phosphate groups in LPS through ionic forces, and displace divalent cations (Ca2+ and Mg2+) that normally function to bridge and stabilize the LPS outer membrane of bacteria. Meanwhile, the N-terminal fatty acyl chain of PMB can insert into the adjacent fatty acyl chains of LPS, causing expansion of the outer membrane of bacteria.33 Due to this combined effect of both the electrostatic and hydrophobic interactions, the PMB molecules in the PMB-HA nanoparticles are expected to competitively bind with LPS in the outer membranes of Gram-negative bacteria, thereby realizing the forced release of PMB from the nanoparticles for the effective inhibition of bacteria growth. This smart LPS-triggered responsive behavior of nanoparticles is designed to ensure both therapeutic effects and biosafety to normal tissues.
In the experiments reported below, we describe the construction of PMB-HA nanoparticles, and characterize their morphology, hydrodynamic size, zeta potential, and LPS-triggered PMB release properties. In cell and animal studies, we show that the nanoparticles possess both outstanding anti-inflammation therapeutic effects on pneumonia caused by multidrug-resistant-Pseudomonas aeruginosa (MDR-P. aeruginosa) and satisfactory biosafety requirements as anticipated.
2 Experimental
2.1 Materials
Polymyxin B (PMB) sulphate, lipopolysaccharides (LPS) and the BCA kit were purchased from Solarbio (Beijing, China). Hyaluronic acid (HA, MW: approximately 8000 Da) was obtained from Guanglong biological company (Shandong, China). Fluorescent dye Cy5.5-NHS was purchased from Aladdin Industrial Corporation (Shanghai, China). Luria–Bertani (LB) broth and LB agar were obtained from Thermo Fisher Scientific (USA). The Annexin V-FITC Apoptosis Detection Kit was purchased from Solarbio (Beijing, China). The ELISA kits of IL-1β, IL-6, IL-10, and TNF-α were purchased from Abcam (Shanghai, China). Human pulmonary alveolar epithelial cells (HPAEpiC), human umbilical vein endothelial cells (HUVECs) and human alveolar basal epithelial cells (A549) were purchased from American Type Culture Collection (ATCC, USA). Multidrug-resistant-Pseudomonas aeruginosa (MDR-P. aeruginosa) was purchased from American Type Culture Collection (ATCC2110, USA) and P. aeruginosa was purchased from BeNa Culture Collection, China (BNCC-186666).2.2 Preparation of PMB-HA nanoparticles
PMB-HA with different molar ratios of HA and PMB was prepared. Typically, PMB was dissolved in 5 mM HEPES buffer at pH 7.4, and a solution of HA (2.5 mM) in 5 mM HEPES buffer at pH 7.4 was also prepared. The PMB solution was then slowly added into the HA solution under constant stirring (400 rpm). After stirring for 24 h, the mixture was filtered through 0.45 μm nylon syringe filters. The resulting PMB-HA was then processed with 3000 molecular weight cut off (MWCO) centrifugal filters to remove the unreacted free PMB. To confirm the complete removal of free PMB, the filtrate obtained after the ultrafiltration was collected to determine the concentration of PMB in it through the BCA kit. The free PMB is considered to be completely removed when the absorbance of the resultant mixture at 562 nm is comparable with PBS background. Finally the resultant nanoparticles were stored at 4 °C for further use. The whole filtrate was collected to calculate the PMB encapsulation rate.The encapsulation efficiency can be calculated through the following equation:
2.3 Nanoparticle characterization
The morphology of the nanoparticles was observed by transmission electron microscopy (TEM) on a HT7700 TEM (Hitachi, Japan) at an acceleration voltage of 80 kV. The hydrodynamic size and zeta potential of the nanoparticles were measured with a dynamic light scattering system (Zetasizer Nano-ZS; Malvern Instruments) equipped with a He–Ne laser (633 nm) at 25 °C. Cell fluorescence images were captured with a Leica Inverted Fluorescence Microscope. The PMB content was measured through absorption spectroscopy (λ = 222 nm) at room temperature with a UV-Vis spectrophotometer (Fig. S4†).2.4 LPS-responsive drug release from PMB-HA nanoparticles
To study the PMB release kinetics of PMB-HA nanoparticles, 2 mL of a PMB-HA nanoparticle (1.2 mg mL−1 with respect to PMB) was added into a dialysis membrane (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO) and dipped into 10 mL of 0.9% NaCl solution (normal saline) with (100 μg mL−1) or without LPS at 37 °C under stirring (300 rpm). Then, the temporal evolution of the PMB content in the dialysate was detected with a BCA kit.
000 MWCO) and dipped into 10 mL of 0.9% NaCl solution (normal saline) with (100 μg mL−1) or without LPS at 37 °C under stirring (300 rpm). Then, the temporal evolution of the PMB content in the dialysate was detected with a BCA kit.
2.5 Fluorescent labeling of nanoparticles
For imaging purposes, PMB was labeled with Cy5.5 fluorescent dye. A Cy5.5-NHS stock solution (0.2% (w/v) in PBS) was added into PMB solution (1.65 mg mL−1 in PBS) at a molar ratio of Cy5.5 : PMB = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The reaction was kept under stirring at 4 °C for 2 h. Then, Cy5.5 labeled PMB-HA nanoparticles (PMBCy5.5-HA) were synthesized following the above procedures for the preparation of PMB-HA nanoparticles. Finally, the resultant nanoparticles were dialyzed against PBS to remove the unconjugated Cy5.5-NHS with a 3000 molecular weight cut off dialysis bag.
1. The reaction was kept under stirring at 4 °C for 2 h. Then, Cy5.5 labeled PMB-HA nanoparticles (PMBCy5.5-HA) were synthesized following the above procedures for the preparation of PMB-HA nanoparticles. Finally, the resultant nanoparticles were dialyzed against PBS to remove the unconjugated Cy5.5-NHS with a 3000 molecular weight cut off dialysis bag.
