Differentiation and classification of bacterial endotoxins based on surface enhanced Raman scattering and advanced machine learning†
Yanjun
Yang
 *a,
Beibei
Xu
b,
James
Haverstick
c,
Nabil
Ibtehaz
*a,
Beibei
Xu
b,
James
Haverstick
c,
Nabil
Ibtehaz
 d,
Artur
Muszyński
d,
Artur
Muszyński
 e,
Xianyan
Chen
b,
Muhammad E. H.
Chowdhury
f,
Susu M.
Zughaier
e,
Xianyan
Chen
b,
Muhammad E. H.
Chowdhury
f,
Susu M.
Zughaier
 *g and
Yiping
Zhao
*g and
Yiping
Zhao
 *c
*c
aSchool of Electrical and Computer Engineering, College of Engineering, The University of Georgia, Athens, GA 30602, USA. E-mail: YanjunYang@uga.edu
bDepartment of Statistics, The University of Georgia, Athens, GA 30602, USA
cDepartment of Physics and Astronomy, The University of Georgia, Athens, GA 30602, USA. E-mail: zhaoy@uga.edu
dDepartment of Computer Science, Purdue University, West Lafayette, IN 47907, USA
eComplex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA
fDepartment of Electrical Engineering, College of Engineering, Qatar University, PO. Box 2713, Doha, Qatar
gDepartment of Basic Medical Sciences, College of Medicine, QU Health, Qatar University, PO. Box 2713, Doha, Qatar. E-mail: szughaier@qu.edu.qa
First published on 17th May 2022
Abstract
Bacterial endotoxin, a major component of the Gram-negative bacterial outer membrane leaflet, is a lipopolysaccharide shed from bacteria during their growth and infection and can be utilized as a biomarker for bacterial detection. Here, the surface enhanced Raman scattering (SERS) spectra of eleven bacterial endotoxins with an average detection amount of 8.75 pg per measurement have been obtained based on silver nanorod array substrates, and the characteristic SERS peaks have been identified. With appropriate spectral pre-processing procedures, different classical machine learning algorithms, including support vector machine, k-nearest neighbor, random forest, etc., and a modified deep learning algorithm, RamanNet, have been applied to differentiate and classify these endotoxins. It has been found that most conventional machine learning algorithms can attain a differentiation accuracy of >99%, while RamanNet can achieve 100% accuracy. Such an approach has the potential for precise classification of endotoxins and could be used for rapid medical diagnoses and therapeutic decisions for pathogenic infections.
Introduction
Bacterial infections in humans account for a significant burden of disease and require rapid detection and treatment. Different approaches have been implemented to detect bacterial infections. The most common method is the gold standard bacterial culture method utilized in clinical and diagnostic microbiology laboratories. Other methods identify bacterial infections indirectly by detecting bacterial biomarkers namely endotoxins, pigments, metabolites, and small molecules, such as pyocyanin and pyoverdine.1,2 Among different bacterial biomarkers, an endotoxin is unique. A Gram-negative bacterium outer membrane contains a glycolipid or an endotoxin known as a lipopolysaccharide (LPS) that is shed during infection or bacterial lysis.3 Bacterial endotoxins are very potent inducers of inflammation by activating toll-like receptor 4 (TLR4)-mediated innate immune responses leading to a cytokine storm usually observed in sepsis.4 Endotoxin circulating in blood, even at low concentrations, is associated with septic shock and mortality.5 Therefore, rapid detection of endotoxin is highly desired and could aid in medical diagnoses and therapeutic decisions.In recent years, with the development of nanotechnology, biosensors based on novel nanostructures have been used to detect and identify trace amounts of endotoxins in human fluid samples based on fluorescence, chemiluminescence, and electrical gradient.6,7 Surface-enhanced Raman spectroscopy (SERS), with the potential to achieve single molecule detection, is very attractive and promising for multiplex detection.8,9 SERS offers a unique “signature” spectral profile with very narrow spectral peaks for individual analytes and has demonstrated the ability to directly detect various biomolecules.10–13 Recently, Wu et al. reported the SERS fingerprint spectra of the LPS KDO2-lipid A and lipid A endotoxin structures of Neisseria meningitidis as well as those of enteric LPSs from E. coli, S. typhimurium, S. Minnesota, V. cholerae, R. CE3, and R. NGR,14,15 and demonstrated the possibility of using SERS for endotoxin detection.
One challenge for SERS endotoxin detection is to identify the unique SERS spectral features. Since the chemical structures of many endotoxins are similar, their SERS spectra are very much alike. Therefore, in order to classify the spectra, different statistical methods are applied, including supervised and unsupervised learning.16–19 Since the SERS spectra can be viewed as multi-variant data, chemometric analysis is often applied to reduce the dimensionality of the spectral data and maximize the variance among spectral fingerprints in order to differentiate bacteria. For unsupervised learning, such as principal component analysis (PCA) and hierarchical cluster analysis (HCA), training data do not have ground truth labels, the model identifies structures such as clusters, and testing data can be assigned to different clusters. For supervised learning, including partial least squares discriminant analysis (PLS-DA), partial least squares regression (PLS), linear discriminant analysis (LDA), support vector machine (SVM), k-nearest neighbor (KNN), random forest (RF), etc., each training sample has a ground truth label. The model learns a decision boundary and replicates the labeling on the testing data. These methods allow successful spectral and image analysis of complex biological samples, such as cell identification,20 disease diagnosis,21 and forensic analysis.22
However, the uses of traditional chemometrics methods to differentiate SERS spectra are increasingly challenged due to several reasons. First, the high dimensionality of the spectra and the multivariate or even megavariate nature, as a result of the inherent complexity of the biological systems, increase the difficulty of data analysis. Second, the vibrational spectra of single cellular or microbial systems suffer from low signal-to-noise ratio (SNR), which further increases the difficulty of data analysis. Third, advanced high-throughput chemical profiling for biological detection can significantly increase data size. This not only leads to the difficulty of calculation but also prevents extraction of any subtle variations of sophisticated hidden features within the big data through a single traditional data processing algorithm. However, deep learning methods have the potential to circumvent the complexity and heterogeneity in data. For example, a convolutional neural network (CNN), which is one of the most popular deep learning architectures, has been widely used and has shown superior performance in analyzing spectroscopic signals including those from SERS spectroscopy of complex biological samples.23–25
In this paper, the SERS spectra of eleven bacterial endotoxins have been measured based on silver nanorod array (AgNR) substrates. The characteristic SERS peaks from these endotoxins have been identified. Different classical machine learning algorithms (MLAs) and a modified CNN model, i.e., RamanNet, have been applied to differentiate and classify endotoxins based on these SERS spectra. It has been shown that with appropriate spectral pre-processing procedures and MLAs, the SERS spectra of endotoxins can be differentiated with 100% accuracy. Such an approach has the potential for rapid detection of endotoxins and could be used in medical diagnoses and therapeutic decisions.
