 Open Access Article
Open Access ArticleFluorescence lifetime microscopy unveils the supramolecular organization of liposomal Doxorubicin†
Paolo
Tentori
ab,
Giovanni
Signore
c,
Andrea
Camposeo
 d,
Annalisa
Carretta
a,
Gianmarco
Ferri
a,
Pasqualantonio
Pingue
ad,
Stefano
Luin
a,
Daniela
Pozzi
d,
Annalisa
Carretta
a,
Gianmarco
Ferri
a,
Pasqualantonio
Pingue
ad,
Stefano
Luin
a,
Daniela
Pozzi
 e,
Enrico
Gratton
f,
Fabio
Beltram
ab,
Giulio
Caracciolo
e,
Enrico
Gratton
f,
Fabio
Beltram
ab,
Giulio
Caracciolo
 e and
Francesco
Cardarelli
e and
Francesco
Cardarelli
 *ad
*ad
aLaboratorio NEST, Scuola Normale Superiore, Pisa, Italy. E-mail:. E-mail: francesco.cardarelli@sns.it
bCenter for Nanotechnology Innovation @NEST, Pisa, Italy
cFondazione Pisana per la Scienza (FPS), Pisa, Italy
dNEST, Istituto Nanoscienze-CNR, Piazza S. Silvestro, 12, I-56127, Pisa, Italy
eDepartment of Molecular Medicine, Sapienza University of Rome, Rome, Italy
fLaboratory for Fluorescence Dynamics, Department of Biomedical Engineering, University of California at Irvine, Irvine, California, USA
First published on 17th June 2022
Abstract
The supramolecular organization of Doxorubicin (DOX) within the standard Doxoves® liposomal formulation (DOX®) is investigated using visible light and phasor approach to fluorescence lifetime imaging (phasor-FLIM). First, the phasor-FLIM signature of DOX® is resolved into the contribution of three co-existing fluorescent species, each with its characteristic mono-exponential lifetime, namely: crystallized DOX (DOXc, 0.2 ns), free DOX (DOXf, 1.0 ns), and DOX bound to the liposomal membrane (DOXb, 4.5 ns). Then, the exact molar fractions of the three species are determined by combining phasor-FLIM with quantitative absorption/fluorescence spectroscopy on DOXc, DOXf, and DOXb pure standards. The final picture on DOX® comprises most of the drug in the crystallized form (∼98%), with the remaining fractions divided between free (∼1.4%) and membrane-bound drug (∼0.7%). Finally, phasor-FLIM in the presence of a DOX dynamic quencher allows us to suggest that DOXf is both encapsulated and non-encapsulated, and that DOXb is present on both liposome-membrane leaflets. We argue that the present experimental protocol can be applied to the investigation of the supramolecular organization of encapsulated luminescent drugs/molecules all the way from the production phase to their state within living matter.
The use of liposomes as drug-delivery carriers for chemotherapeutic agents, proposed originally by Gregoriadis in 1981,1 offers a potential means of modulating drug distribution to increase drug efficacy and reduce cytotoxicity.2 A paradigmatic case is encapsulated Doxorubicin in its prototypical form, i.e. Doxil®, the first FDA-approved nano-drug (1995), currently used for the treatment of a number of pathologies including AIDS-related Kaposi's sarcoma, recurrent ovarian cancer, metastatic breast cancer, multiple myeloma.3 Doxil® consists of a formulation of 85 nm-diameter liposomes with 2000 Da segments of poly-(ethylene glycol) (PEG) engrafted onto the liposome surface and loaded with DOX. Doxil® performance proved superior with respect to isolated DOX thanks to three peculiar properties: (i) prolonged drug-circulation time (with avoidance of the reticuloendothelial system), (ii) a protective lipid bilayer in the “liquid ordered” phase composed of high-melting-temperature (Tm) phosphatidylcholine and cholesterol, (iii) stable loading of a high concentration of DOX (∼2 mg ml−1). In spite of such a rewarding path, almost 10 years after Doxil®-related patents expired, no FDA- or EMA-approved generic “Doxil” was still available. As clearly pointed out by Y. Barenholz (one of Doxil®'s inventors),3 a major cause, at the very bottom, is the limited understanding of the in-cuvette “synthetic identity” of this DOX liposomal formulation, i.e. the set of physicochemical properties resulting from production. This largely stems from the lack of analytical tools that can quantitatively dissect the molecular organization of the drug within the intact liposomal formulation. To date, high-resolution techniques (e.g. cryo-EM, SAXS, WAXS) provided demonstration that a nanorod-shaped crystal of ammonium-sulfate DOX is present within Doxil® liposomes4 (see e.g.Fig. 1A), as expected based on the drug remote-loading mechanism.5–8 However, the semi-quantitative nature of such investigations is not sufficient to distinguish possible coexisting phase-separated drug pools in the formulation (i.e. the drug supramolecular organization), even less to quantify the fractional amount of each drug sub-population. These limitations affect our ability to control the performance of encapsulated DOX in delivery applications, and to improve by rational design the efficacy of new formulations.