2.6 Cell culture
HUVECs were cultured in endothelial cell medium (ECM) with 10% fetal bovine serum (FBS), 100 U mL−1 penicillin, and 0.1 mg mL−1 streptomycin at 37 °C under a 5% CO2 atmosphere. HPAEpiC cells and A549 cells were cultured in RPMI-1640 medium with 10% FBS, 100 U mL−1 penicillin, and 0.1 mg mL−1 streptomycin at 37 °C under a 5% CO2 atmosphere.2.7 Western blot analysis for CD44 protein expression
The protein samples were extracted from HUVECs incubated with the cell culture medium in the presence (1 μg mL−1) or absence of LPS for 24 h. HUVECs were washed with PBS and lysed with a lysis buffer freshly supplemented. Then, the BCA kit was used to determine the protein concentration of samples. Next, the extracted protein supernatant and protein loading buffer solution were placed in a boiling water bath for 8 min to complete protein denaturation. Subsequently, protein samples and markers were added to the loading wells and electrophoresis was performed. Finally, the gel was removed and transferred in a transfer tank, and then developed by immunoblotting.2.8 Cell binding assays
HUVECs were seeded in a 12-well cell culture plate and then cultured in an atmosphere containing 5% CO2 overnight at 37 °C to allow firm adherence. Then, the cells were activated by being cultured in ECM containing 1 μg mL−1 LPS for 24 h in the absence of FBS. Typically, 1 mL of ECM containing PMBCy5.5-HA nanoparticles (100 μg mL−1 with respect to PMB) was added to the wells and incubated with the adherent cells for 6 h. After the supernatant was decanted, the cells were rinsed three times with PBS to remove the unbound particles. After that, the cells were fixed by adding 4% paraformaldehyde. After staining the nucleus with Hoechst 33342, fluorescence images were captured on a fluorescence microscope (Leica DMI 3000B).2.9 Determination of minimum inhibitory concentrations (MIC)
P. aeruginosa was cultured in LB media at 30 °C with rotation (180 rpm) overnight. MIC for antimicrobial susceptibility testing of PMB-HA nanoparticles and PMB was performed through the broth dilution method according to the guidelines of the Clinical and Laboratory Standards Institute.34 The bacterial concentration was adjusted in a McBride turbidimetric tube to 106 CFU mL−1. Approximately 104P. aeruginosa were suspended and cultured with PMB-HA nanoparticles or PMB solutions at different concentrations in 96-well plates. In particular, the bacteria were incubated in liquid medium at a series of initial drug concentrations. After 24 h of incubation, the bacterial suspension of each well was centrifuged, and the optical density of the supernatant of each well at 600 nm was recorded with a microplate reader (FilterMax F5, USA). The MIC was determined as the lowest antibiotic concentration at which the growth of bacteria was totally inhibited.2.10 Determination of the diameter of the bacteriostatic zone
P. aeruginosa was inoculated into LB medium and placed in an incubator at 30 °C overnight (180 rpm). A sample (20 μL) of the bacterial suspension (106 CFU mL−1) was added to 100 mL of sterile LB medium containing melted agar at 40 °C, and the mixture was poured into a sterile Petri dish. After the culture medium solidified, sterilized Oxford cups were placed symmetrically on the prepared plate, and samples (150 μL) of PMB-HA or PMB nanoparticles (containing the equivalent of 20 μg mL−1 PMB) were added into the two Oxford cups. After incubating for 24 h at 30 °C, the diameters of inhibition zones were measured with the cross-sectional method with a Vernier caliper.352.11 In vitro bactericidal activity of nanoparticles by standard plate counting assays
Colonies of P. aeruginosa were transferred to 6 mL of LB medium and cultivated in a rotating shaker (180 rpm) at 30 °C overnight to obtain bacteria in the exponential growth phase. The bacterial suspensions were then centrifuged at 5000 rpm for 5 min to remove the medium and resuspended in sterile PBS. The bacterial suspensions were adjusted to an initial concentration of 2.5 × 107 CFU mL−1. P. aeruginosa was co-incubated with 100 μL PMB-HA nanoparticles or PMB at 1 μg mL−1, 2 μg mL−1, 4 μg mL−1 and 8 μg mL−1 (equivalent to PMB) in 96-well plates for 12 h. After dilution with the same multiple, the suspensions were transferred to LB plates and incubated for 20 h at 30 °C. Through counting the colonies, the antibacterial ratio was determined using the following formula:2.12 Cytotoxicity assays
CCK-8 assays of HPAEpiC cells, A549 cells, and HUVECs were carried out as follows. Cells were seeded into 96-well cell culture plates at 5 × 103 cells per well under 100% humidity and then cultured at 37 °C in an atmosphere containing 5% CO2 for 24 h. Culture medium containing PMB-HA nanoparticles or PMB at a series of concentrations was added to the wells, and the cells were incubated for 24 h at 37 °C under 5% CO2. Subsequently, the supernatant containing the excess particles or free PMB was decanted, and the cells were incubated for another 24 h at 37 °C under 5% CO2. CCK-8 (100 μL, 10% v/v) was then added to each well, and the plates were incubated for 2 h at 37 °C. The optical density of each well at 450 nm was recorded on a microplate reader (Thermo Fisher, USA).The cell viability of each well was calculated according to the following equation.
 ,
,  , and
, and  are the average optical density of test wells (cells + samples), control wells (healthy cells), and blank wells (culture medium), respectively.
are the average optical density of test wells (cells + samples), control wells (healthy cells), and blank wells (culture medium), respectively.
2.13 Live/dead cell staining
In order to differentiate dead cells from viable cells after co-incubation with PMB-HA nanoparticles or PMB (150 μg mL−1 with respect to PMB for 24 h), the live/dead staining reagents 3′,6′-di(O-acetyl)-4′,5′-bis[N,N-bis(carboxymethyl) aminomethyl] fluorescein, tetraacetoxymethyl ester (calcein-AM), and propidium iodide (PI) were used to stain the viable cells green (λex = 490 nm, λem = 515 nm) and dead cells red (λex = 535 nm, λem = 617 nm). In particular, portions (100 μL) of calcein-AM and PI solution were added after the removal of the culture medium and rinsing of the disks. After 30 min of incubation, the staining solution was removed and cells were rinsed with PBS twice prior to examination with a fluorescence microscope (Leica DMI 3000B, Germany).2.14 Cell apoptosis assays
HPAEpiC cells were cultured in 6-well plates at a density of 1 × 105 cells per well. When the cells reached 70 to 80% confluence, the culture medium was replaced with 2 mL of fresh culture medium containing PMB-HA nanoparticles or PMB (150 μg mL−1 with respect to PMB), or HA (10 mg mL−1). The control group was treated with the same amount of culture medium. After incubation for another 24 h, cell apoptosis was detected by using the Annexin V-FITC/PI Apoptosis Detection Kit.2.15 Treatment of MDR-P. aeruginosa infectious pneumonia with PMB-HA nanoparticles
The ICR mice (6 weeks of age, 20–23 g, male) were anesthetized with isoflurane and then infected with 5 × 107 CFU of MDR-P. aeruginosa in 50 μL through intranasal drips. The mouse pneumonia model was considered to be established at 2 h post-injection of the bacteria. Mice with pneumonia (n = 30) were randomly allocated into 3 groups (n = 10), and intravenously injected with PMB-HA nanoparticles, PMB (3 mg kg−1 with respect to PMB) or PBS at the same volume (100 μL) at 2 h and 8 h post-modeling. Ten healthy mice were set as the negative controls. Mice were sacrificed 32 h post-modeling. Their lung tissue was extracted and weighed to calculate the lung index inhibition rate, followed by morphological observation and histological analysis. After observation, 3 lung samples were homogenized in 10 mL of PBS and then diluted 10-fold prior to counting of colony-forming units. The other 3 lung tissues were also homogenized, and their supernatant was collected to analyze the level of inflammatory-related factors, including IL-1β, IL-6, IL-10, and TNF-α with ELISA kits. The rest of the lung tissue was analysed by H&E staining. Samples of blood from all mice were collected after sacrificing for biochemical and routine blood tests.The lung index can be expressed as the ratio of the mean lung weight to the mean body weight. And the lung index inhibition rate can be calculated according to the following equations:
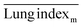 ,
, 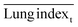 , and
, and 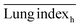 are the average lung index of model group, treatment group, and healthy group.
are the average lung index of model group, treatment group, and healthy group.