Experimental section
General detection and classification strategy
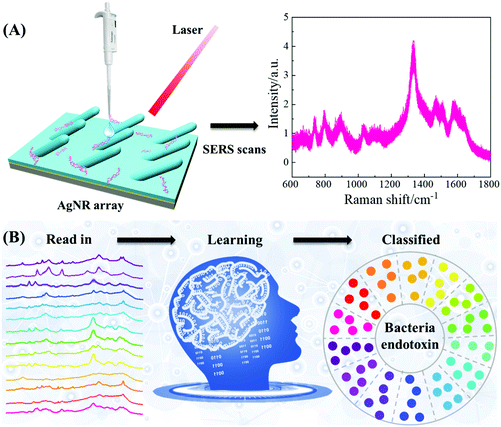
The procedure to use SERS and MLAs to differentiate and classify eleven bacterial endotoxins is illustrated in Fig. 1. First, an extensive SERS spectral database of bacterial endotoxins is produced by collecting spectra from highly sensitive silver nanorod array (AgNR) substrates. Then, according to the spectral feature, a simple and reliable baseline correction method is developed to obtain highly reproducible spectra. Finally, by applying a classical MLA, such as SVM, RF, KNN, PLS-DA, and LDA, or a novel deep learning model (RamanNet, based on CNN), bacterial endotoxins can be accurately distinguished based on their SERS spectra.Materials
Sulfuric acid (Fisher Scientific, 98%), ammonium hydroxide (Fisher Scientific, 98%), hydrogen peroxide (Fisher Scientific, 30%), and ethyl alcohol (EtOH, reagent grade) were used to clean glass slides (Gold Seal, Part# 3010). Silver (Kurt J. Lesker, 99.99%) and titanium pellets (Kurt J. Lesker, 99.995%) were purchased as the evaporation materials. Pure water (Sigma-Aldrich) was used throughout all the experiments.AgNR substrate fabrication
AgNR arrays prepared by oblique angle deposition (OAD) are excellent SERS substrates as reported previously.26–29 Briefly, clean glass slides (0.5 inch × 0.5 inch) were loaded into a vacuum deposition chamber with the substrate normal antiparallel to the incident vapor direction. A layer of 20 nm-thick Ti film and a layer of 200 nm-thick Ag film were deposited in sequence at a rate of 0.2 nm s−1 and 0.3 nm s−1, respectively. Then, the substrate normal was rotated to 86° relative to the incident vapor direction, and a layer of 2000 nm-thick Ag film was deposited at a rate of 0.3 nm s−1 to obtain AgNR arrays. The entire evaporation process was conducted under high vacuum conditions with a pressure <3 × 10−6 Torr. A typical SEM image of a AgNR substrate is shown in Fig. S1,† and the detailed deposition procedure and conditions can be found in ref. 30. According to previous extensive studies, AgNR substrates have been demonstrated to possess good SERS reproducibility with <10% relative standard deviation (RSD), high SERS enhancement factors up to 109, and large area uniformity.26,27,31,32Preparation and purification of bacterial endotoxins
Eleven different kinds of LPSs were prepared for this study. These 11 kinds of LPSs are representative of the most common bacteria that cause disease in humans (see section S1 in the ESI† for details). The lipopolysaccharides were extracted from the bacterial cells by the hot phenol-water extraction procedure.33 The water phases were dialyzed (12–14 kDa cutoff membrane), freeze-dried, and washed in 90% EtOH to remove traces of phospholipids. The detailed purification procedures of specific LPS samples are in the corresponding references: Francisella tularensis LVS;34Moraxella catarrhalis LOS;35Pseudomonas aeruginosa;36 and S. meliloti (this work). The endotoxin of Salmonella enterica serovar, typhimurium (S-type LPS), Salmonella enterica serovar, minnesota Re595 (R-type LPS; Re), E. coli-EH100 (R-type LPS; Ra), LPS E. coli-O128:B12, E. coli-O11:B4, and E. coli-J5 (R-type LPS; Rc) were obtained from Sigma Aldrich, and the LPS of Helicobacter pylori GU2 was obtained from Wako Pure Chemicals (Tokyo, Japan). Table S1 in the ESI† lists the general structures and properties of these LPSs. Of note, all LPSs used in this study were derived from bacteria that can cause disease in humans except for S. meliloti, which is a Gram-negative bacteria found in soil and does not cause infections in humans. The LPS structure of S. meliloti (also known as Rhizobium meliloti) is very different from those of other bacteria as it harbors long fatty acyl chains.37 LPSs with extended length fatty acyl chains are also found in gut microbiota species such as Bacteroides and Prevotella, which harbor the biologically inactive LPS as a TLR4 ligand and therefore would not lead to innate immune activations.38 Three different macromolecules were used as control samples since they are structurally very different from the LPS: peptidoglycan (PGN) from Gram-positive S. aureus (Sigma-Aldrich), lipoteichoic acid (LTA) from B. subtilis (Sigma-Aldrich) and chitin from crab shell (Sigma-Aldrich). In rare mixed infections of Gram-positive and Gram-negative bacteria, PGN and LTA could be present along with the LPS in a clinical sample. The SERS spectra may look similar. Chitin, however, is not present in bacteria that cause disease in humans but is used as a control since it is a repeating sugar polymer found in plants and fungi. Note that the LPS structure contains O-antigen, which is a repeating sugar chain linked to the lipid A part of the LPS molecule. PGN purified from S. aureus and LTA purified from B. subtilis originated from Gram-positive bacteria. PGN is the main membrane component in Gram-positive bacteria, which is in contrast to Gram-negative bacteria that also contain a quite thin layer of PGN embedded in the membrane beneath the LPS layer. Chitin is not found in bacteria and characteristic of fungi and insects.39SERS characterization
Bacterial endotoxin samples or negative controls were diluted into 100 μg mL−1 using pure water. 2 μL of the diluted sample was dispensed onto the AgNR substrate and air dried at 20 °C. The average spreading area was estimated to be 3.5 mm2. The SERS spectra were recorded on a confocal Raman microscope (Renishaw, InVia) using a 785 nm excitation laser. To reduce the florescence background signal from the targeted analytes, a 785 nm excitation wavelength was selected. Additionally, 785 nm excitation can generate a large SERS enhancement from AgNR substrates compared to other short wavelength excitations. Unless otherwise specified, the laser power was set to 9 mW at the sample position with a 5× objective lens and 10 s acquisition time. The excitation laser spot size was 1875 μm2 and the amount of analyte (LPS or reference molecules) in a single SERS measurement was estimated to be 8.75 pg. In order to obtain sufficient SERS spectra for MLAs, discrete SERS mappings were taken from 4 to 5 AgNR substrates with at least 300 μm spacing among the sampling spots to avoid mapping overlap. About 100 spectra were taken for each mapping and some outlier spectra with obvious inconsistencies (such as multiple spike peaks presented in the spectra, featureless spectra, etc.) were removed from the spectral data set. The final number of SERS spectra taken for each endotoxin is between 338 and 440, and the detail is listed in Table S2.†Data pre-processing