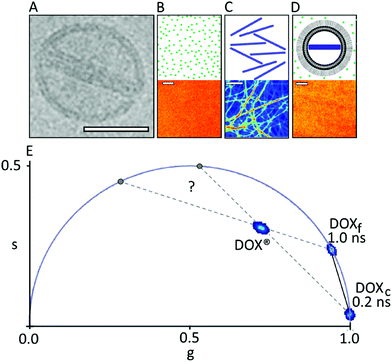 | ||
| Fig. 1 Phasor-FLIM fingerprint of DOX®. (A) Cryo-EM image of a single DOX® nanoparticle (adapted from ref. 9 scale bar: 50 nm). (B–D) Schematic representation of DOXf in aqueous solution, isolated DOXc, and DOX® in solution, respectively (top panels) with the corresponding confocal images (bottom panels). Scale bars: 5 μm. (E) Phasor plot containing the characteristic lifetime data from the three samples described above, namely: DOXf (cluster on the universal circle at ∼1 ns), DOXc (cluster on the universal circle at ∼0.2 ns), and DOX® (cluster within the universal circle). | ||
To tackle these issues, we exploit DOX intrinsic fluorescence as a source of signal and fluorescence lifetime imaging microscopy (FLIM) as a tool with exquisite sensitivity to the nanoscale supramolecular organization of the emitter.
The phasor approach to FLIM data is used here as a fast graphical method to extract the information encrypted in lifetime measurements.10,11 In brief, the fluorescence lifetime decay spectra measured at each pixel of the image are mapped onto a “phasor” plot whose polar coordinates (‘s’ and ‘g’ in Fig. 1, amplitude and phase, respectively) are derived by the Fourier transform of the fluorescence decay in time at the angular repetition frequency of the measurement (eqn (S2) and (S3) in ESI†). Thus, pixels with similar decay curves will have similar coordinates in the phasor plot; also, pixels containing a combination of two (or more) distinct lifetime decays will be mapped according to the weighted linear combination of these contributions.12 As a consequence, if all the distinct contributing lifetimes are known, the fractional contribution of each decay can be retrieved by simple linear algebra or even graphically13 (for more details refer to ESI, eqn (S4)–(S7) and Fig. S1†). As a model system of encapsulated DOX we analyzed Doxoves® (or DOX®), a research-grade product of PEGylated liposomal DOX whose physical characteristics and pharmacokinetics are comparable to those of Doxil®.9 Based on manufacturer's indications, DOX® formulation comes with most of the drug molecules (typically >98%) encapsulated within the aqueous liposome lumen (and presumably all within a nanorod-shaped crystal, hereafter: DOXc), while the remaining minor fraction of molecules (<2%) are supposed to be free in solution (hereafter: DOXf), presumably non-encapsulated. Our analysis started by analyzing the phasor-FLIM signature of the two pure species, DOXf and DOXc, and that of intact DOX® (Fig. 1B–D). DOXf in aqueous solution yields a phasor plot characteristic of a mono-exponential decay with ∼1 ns lifetime (on the “universal circle”,10Fig. 1E), in keeping with previous reports.14,15 Ribbon-shaped DOX crystals, produced following the protocol by Wei and collaborators,16 are characterized by a phasor plot centered on a spot on the universal circle corresponding to a single lifetime of ∼0.2 ns. Results from both DOXf and DOXc samples are highly reproducible, as shown by the low variability reported on their lifetimes (SD < 1%, Table 1). The phasor-FLIM signatures of these pure species identify a segment in the phasor plot (solid black line in Fig. 1E) on which all the possible mixtures of DOXf and DOXc are expected to appear. Measured DOX® phasors, however, do not lie on this segment (Fig. 1E, see also a representative fluorescence decay in