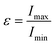Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Impact of drug aggregation on the structural and dynamic properties of Triton X-100 micelles†
Hrachya
Ishkhanyan
a,
Natasha H.
Rhys
a,
David J.
Barlow
b,
M. Jayne
Lawrence
 b and
Christian D.
Lorenz
b and
Christian D.
Lorenz
 *a
*a
aBiological & Soft Matter Research Group, Department of Physics, Faculty of Natural, Mathematical & Engineering Sciences, King's College London, London, UK. E-mail: chris.lorenz@kcl.ac.uk
bDivision of Pharmacy and Optometry, School of Health Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Stopford Building, Oxford Road, Manchester, UK
First published on 16th March 2022
Abstract
Surfactants are used in a wide range of chemical and biological applications, and for pharmaceutical purposes are frequently employed to enhance the solubility of poorly water soluble drugs. In this study, all-atom molecular dynamics (MD) simulations and small-angle neutron scattering (SANS) experiments have been used to investigate the drug solubilisation capabilities of the micelles that result from 10 wt% aqueous solutions of the non-ionic surfactant, Triton X-100 (TX-100). Specifically, we have investigated the solubilisation of saturation amounts of the sodium salts of two nonsteroidal anti-inflammatory drugs: ibuprofen and indomethacin. We find that the ibuprofen-loaded micelles are more non-spherical than the indomethacin-loaded micelles which are in turn even more non-spherical than the TX-100 micelles that form in the absence of any drug. Our simulations show that the TX-100 micelles are able to solubilise twice as many indomethacin molecules as ibuprofen molecules, and the indomethacin molecules form larger aggregates in the core of the micelle than ibuprofen. These large indomethacin aggregates result in the destabilisation of the TX-100 micelle, which leads to an increase in the amount of water inside of the core of the micelle. These combined effects cause the eventual division of the indomethacin-loaded micelle into two daughter micelles. These results provide a mechanistic description of how drug interactions can affect the stability of the resulting nanoparticles.
Introduction
Without a drug delivery vehicle (DDV), consistent in vivo drug concentrations would be difficult to ensure which then would lead to unpredictable effectiveness and safety of the therapeutic treatment.1,2 With the advances of nanomedicine, more stable, controllable and precise drug delivery is possible.3–6 Increasingly drugs are loaded into nano-sized vesicles, which are constructed and optimised for the nature of the drug and its biological target.7,8 DDVs including nano-sized carriers offer a wide range of advantages including greater drug solubility, lower toxicity and more targeted delivery. With their high surface area to volume ratio, nano-carriers are used to solubilise small drug molecules, nucleic acids, and proteins with good efficiency and are used for targeted as well as non-targeted therapy.9A popular family of nano-DDV are self-assembled structures, which are cost efficient and simple to make. Self-assembly is the process of the organisation of materials into structures based on non-covalent interactions. Alongside the self-assembled structures abundant in nature, artificially designed ones are of great interest in biomedical applications, particularly as antimicrobial and detection agents10 or drug delivery vesicles.11–13 There is a vast range of materials such as biopolymers, peptides, nucleic acids and inorganic compounds used as building blocks for self-assembled structures.14–16 Peptides can be self-assembled into nanotubes preserving their biological recognition properties and used in microelectronics to easily construct electrical circuits17 while nucleic acids and surfactants can be self-assembled into structures suitable for drug delivery.18,19
Surfactants are surface active molecules widely used as solubilising agents, emulsifiers, antimicrobial agents, and drug and gene delivery systems.20,21 Traditional surfactants are low molecular weight amphiphiles that in aqueous solution can self-assemble into various structures such as micelles or liposomes. There are four main categories of surfactants classified on the basis of their charge: non-ionic, cationic, anionic and zwitterionic. Due to their ability to self-assemble, surfactant micelles are easy to produce and consequently have a low production cost. Additionally, micelles have a large drug solubilisation capacity and can increase the bioavailability of poorly soluble drug molecules. Therefore surfactants, particularly the non-ionic variety, are attractive candidates when forming drug delivery vesicles.
In this work, we investigate the structural properties and drug solubilisation capabilities of Triton X-100 (TX-100) (Fig. 1c) micelles using a combination of all-atom molecular dynamics (MD) simulations and small-angle neutron scattering (SANS) experiments. TX-100 is a non-ionic surfactant with a hydrophilic polyethylene oxide headgroup and a hydrophobic tail. TX-100 is widely used for protein and cell organelle extraction, cell lysation, and membrane permeabilisation.22,23 More recently, TX-100 has become increasingly investigated for its application in drug delivery.24–27
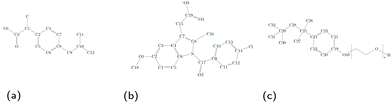 | ||
| Fig. 1 Structures of molecules in simulations, including (a) the ibuprofen ion, (b) the indomethacin ion and (c) the TX-100 surfactant molecule. | ||
The self-assembly of TX-100 surfactant molecules into micelles in an aqueous environment has been extensively investigated experimentally.28–30 The first average aggregation number and molecular weight of the resulting TX-100 micelles was determined using static light scattering by Kushner and Hubbard in 1954.31 The size and shape of the TX-100 micelles in different solvent environments and at different temperatures has also been investigated in numerous studies.29–34 However, to date there is still no consensus about the shape of TX-100 micelles in an aqueous environment. For example, Robson and Dennis used the results of hydrodynamic and viscosity measurements to argue that TX-100 micelles are non-spherical in shape.30 Paradies29 and Podo et al.35 used the results of Nuclear Overhauser Effect SpectroscopY (NOESY) NMR and hydrodynamic measurements to suggest that at low aggregation numbers the shape of TX-100 micelles is slightly non-spherical, and as the aggregation numbers increase the micelles become increasingly non-spherical.
While there has been a significant amount of experimental investigation of the structural properties of self-assembled TX-100 micelles there have been comparatively few investigations of these micelles using MD simulations. Yordanova et al. reported the CHARMM force field parameters for Triton X series surfactants.36 Also, Milano et al. have used all-atom and coarse-grain simulations, as well as their MD-self-consistent field theory (MD-SCF) approach, to explore the self-assembly of TX-100 and the structure and shape of the resultant micelles.37,38 In their original paper, they report on numerous structural properties of TX-100 micelles in aqueous solutions and show that the shape of the micelles become increasingly non-spherical as the aggregation number of the micelle increases. While in their more recent paper, they show that the transition from the more spherical micelles to the more non-spherical micelles is a result of the energetic cost due to the growth of the interfacial region, and the resulting larger hydrophobic surface exposed to the aqueous environment, as the aggregation number increases.