2.16 Hemolysis assays
Hemolysis assays were conducted according to a previous report.36 Briefly, a 2 mL fresh blood sample of mice was mixed with 6 mL of normal saline (NS) and centrifuged at 1500 rpm for 15 min. The supernatant was drawn off, and three subsequent washes were carried out before diluting the sample to a final volume of 4 mL in PBS. The red blood cells were then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 into NS solutions (negative control); water (positive control); or nanoparticle solutions (in NS) at varying concentrations (tested samples). The samples were left at room temperature in the dark for 4 h at 37 °C and then centrifuged at 3000 rpm for 5 min. After centrifuging, the supernatant was transferred to a 96-well plate, and the absorbance at 541 nm was recorded.
4 into NS solutions (negative control); water (positive control); or nanoparticle solutions (in NS) at varying concentrations (tested samples). The samples were left at room temperature in the dark for 4 h at 37 °C and then centrifuged at 3000 rpm for 5 min. After centrifuging, the supernatant was transferred to a 96-well plate, and the absorbance at 541 nm was recorded.
The hemolysis rate was calculated with the following equation:
2.17 Acute toxicity of PMB-HA nanoparticles
Ten healthy male ICR mice (28–32 g) were randomly divided into 2 groups (n = 5), and intravenously injected with PBS containing PMB-HA nanoparticles or free PMB at the dosage of 10 mg kg−1 with respect to PMB. The survival curves of the two groups were plotted, and the survival rates were analyzed dynamically.2.18 Long-term toxicity of PMB-HA nanoparticles
Fifteen healthy male ICR mice (28–32 g) were randomly divided into 3 groups (n = 5). Two group of mice were intravenously injected with PBS containing PMB-HA nanoparticles or free PMB every 8 h for 3 days at the dosage of 6 mg kg−1 with respect to PMB. The third group was set as the negative control, in which the mice were intravenously injected with PBS at the same administration volume and frequency. After 3 d of injection, animals were sacrificed at 4 h after the last injection. The major organs, including heart, liver, spleen, lungs, kidneys and brain, were extracted and subjected to hematoxylin and eosin (H&E) staining. In addition, one slice of the kidney and brain adjacent to an HE-stained slice was subjected to TUNEL staining to identify apoptotic cells. When the mice were sacrificed, blood was also collected for routine blood and blood-chemistry analyses.All animal experiments reported herein were performed according to a protocol approved by the Peking University Institutional Animal Care and Use Committee (LA2019083).
2.19 Statistical analysis
Data were analyzed with GraphPad Prism 6 (GraphPad Software, Inc.), and all biological experiments were performed at least in triplicate. The results are presented as mean ± standard deviation. Paired t-tests and one-way ANOVA were used for multiple comparisons (when more than two groups were compared). Differences for which p values were less than 0.05 were considered to be significant, differences for which p values were less than 0.01, 0.001, or 0.0001 were considered to be highly significant.3 Results and discussion
3.1 Synthesis and characterization of PMB-HA nanoparticles
The construction procedure and the structure of PMB-HA nanoparticles are illustrated in Scheme 1. According to a previous report, the toxicity of PMB is mainly attributed to its positive charge,37 which leads to adverse side effects and largely hampers its clinical applications. Therefore, it can be inferred that neutralizing the positive charge of PMB might be a feasible strategy to improve the biocompatibility and biosafety of PMB. Based on this design concept, negatively charged HA molecules were introduced. Owing to the electrostatic interaction between the amino residues of PMB and the carboxyl groups of HA, the PMB-HA nanoparticles were predicted to be readily formed under physiological pH, where HA molecules can effectively neutralize and shielding the positive charges of PMB.To optimize the size and stability of the self-assembly system, the mixing of PMB and HA at different molar ratios was explored. For adequately shielding the positive charge of PMB, the excess HA molecules were added. The surface charge variation of the resultant mixtures with different HA contents were first investigated. As shown in Fig. 1A, when the molar ratio of HA to PMB is 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the zeta potential of the resultant PMB-HA mixture is close to zero, indicating that the positive charge of PMB was fully neutralized. In addition, with the proportion of HA continually increased, i.e., from 6.25
1, the zeta potential of the resultant PMB-HA mixture is close to zero, indicating that the positive charge of PMB was fully neutralized. In addition, with the proportion of HA continually increased, i.e., from 6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8.3
1 to 8.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the zeta potential of mixtures decreased from −7.3 mV to −12.3 mV. Thereafter, the hydrodynamic size of these mixtures was measured through dynamic light scattering analysis. Accordingly, the hydrodynamic diameter (Dh) of the mixture decreased from 334.7 nm to 196.5 nm with the molar ratio of HA to PMB increasing from 5
1, the zeta potential of mixtures decreased from −7.3 mV to −12.3 mV. Thereafter, the hydrodynamic size of these mixtures was measured through dynamic light scattering analysis. Accordingly, the hydrodynamic diameter (Dh) of the mixture decreased from 334.7 nm to 196.5 nm with the molar ratio of HA to PMB increasing from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 6.25
1 to 6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and further decreased to 113.4 nm when the ratio reached 8.3
1, and further decreased to 113.4 nm when the ratio reached 8.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 1B and S1†).
1 (Fig. 1B and S1†).
To further reveal the morphology of the PMB-HA nanoparticles, the transmission electron microscopy (TEM) images were obtained. As shown in Fig. 1C, uniform nanoparticles with ∼180 nm diameter can be observed in TEM image when the HA to PMB ratio is 6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. In comparison, larger nanoparticles and smaller nanoparticles with inhomogeneous sizes could also formed when the ratio are 5
1. In comparison, larger nanoparticles and smaller nanoparticles with inhomogeneous sizes could also formed when the ratio are 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 8.3
1 and 8.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. However, the uniformity of these resultant nanoparticles in TEM images were not as good as the nanoparticles with the ratio of 6.25
1, respectively. However, the uniformity of these resultant nanoparticles in TEM images were not as good as the nanoparticles with the ratio of 6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. S2 and S3†). Taking the DLS size distribution profile and the TEM images of the resultant PMB-HA nanostructures together, the HA to PMB ratio of 6.25
1 (Fig. S2 and S3†). Taking the DLS size distribution profile and the TEM images of the resultant PMB-HA nanostructures together, the HA to PMB ratio of 6.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was selected for the most suitable ratio for the PMB-HA construction.
1 was selected for the most suitable ratio for the PMB-HA construction.