Usually, SERS spectra contain unwanted spectral features such as spikes (noise) and baselines that need to be removed before further data analysis. The baselines of SERS spectra originate from different sources, which one could catalogue as being of intrinsic or extrinsic origin. The intrinsic baseline is due to the fluorescence and Rayleigh scattering from the analyte molecules, especially biomolecules.40,41 Extrinsic baselines include the scattering/absorption from the nanostructures in the SERS substrates and the instrument responses. The targeted analyte molecules or other background molecules could randomly adsorb onto the hot spots where strong SERS signals are produced. Such a process not only makes the entire spectral signal move up or down, but also introduces noise in the spectra. Since the intrinsic baseline is directly associated with the targeted analytes, in terms of spectral classification, it is important information that should not be removed; however, the extrinsic baseline, and the contributions from SERS substrates and instruments should be removed since they are irrelevant to the targeted analytes and cause problems in future data analysis. According to the mechanisms of extrinsic baselines, both should contribute to the baseline of a monotonic function. Therefore, the criteria for a good baseline correction method should be: (1) the removal of the baseline should, in most cases, decrease the variation in spectra from measurement to measurement for the same sample and SERS substrate; (2) by removing the baseline, one should remove information that is similar to all spectra; and (3) after baseline removal, spectra from the same sample and SERS substrate should be highly correlated. A typical raw SERS spectrum is shown in Fig. S2,† and in the wavenumber ranges of 300–400 cm−1 and 1800–2500 cm−1, there are no Raman peaks signifying the fingerprint features of the analytes. All the experimental spectra share similar features in these two wavenumber regions, indicating that these are common features in all our SERS measurements and independent from the target analytes used, thus these extrinsic features need to be removed. Since the spectral feature in the 300–400 cm−1 region shows a rapid decay while in the 1800–2500 cm−1 region it exhibits a slow decrease, we chose to use a mixed Gaussian and Lorentzian function to fit the baseline in the wavelength ranges of 300–400 cm−1 and 1800–2500 cm−1 (other math functions that can best fit these two spectral features shall also work well),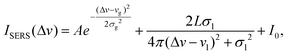 | (1) |
Machine learning model
As shown in Table S2,† there is a total of 5624 SERS spectra obtained from 11 bacterial endotoxins, peptidoglycan (PGN), lipoteichoic acid (LTA) and chitin. After baseline correction and normalization following the above mentioned procedure (see Fig. S3†), 3936 and 1688 SERS spectra were randomly chosen as the training spectrum set and testing spectrum set (a ratio of 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and analyzed by different MLAs, including SVM, RF, KNN, PLS-DA, and LDA with a machine learning library scikit-learn in Python 3.8.3. Only the training spectrum set was used to train the model, while the testing set was used to obtain the prediction performance of the trained model. During the training process, a five-fold cross validation was employed to tune hyperparameters, such as C (regularization parameter) and γ (kernel coefficient) in the SVM algorithm, and k (number of neighbors) in the KNN algorithm. At the end of cross validation, the best hyperparameters were chosen and the unbiased model performance was obtained using seven measures: accuracy, micro and macro precision, micro and macro recall, and micro and macro F-score on validation sets. The models were further evaluated by the seven model performance measures on testing data for external validation. Feature importance was obtained by applying the models with best hyperparameters on the whole training set. The confusion matrix and receiver operating characteristic (ROC) curve were obtained by implementing the models on the testing spectrum set.
3) and analyzed by different MLAs, including SVM, RF, KNN, PLS-DA, and LDA with a machine learning library scikit-learn in Python 3.8.3. Only the training spectrum set was used to train the model, while the testing set was used to obtain the prediction performance of the trained model. During the training process, a five-fold cross validation was employed to tune hyperparameters, such as C (regularization parameter) and γ (kernel coefficient) in the SVM algorithm, and k (number of neighbors) in the KNN algorithm. At the end of cross validation, the best hyperparameters were chosen and the unbiased model performance was obtained using seven measures: accuracy, micro and macro precision, micro and macro recall, and micro and macro F-score on validation sets. The models were further evaluated by the seven model performance measures on testing data for external validation. Feature importance was obtained by applying the models with best hyperparameters on the whole training set. The confusion matrix and receiver operating characteristic (ROC) curve were obtained by implementing the models on the testing spectrum set.