In our study, we investigate how the solubilisation of two non-steroidal anti-inflammatory drugs (NSAIDs), ibuprofen (C13H18O2) and indomethacin (C19H16ClNO4) (chemical structures shown in Fig. 1),39–41 affects the structural properties of TX-100 micelles. Both drug molecules contain a benzene ring and carboxyl group, which has a low pKa![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 42 resulting in both drugs being deprotonated at neutral pH. Despite being deprotonated, both molecules are poorly soluble in aqueous solutions, and therefore in order to increase their bioavailability they are best solubilised in some kind of a drug delivery vehicle. Previously, Bahadur et al. have investigated the effect of the solubilisation of small molecules within TX-100 micelles.43,44 In both publications, they have shown that the solubilisation of more hydrophobic small molecules results in the growth of the TX-100 micelles, while the solubilisation of more hydrophilic small molecules results in the micelles staying approximately the same size.
42 resulting in both drugs being deprotonated at neutral pH. Despite being deprotonated, both molecules are poorly soluble in aqueous solutions, and therefore in order to increase their bioavailability they are best solubilised in some kind of a drug delivery vehicle. Previously, Bahadur et al. have investigated the effect of the solubilisation of small molecules within TX-100 micelles.43,44 In both publications, they have shown that the solubilisation of more hydrophobic small molecules results in the growth of the TX-100 micelles, while the solubilisation of more hydrophilic small molecules results in the micelles staying approximately the same size.
We show here that the sodium salts of both ibuprofen and indomethacin are solubilised within TX-100 micelles. We find that TX-100 micelles consisting of approximately 147 surfactant molecules are able to solubilise approximately twice as much indomethacin than ibuprofen. Once solubilised within the core of the micelles, the drug molecules reorient in order to allow their carboxyl groups to remain hydrated by the water in the aqueous environment surrounding the core of the micelles. We find that there are more extensive interactions between indomethacin molecules than between ibuprofen molecules. As a result, the indomethacin molecules form larger aggregates within the core of the micelle than ibuprofen. Furthermore, we observe that the indomethacin-loaded micelle divided into two after reaching its maximum loading. The division of the indomethacin-loaded micelle is a result of the growth of the solubilised indomethacin aggregates which destabilise the core of the micelle. In the following sections of the manuscript, we present the methods we have used in order to conduct this investigation, and then report on the structural properties of the TX-100 micelles with and without drugs, as well as the drug solubilisation process.
Methods
Molecular dynamics simulations
In this study, three systems (summarised in Table 1) have been investigated with all-atom molecular dynamics simulations using the GROMACS 2018.2 simulation engine.45–47 The CHARMM36 force-field48 was used to describe the interactions of the TX-100 molecules36 and the Na+ ions in solution. The drug molecules were parameterised using the CHARMM general force field.49 The interactions of the water molecules were modelled using the CHARMM-modified version of the TIP3P potential.| System | Drug | Triton X-100 | Water |
|---|---|---|---|
| TX-100 | 0 | 150 (![[1 with combining low line]](https://www.rsc.org/images/entities/char_0031_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[. with combining low line]](https://www.rsc.org/images/entities/char_002e_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[w with combining low line]](https://www.rsc.org/images/entities/char_0077_0332.gif) ![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) %) %) |
52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 266 266 |
| TX-100-IBUP | 100 | 150 (![[7 with combining low line]](https://www.rsc.org/images/entities/char_0037_0332.gif) ![[. with combining low line]](https://www.rsc.org/images/entities/char_002e_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[w with combining low line]](https://www.rsc.org/images/entities/char_0077_0332.gif) ![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) %) %) |
75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 181 181 |
| TX-100-INDO | 100 | 150 (![[8 with combining low line]](https://www.rsc.org/images/entities/char_0038_0332.gif) ![[. with combining low line]](https://www.rsc.org/images/entities/char_002e_0332.gif) ![[0 with combining low line]](https://www.rsc.org/images/entities/char_0030_0332.gif) ![[w with combining low line]](https://www.rsc.org/images/entities/char_0077_0332.gif) ![[t with combining low line]](https://www.rsc.org/images/entities/char_0074_0332.gif) %) %) |
65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 673 673 |
In order to build an initial configuration of the TX-100 system, we pre-assembled a spherical micelle containing 150 TX-100 surfactant molecules using Packmol.50 The input code is presented in the ESI.† Then for the systems containing ibuprofen (TX-100-IBUP) and indomethacin (TX-100-INDO) we placed 100 drug molecules randomly around the pre-assembled micelle. In each system, water molecules are then added to fill the simulation box with dimensions of 120 Å × 120 Å × 120 Å for the TX-100 system and 130 Å × 130 Å × 130 Å for the TX-100-IBUP and TX-100-INDO systems (Table 1). In the drug containing systems, we also added 100 sodium (Na+) ions as counterions to the negatively charged drug molecules.
Each system was simulated using the same protocol. First the system underwent 5000 steps of minimisation with the steepest descent method. Then a 50 ps NVT simulation using the V-rescale thermostat and a 200 ps NPT simulation using the same thermostat and the Berendsen barostat were conducted in order to equilibrate the temperature and pressure of the system, respectively. Finally, a production simulation using the NPT ensemble with the Nosé–Hoover thermostat and Parrinello–Rahman barostat was conducted. The production simulation was run for long enough for the size and shape of the micelle to equilibrate. Therefore, the production simulations were run for 200 ns (TX-100), 300 ns (TX-100-IBUP) and 600 ns (TX-100-INDO) using a timestep of 2 fs. The resulting simulation box sizes were 119.4 Å × 119.4 Å × 119.4 Å, 134.2 Å × 134.2 Å × 134.2 Å and 129.0 Å × 129.0 Å × 129.0 Å for the TX-100, TX-100-IBUP and TX-100-INDO systems, respectively.