Fig. S2†). In order to rationalize the experimental DOX® lifetime, at least one third species must be present in the mixture. This ‘third-species’ hypothesis can be specifically tested in an experiment in which either one species between DOXf and DOXc is selectively removed from the liposomal formulation. To this end, we used the protocol at low molar concentration of ammonium sulfate by Wei and collaborators in order to produce a variant of DOX® (hereafter ‘DOX®-like nanoparticle’, or DLN) that does not contain the DOXc species (Fig. 2A, left). The multi-exponential nature of the corresponding experimental lifetime (phasor denoted with ‘*’ in Fig. 2B) confirms the presence of at least one additional species mixing with DOXf in DLN. An obvious candidate as additional species in DOX® is represented by drug molecules associated/bound to the liposome membrane (hereafter: DOXb). To test this hypothesis we performed an experiment in which DLNs were spin-coated onto a glass surface (Fig. 2A, middle). This procedure mechanically destroyed the liposomal particles while recovering the liposomal membranes on the glass (DOXf is washed out). Of note, phasor-FLIM analysis of the signal associated to these membrane patches yielded a highly-reproducible, nearly mono-exponential lifetime (∼4.5 ns, phasor cluster denoted as ‘**’ in Fig. 2B and Table 1). This closely resembles data obtained for DOX molecules attached chemically, via imine bonds, onto the polymeric surface on iron-oxide nanoparticles.17 Interestingly, if DOX® is spin-coated on the glass (Fig. 2A, right), separate patches of DOXc and DOXb species are observed, yielding two separate clusters in the phasor plot, which are close to spots corresponding to 4.5 and 0.2 ns lifetimes (both marked ‘***’ in Fig. 2C), respectively. Overall, the experiments performed on spin-coated compounds validate the hypothesis that DOXf, DOXc, and DOXb species are all co-existing within DOX® nanoparticles. The fractional-intensity contribution of each species can be determined from the position of the DOX® phasor-plot cluster in the triangle with the three pure species as vertices (Fig. 3A), using algebraic rules13,18 (eqn (S7) and Fig. S1†). Our results are reported in the pie chart of Fig. 3B and in Table 1 (column ‘DOX®FLIM’). Please note that, at this level, the procedure can already be used to quantitatively compare different datasets (e.g. distinct drug preparations; see data from a second batch reported in Fig. S3†).
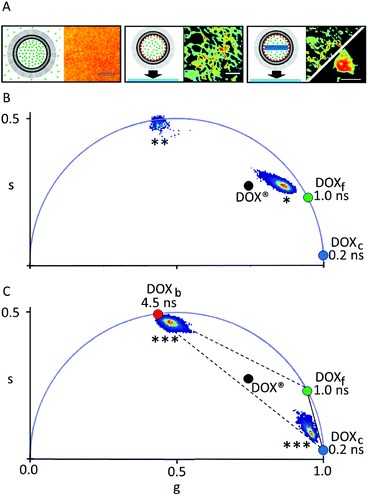 | ||
| Fig. 2 Phasor-FLIM analysis of DOX® synthetic identity. (A) Schematic representation of the sample and the corresponding confocal image for DLN in solution (left), DLN spin-coated on glass (middle) and DOX® spin-coated on glass (right). Scale bars: 3 μm. (B) Phasor plot containing the clusters measured from DLN in solution (‘*’) and DLN after spin-coating (‘**’). (C) Phasor plot containing the two clusters obtained if DOX® is spin-coated on glass, corresponding to the membrane-bound and crystal-like components of DOX® (both marked ‘***’). The two components were measured separately (see example in 2A, right) as DOXc-enriched patches on the glass were sensibly less fluorescent than DOXb-enriched patches. In panels B and C, the black dot corresponds to the centroid of the cluster “DOX®” in Fig. 1E. | ||
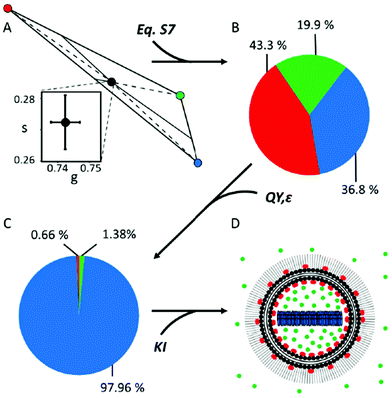 | ||