Upon employing of this ratio, the PMB encapsulation efficiency of the PMB-HA nanoparticle was determined. The concentration of PMB in the ultrafiltrate was determined with UV spectroscopy (222 nm), with reference to a standard curve (Fig. S4†). The PMB encapsulation efficiency of the PMB-HA nanoparticle was calculated as 60.3 ± 0.12%, which is relatively high compared with those previous reported for other nanoparticulate drug delivery systems.35,38,39
Based on the design rationale, the amino groups in PMB can bind with phosphate groups in LPS through ionic forces, and the N-terminal fatty acyl chain of PMB can attach with the fatty acyl chains of LPS through the hydrophobic interaction. Therefore, the PMB-HA nanoparticles were expected to remain stable under normal physiological conditions, but to specifically release the PMB molecules in an inflammatory area through the competitive binding between PMB and the LPS in bacterial cell walls, thereby promoting the efficacy and biosafety of PMB. To confirm the feasibility of this hypothesis, the PMB release kinetics under different circumstances were assessed in vitro, where the amount of released PMB by PMB-HA nanoparticles was determined through the BCA kit. As shown in Fig. 1D, in the presence of 100 μg mL−1 LPS, which mimics the bacterial wall, a fast PMB release was observed during the initial stage, and 39.3% of the loaded PMB was released within the initial 4 h. After the initial burst release, the PMB molecules continued to release until reaching 69.0% at 12 h. By contrast, the PMB molecules in PMB-HA nanoparticles released much more slowly in the absence of LPS, and only 39.2% of PMB was released in normal saline after 12 h incubation (p < 0.001). With this LPS-triggered PMB release property, the PMB-HA nanoparticles are expected to smartly release more PMB on the surface of bacteria, thereby minimizing the acute nephrotoxicity and neurotoxicity.
3.2 The pneumonia targeting ability and the antibacterial effect of PMB-HA nanoparticles in vitro
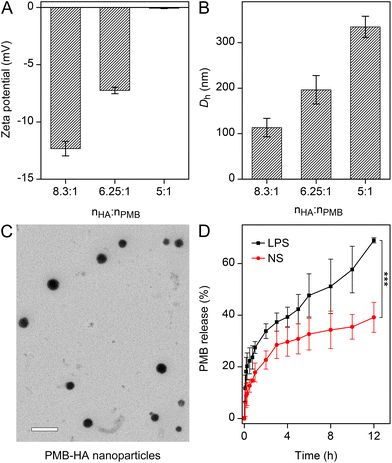
The pneumonia-specific targeting ability of the nanoparticle would be desired with regard to precise treatments. According to the design concept, the HA residues on the outer surface of the PMB-HA nanoparticles should bind with CD44 receptors over-expressed on the membranes of endothelial cells after inflammatory injury,40 including the LPS-activated HUVECs (Fig. S5†). This binding would endow the nanoparticles with a specific inflammatory site targeting ability. Therefore, this potential inflammatory targeting ability of the PMB-HA nanoparticles was investigated using HUVECs.For the locations of the PMB-HA nanoparticles, Cy5.5-NHS ester was employed to label the PMB molecules through the amino groups. According to the DLS and zeta potential results, the resultant PMBCy5.5-HA nanoparticles possess similar properties to the original PMB-HA nanoparticles (Fig. S6†). The fluorescence microscopy images in Fig. 2A clearly reveal that the PMBCy5.5-HA nanoparticles specifically targeted LPS-stimulated HUVECs, which represent inflamed endothelial cells, in which CD44 was significantly up-regulated.29 Meanwhile, non-activated HUVECs only showed weak fluorescence signal after incubation with PMB-HA under the same conditions. Therefore, it can be reasonably inferred that PMB-HA can successfully target inflammatory sites owing to the binding affinity between HA and CD44.
A prerequisite for an ideal antibacterial agent is the ability to efficiently suppress the growth of commonly seen multidrug-resistant bacteria.41 Therefore, the in vitro antibacterial effect of the current nanoparticles was evaluated on P. aeruginosa. A series of concentrations of PMB-HA nanoparticles or free PMB were added to well plates containing cultures with equivalent initial amounts of this resistant strain, and the suppression effect was determined by detecting the OD600 of the supernatant of each well. As shown in Fig. S7,† comparable antibacterial effects could be observed for both free and nanoparticulate PMB, with MICs as low as 0.541 μg mL−1.
In addition to the MIC determination, the effects against P. aeruginosa were also characterized with plate antagonism tests (Fig. 2B and C). The diameters of the zones of inhibition of PMB-HA and free PMB were found to be 13.8 ± 0.12 mm and 14.3 ± 0.07 mm, respectively, without significant difference (p > 0.05). This result indicated that the PMB can be readily released from the PMB-HA nanoparticles in the presence of bacteria. In addition, the efficiencies of PMB-HA nanoparticles and free PMB were also calculated with a colony counting assay (Fig. S8†). The antibacterial efficiency of these drugs was calculated via 100% minus the percentage of colony forming unit (CFU) number in treatment groups relative to the control group, as shown in Fig. 2D. The antibacterial efficiency of PMB-HA nanoparticles against P. aeruginosa was more than 70% at 4 μg mL−1 (with respect to PMB in the PMB-HA nanoparticles), and reached nearly 100% when the concentration was higher than 8 μg mL−1 (with respect to PMB in the PMB-HA nanoparticles), which is comparable to that of the free PMB. According to the results of MIC, inhibition zone, and colony counting assays, it can be concluded that PMB-HA nanoparticles exhibited excellent performance in combating drug-resistant bacteria, and that the HA molecules did not impair the antibacterial effect of the loaded PMB.
3.3 The cytotoxicity of PMB-HA nanoparticles
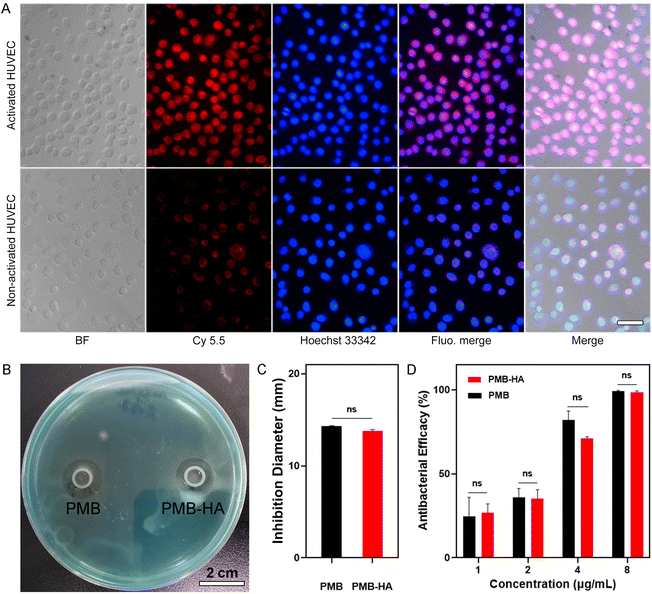
An ideal antibacterial drug should selectively kill bacteria, but cause minimal harm to normal human cells. Due to the encapsulation of PMB by HA molecules, which shielded the positive charge, the cytotoxicity of PMB should be largely reduced. Therefore, a Calcein-AM and PI co-staining assay was employed to distinguish live and dead cells in A549, HUVEC, and HPAEpiC (Fig. S9†) after incubation with free PMB, HA, and PMB-HA nanoparticles. As shown in Fig. 3A, in comparison with the control groups, the number of dead cells in all three cell lines increased significantly after incubation with free PMB, as indicated by the increased PI-stained cells (red fluorescent signals) in the field of view. In contrast, fewer cells were damaged after incubation with the PMB-HA nanoparticles at the same PMB concentration, which suggested that the cytotoxicity of PMB-HA nanoparticles is negligible. These results can be further quantitatively characterized through the proportion of dead cells. As displayed in Fig. S10,† only 0.82%, 0.46% and 0.14% of HPAEpiC, HUVEC and A549 cells were dead after co-incubating with PMB-HA nanoparticles, in contrast to the 10.82%, 1.71% and 8.27% of these kind of cells after treating with free PMB at the same concentration.For further quantitative analysis, the CCK-8 cell proliferation assay was employed in the aforementioned three cell lines. As shown in Fig. 3B–D, the PMB-HA nanoparticles exhibited good biosafety relative to that of PMB. More than 90% of cells in all three cell lines were viable when treated with PMB-HA with a PMB equivalent as high as 250 μg mL−1. By contrast, free PMB exhibited evident cytotoxicity with increasing concentrations. Treatment with 250 μg mL−1 of PMB was associated with the non-viability of 27.99 ± 1.26%, 45.31 ± 1.73%, and 26.00 ± 2.54% of HPAEpiC, HUVEC and A549 cells, respectively.