In all simulations, the target pressure and temperature were set to 1 atm and 303.15 K respectively. Cut-off distances for Coulomb and Lennard-Jones interactions were set to 1.2 nm. Long-range electrostatic interactions were calculated using the particle-mesh Ewald method. Hydrogen containing bonds were constrained using the LINCS algorithm.51
Analysis of MD simulations
The physical properties of each simulated system were analysed using in-house Python scripts and the MDAnalysis package.52,53 In order to characterise the shape of the micelles, we have calculated the ellipticity of the entire micelle, defined as:where Imax and Imin are the largest and smallest terms in a diagonal inertia tensor. For a sphere, ε is equal to 1, and ε increases as the shape becomes increasingly elliptical. The shape of the micelle can be further characterised by comparing the diagonal terms in the inertia tensor. If two axes of the ellipsoid are equal and greater or less than the third axes, the ellipsoid is oblate or prolate respectively. Otherwise, when all three axes are equal, the structure is spherical.
The solvent-accessible surface area (SASA) algorithm represents the surfaces of our micelles as overlapping spheres with van der Waals radii of the corresponding atoms, and uses a probe with a small radius, that is typically 1.4 Å to represent a water molecule, to determine the maximum permitted contact with the surface.54 In this study, the FreeSASA55 module was used to calculate the SASA of the various micelles.
In order to determine whether a drug molecule was solubilised in a micelle, we first calculated the distance between the center of mass of drug molecules and the micelle. If the distance is less than 5 Å, the drug is considered to be solubilised. A cut-off distance of 5 Å is chosen as it is roughly the distance to the first neighbouring atom. This value is used throughout the rest of the analysis.
Further analysis was performed to characterise the hydration of drugs throughout the simulation. The number of water molecules around each drug has been plotted against Δr, which is defined as:
| Δr = rd − rs |
Finally, drug–drug and drug-TX-100 interactions were characterised by creating contact maps. The distance between the heavy (non-hydrogen) atoms of the molecules of interest that were 5 Å away from one another were considered in contact. The number of contacts were counted between each pair of heavy (non-hydrogen) atoms on the molecules of interest, and then the maximum number of contacts between any two pair of atoms was used to normalise the various values, such that the pair of atoms most frequently in contact had a value of 1.0 in the contact maps.
Small-angle neutron scattering
Small angle neutron scattering (SANS) studies were performed on the SANS2D small angle diffractometer at the ISIS pulsed neutron source (ISIS, Rutherford Appleton Laboratory, STFC, Didcot, Oxford). The SANS2D diffractometer was configured to provide a scattering vector Q = (4πλ)sin(θ/2) in the range of 0.0045 Å−1 < Q < 0.4 Å−1. All the protiated surfactant solutions were prepared using D2O as the solvent. When preparing the solutions with D2O, the weight ratio of surfactant to D2O was re-calculated to ensure the molar ratio of surfactant to D2O was the same as in H2O. The samples were measured in quartz banjo cells of 2 mm path length. All measurements were performed at 298 ± 0.1 K. The measured SANS data were model-fitted using SasVView.56 A range of models/scattering form factors (P(Q)) including core–shell spheroids (namely sphere, oblate, prolate) and triaxial micelles were considered. As the TX-100 concentrations used were large, it was necessary to account for any interparticulate interactions (S(Q)), which was achieved using a hard sphere model. The best model found to fit the data was a core–shell spheroid model. The modelling of the SANS data assumed a flat background correction to allow for any mismatch in the incoherent and inelastic scattering between the samples and solvent and the fitted background levels were checked to ensure they were of a physically reasonable magnitude.Results
In the following sections we summarise the results from our all-atom MD simulations and compare them to the results of our neutron scattering experiments in order to provide a detailed description of the size, shape and internal structure of a TX-100 micelle. We also provide a detailed description of how these properties change when ibuprofen and indomethacin are loaded within the micelle.Structural properties of pure Triton X-100 micelles
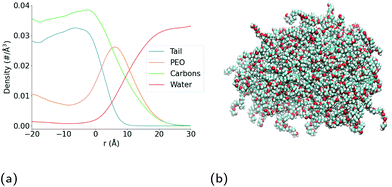
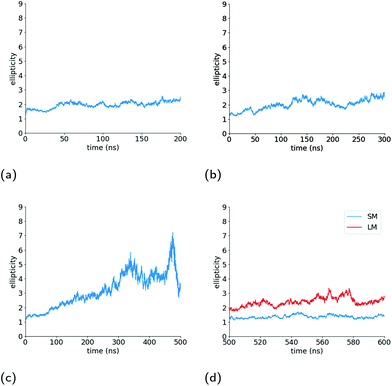
As can be seen in the snapshot shown in Fig. 2b, the pure TX-100 micelle relaxes into a non-spherical structure. The equilibrated micelle contains an average of 147 TX-100 molecules, with the remaining three molecules in a dynamic equilibrium of joining and leaving the micelle. In order to quantify the shape of this micelle (TX-100), we calculated the ellipticity as a function of time (Fig. 3a). The shape of the TX-100 micelle was found to remain slightly non-spherical throughout the production simulation (ε ∼ 1.48) with a prolate shape and a length of 107.8 ± 0.8 Å along the primary axis. Likewise, the measured SASA of the micelle was found to remain nearly constant over time (Fig. 4a), averaging 6.01 × 104 (±0.01 × 104) Å2.In order to characterise the internal structure of the micelle, we first needed to identify the intrinsic surface of the hydrophobic core of the micelle. In order to identify this intrinsic surface, we used the NANOCISC code.57 We chose the carbon atom in the aromatic ring of the surfactant (atom C19 in Fig. 1c) bonded to the first oxygen in the polyethyllene oxide (PEO) as the anchor point, which was then used to define the surface of the hydrophobic core of the micelle and was therefore defined as where r = 0 in Fig. 2a and 6. Furthermore, the intrinsic densities of water (using the oxygen atoms in the water molecules) and the TX-100 molecules were calculated (Fig. 2a). At large distances, the number density of the oxygen atoms in the water molecules is 0.033 Å−3, which is consistent with the bulk water density found in our previous studies.57–59 The water density begins to decrease slightly at a distance of 25 Å from the micelle's core surface and continues to decrease until it reaches a minimum of 0.02 Å−3 at a distance of r ∼ −9 Å (e.g., 9 Å inside of the hydrophobic core of the micelle). Then in the centre of the micelle (r ∼ −20 Å), the number density of water is 0.002 Å−3, indicating the existence of a small amount of water inside of the micelle core.