| Fig. 3 Quantification of the molar fractions of the three DOX species within DOX®. The fractional-intensity contributions of the three DOX species within Doxoves® represented both by a schematic phasor representation (A) and by the corresponding pie-chart (B). (C) Pie chart representing the molar fractions of the different species after correction by QY and ε (see ESI;† uncertainties are reported in Table 1). (D) Schematic representation of DOX® nanoparticle based on phasor-FLIM results, including KI-based ones: DOXf (green) is present both as non-encapsulated and encapsulated; DOXb (red) is associated with both membrane leaflets; DOXc (blue) is buried in the liposome core. | ||
| Pure species | Composite species | |||||||
|---|---|---|---|---|---|---|---|---|
| DOXf | DOXc | DOXb | DLNFLIM | DLNmol | DOX®FLIM | DOX®mol | DOX®mol-ex/inc | |
| All values are expressed as mean ± SD except for QY and ε470 which are expressed as mean ± SE. For derivation of the uncertainties, see ESI.† ND: not determined. ‘—’: not pertinent.a Derived using the DOXb/DOXf lifetime ratio, as described in ESI.†b Same as DOXf, see main text.c Values derived using the fractional contributions of external (‘Ex’) and internal (‘In’) DOXf and DOXb reported in Table S1.†d Calculated by eqn (S18).† | ||||||||
| N | 12 | 7 | 4 | 7 | 7 | 24 | 24 | — |
| τ m (ns) | 1.002 ± 0.008 | 0.200 ± 0.001 | 4.54 ± 0.04 | 3.45 ± 0.05 | — | 2.32 ± 0.06 | — | — |
| DOXf (%) | 100 | — | — | 77.2 ± 4.5 | 93.9 ±1.5 | 19.9 ± 1.8 | 1.37 ±0.22 | Ex: 0.89 ± 0.16 |
| In: 0.48 ± 0.10 | ||||||||
| DOXb (%) | — | — | 100 | 22.8 ± 4.5 | 6.1 ± 1.5 | 43.3 ± 1.9 | 0.66 ± 0.08 | Ex: 0.25 ± 0.06 |
| In: 0.41 ±0.07 | ||||||||
| DOXc (%) | — | 100 | — | — | — | 36.8 ± 2.4 | 97.98 ±0.29 | 97.98 ±0.29 |
| QY488 (%) | 4.23 ± 0.09 | 0.150 ± 0.004 | 19.17 ± 0.47a | ND | 5.14 ± 0.25d | ND | 0.40 ± 0.03d | — |
| ε 470 (M−1 cm−1) | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340± 35 340± 35 |
7510 ± 490 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340± 35b 340± 35b |
ND | — | ND | — | — |
Still, the fractional-intensity contribution of a species will coincide with its actual molar fraction only if the distinct pure species have the same brightness (given by the product of their quantum yield QY and their molar absorption coefficient ε) under the experimental conditions used. We tested this experimentally. First, the QY of DOXf was measured exciting at 488 nm by using an integration sphere, following the methodology reported by de Mello and co-workers,19 and obtained a QY ∼4,2% (Table 1), a value in good agreement with available literature.14 Similarly, the molar absorption coefficient of DOXf at 470 nm (ε470) was derived from the Lambert–Beer relation by using the measured absorbance, 1 cm optical path, and DOXf concentration: it resulted being ∼10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 340 M−1 cm−1 (Table 1), a value again in line with previous estimates.20 These measurements on DOXf served as calibration for similar quantifications on the DOXc sample. QY of DOXc at 488 nm was found to be significantly decreased with respect to the free drug, that is: ∼0.15% (Table 1).
340 M−1 cm−1 (Table 1), a value again in line with previous estimates.20 These measurements on DOXf served as calibration for similar quantifications on the DOXc sample. QY of DOXc at 488 nm was found to be significantly decreased with respect to the free drug, that is: ∼0.15% (Table 1).