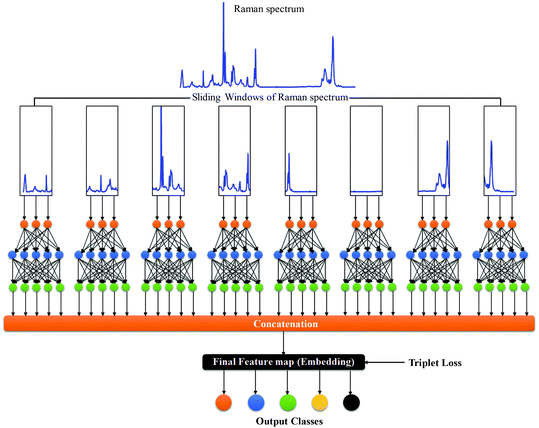
RamanNet model
RamanNet is a novel neural network architecture designed to focus on the unique properties of Raman spectra.42 The state-of-the-art CNN models, despite being able to generalize better and extract the complex and novel pattern from the signal or image data, are not suitable for Raman spectral analysis. Since the horizontal axis in a Raman spectrum represents Raman shift, not time or any other independent variable, the equivariance to translation property in CNN proves to be troublesome, as it will treat pattern signatures at different Raman shifts in an identical manner. In contrast, for traditional MLAs, one faces the curse of dimensionality and at the same time disregards any correlation between intensities at neighboring Raman shifts, both of which could have been solved by the sparse connectivity found in CNN.RamanNet attempts to be at the middle ground between both of the two approaches, by ensuring sparse connectivity and by disabling temporal invariance. This is performed by employing the traditional densely connected blocks in a novel manner. We consider overlapping windowed segments from the Raman spectrum and analyze them in shifted densely connected blocks as shown in Fig. 2. This effectively mimics a 1D convolutional operation, but with context localization or without translation. Therefore, we can extract features in the same fashion as a 1D CNN. Mathematically, a typical 1D CNN operation can be simplified as,
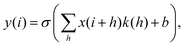 | (2) |
In RamanNet, the proposed modification is to use the shifted densely connected blocks,
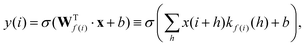 | (3) |
RamanNet analyses were conducted in a server computer with Intel Xeon @2.2 GHz CPU, 24 GB RAM, and NVIDIA TESLA P100 (16 GB) GPU. The RamanNet architecture was implemented using Tensorflow. In order to compare the performance of the RamanNet model with the classical machine learning models in the classification of bacterial endotoxin Raman spectra, a similar five-fold cross-validation scheme was used.
Results and discussion
Characteristics of LPS SERS spectra
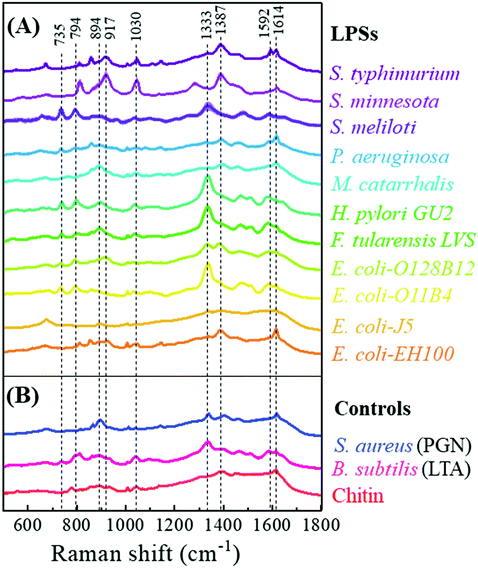
Fig. 3 shows the characteristic average SERS spectra of the 11 LPS samples and 3 reference samples. The detailed SERS peak assignments based on the molecular vibrational modes for these LPS samples are summarized in Table S6.† Intact bacterial LPSs are amphiphilic macromolecules with a molecular mass of 10–20 kDa and having three structural components: (1) a hydrophobic lipid section, lipid A, which is responsible for the toxic properties of the molecule, (2) a hydrophilic core polysaccharide chain, and (3) a repeating hydrophilic O-antigenic oligosaccharide polymer that is specific to the bacterial serotype.44 Therefore, many LPSs share similar spectral features with similar vibrational modes. Table S7† rearranges the SERS peak assignments and shows the common peaks among different LPSs. The number of LPS samples with shared common peaks varies from 1 to 11. The three most common peaks are the peaks at Δv = 1614 cm−1 and Δv = 1003 cm−1 corresponding to both v(C–O) and v(C–C) modes, and the peak at Δv = 1333 cm−1 resulting from the δ(C–H) mode. Other obvious common peaks are the Δv = 735 cm−1 peak corresponding to the β(C–O–C) mode, the peaks at Δv = 794 cm−1 and 1592 cm−1 due to v(C–O) modes, and the peaks at Δv = 894 cm−1 and 917 cm−1 resulting from both the δ(C–C–H) and δ(C–O–H) modes. Also, there are some unique peaks only belonging to the specific LPS, e.g., Δv = 531 cm−1, 568 cm−1, 577 cm−1, 648 cm−1, 759 cm−1, 775 cm−1, 1174 cm−1, 1304 cm−1, 1326 cm−1, 1578 cm−1, and 1642 cm−1, respectively. However, their relative peak intensities are very weak. The similarity and difference in the SERS peaks are due to the intrinsic molecular structure similarity and difference in these LPSs.Since the reference samples PGN and LTA are obtained from Gram-positive bacteria while chitin is not from any bacterium, their molecular structures are very different from LPSs. Thus, it is expected that their SERS spectra should be significantly different from those of the LPSs. As shown in Fig. 3, generally the SERS spectra of the three reference samples have fewer characteristic peaks, and their overall spectral shapes are very similar. Their detailed SERS peak assignments are shown in Table S6.† These three samples have only one common peak at Δv = 1614 cm−1, which is due to the v(C–O) and v(C–C) modes. The SERS spectra of PGN and LTA have a common peak at Δv = 1333 cm−1, corresponding to the δ(C–H) deformation mode, while LTA and chitin have a common peak at Δv = 894 cm−1, resulting from the δ(C–C–H) and δ(C–O–H) modes. Chitin has a notable unique peak at Δv = 775 cm−1, which results from the v(C–O) modes.