Additionally, we have determined the intrinsic densities of the carbon atoms in the whole micelle (green curve), the poly(ethylene oxide) chain (blue curve) and the hydrophobic tail of the surfactant (orange curve) as shown in Fig. 2a. Some of the PEO chains extend up to a distance of r ∼ 25 Å, which corresponds to the distance at which the density of water begins to decrease. The thickness of the PEO headgroup, which we define as the distance from the surface of the hydrophobic core of the micelle at which the density of the PEO chains becomes larger than the density of the water, is r ∼ 10.7 Å. The peak density of the PEO chains is found at a distance of r ∼ 7 Å from the surface of the hydrophobic core of the micelle. In the hydrophobic core of the micelle, the density of PEO decreases until it reaches a minimum at r ∼ −7.5 Å and then increases to a density of ∼0.011 Å−3 at r = −20 Å. In the core of the micelle, PEO makes up approximately one-third of the surfactant's density.
Meanwhile the hydrophobic tails of the Triton X-100 molecules, which consist of the benzene ring and the attached short hydrocarbon chain, are found primarily within the core of the micelle (r < 0 Å). The hydrophobic tails take a variety of orientations at the interface of the hydrophobic core and the surrounding aqueous environment which results in the small density of these groups found within 7.5 Å of this interface (the end-to-end distance of the hydrophobic tail is ∼7.5 Å). Within the core of the micelle, the density of the surfactant tails begins to decrease at r = −7.5 Å, which is the distance within the core where we observe the increase in the PEO density. The hydrophobic tails of the Triton X-100 molecules interact via a combination of hydrophobic interactions of the benzene rings and the attached methyl groups, as can be seen in Fig. S4.†
Triton X-100 micelles loaded with ibuprofen
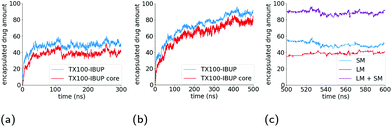
The surface area of the micelle initially increases as the ibuprofen molecules are solubilised within (Fig. 4b), as can be seen by an increase in the SASA value from an initial value of 6.17 × 104 Å2 SASA to a value of 6.4 ± 0.006 × 104 Å2. As the SASA values plateau after approximately 75 ns and the ellipticity after 150 ns, it appears that the micelle continues to become increasingly rod-like in such a manner that it maintains a constant surface area.
The structure of the micelle when loaded with ibuprofen is similar to that found for the TX-100 micelle. As such, the PEO chains of the surfactant molecules are found on the surface of the hydrophobic core of the micelle, extending into the surrounding aqueous environment, while the benzene ring and corresponding short hydrocarbon chain of the surfactant molecules are generally hidden from exposure to the surrounding aqueous environment.
Solubilisation of ibuprofen
As shown in Fig. 5a, the number of ibuprofen molecules solubilised within the micelle converges to an average of 49 after 50 ns. On average, 39 of these ibuprofen molecules are solubilised within the core of the micelle, and interestingly the number of ibuprofen molecules within the core stabilises at approximately the same point in time.In order to assess how the hydration of the ibuprofen molecules changes during the solubilisation process, we first measured the radial distribution functions (rdfs) (Fig. S1†) of the polar oxygen atoms within the ibuprofen molecule and the oxygen atom in surrounding water molecules.
From these rdfs, we determined a first neighbour distance between the water molecules and these oxygen atoms of 2.8 Å. We then counted the number of water molecules within 2.8 Å of the polar oxygen atoms within each ibuprofen during the course of the simulation, and have plotted the average number of water molecules hydrating each of the two oxygen atoms as a function of the intrinsic distance from the interface of the hydrophobic core of the micelle (Fig. 6(a) & (b)), which is defined by the location of the benzene ring of the TX-100 molecules.
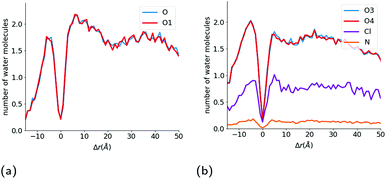 | ||
| Fig. 6 Hydration of drug molecules during the solubilisation process. Number of water molecules hydrating the (a) O1 and O atoms in the ibuprofen molecules and (b) O3, O4, N and Cl atoms in indomethacin as a function of the distance of each atom from the interface of the core of the micelle. Atom labels are the same as shown in Fig. 1. | ||
As can be seen in Fig. 6b, the number of water molecules around the O1 and O atoms of the ibuprofen molecules remain approximately constant until the drug molecules get within 25 Å of the surface of the hydrophobic core of the micelle (2.18 ± 0.02 water molecules around O1 and O). Then at a distance of 25 Å, which corresponds to the maximum extent of the PEO chains of the TX-100 molecules from the surface of the micelle's core, the number of water molecules around each oxygen decreases slightly. Then at a distance of 20 Å the number of water molecules around each atom increases and reaches a peak at ∼7.5 Å which corresponds to the distance which the hydrophobic tails of TX-100 extend from the surface of the core of the micelle into the aqueous environment. Both oxygen atoms of the ibuprofen molecules then become significantly dehydrated as the ibuprofen molecule crosses from the corona of the micelle into the core of the micelle. This dehydration of the oxygens at the interface of the hydrophobic core of the micelle is likely due to the large steric barrier that exists for the drug molecules when they cross into the core of the micelle. When the ibuprofen molecules enter into the core of the micelle, they generally reorientate such that the oxygens in the ibuprofen molecules are at the interface of the core with the aqueous environment and become hydrated to nearly the same level as in the bulk aqueous environment, which is demonstrated by the peak in hydration of the oxygen atoms observed at Δr ∼ −7.5 Å in Fig. 6b. The oxygen atoms on the drug molecules deeper into the core of the micelle become significantly dehydrated once again.
In Fig. S7b,† we show the number of water molecules as a function of Δr at various time points during the drug solubilisation process. This figure shows that there is a very slight increase in the number of water molecules deep within the core of the micelle (Δr < −10 Å) as time passes, which demonstrates that the ibuprofen molecules do not bring a significant amount of water into the micelles with them. In comparison to the TX-100 micelle (Fig. S7a†) formed in the absence of drug, there is very little difference in the number of water molecules within the core micelle of over the course of the two simulations.