The cytotoxicity profiles were further investigated in HPAEpiC cells by measuring apoptosis with an Annexin V-FITC/PI staining flow cytometry-based detection kit, as displayed in Fig. S11.† The Annexin V can label the phosphatidylserine exposed on the outer membrane of the early apoptotic cells, while the PI can only cross the destroyed cell membrane with increased permeability of late apoptotic cells. Apparently, after incubation with PMB-HA nanoparticles or HA (150 μg mL−1 with respect to PMB and 10 mg mL−1 with respect to HA), the proportion of early apoptotic cells (Annexin V+/PI− staining) was only 2.18% and 1.66%, which did not significantly increase in comparison with that of PBS-treated cells (1.42%). However, 34.65% of cells underwent early apoptosis after incubation with free PMB (150 μg mL−1), indicating the higher cytotoxicity. In addition, 1.49% of late apoptotic cells (Annexin V+/PI+ staining) can be measured after treated with free PMB, which is also higher than the PMB-HA nanoparticles treated cells (0.14%). Collectively, the above results confirmed that the toxicity of PMB was dramatically reduced upon HA encapsulation, laying a foundation for further in vivo experiments.
3.4 Pneumonia treatment of PMB-HA nanoparticles in vivo
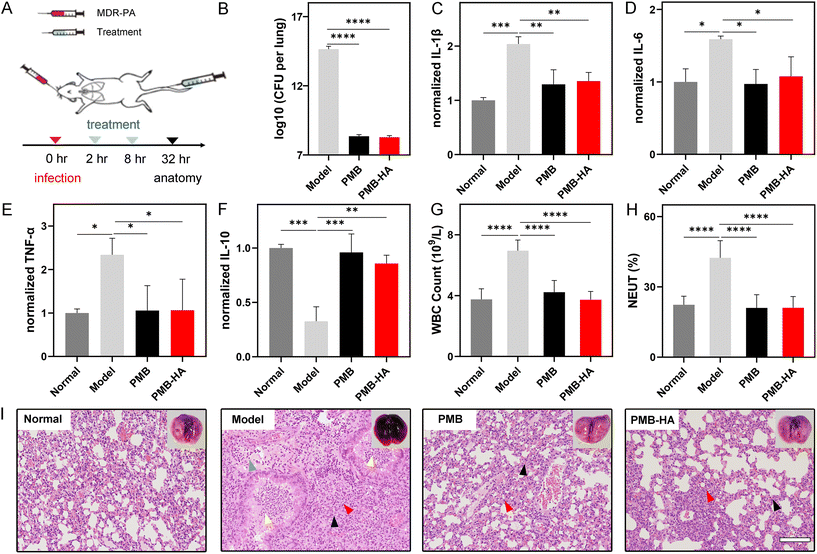
Considering the outstanding MDR-P. aeruginosa inhibition activity and satisfactory biosafety profiles of the PMB-HA nanoparticles as demonstrated in vitro, the antibacterial effect of PMB-HA nanoparticles was further evaluated on MDR-P. aeruginosa-induced pneumonia mouse models in vivo, in which free PMB was set as the control. A total of 40 healthy ICR mice were randomly divided into 4 groups (n = 10). Three groups of mice were infected with MDR-P. aeruginosa by nasal drips to build the MDR-P. aeruginosa pneumonia mice models. After 2 h infection, the mice lungs were adequately infected. At 2 h and 8 h post-infection, two groups of model mice were intravenously injected with PMB-HA nanoparticles or PMB, respectively, at 3 mg kg−1 PMB equivalent (Fig. 4A). After treatment, the mice from each group were sacrificed and then the lung tissues were harvested. The morphological changes in the lung tissues of the mice were first investigated according to the representative photographs of the lungs from each group. As shown in Fig. S12,† in comparison with the healthy pink lung tissue without any congestion, the MDR-P. aeruginosa infected mice exhibited a large area of congestion in the lung tissues, which presented as an abnormal dark red color. By contrast, after treatment with PMB-HA or free PMB, the color of lung tissue of pneumonia mice was recovered to the normal pink color, and no congestion could be observed on the lungs, which indicated that the pulmonary inflammation was successfully eliminated.As is known, after the infection with P. aeruginosa, the inflammatory edema will significantly enhance the wet weight of lungs, thus inducing the increase of the lung index expressed as the ratio of the mean lung weight to the mean body weight. For further quantitatively assessment of the therapeutic efficacy of the PMB-HA nanoparticles, the lung indexes after different treatments were calculated.42 As shown in Table S1,† the lung index was significantly higher in the MDR-P. aeruginosa-infected mice than in the healthy control mice (p < 0.0001), which suggested the successful establishment of the pneumonia model. More importantly, treatments with both PMB-HA nanoparticles and free PMB significantly decreased the lung index (p < 0.01); the inhibition rate according to the lung index for PMB-HA nanoparticles and free PMB was calculated as 64.80% ± 18.14% and 41.87% ± 29.31%, respectively (p < 0.05). These results indicated that the PMB-HA nanoparticles exhibited superior treatment outcomes for pneumonia as compared with free PMB. This superiority can be reasonably attributed to the long-term retention of nanoparticles and LPS-triggered release of PMB.
After calculating the lung index, three lungs from each group were homogenized and subjected to bacterial culturing to quantitatively identify the number of pathogenic bacteria that invaded into the lungs. As shown in Fig. 4B, the average number of CFU formed by the infected lung that received PMB-HA treatment was less than 109, which was comparable to that of free PMB-treated lung, but remarkably smaller than that of the infected lungs without any treatment, which nearly reached the order of 1015 CFU.