Finally, quantification of the ε470 of the crystal was obtained by combining the absorbance (measured by the integration sphere method) with a careful experimental estimate of the effective optical path in the DOXc sample (see Fig. S4† for further details). The value of ε470 obtained for the DOXc sample is ∼7510 M−1 cm−1 (Table 1). The decreased ε470 of DOXc as compared to DOXf is not surprising, also in light of the measured red-shift in the absorption spectrum of the crystal with respect to the free drug (Fig. S5†). Concerning DOXb, based on available literature (e.g. DOX embedded into PVA films14), we assumed that the QYs of DOXb and DOXf are in the same ratio as their lifetimes (yielding an estimated QY for DOXb of about 19%, Table 1) and that ε470 is the same for the two forms. We could then derive the molar fractions of DOXb and DOXf within DLN directly from FLIM data and use them to estimate the QY of DLN (QYDLN = 5.14 ± 0.25%, Table 1, eqn (S18)†). Importantly, this estimate is in good agreement with the QY of DLN independently measured by in-cuvette absorption/fluorescence spectroscopy (5.03 ± 0.25%, details in ESI Methods and Fig. S6†). This result prompted us to use all the spectroscopic parameters at our disposal to convert the fractional-intensity contributions of the three pure species within DOX® into their actual molar ones (Fcorri, eqn (S15),† results in Fig. 3C and Table 1, column ‘DOX®mol’). The resulting dominance of the DOXc component (∼98%) well agrees with expectations based on the drug active-loading procedure used.3,4 By contrast, the presence of a minor fraction of membrane-bound drug (∼0.7%) is revealed here, although this does not contradict previous reports on the nature of DOX-membrane interaction.21 Finally, the retrieved molar fraction of DOXf (∼1,4%) appears within the manufacturer's expectations for non-encapsulated drug (i.e. <2%). It remains unclear, however, if a fraction of DOXf is trapped within the aqueous lumen of DOX®. In order to investigate this point, DOX® lifetime was measured before and after incubation with 370 mM KI, an effective dynamic quencher of non-encapsulated DOX (see Fig. S7†). Assuming that DOXc cannot be affected by KI, lifetime data from N = 3 experiments suggest that a fraction of DOXf must be contained in nanoparticles (∼35% of total DOXf, calculated using eqn (S19) and (S20) in ESI, data reported in Table S1†), with the remaining fraction (∼65%) being non-encapsulated DOXf. Interestingly, data also indicate that a fraction of DOXb (∼38% of total DOXb, Table S1†) is affected by KI, suggesting that DOX molecules can associate to both leaflets of the membrane during loading, with a final prevalence of molecules on the internal leaflet (∼62% of total DOXb). Data from this experiment can be used to re-calculate the molar fractions of the three species, according to their actual localization (Table 1, column ‘DOX®mol-ex/in’). The final picture on DOX® ‘synthetic identity’ is represented schematically in Fig. 3D. Citing again the words by Y. Barenholz: “in Doxil® each component matters and contributes to the optimized performance”.3 Based on present results, we argue that quantifying the abundance of each component is a fundamental step towards understanding the performances (in vitro and in vivo) of encapsulated DOX as a whole drug. In fact, the form in which the drug is administered, and the one in which it then reaches the tumor cell, are important factors in defining its therapeutic efficacy. For instance, it appears obvious that non-monomeric and non-isolated DOX molecules, such as DOXc and DOXb, cannot interact with DNA with the same efficacy of the monomeric, isolated ones (i.e. DOXf). Based on our results, the putative low amount of free drug within Doxil® (∼1%) would apparently contrast with the superior performances of the latter in in vivo applications. However, Doxil® increased efficacy, compared to the free drug, may stem from a complex combination of (i) its demonstrated ability to accumulate a large amount of intact drug preferentially at the tumor site and (ii) an hitherto elusive ability, upon reaching the target site, to gradually release free drug from the crystalline reservoir. In this regard, it should be noted that the use of visible light makes the proposed platform particularly promising for investigating how Doxil® ‘synthetic identity’ may change upon interaction with living matter, at any level, from bodily fluids (e.g. by the adsorption of proteins from the blood serum22) to the intracellular environment.