Internal structure of ibuprofen-loaded Triton X-100 micelles
To better understand the surfactant–drug and drug–drug interactions, we have measured the amount of contact between the various regions of the molecules. In doing so, we have constructed contact maps, which are generated by calculating the distance between each heavy (non-hydrogen) atom of a TX-100 (or ibuprofen) molecule and a neighbouring ibuprofen molecule. In order to determine a distance at which the molecules are in contact, we first calculated the minimum distance between a TX-100 (or ibuprofen) molecule and an ibuprofen molecule when they interact with each other. From these measurements, we observed that the minimum distance plateaus to ∼5 Å when two molecules aggregate with one another. Therefore, if any two heavy (non-hydrogen) atoms are within 5 Å of one another then we count that as a contact between the two molecules.The regions of the TX-100 surfactant molecules that have the most contact with the ibuprofen molecules are the hydrophobic tails, which correspond to atoms C19–C32 on horizontal axis on Fig. 7a. Meanwhile, the regions of the ibuprofen molecule which are in contact with the surfactant molecules are the two terminal methyl carbon atoms, C11–C12, as highlighted in Fig. 7b. Also we find that the carboxyl group on the ibuprofen molecules makes no contact with the surfactant molecules. Thus the ibuprofen molecules are solubilised within the micelle via hydrophobic interactions between themselves and the surfactants while the oxygen atoms remain solvated by the aqueous environment surrounding the core of the micelle.
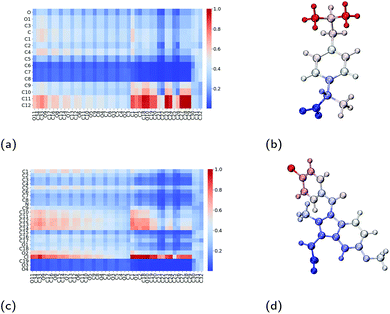 | ||
| Fig. 7 Interactions between Triton X-100 surfactants and drug molecules. Contact maps which show the amount of contact between an atom on the surfactant molecule (x-axis) and an atom on the drug molecule (y-axis) for (a) ibuprofen and (c) indomethacin. Diagrams of (b) ibuprofen and (d) indomethacin molecules which have been coloured by their amount of contact with neighbouring Triton X-100 molecules, where the colours are consistent with those used in the contact maps. Atom labels used within the contact maps are those shown in Fig. 1. | ||
Within the core of the micelle, the ibuprofen molecules aggregate with one another. In doing so, we find that methyl carbon atoms and the carbon in between them (C10, C11 and C12) form contacts with one another, as shown in Fig. 8a and visualised in Fig. 8b. Representative aggregates of ibuprofen molecules formed within the core of the micelle can be seen in Fig. S5.† We have also investigated the contacts which exist within the crystalline form of ibuprofen, which are shown in Fig. S2a.† By comparing these two contact maps (Fig. S3a†), we find that the packing of the drugs within the micelle are significantly different than within their crystalline form. This is largely due to the reorientation of the drugs in the micelle such that the carboxyl group can interact with the surrounding aqueous environment.
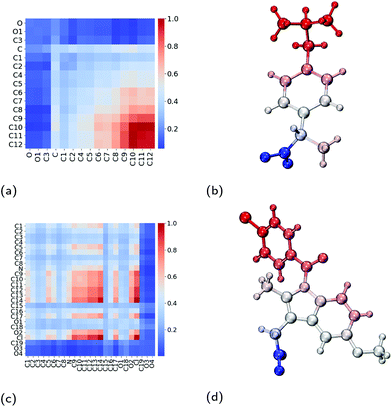 | ||
| Fig. 8 Interactions between pairs of neighbouring drug molecules. Contact maps which show the amount of contact between two atoms on two neighbouring drug molecules for (a) ibuprofen and (c) indomethacin. Diagrams of (b) ibuprofen and (d) indomethacin molecules which have been coloured by their amount of contact with neighbouring drug molecules, where the colours are consistent with those used in the contact maps. Atom labels used within the contact maps are those shown in Fig. 1. | ||
Triton X-100 micelles loaded with indomethacin
The SASA of the TX-100 micelle loaded with indomethacin grows until it reaches a maximum value of ∼8.0 × 104 Å2 (Fig. 4c). After the micelle splits into two, the SASA values of the daughter micelles remain fairly constant during the rest of the simulation, as can be seen in Fig. 4d. Interestingly the sum of the SASA values of the two daughter micelles (3.48 ± 0.01 × 104 Å2 & 4.53 ± 0.01 × 104 Å2) is approximately the same as the SASA value found for parent micelle before it split (∼8.3 × 104 Å2).
Solubilisation of indomethacin
Fig. 5b shows the number of indomethacin molecules that have been solubilised within the micelle and the core of the micelle, respectively, as a function of time. While the amount of ibuprofen loaded into the micelle saturated after approximately 100 ns, the amount of solubilised indomethacin continues to increase until approximately 400 ns. At which point, the number of indomethacin molecules solubilised in the micelle and the core of the micelle reach values of 87 ± 1 and 77 ± 2, respectively. The amount of indomethacin that is solubilised into the core of the micelle is approximately twice the number of ibuprofen molecules solubilised in the core of the micelle.The number of solubilised indomethacin molecules continues to increase after 400 ns and as a result the values of the ellipticity and the solvent-accessible surface area for the parent micelle continue to increase until the micelle splits. The parent micelle divides into two after it solubilises ∼80 indomethacin molecules into the core of the micelle which results in the micelle continuing to elongate until it becomes destabilised and finally divides. After the parent indomethacin-loaded TX-100 micelle divides into the two daughter micelles, the amount of drug in each of the two new micelles is constant and none of the drugs that remained in solution were observed to solubilise into the micelles (Fig. 5c). The smaller micelle which consists of 62 TX-100 molecules, solubilises 40 ± 1 indomethacin molecules, whereas the larger micelle, which consists of 88 surfactant molecules, solubilises 50 ± 3 drug molecules. The ratio of indomethacin to TX-100 in the two daughter micelles (0.65 & 0.57) is similar to that found in the parent indomethacin-loaded micelle before it divided (0.61). This ratio for the daughter micelles (and the parent micelle) of the indomethacin-loaded systems, however, is twice that found for the ibuprofen-loaded micelle (0.34).