An additional 3 lungs from each group were also homogenized. The resultant supernatants were collected, and the level of pro-inflammatory cytokines, including IL-1β, IL-6, and TNF-α, was determined by ELISA. As shown in Fig. 4C–E, after infected by the bacteria, these pro-inflammatory were dramatically up-regulated, which exhibited 2.04, 1.59, and 2.34 times increase, respectively, indicating that the inflammatory response in lungs was serious. In contrast, the high levels of all three pro-inflammatory cytokines were significantly down-regulated after treatment with both PMB and PMB-HA nanoparticles, which implied that the PMB-HA nanoparticles possess a comparable ability to promote the remission of the inflammation with free PMB.
In addition, the anti-inflammatory cytokine IL-10, which can activate macrophages to inhibit the expression of inflammatory factors, was also quantitatively measured. As shown in Fig. 4F, the level of IL-10 in lung tissues of pneumonia model mice was down-regulated to only one third (0.33) of that in the normal group, and this factor up-regulated to 0.86 after treatment with PMB-HA, which suggested that the inflammation was on the mend.
To evaluate the alleviation of systemic inflammation after treatment, the blood samples of these sacrificed mice were also collected for routine blood cell and blood chemical testing. As shown in Fig. 4G and H, after the treatment of PMB-HA nanoparticles, the increased white blood cell and neutrophil counts in peripheral blood of the model mice were significantly decreased to levels that were comparable to those of mice treated with free PMB. These levels were almost recovered to the normal level. These results suggested that the systemic inflammation reaction was relieved upon the treatment with PMB-HA nanoparticles.
Overall, according to the 4C–4H, the inflammation- and blood-related index values of the P. aeruginosa-infected mice were abnormally up- or down-regulated with a statistical difference compared with the healthy mice, indicating that these indexes were affected by the inflammation. In addition, all these indexes of treated-mice were recovered to varying degrees, which were closer to those of health mice. All these variations highlight the therapeutic efficacy of the PMB-HA nanoparticles.
To further visualize the pathological changes in the lungs of the treated and untreated mice, the remaining lung tissues from each group were cut into slices and subjected to H&E staining for histopathological analysis. As shown in Fig. 4I and the locally enlarged frame in Fig. S13,† in comparison with healthy lungs, the lungs of MDR-P. aeruginosa-infected mice displayed marked inflammatory infectious features, including the lung consolidation (black arrows), granulocyte infiltrating (red arrows), inflammatory exudates and granulocytes within bronchial lumen (yellow arrows), and lymphocyte infiltration (blue arrows). By contrast, these histopathological features were largely eliminated upon the treatment with PMB-HA, and also significantly reduced after treatment with free PMB. These results confirmed that PMB-HA possesses outstanding antibacterial ability and can effectively ameliorate the lung damage in vivo, and that this preparation achieved a better curative effect than did free PMB, owing to their nanostructure.
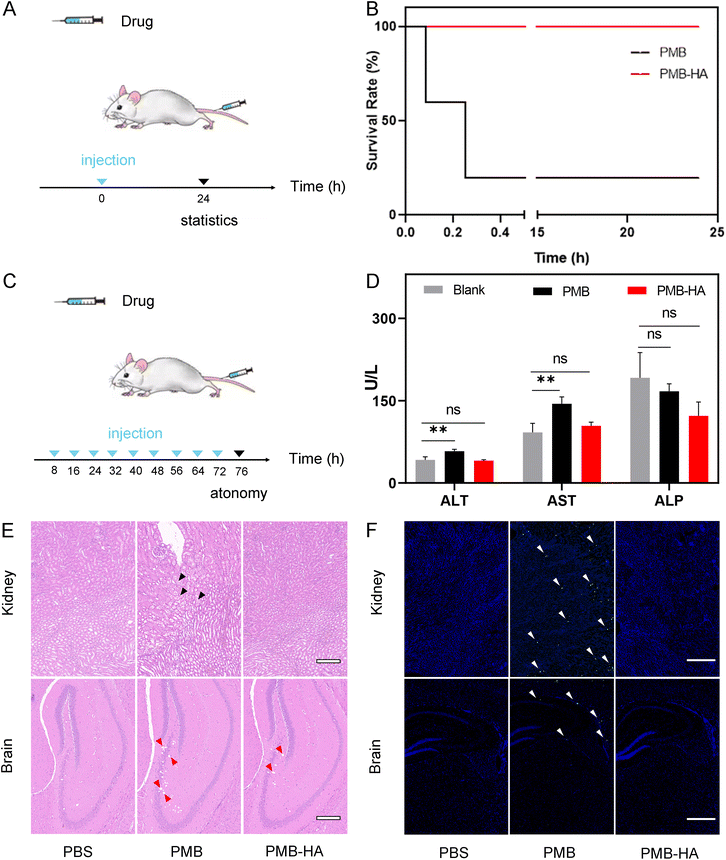
3.5 The evaluation of the biosafety of PMB-HA nanoparticles in vivo
Given that the toxicity of PMB was significantly reduced after encapsulation by HA, as suggested by the aforementioned in vitro experiments, the biosafety of PMB-HA nanoparticles was further evaluated in vivo by treatment of healthy mice. The hemolysis rate of the mice was investigated prior to the initial intravenous injection. As shown in Fig. S14,† the positive control, pure water, and negative control, normal saline (NS), showed nearly 100% and 0% hemolysis rates, respectively. With respect to the PMB-HA nanoparticles in the NS solution, the hemolysis rates were less than 3% at concentrations ranging from 32 to 1024 μg mL−1. These rates are lower than the threshold hemolysis rate of 5% set by the International Organization for Standardization and the American Society for Testing and Materials.43 In significant contrast, free PMB exhibited a considerable hemolytic toxicity even at the low concentration of 32 μg mL−1, with the hemolysis rates of 7.55%. These results provided a prerequisite indication of safety for the further intravenous injection of the nanoparticles.For evaluating the acute toxicity of the PMB-HA nanoparticles, ten healthy mice were randomly divided into 2 groups (n = 5), and these mice were intravenously injected with high doses of PMB-HA nanoparticles or free PMB (10 mg kg−1 with respect to PMB, 3-fold higher than the therapeutic dose) (Fig. 5A). After injection of free PMB, all mice were found to be acutely poisoned, in which serious adverse side effects, including tachycardia, convulsion and respiratory inhibition, were observed. In contrast, no apparent adverse reactions were observed in the mice receiving PMB-HA nanoparticles, which confirmed that the biosafety of PMB-HA nanoparticles was improved compared with free PMB. The survival rate of these two groups of mice was then calculated, as shown in the Fig. 5B. After 24 h of medication, all of the mice in PMB-HA groups were still alive, whereas 4 out of 5 mice treated with free PMB died within the first 20 minutes.