SERS spectral classification using different MLAs
Simple PCA showed little discrimination between different types of LPSs and control samples as shown in Fig. S6.† For MLA or deep learning analysis, sufficient spectra are needed for the training spectrum set. As shown in Table S2,† the total SERS spectra collected from 11 bacterial LPS samples and 3 control samples on the AgNR substrates are 5624. After the baseline correction and normalization shown in Fig. S3,† most SERS spectra obtained from the same sample have a correlation coefficient larger than 0.98. Only a few, i.e., E. coli-O11:B4 (0.954), P. aeruginosa (0.942), and S. meliloti Rm1021 (0.900), have smaller correlation coefficients, but all are larger than 0.9. Therefore, the pre-processed spectra are good for MLA or deep learning analysis.Different machine learning models were applied for the classification of the SERS spectra of the LPSs. For the SVM classifier, different kernel functions, including linear, polynomial, radial basis function (RBF), and sigmoid, were used to optimize SVM, and their corresponding validation accuracies are shown in Table S4.† SVM classifiers with linear, polynomial, and RBF kernels show comparable accuracies of >99%, while the sigmoid kernel function gives the lowest accuracy of 93.1% ± 0.9%. The polynomial and sigmoid kernels are nonlinear kernels, which are more complex and may increase the chance of overfitting the model and decrease accuracy on the testing set. The comparable accuracies of linear and RBF kernels suggest that the dataset is linearly separable, thus the linear kernel was chosen for the SVM classifier. The hyperparameters for the SVM classifier were chosen as C = 1 (γ is a constant for the linear kernel function) by cross validation. For the KNN classifier, the number of neighbors k was chosen as 4. Table 1 shows the performance of five classic MLAs based on the 5-fold cross-validation. All models perform well, with accuracies, precisions, recalls and the F1-score greater than 98% (the definitions of all the parameters can be found in Table S8†). Among all five models, SVM performs the best, giving almost 100% for almost all the parameters, including the accuracy. The small variability for all the performance measures demonstrates the good stability of the predictive model.
| SVM | RF | KNN | LDA | PLS-DA | |
|---|---|---|---|---|---|
| Accuracy | 0.9998 ± 0.0005 | 0.995 ± 0.001 | 0.992 ± 0.003 | 0.998 ± 0.001 | 0.98 ± 0.02 |
| Micro precision | 0.9998 ± 0.0005 | 0.995 ± 0.001 | 0.992 ± 0.003 | 0.998 ± 0.001 | 0.98 ± 0.02 |
| Macro precision | 0.9998 ± 0.0004 | 0.995 ± 0.002 | 0.992 ± 0.002 | 0.998 ± 0.002 | 0.98 ± 0.01 |
| Micro recall | 0.9998 ± 0.0005 | 0.995 ± 0.001 | 0.992 ± 0.003 | 0.998 ± 0.001 | 0.98 ± 0.02 |
| Macro recall | 0.9997 ± 0.0006 | 0.995 ± 0.001 | 0.992 ± 0.003 | 0.998 ± 0.002 | 0.98 ± 0.01 |
| Micro F1-score | 0.9998 ± 0.0005 | 0.995 ± 0.001 | 0.992 ± 0.003 | 0.998 ± 0.001 | 0.98 ± 0.02 |
| Macro F1-score | 0.9998 ± 0.0005 | 0.995 ± 0.002 | 0.992 ± 0.003 | 0.998 ± 0.002 | 0.98 ± 0.02 |
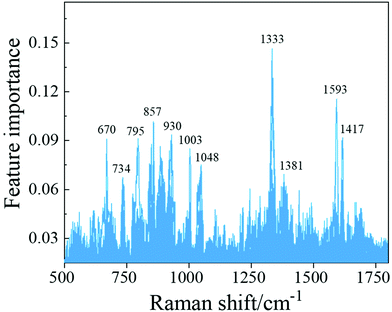
To investigate the important structures/peaks in the SERS spectra that contribute most to a classifier, we applied the trained SVM model with the best hyperparameters to the whole training spectrum set and obtained the feature importance of the Raman shift. As shown in Fig. 4, the peaks at Δv = 857 cm−1, 1333 cm−1 and 1593 cm−1 were found to be the most prominent features, which correspond to the δ(C–C–H) and δ(C–O–H) deformation modes, the δ(C–H) deformation mode and the v(C–O) stretching mode, respectively. Table S9† lists all these importance features, which include peak assignments and peak distributions.
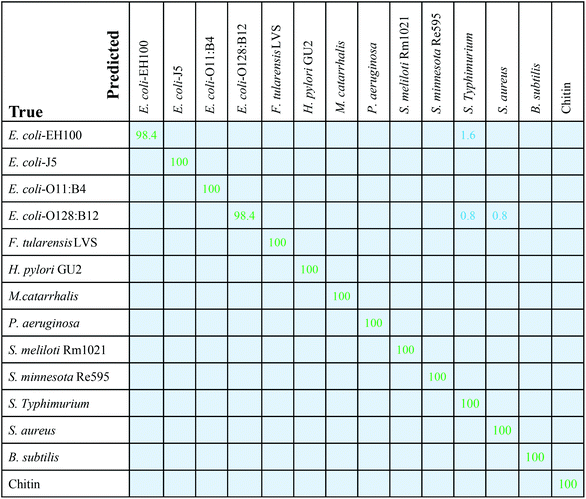
Applying the trained SVM classifiers on the testing spectrum set, the performance of SVM on the testing spectrum set was obtained, and the corresponding confusion matrix is shown in Fig. 5. Among the 11 LPSs and 3 reference samples, 12 are classified with 100% accuracy, with a few spectra from E. coli-EH100 and E. coli-O128:B12 being misclassified. The accuracy for E. coli-EH100 is 98.4%; 1.6% spectra were recognized as S. typhimurium. The accuracy for E. coli-O128:B12 is 98.4%, with 0.8% spectra recognized as S. typhimurium, and 0.8% spectra recognized as S. aureus. The corresponding ROC curve is shown in Fig. S8.† For all 14 samples, the mean value of the areas under the ROC curves (AUC) is greater than 0.99, which suggests that the SVM model classifies different LPSs and reference samples with a very high specificity and sensitivity based on the SERS spectra.