As was done for the ibuprofen molecules, we have calculated the hydration of the indomethacin molecules as they approach the surface of the hydrophobic core of the micelle (Fig. 6b). For the indomethacin molecules, we have used the chlorine atom, nitrogen atom and the two oxygen atoms in the carboxyl group in order to characterise the hydration of the drug molecules. At large distances (r > 40 Å) from the hydrophobic core of the micelle, there are 0.80 ± 0.01, 1.86 ± 0.05, 1.85 ± 0.05 and 0.15 ± 0.01 water molecules around the chlorine, O3, O4 and nitrogen atoms, respectively. The hydration of all of the atoms remains constant until a distance of ∼15 Å from the surface of the hydrophobic core of the micelle. Then the hydration of the N and Cl atoms increase slightly until the drugs are approximately 5 Å from the surface of the micelle's hydrophobic core, and then are dehydrated as the drugs get close to the interface of the core of the micelle. Then the oxygens, nitrogen and chlorine atoms are significantly dehydrated as they cross into the hydrophobic core of the micelle.
After the drug molecules pass the interface of the core of the micelle, all of the atoms are hydrated again to approximately the same amount as in the bulk aqueous environment. Also the hydration of all four of the atoms we investigated for the indomethacin molecules deeper inside the core of the micelle (Δr < −10 Å) is larger than that found for the ibuprofen molecules in the same region of the micelle.
We then looked at the number of water molecules as a function of Δr at various time points through the solubilisation process (see Fig. S7c†). As time increases, which in the case of the indomethacin-loaded micelle also corresponds to more and more indomethacin in the core of the micelle (see Fig. 5b), we observe an increase in the amount of water within the core of the micelle. At times larger than 400 ns, we observe that the amount of water in the core of the micelle exceeds that found in either the TX-100 micelle (Fig. S7a†) or the ibuprofen-loaded micelles (Fig. S7b†).
Internal structure of indomethacin-loaded Triton X-100 micelles
As was calculated for the ibuprofen-loaded micelle, we again determined the contacts between indomethacin and the surfactant molecules, and between the indomethacin molecules. The indomethacin molecules interact with the TX-100 surfactant molecules primarily via its chlorine atom (Cl), which interacts with the benzene ring and the hydrocarbon chain (atoms C19–C32) which form the hydrophobic tail of the surfactants (Fig. 7c & d). Meanwhile, the indomethacin molecules in the core of the micelle bind to one another primarily via their chlorobenzene group (atoms C9–C14 & Cl). Representative clusters of indomethacin molecules within the core of the micelle are shown in Fig. S6.† The carboxyl group (atoms C19, O3 & O4) of the indomethacin molecules are not interacting with either the surfactants or other indomethacin molecules. Instead the carboxyl groups are oriented such that they are interacting with the aqueous environment surrounding the micelle.As a point of comparison, we have determined the contact map between neighbouring indomethacin molecules in its crystalline form, as shown in Fig. S2b.† When comparing the contact maps for the indomethacin in the core of the micelle and that found for the drug in its crystalline form (Fig. S3a†), we find that the chlorobenzene group plays a significant role in the interactions between the drugs in both cases. In the micelle, because the indomethacin molecules are oriented such that their carboxyl group remains hydrated by the surrounding aqueous environment, there is more disorder in the respective orientations of the drug molecules than is found in the crystalline form.
Discussion & conclusions
All-atom molecular dynamics simulations have been used to investigate the internal structure of Triton X-100 micelles with an aggregation number of ∼147 molecules. We find that there the PEO chains of the Triton X-100 molecules are present alongside the hydrophobic tails of the surfactant inside of the core of the micelle. While we have not observed such behaviour in our investigations of surfactants with smaller hydrophilic headgroups,58,60 the intrusion of PEO chains into the hydrophobic core of micelles has been previously observed experimentally Backspace in other PEO containing surfactant molecules.61–64We find that the TX-100 micelles without any drug are non-spherical, which is consistent with the findings of Milano et al.38 for micelles of a similar aggregation number. When investigating the solubilisation of ibuprofen and indomethacin into these micelles, we found that nearly twice as many indomethacin as ibuprofen molecules are solubilised within the micelles. After becoming saturated with drug molecules, we find that the size and shape of the equilibrated ibuprofen-loaded micelles were elongated compared to the TBackspaceX-100 micelles without any drug. In comparison, upon becoming saturated with drug molecules, the indomethacin-loaded micelles become increasingly asymmetric eventually spontaneously dividing into two daughter micelles – a smaller, slightly asymmetric one and a larger, significantly asymmetric one. In addition, we have determined the ellipticity of the hydrophobic core of the two daughter micelles over time, which shows that the core of the larger of the daughter micelles is more dynamic than the smaller micelle.
The SANS experiments that we have conducted in parallel to these simulations show that the TX-100 micelle is non-spherical being model-fitted as a core–shell prolate ellipsoid when it is not loaded with any drug molecules (Fig. 10a & Table 2), which is consistent with what we find via our MD simulations. We, also, find that the average maximum diameter of our simulated micelle is 107.8 Å, which is in reasonable agreement with the longest dimension of the micelle as suggested by the fitting of the SANS data (((20.4 Å × 3.8) + 6.7 Å) = 168.8 Å). It is worth commenting that the aggregation numbers obtained from SANS and the MD simulations are not exactly the same because of the limitation of using only one contrast (i.e. protiated drug and surfactant dispersed in D2O) for the SANS data which does not allow us to establish whether any PEO is intruding into the core of the micelle. To establish this, it would be necessary to use additional contrasts using deuterated drug and surfactant (neither of which are readily available) and then to perform a simultaneous constrained model fit across all of the measured data. However, the SANS data shows that the ibuprofen-loaded TX-100 micelles are more asymmetric than the indomethacin-loaded TX-100 micelles, which in turn are more asymmetric than the micelles with no drug. When considering the MD simulations, and in particular the average ellipticity of the two daughter micelles which form after the unstable, indomethacin-saturated micelle divides in two, the results are reassuringly consistent with the SANS results. (It should be noted here that the measured SANS profiles, recorded over several minutes, will show the scattering arising from individual aggregates weighted according to their volume.) The ibuprofen-loaded micelle is more asymmetric than the average of the two daughter, indomethacin-loaded, micelles, which are both more asymmetric than the micelle in the absence of any drug.