More in general, the present approach can be fruitfully applied to investigation of the supramolecular organization of a broad range of drugs transported by carriers, with significant advantages over standard methods, namely: (i) it is a label-free procedure, i.e. it does not require chemical modification of the molecule under study but exploits intrinsic signals in native conditions; (ii) it exploits a fast, fit-free data-analysis procedure; (iii) it affords exquisite nanoscale sensitivity in standard optical setups. Finally, we envision similar applications in adjacent fields to provide fast readouts in quality tests along the production line of substances such as agrochemicals (e.g. controlled-release pesticides), industrial chemicals (e.g. paints, adhesives, inks, anti-counterfeiting inks, cosmetics), textiles, nutraceutical/dietary supplements.
Author contributions
Investigation: P. T., G. S., A. Cam., A. Car., G. F., P. P., S. L., D. P.; formal analysis: P. T., G. S., A. Cam., A. Car., G. F., S. L.; methodology: P. T., D. P., G. C., F. B., F. C.; software: E. G.; conceptualization: F. B., G. C., F. C.; writing-original draft: P. T., F. C.; writing-review & editing: E. G., G. C., F. B., F. C. supervision: F. C.Conflicts of interest
No competing financial interests have been declared.Acknowledgements
The authors acknowledge D. Pisignano for support with the instrumentation, L. Lanzanò for useful discussions. This research was partly funded by MIUR PRIN Grant (2017YF9FBS) to F. C.References
- G. Gregoriadis, Targeting of drugs: implications in medicine, Lancet, 1981, 318(8240), 241–247 CrossRef.
- T. Safra, F. Muggia, S. Jeffers, D. D. Tsao-Wei, S. Groshen, O. Lyass, R. Henderson, G. Berry and A. Gabizon, Pegylated Liposomal Doxorubicin (Doxil): Reduced Clinical Cardiotoxicity in Patients Reaching or Exceeding Cumulative Doses of 500 Mg/M2, Ann. Oncol., 2000, 11(8), 1029–1034 CrossRef CAS PubMed.
- Y. Barenholz, (Chezy). Doxil®—The First FDA-Approved Nano-Drug: Lessons Learned, J. Controlled Release, 2012, 160(2), 117–134 CrossRef CAS PubMed.
- Y. Schilt, T. Berman, X. Wei, Y. Barenholz and U. Raviv, Using Solution X-Ray Scattering to Determine the High-Resolution Structure and Morphology of PEGylated Liposomal Doxorubicin Nanodrugs, Biochim. Biophys. Acta, Gen. Subj., 2016, 1860(1), 108–119 CrossRef CAS PubMed.
- G. Haran, R. Cohen, L. K. Bar and Y. Barenholz, Transmembrane Ammonium Sulfate Gradients in Liposomes Produce Efficient and Stable Entrapment of Amphipathic Weak Bases, Biochim. Biophys. Acta Biomembr., 1993, 1151(2), 201–215 CrossRef CAS.
- E. M. Bolotin, R. Cohen, L. K. Bar, N. Emanuel, S. Ninio, Y. Barenholz and D. D. Lasic, Ammonium Sulfate Gradients for Efficient and Stable Remote Loading of Amphipathic Weak Bases into Liposomes and Ligandoliposomes, J. Liposome Res., 1994, 4(1), 455–479 CrossRef.
- D. D. Lasic, B. Čeh, M. C. A. Stuart, L. Guo, P. M. Frederik and Y. Barenholz, Transmembrane Gradient Driven Phase Transitions within Vesicles: Lessons for Drug Delivery, Biochim. Biophys. Acta Biomembr., 1995, 1239(2), 145–156 CrossRef.
- Y. Barenholz, Amphipathic Weak Base Loading into Preformed Liposomes Having a Transmembrane Ammonium Ion Gradient: From the Bench to Approved Doxil, in Liposome Technology, ed. G. Gregoriadis, Informa Healthcare, 2006, vol. II, pp. 1–25 Search PubMed.
- S. Zhu, L. Ma, S. Wang, C. Chen, W. Zhang, L. Yang, W. Hang, J. P. Nolan, L. Wu and X. Yan, Light-Scattering Detection below the Level of Single Fluorescent Molecules for High-Resolution Characterization of Functional Nanoparticles, ACS Nano, 2014, 8(10), 10998–11006 CrossRef CAS PubMed.