RamanNet analysis
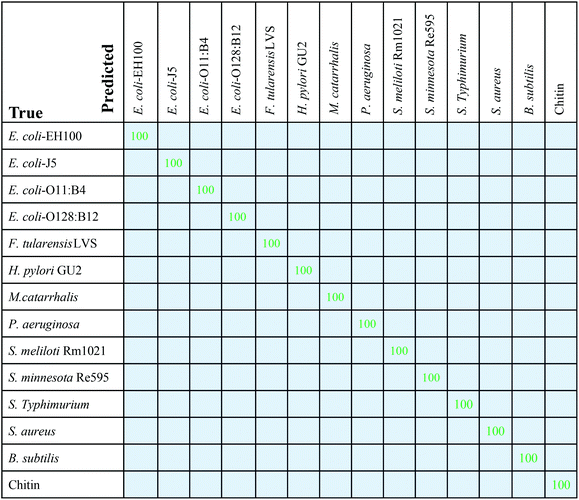
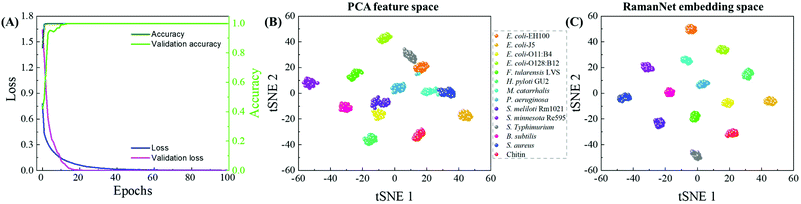
Similar to the evaluation procedure followed for the classical MLAs, 5-fold cross-validation was performed with RamanNet. RamanNet manages to classify all the samples with perfect 100% accuracy in all the folds as shown in Fig. 6. Although this increase is apparently insignificant compared to the result reported in Fig. 5, it should be noted that this performance is achieved by a shallow 3 layers’ network with only 1 M parameters. The summarized results from the 5-fold tests are presented in Fig. 6 as a confusion matrix.Fig. 7A shows the training and validation loss and accuracy plots, where it can be observed that the model was trained for 100 epochs, but the model quickly achieved convergence within 40 epochs. This makes the model not only accurate (100% multiclass accuracy) but also fast (2 M FLOPs) in training. Moreover, the inference time is very small as the model is very shallow.
In addition to the improved performance, another feature of RamanNet is the lack of reliance on dimensionality reduction algorithms, whereas traditional methods have to rely on the feature compression scheme like PCA, and RamanNet is capable of inherently extracting a lower dimensional representation from the data. Furthermore, the use of triplet loss in the hidden layers of RamanNet provides a better class separability compared to PCA. For example, the compressed feature spaces obtained from PCA and RamanNet are presented. A 256-dimensional feature space using PCA and extract embedding of similar dimensions from RamanNet were computed. Then the 256-dimensional feature space is projected into a 2 dimensional-map using t-distributed stochastic neighbor embedding (t-SNE).45 As shown in Fig. 7B and C, though the t-SNE plot derived from PCA shows separated clusters for individual LPSs and reference samples, the t-SNE plot using the embedding learnt in RamanNet shows that the clusters are free from overlaps, and all the classes are distributed properly with minimizing intraclass distance and maximizing interclass distance. This ensures better distinction among the classes, which increases the classification performance as shown in Fig. 6. In addition, as shown in the RamanNet embedding space plotted in Fig. S5† for different data pre-processing, the RamanNet model enables most clusters to distribute properly without overlap and maximize the interclass distance for raw spectra (Fig. S5B†), normalized raw spectra (Fig. S5D†), and the WiRE correction (Fig. S5F†); only a few spectra were misclassified. Therefore, we expect that combining our baseline correction method with the RamanNet machine learning model could achieve a higher accuracy for more complex spectral sets measured from real patient samples.
Classification of the LPS mixture
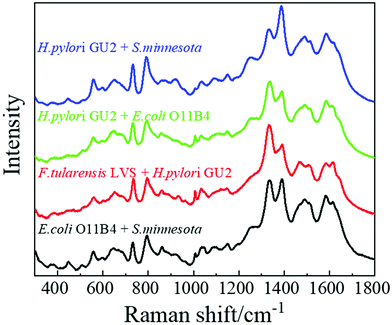
Occasionally a patient may be infected by more than one type of bacterium, and multiple LPSs may co-exist in the clinic specimen. Detection and differentiation of multiple analytes from a single specimen using SERS and machine learning is still a challenging topic and is beyond the scope of this paper. One of the challenges is how to obtain the training spectrum set. Several publications used a data augmentation strategy to map out all the possible combinations of different mixed analytes to build the training spectrum set based on spectral linear combinations,46,47i.e., for a given mixture, the SERS spectrum (Sm) of a mixture is a linear combination of the SERS spectra of the individual analyte 1 (S1) and analyte 2 (S2), Sm = aS1 + bS2, where a and b represent the relative contribution of each analyte in the mixture. According to ref. 46 and 47, such a strategy seems to work quite well. Here, as a proof of concept, four kinds of two-LPS mixtures, E. coli-O11:B4 and S. minnesota Re595 (ES), F. tularensis LVS and H. pylori GU2 (FH), E. coli-O11:B4 and H. pylori GU2 (EH), as well as S. minnesota Re595 and H. pylori GU2 (SH), with 50 μg mL−1 each for each analyte, were prepared for SERS measurements. Their corresponding biological significance is listed in Table S10.†Fig. 8 shows the typical average SERS spectra of these four mixtures. The most important SERS peaks listed in Table S9† can be observed. Combining spectra from single LPSs and mixtures for machine learning analysis, Table S11† shows the predicted accuracies of six MLAs based on the 5-fold cross-validation. The PLS-DA model shows a low accuracy of 0.928, while the other five models perform quite well, with accuracies greater than 99%. Still, RamanNet enables the classification of all the samples with a perfect 100% accuracy as shown in an sample confusion matrix (Fig. S9†). The t-SNE plot shows that all the clusters are free from overlaps and distributed properly while minimizing the intraclass distance and maximizing the interclass distance (Fig. S10†). These results show that the SERS spectra of mixtures are very different from those of single LPSs, and in principle, they should be able to be used for machine learning to identify the mixed species and their relative concentrations.However, we find that it is not suitable to use the linear combinations of the SERS spectra of single LPSs to construct the training spectrum set for multiplex detection. Fig. S11† shows the experimentally obtained SERS spectra from LPS mixtures (red curves) and the best linear combinations (black curves) of spectra from two LPSs in the mixture through a least-squares fitting.48 There are significant differences between the experimental and linearly combined spectra. For example, in a S. minnesota Re595 and H. pylori GU2 (SH) mixture, the experimental result shows obvious SERS peaks at Δv = 558 cm−1 and 1387 cm−1, while SERS peaks at Δv = 898 cm−1, 1034 cm−1, and 1333 cm−1 are distinctive in the fitting result. The possible reasons are currently under investigation. In fact, for ref. 46 and 47, the linear combination hypothesis may not be reasonable from the SERS mechanism point of view. There are many underlying assumptions for this hypothesis: (1) the SERS spectrum of each analyte does not change; (2) there is no interaction between the two (or more) target analytes; (3) each analyte has the same enhancement; and (4) the adsorption mechanism of each analyte to the SERS hot spots does not change. Unfortunately, our results show that some of the above assumptions do not hold. Therefore, more investigations are needed to delineate SERS spectra from LPS mixtures for practical applications.