In addition to the acute toxicity, the long-term toxicity of the nanoparticles was also investigated. As illustrated in Fig. 5C, six healthy mice were randomly divided into 2 groups (n = 3), and successively intravenously injected with PMB-HA nanoparticles or free PMB (6 mg kg−1 with respect to PMB, 2-fold greater than therapeutic dose) every 8 h for 3 d. The mice were then sacrificed 96 h after the first treatment. The blood samples were collected for testing, and the main organs, including heart, liver, spleen, lungs, kidneys and brain, were extracted for further histological analysis. As shown in Fig. S15,† according to the routine blood test, after the successive intravenous administration of free PMB, the amount of WBC and lymphocyte was significantly reduced, implying that the immune systems of mice were impaired by PMB. In contrast, there was no abnormal variation in these two indexes after the administration of PMB-HA nanoparticles. In addition, the serum biochemical test showed that after the intravenous injection of free PMB, the level of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) of mice significantly up-regulated compared with that of healthy mice (p < 0.01), implying that free PMB may lead to the hepatic injury. In contrast, PMB-HA nanoparticles with the same PMB dosage would not cause the significant changes in these liver biochemical indexes (Fig. 5D).
Furthermore, according to the representative field of view of H&E stained kidney and brain slices shown in Fig. 5E (the locally enlarged frames are shown in Fig. S16 and S17†), the swelling of renal tubule epithelial cells with the distributed eosinophilic granular (black arrows) could be observed in the kidneys of mice treated with free PMB. In contrast, such histopathological features were not observed in kidney slices from mice treated with PMB-HA nanoparticles. As for the brain slices, free PMB led to the significant cavitation in hippocampus (red arrows), and a small number of atrophic neurons were observed in the cortex, with deep staining (yellow arrows in Fig. S17†), while these abnormalities in the brain slices of the mice that received PMB-HA nanoparticles were not serious. In addition, no noticeable inflammation or damage was observed in any other major organs of the mice receiving PMB-HA nanoparticles, as shown in Fig. S18.† The above evaluation results confirmed that even at high doses, PMB-HA nanoparticles can be administered intravenously without obvious side effects, which highlights their biosafety.
4 Conclusions
In summary, a targeted nano-based PMB delivery system, PMB-HA nanoparticle, was successfully constructed in pursuit of a safe and effective treatment of drug-resistant pneumonia. These particles were produced through the electrostatic self-assembly of HA and PMB molecules. Through shielding the cationic nature of PMB, PMB-HA nanoparticles exhibited outstanding biosafety performance both in vitro and in vivo in comparison to free PMB. More importantly, systematic studies clearly revealed that after intravenous injection, PMB-HA nanoparticles can precisely target the CD44 receptors over-expressed on the membrane of activated endothelial cells in inflammatory sites, and come into contact with the bacteria surrounding the damaged alveolar-capillary membrane. Due to both the electrostatic and hydrophobic interactions of PMB and LPS, the PMB molecules in the PMB-HA nanoparticles competitively bind with LPS in the cell walls of Gram-negative bacteria, thereby escaping from the nanoparticles for the effective inhibition of bacterial growth. This smart LPS-triggered responsive behavior of nanoparticles enables both increased therapeutic effects and enhanced biosafety. Given the FDA-approved status of HA, it is believed that this smart delivery system may pave a new way for the resurrection of PMB for the future clinical treatment of bacterial inflammatory diseases.Author contributions
Peisen Zhang, Qiuhong Ouyang and Tianshu Zhai contributed to the work equally. Peisen Zhang – Data collections, first draft writer and editing. Qiuhong Ouyang – Data collections, first draft writer and submission. Tianshu Zhai – Data collections, resources and validation. Jing Sun – Data collections and resources. Jun Wu – Data collections and formal analysis. Feng Qin – Formal analysis. Ni Zhang – Formal analysis. Saisai Yue – Data collections. Xinchen Yang – Data collections. Hanyi Zhang – Data collections. Yi Hou – Supervision. Li Deng – Supervision. Fang Wang – Supervision. Qingyuan Zhan – Investigation, supervision and draft reviewing. Qingsong Yu – Investigation, supervision and draft reviewing. Meng Qin – Investigation, supervision and draft reviewing. Zhihua Gan – Methodology and project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 51973014, 21774008, and 82102679).References
- L. Armand-Lefevre, C. Angebault, F. Barbier, E. Hamelet, G. Defrance, E. Ruppe, R. Bronchard, R. Lepeule, J. C. Lucet and A. El Mniai, Antimicrob. Agents Chemother., 2013, 57, 1488–1495 CrossRef CAS PubMed.
- E. Schulz, A. Goes, R. Garcia, F. Panter and G. Fuhrmann, J. Controlled Release, 2018, 290, 46–55 CrossRef CAS.
- D. Y. Feng, Y. Q. Zhou, X. L. Zou, M. Zhou, W. B. Wu, X. X. Chen, Y. H. Wang and T. T. Zhang, J. Infect. Public Health, 2019, 12, 630–633 CrossRef PubMed.
- N. Thundon, B. Adhiratha, C. Lantharita, N. Sireethorn, L. Narisorn, W. Luksame and T. Visanu, Infect. Drug Resist., 2018, 11, 1219–1224 CrossRef PubMed.
- A. Mahableshwar, N. Alan, B. Karen and M. G. Alasdair, J. Antimicrob. Chemother., 2014, 69, 1432–1434 CrossRef PubMed.
- M. U. Ahmed, T. Velkov, Y. W. Lin, B. Yun, C. J. Nowell, F. Zhou, Q. T. Zhou, K. Chan, M. Azad and J. Li, Antimicrob. Agents Chemother., 2017, 61, AAC.02690-02616 CrossRef PubMed.
- W. Sun, B. Hu, X. Zhang, Y. Wang and G. Lin, Drug Des., Dev. Ther., 2021, 15, 611–616 CrossRef.
- C. Dai, X. Xiao, J. Li, G. D. Ciccotosto, R. Cappai, S. Tang, E. K. Schneider-Futschik, D. Hoyer, T. Velkov and J. Shen, ACS Chem. Neurosci., 2019, 10, 120–131 CrossRef CAS PubMed.
- S. A. Yuk, H. Kim, N. S. Abutaleb, A. M. Dieterly, M. S. Taha, M. D. Tsifansky, L. T. Lyle, M. N. Seleem and Y. Yeo, Sci. Adv., 2021, 7, 1577 CrossRef.
- I. Insua, S. Majok, A. Peacock, A. M. Krachler and F. Fernandez-Trillo, Eur. Polym. J., 2017, 87, 478–486 CrossRef CAS PubMed.
- Y. H. Liu, S. C. Kuo, B. Y. Yao, Z. S. Fang, Y. T. Lee, Y. C. Chang, T. L. Chen and C. M. Hu, Acta Biomater., 2018, 82, 133–142 CrossRef CAS.
- P. Temboot, S. Kaewpaiboon, K. Tinpun, T. Nakpeng, R. Khalil, Z. Ul-Haq, V. Thamlikitkul, S. Tiengrim and T. Srichana, J. Drug Delivery Sci. Technol., 2020, 58, 101779 CrossRef CAS.