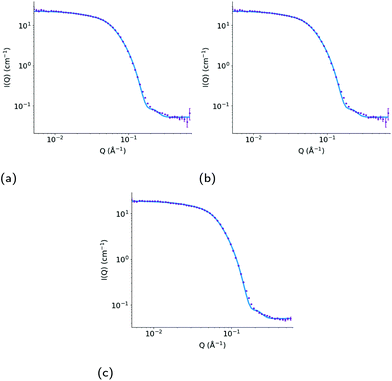 | ||
| Fig. 10 Measured SANS profile for (a) a 10 wt% dispersion of Triton X-100 in D2O and for a 7.5 wt% dispersion of Triton X-100 in D2O with added (b) ibuprofen and (c) indomethacin. Error bars show the standard errors on the measured data, and the model fitted curve in each plot (red) shows the calculated scattering intensity (I(Q), cm−1) as a function of momentum transfer (Q, Å−1), assuming the parameters presented in Table 2. | ||
| System | Core equatorial radius (Å) | Core axial ratio | Shell thickness (Å) | Overall micelle dimensions (Å) | χR 2 |
|---|---|---|---|---|---|
| No drug | 20.4 ± 0.1 | 3.8 ± 0.1 | 6.7 ± 0.1 | 168 × 84 | 1.9 |
| Ibuprofen | 20.1 ± 0.2 | 5.3 ± 0.1 | 8.1 ± 0.1 | 229 × 56 | 1.3 |
| Indomethacin | 18.6 ± 0.1 | 5.0 ± 0.1 | 8.8 ± 0.1 | 207 × 55 | 1.7 |
As the drug molecules are solubilised in the micelle, we find that the polar atoms are significantly dehydrated as they pass from the hydrated PEO rich corona of the micelle to the micelle's core. Once they have penetrated into the micelle core, the drug molecules reorient such that their carboxyl groups are rehydrated by the water in the aqueous environment. As the interactions between the indomethacin molecules were more extensive than those between ibuprofen molecules, this resulted in the formation of several larger aggregates within the core of the micelle than is found with ibuprofen. The formation of these larger aggregates of indomethacin within the drug-saturated micelle result in the destabilisation of the initial micelle such that it divides into two smaller micelles. Significantly, we do not observe any such destabilisation of the ibuprofen-loaded micelle which is considered to be due to the fact that the ibuprofen molecules do not interact as extensively with one another in the micelle and therefore do not aggregate as much. A previous experimental investigation of the solubilisation of various drug molecules in sodium lauryl sulfate found more loading of indomethacin than ibuprofen in their resulting micelles,65 which is the same trend we observe with our simulations. The results in our study provide details of the molecular scale mechanisms which lead to this trend and in our case the disruption of the micelles that form when solubilising the indomethacin. These results show that when designing drug-delivery vehicles the interactions between all of the components present (drugs and surfactant(s)) will play a significant role in the stability of the formulation and the size and shape of the nanoparticles that result.
Author contributions
HI – data curation, formal analysis, investigation, methodology, software, validation, visualisation, writing – original draft. NHR – supervision, writing–review & editing. DJB – conceptualisation, supervision, data curation, formal analysis, methodology, resources, writing–review & editing. MJL – conceptualisation, supervision, funding acquisition, resources, writing–review & editing. CDL – conceptualisation, supervision, funding acquisition, resources, project administration, writing–review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Through C. D. L.'s membership within the UK HPC Materials Chemistry Consortium, which is funded by the Office of Science and Technology through the EPSRC High End Computing Programme (grant no. EP/L000202, EP/R029431), the use of ARCHER, the UK National Supercomputing Service (https://www.archer.ac.uk) and the UK Materials and Molecular Modelling Hub (MMM Hub), which is partially funded by the EPSRC (EP/P020194/1, EP/T022213), was used for the molecular dynamics simulations presented in this work. A special thanks to “Tekeyan Trust London” whose funding has made H. I.'s research at KCL possible. N.H.R. is supported by a King's Prize Fellowship funded by a Wellcome Trust (https://wellcome.ac.uk/) Institutional Strategic Support Fund grant to King's College London (204823/Z/16/Z). Experiments at the ISIS Neutron and Muon Source were supported by beamtime allocation from STFC, and the SANS data are available at https://doi.org/10.5286/ISIS.E.RB1710299. This work benefited from the use of the SasView application, originally developed under National Science Foundation award DMR-0520547. SasView also contains acode developed with funding from the European Union's Horizon 2020 research and innovation program under the SINE2020 project, grant no. 654000.Notes and references
- V. Patravale, P. Dandekar and R. Jain, Nanoparticulate Drug Delivery, Woodhead Publishing, 2012, pp. 29–85 Search PubMed.
- R. H. Muller and C. M. Keck, J. Biotechnol., 2004, 113, 151–170 CrossRef CAS PubMed.
- A. Puri, K. Loomis, B. Smith, J. H. Lee, A. Yavlovich, E. Heldman and R. Blumenthal, Crit. Rev. Ther. Drug Carrier Syst., 2009, 26, 523–580 CrossRef CAS PubMed.
- K. Möller and T. Bein, Chem. Mater., 2019, 31, 4364–4378 CrossRef.
- V. Karthika, P. Kaleeswarran, K. Gopinath, A. Arumugam, M. Govindarajan, N. S. Alharbi, J. M. Khaled, M. N. Al-anbr and G. Benelli, Mater. Sci. Eng., C, 2018, 90, 589–601 CrossRef CAS PubMed.
- M. Medina-Sánchez, H. Xu and O. G. Schmidt, Ther. Delivery, 2018, 9, 303–316 CrossRef PubMed.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Chem. Rev., 2015, 207, 18–30 CAS.
- K. Cho, X. Wang, S. Nie, Z. Chen and D. Shin, Clin. Cancer Res., 2008, 14, 1310–1316 CrossRef CAS PubMed.
- F. U. Din, W. Aman, I. Ullah, O. S. Qureshi, O. Mustapha, S. Shafique and A. Zeb, Int. J. Nanomed., 2017, 12, 7291–7309 CrossRef PubMed.
- L.-L. Li, H.-W. An, B. Peng, R. Zheng and H. Wang, Mater. Horiz., 2019, 6, 1794–1811 RSC.
- G. S. Kwon and T. Okano, Pharm. Res., 1999, 16, 597–600 CrossRef CAS PubMed.
- C. J. Drummond and C. Fong, Curr. Opin. Colloid Interface Sci., 1999, 4, 449–456 CrossRef CAS.
- Y. Jin, L. Tong, P. Ai, M. Li and X. Hou, Int. J. Pharm., 2006, 309, 199–207 CrossRef CAS PubMed.