Conclusions
In summary, SERS spectra of eleven bacterial endotoxins at a very low amount (8.75 pg) have been obtained from AgNR substrates, and the characteristic SERS peaks have been identified. Different classical machine learning algorithms and a deep learning algorithm RamanNet have been applied to differentiate and classify various endotoxins. After implementing appropriate spectral pre-processing procedures and machine learning algorithms, it has been found that most conventional machine learning algorithms can obtain a differentiation accuracy of >99%, while RamanNet can achieve 100% accuracy. Such an approach has the potential for rapid detection of endotoxins and could aid in medical diagnosis, such as sepsis, and in making therapeutic decisions. In addition, a patient may occasionally be infected by more than one type of bacterium, and our results indicate that the SERS spectra of endotoxin mixtures can also be classified with 100% accuracy using the RamanNet model. However, a practical multiplex detection strategy to determine the possible species and relative compositions in a mixture based on SERS spectra is still under investigation. The challenges originate from the possible changes in the SERS spectra of individual analytes and the way to establish a reliable training spectrum set for MLAs.Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
YY, SMZ, and YZ were supported by the Qatar National Research Fund (grant number: NPRP12S-0224-190144). Work at the Complex Carbohydrate Research Center was supported by the US Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES), under Award DE-SC0015662, and by the NIH grant R24GM137782-01 to Parastoo Azadi.References
- X. Wu, J. Chen, X. Li, Y. Zhao and S. M. Zughaier, Nanomedicine, 2014, 10, 1863–1870 CrossRef CAS PubMed.
- A. Gopal, L. Yan, S. Kashif, T. Munshi, V. A. L. Roy, N. H. Voelcker and X. Chen, Adv. Healthcare Mater., 2022, 11, 2101546 CrossRef CAS PubMed.
- B. S. Park, D. H. Song, H. M. Kim, B.-S. Choi, H. Lee and J.-O. Lee, Nature, 2009, 458, 1191–1195 CrossRef CAS PubMed.
- S. M. Zughaier, S. M. Zimmer, A. Datta, R. W. Carlson and D. S. Stephens, Infect. Immun., 2005, 73, 2940–2950 CrossRef CAS PubMed.
- P. Brandtzaeg, R. Ovstebø and P. Kierulf, Prog. Clin. Biol. Res., 1995, 392, 219–233 CAS.
- R. Seth, M. Ribeiro, A. Romaschin, J. A. Scott, M. Manno, J. A. Scott, G. M. Liss and S. M. Tarlo, J. Allergy Clin. Immunol., 2011, 127, 272–275 CrossRef CAS PubMed.
- S.-E. Kim, W. Su, M. Cho, Y. Lee and W.-S. Choe, Anal. Biochem., 2012, 424, 12–20 CrossRef CAS PubMed.
- K. Kneipp, Y. Wang, H. Kneipp, L. T. Perelman, I. Itzkan, R. R. Dasari and M. S. Feld, Phys. Rev. Lett., 1997, 78, 1667–1670 CrossRef CAS.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- Y. Yang, X. Jiang, J. Chao, C. Song, B. Liu, D. Zhu, Y. Sun, B. Yang, Q. Zhang, Y. Chen and L. Wang, Sci. China Mater., 2017, 60, 1129–1144 CrossRef CAS.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, Nanomedicine, 2010, 6, 214–226 CrossRef CAS.
- S. P. Mulvaney, M. D. Musick, C. D. Keating and M. J. Natan, Langmuir, 2003, 19, 4784–4790 CrossRef CAS.
- M. D. Porter, R. J. Lipert, L. M. Siperko, G. Wang and R. Narayanan, Chem. Soc. Rev., 2008, 37, 1001–1011 RSC.
- X. Wu, Y. Zhao and S. M. Zughaier, Biosensors, 2021, 11, 234 CrossRef CAS PubMed.
- X. Wu, J. Chen, B. Park, Y.-W. Huang and Y. Zhao, Advances in Applied Nanotechnology for Agriculture, American Chemical Society, 2013, ch. 5, vol. 1143, pp. 85–108 Search PubMed.
- O. Adir, M. Poley, G. Chen, S. Froim, N. Krinsky, J. Shklover, J. Shainsky-Roitman, T. Lammers and A. Schroeder, Adv. Mater., 2020, 32, 1901989 CrossRef CAS PubMed.
- N. M. Ralbovsky and I. K. Lednev, Chem. Soc. Rev., 2020, 49, 7428–7453 RSC.
- H. He, S. Yan, D. Lyu, M. Xu, R. Ye, P. Zheng, X. Lu, L. Wang and B. Ren, Anal. Chem., 2021, 93, 3653–3665 CrossRef CAS PubMed.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS.