- M. A. Digman, V. R. Caiolfa, M. Zamai and E. Gratton, The Phasor Approach to Fluorescence Lifetime Imaging Analysis, Biophys. J., 2008, 94(2), L14–L16 CrossRef CAS PubMed.
- S. Ranjit, L. Malacrida, D. M. Jameson and E. Gratton, Fit-Free Analysis of Fluorescence Lifetime Imaging Data Using the Phasor Approach, Nat. Protoc., 2018, 13(9), 1979–2004 CrossRef CAS PubMed.
- G. Weber, Resolution of the Fluorescence Lifetimes in a Heterogeneous System by Phase and Modulation Measurements, J. Phys. Chem., 1981, 85(8), 949–953 CrossRef CAS.
- S. Ranjit, R. Datta, A. Dvornikov and E. Gratton, Multicomponent Analysis of Phasor Plot in a Single Pixel to Calculate Changes of Metabolic Trajectory in Biological Systems, J. Phys. Chem. A, 2019, 123(45), 9865–9873 CrossRef CAS PubMed.
- S. Shah, A. Chandra, A. Kaur, N. Sabnis, A. Lacko, Z. Gryczynski, R. Fudala and I. Gryczynski, Fluorescence Properties of Doxorubicin in PBS Buffer and PVA Films, J. Photochem. Photobiol., B, 2017, 170, 65–69 CrossRef CAS PubMed.
- T. Zhou, T. Luo, J. Song and J. Qu, Phasor–Fluorescence Lifetime Imaging Microscopy Analysis to Monitor Intercellular Drug Release from a PH-Sensitive Polymeric Nanocarrier, Anal. Chem., 2018, 90(3), 2170–2177 CrossRef CAS PubMed.
- X. Wei, D. Shamrakov, S. Nudelman, S. Peretz-Damari, E. Nativ-Roth, O. Regev and Y. Barenholz, Cardinal Role of Intraliposome Doxorubicin-Sulfate Nanorod Crystal in Doxil Properties and Performance, ACS Omega, 2018, 3(3), 2508–2517 CrossRef CAS PubMed.
- J. S. Basuki, H. T. T. Duong, A. Macmillan, R. B. Erlich, L. Esser, M. C. Akerfeldt, R. M. Whan, M. Kavallaris, C. Boyer and T. P. Davis, Using Fluorescence Lifetime Imaging Microscopy to Monitor Theranostic Nanoparticle Uptake and Intracellular Doxorubicin Release, ACS Nano, 2013, 7(11), 10175–10189 CrossRef CAS PubMed.
- A. Vallmitjana, A. Dvornikov, B. Torrado, D. M. Jameson, S. Ranjit and E. Gratton, Resolution of 4 Components in the Same Pixel in FLIM Images Using the Phasor Approach, Methods Appl. Fluoresc., 2020, 8(3), 035001 CrossRef CAS PubMed.
- J. C. de Mello, H. F. Wittmann and R. H. Friend, An Improved Experimental Determination of External Photoluminescence Quantum Efficiency, Adv. Mater., 1997, 9(3), 230–232 CrossRef CAS.
- Y. Tian, L. Bromberg, S. N. Lin, T. A. Hatton and K. C. Tam, Complexation and Release of Doxorubicin from Its Complexes with Pluronic P85-b-Poly(Acrylic Acid) Block Copolymers, J. Controlled Release, 2007, 121(3), 137–145 CrossRef CAS PubMed.
- A. Martí, X. Armengol, J. Estelrich and J. Hernández-Borrell, Encapsulation of Doxorubicin in Neutral Liposomes by Passive Methods: Evidence of Drug-Lipid Interaction at Neutral PH, J. Microencapsulation, 1991, 9(2), 191–200 CrossRef PubMed.
- G. Caracciolo, S. Palchetti, L. Digiacomo, R. Z. Chiozzi, A. L. Capriotti, H. Amenitsch, P. M. Tentori, V. Palmieri, M. Papi, F. Cardarelli, D. Pozzi and A. Laganà, Human Biomolecular Corona of Liposomal Doxorubicin: The Overlooked Factor in Anticancer Drug Delivery, ACS Appl. Mater. Interfaces, 2018, 10(27), 22951–22962 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods. Fig. S1–S7, Table S1. See DOI: https://doi.org/10.1039/d2nr00311b |
| This journal is © The Royal Society of Chemistry 2022 |