- S. M. Wu, C. Xu, Y. W. Zhu, L. Zheng, L. D. Zhang, Y. Hu, B. R. Yu, Y. G. Wang and F. J. Xu, Adv. Funct. Mater., 2021, 31, 2103591 CrossRef CAS.
- P. S. Zhang, Y. Y. Li, W. Tang, J. Zhao, L. H. Jing and K. J. McHughc, Nano Today, 2022, 42, 101335 CrossRef CAS.
- P. S. Zhang, J. L. Meng, Y. Y. Li, C. Yang, Y. Hou, W. Tang, K. J. McHugh and L. H. Jing, Innovation, 2021, 2, 100174 CAS.
- Y. Yang, S. S. Yue, Y. Y. Qiao, P. S. Zhang, N. Jiang, Z. B. Ning, C. Y. Liu and Y. Hou, Front. Chem., 2021, 8, 572471 CrossRef PubMed.
- Y. R. Ren, Y. C. Feng, K. Y. Xu, S. S. Yue, T. T. Yang, K. L. Nie, M. Xu, H. J. Xu, X. Xiong, F. Körte, M. Barbeck, P. S. Zhang and L. Liu, Front. Pharmacol., 2021, 12, 721988 CrossRef CAS PubMed.
- P. S. Zhang, Y. Hou, J. F. Zeng, Y. Y. Li, Z. H. Wang, R. Zhu, T. C. Ma and M. Y. Gao, Angew. Chem., Int. Ed., 2019, 58, 11088–11096 CrossRef CAS PubMed.
- M. Y. Chai, Y. F. Gao, J. Liu, Y. Y. Deng, D. F. Hu, Q. Jin and J. Ji, Adv. Healthcare Mater., 2020, 9, 1901542 CrossRef CAS PubMed.
- J. J. Wang, Q. K. Ni, Y. F. Wang, Y. X. Zhang, H. Y. He, D. W. Gao, X. W. Ma and X. J. Liang, J. Controlled Release, 2020, 331, 282–295 CrossRef.
- J. Zhou, M. H. Li, Y. H. Hou, Z. Luo, Q. F. Chen, H. X. Cao, R. L. Huo, C. C. Xue, L. Sutrisno, L. Hao, Y. Cao, H. Ran, L. Lu, K. Li and K. Y. Cai, ACS Nano, 2018, 12, 2858–2872 CrossRef CAS.
- H. R. Jia, Y. X. Zhu, X. Liu, G. Y. Pan and F. G. Wu, ACS Nano, 2019, 13, 11781–11792 CrossRef CAS PubMed.
- S. Z. F. Phua, G. B. Yang, Q. L. Wei, A. Verma, H. Z. Chen, T. Thanabalu and Y. L. Zhao, ACS Nano, 2019, 13, 4742–4751 CrossRef CAS PubMed.
- H. Kim, J. Cha, M. Jang and P. Kim, Biomater. Sci., 2019, 7, 2264–2271 RSC.
- Q. Shi, L. Zhao, C. M. Xu, L. F. Zhang and H. Zhao, Molecules, 2019, 24, 1766 CrossRef CAS PubMed.
- Y. Kudo, S. Kohi, K. Hirata, M. Goggins and N. Sato, Oncotarget, 2019, 10, 5592–5604 CrossRef PubMed.
- I. Avar and D. Kero, ST-OPEN, 2020, 1, 1–16 Search PubMed.
- A. G. Tavianatou, C. Ilaria, F. Marco, P. Zoi, G. Devis and N. K. Karamanos, FEBS J., 2019, 286, 2883–2908 CrossRef CAS.
- C. Yang, M. Cao, L. Hua, Y. He and G. Feng, J. Biol. Chem., 2012, 287, 43094–43107 CrossRef CAS.
- K. L. Schwertfeger, M. K. Cowman, P. G. Telmer, E. A. Turley and J. B. Mccarthy, Front. Immunol., 2015, 6, 236 Search PubMed.
- X. Y. Hou, H. Lin, X. D. Zhou, Z. T. Cheng, Y. Li, X. Liu, F. Zhao, Y. P. Zhu, P. Zhang and D. Q. Chen, Carbohydr. Polym., 2020, 232, 115787 CrossRef CAS PubMed.
- D. H. Jiang, J. R. Liang, J. Fan, S. Yu, S. P. Chen, Y. Luo, G. D. Prestwich, M. M. Mascarenhas, H. G. Garg, D. A. Quinn, R. J. Homer, D. R. Goldstein, R. Bucala, P. J. Lee, R. Medzhitov and P. W. Noble, Nat. Med., 2005, 11, 1173–1179 CrossRef CAS.
- T. Velkov, P. E. Thompson, R. L. Nation and J. Li, J. Med. Chem., 2010, 53, 1898–1916 CrossRef CAS PubMed.
- J. J. Cao, Y. Zhao, Y. Liu, S. Tian, C. X. Zheng, C. H. Liu, Y. Zhai, Y. L. An, H. J. Busscher, L. Q. Shi and Y. Liu, ACS Macro Lett., 2019, 8, 651–657 CrossRef PubMed.
- A. Omri, Z. E. Suntres and N. S. Pang, Biochem. Pharmacol., 2002, 64, 1407–1413 CrossRef CAS.
- N. Zhang, L. C. Zhu, Q. H. Ouyang, S. S. Yue, Y. C. Huang, S. Qu, R. W. Li, Y. Y. Qiao, M. Xu, F. F. He, B. Zhao, L. Wei, X. A. Wu and P. S. Zhang, Front. Pharmacol., 2021, 12, 784864 CrossRef CAS PubMed.
- G. H. C. Furtado, P. A. d’Azevedo, A. F. Santos, A. C. Gales, A. C. C. Pignatari and E. A. S. Medeiros, Int. J. Antimicrob. Agents, 2007, 30, 315–319 CrossRef CAS PubMed.
- P. Severino, E. F. Silveira, K. Loureiro, M. V. Chaud, D. Antonini, M. Lancellotti, V. H. Sarmento, C. F. D. Silva, M. H. A. Santana and E. B. Souto, Eur. J. Pharm. Sci., 2017, 106, 177–184 CrossRef CAS PubMed.
- M. Alipour, M. Halwani, A. Omri and Z. E. Suntres, Int. J. Pharm., 2007, 355, 293–298 CrossRef.
- H. Frey, N. Schroeder, T. Manon-Jensen, R. V. Lozzo and L. Schaefer, FEBS J., 2013, 280, 2165–2179 CrossRef CAS PubMed.
- J. V. Laura, Nat. Rev. Microbiol., 2017, 15, 639–640 CrossRef PubMed.
- Q. H. Ma, Q. T. Yu, X. F. Xing, S. N. Liu, C. Y. Shi and J. B. Luo, Viruses, 2018, 10, 117 CrossRef.
- E. P. Rondon, H. A. Benabdoun, F. Vallières, M. S. Petrnio and J. C. Fernandes, Int. J. Nanomed., 2020, 15, 6183–6200 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr02026b |
| ‡ These authors contributed to the work equally. |
| This journal is © The Royal Society of Chemistry 2022 |