- J. Hung, A. Goodman, D. Ravel, S. C. P. Lopes, G. W. Rangel, O. A. Nery, B. Malleret, F. Nosten, M. V. G. Lacerda, M. U. Ferreira, L. Rénia, M. T. Duraisingh, F. T. M. Costa, M. Marti and A. E. Carpenter, BMC Bioinf., 2020, 21, 300 CrossRef.
- T. O'Connor, A. Anand, B. Andemariam and B. Javidi, Biomed. Opt. Express, 2020, 11, 4491–4508 CrossRef PubMed.
- M. A. Neaimi, H. A. Hamadi, C. Y. Yeun and M. J. Zemerly, 2020 3rd International Conference on Signal Processing and Information Security (ICSPIS), 2020, pp. 1–4.
- M. Erzina, A. Trelin, O. Guselnikova, B. Dvorankova, K. Strnadova, A. Perminova, P. Ulbrich, D. Mares, V. Jerabek, R. Elashnikov, V. Svorcik and O. Lyutakov, Sens. Actuators, B, 2020, 308, 127660 CrossRef CAS.
- J. Ding, Q. Lin, J. Zhang, G. M. Young, C. Jiang, Y. Zhong and J. Zhang, Anal. Bioanal. Chem., 2021, 413, 3801–3811 CrossRef CAS PubMed.
- N. Cheng, D. Chen, B. Lou, J. Fu and H. Wang, Biosens. Bioelectron., 2021, 186, 113246 CrossRef CAS PubMed.
- Y. J. Liu and Y. P. Zhao, Phys. Rev. B: Condens. Matter Mater. Phys., 2008, 78, 075436 CrossRef.
- J. D. Driskell, S. Shanmukh, Y. Liu, S. B. Chaney, X. J. Tang, Y. P. Zhao and R. A. Dluhy, J. Phys. Chem. C, 2008, 112, 895–901 CrossRef CAS.
- C. Song, B. Yang, Y. Zhu, Y. Yang and L. Wang, Biosens. Bioelectron., 2017, 87, 59–65 CrossRef CAS.
- J. Zhang, Y. Yang, X. Jiang, C. Dong, C. Song, C. Han and L. Wang, Biosens. Bioelectron., 2019, 141, 111402 CrossRef CAS PubMed.
- C. Song, Y. Yang, B. Yang, Y. Sun, Y. P. Zhao and L.-H. Wang, Nanoscale, 2016, 8, 17365–17373 RSC.
- Y. J. Liu, H. Y. Chu and Y. P. Zhao, J. Phys. Chem. C, 2010, 114, 8176–8183 CrossRef CAS.
- Y. J. Liu, Z. Y. Zhang, Q. Zhao, R. A. Dluhy and Y. P. Zhao, J. Phys. Chem. C, 2009, 113, 9664–9669 CrossRef CAS.
- O. Westphal, Methods Carbohydr. Chem., 1965, 5, 83 CAS.
- S. Soni, R. Ernst, A. Muszynski, N. Mohapatra, M. Perry, E. Vinogradov, R. Carlson and J. Gunn, Front. Microbiol., 2010, 1, 129 CAS.
- S. Gao, D. Peng, W. Zhang, A. Muszyński, R. W. Carlson and X.-X. Gu, FEBS J., 2008, 275, 5201–5214 CrossRef CAS.
- M. R. Davis, A. Muszyński, I. V. Lollett, C. L. Pritchett, R. W. Carlson and J. B. Goldberg, J. Bacteriol., 2013, 195, 1504–1514 CrossRef CAS PubMed.
- A. F. Haag, S. Wehmeier, A. Muszyński, B. Kerscher, V. Fletcher, S. H. Berry, G. L. Hold, R. W. Carlson and G. P. Ferguson, J. Biol. Chem., 2011, 286, 17455–17466 CrossRef CAS.
- A. N. Jacobson, B. P. Choudhury and M. A. Fischbach, mBio, 2018, 9, e02289-17 CrossRef PubMed.
- L. Steinfeld, A. Vafaei, J. Rösner and H. Merzendorfer, in Targeting Chitin-containing Organisms, ed. Q. Yang and T. Fukamizo, Springer Singapore, Singapore, 2019, pp. 19–59 Search PubMed.
- J. Li, L.-P. I. Choo-Smith, Z. Tang and M. G. Sowa, J. Raman Spectrosc., 2011, 42, 580–585 CrossRef CAS.
- B. Li, N. M. S. Sirimuthu, B. H. Ray and A. G. Ryder, J. Raman Spectrosc., 2012, 43, 1074–1082 CrossRef CAS.
- N. Ibtehaz, M. E. Chowdhury, A. Khandakar, S. M. Zughaier, S. Kiranyaz and M. S. Rahman, RamanNet: A generalized neural network architecture for Raman Spectrum Analysis, 2022, https://doi.org/10.48550/arXiv.2201.09737 Search PubMed.
- N. Ibtehaz, M. E. H. Chowdhury, A. Khandakar, S. Kiranyaz, M. S. Rahman, A. Tahir, Y. Qiblawey and T. Rahman, IEEE Trans. Emerg. Top. Comput. Intell., 2021, 1–13, DOI:10.1109/TETCI.2021.3131374.
- E. T. Rietschel, T. Kirikae, F. U. Schade, U. Mamat, G. Schmidt, H. Loppnow, A. J. Ulmer, U. Zähringer, U. Seydel, F. Di Padova, M. Schreier and H. Brade, FASEB J., 1994, 8, 217–225 CrossRef CAS PubMed.
- G. Hinton and S. Roweis, presented in part at the Proceedings of the 15th International Conference on Neural Information Processing Systems, 2002, pp. 857–864.
- J. Li and T. Vo-Dinh, Proceedings of SPIE, Plasmonics in Biology and Medicine XIX, 2022, p. 1197805-1 Search PubMed.
- M. H. Mozaffari and L. L. Tay, Raman spectral analysis of mixtures with one-dimensional convolutional neural network, 2021, https://doi.org/10.48550/arXiv.2106.05316 Search PubMed.
- J. L. Abell, J. M. Garren, J. D. Driskell, R. A. Tripp and Y. Zhao, J. Am. Chem. Soc., 2012, 134, 12889–12892 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nr01277d |
| This journal is © The Royal Society of Chemistry 2022 |