- S. Zhang, Nat. Biotechnol., 2003, 21, 1171–1178 CrossRef CAS PubMed.
- I. Marzuoli, C. H. B. Cruz, C. D. Lorenz and F. Fraternali, Nanoscale, 2021, 13, 10342–10355 RSC.
- I. E. Kepiro, I. Marzuoli, K. Hammond, X. Ba, H. Lewis, M. Shaw, S. B. Gunnoo, E. D. Santis, U. Lapriska, S. Pagliara, M. A. Holmes, C. D. Lorenz, B. W. Hoogenboom, F. Fraternali and M. G. Ryadnov, ACS Nano, 2020, 14, 1609–1622 CrossRef CAS PubMed.
- X. Gao and H. Matsui, Adv. Mater., 2005, 17, 2037–2050 CrossRef CAS PubMed.
- V. Linko, A. Ora and M. A. Kostiainen, Trends Biotechnol., 2015, 33, 586–594 CrossRef CAS PubMed.
- I. F. Uchegbu and S. P. Vyas, Int. J. Pharm., 1998, 172, 33–70 CrossRef CAS.
- M. J. Lawrence, Eur. J. Drug Metab. Pharmacokinet., 1994, 19, 257–269 CrossRef CAS PubMed.
- A. Makhlouf, I. Hajdu and I. Badea, Organic Materials as Smart Nanocarriers for Drug Delivery, William Andrew Publishing, 2018, pp. 561–600 Search PubMed.
- A. Rajagopal, A. C. Pant, S. M. Simon and Y. Chen, Cancer Res., 2002, 62, 391–396 CAS.
- D. Hipfner, S. Gauldie, R. Deeley and S. Cole, Cancer Res., 1994, 54, 5788–5792 CAS.
- A. Solomonov, Y. Marfin, E. Rumyantsev, E. Ragozin, T. Shekhter, G. Gellerman, A. Tesler, F. Muench, A. Kumagai and A. Miyawaki, Mater. Sci. Eng., C, 2019, 99, 794–804 CrossRef CAS PubMed.
- A. Mukhija and N. Kishore, J. Mol. Liq., 2018, 265, 1–15 CrossRef CAS.
- A. Das and R. K. Mitra, Colloid Polym. Sci., 2013, 292, 635–644 CrossRef.
- A. Kalatizaki, M. Poulopoulou, A. Xeankis and V. Papadimitriou, Colloids Surf., A, 2014, 442, 80–87 CrossRef.
- E. A. Dennis, Arch. Biochem. Biophys., 1974, 165, 764–773 CrossRef CAS PubMed.
- H. H. Paradies, J. Phys. Chem., 1980, 84, 599–607 CrossRef CAS.
- R. J. Robson and E. A. Dennis, J. Phys. Chem., 1977, 81, 1075–1078 CrossRef CAS.
- L. M. Kushner and W. D. Hubbard, J. Phys. Chem., 1954, 58, 1163–1167 CrossRef CAS.
- K. Streletzky and G. D. J. Phillies, Langmuir, 1995, 11, 42–47 CrossRef CAS.
- A. S. Sadaghiani and A. Khan, Langmuir, 1991, 7, 898–904 CrossRef CAS.
- A. Mandal, S. Ray, A. Biswas and S. Moulik, J. Phys. Chem., 1980, 84, 856–859 CrossRef CAS.
- F. Podo, A. Ray and G. Nemethy, J. Am. Chem. Soc., 1973, 85, 6164–6171 CrossRef.
- D. Yordanova, I. Smirnova and S. Jakobtorweihen, J. Chem. Theory Comput., 2015, 11, 2329–2340 CrossRef CAS PubMed.
- A. De Nicola, T. Kawakatsu, C. Rosano, M. Celino, M. Rocco and G. Milano, J. Chem. Theory Comput., 2015, 11, 4959–4971 CrossRef CAS PubMed.
- W. Murakami, A. D. Nicola, Y. Oya, J.-I. Takimoto, M. Celino, T. Kawakatsu and G. Milano, ACS Appl. Nano Mater., 2021, 4, 4552–4561 CrossRef CAS.
- T. G. Kantor, Ann. Intern. Med., 1979, 91, 877–882 CrossRef CAS PubMed.
- N. M. Davies, Clin. Pharmacokinet., 1998, 34, 101–154 CrossRef CAS PubMed.
- G. Alván, M. Orme, L. Bertilsson, R. Ekstrand and L. Palmér, Clin. Pharmacol. Ther., 1975, 18, 364–373 CrossRef PubMed.
- National Center for Biotechnology Information (2021). PubChem Compound Summary for CID 3715, Indomethacin. Retrieved April 28, 2021 from https://pubchem.ncbi.nlm.nih.gov/compound/Indomethacin .
- U. Patel, N. Dharaiya and P. Bahadur, J. Mol. Liq., 2016, 216, 156–163 CrossRef CAS.
- N. Dharaiya and P. Bahadur, Colloids Surf., A, 2012, 410, 81–90 CrossRef CAS.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- E. Lindahl, Solving Software Challenges for Exascale, 2015, pp. 3–27 Search PubMed.
- S. Pronk, S. Páll, R. Schulz, P. Larsson, P. Bjelkmar, R. Apostolov, M. R. Shirts, J. C. Smith, P. M. Kasson, D. van der Spoel, B. Hess and E. Lindahl, Bioinformatics, 2013, 29, 845–854 CrossRef CAS PubMed.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. de Groot, H. Grubmüller and A. D. MacKerell, Nat. Methods, 2017, 14, 71–73 CrossRef CAS PubMed.
- K. Vanommeslaeghe, E. Hatcher, C. Acharya, S. Kundu, S. Zhong, J. Shim, E. Darian, O. Guvench, P. Lopes, I. Vorobyov and A. D. Mackerell, J. Comput. Chem., 2010, 31, 671–690 CAS.
- L. Martínez, R. Andrade, E. G. Birgin and J. M. Martínez, J. Comput. Chem., 2009, 30, 2157–2164 CrossRef PubMed.
- B. Hess, H. Bekker, H. J. C. Berendsen and J. G. E. M. Fraaije, J. Comput. Chem., 1997, 18, 1463–1472 CrossRef CAS.
- R. J. Gowers, M. Linke, J. Barnoud, T. J. E. Reddy, M. N. Melo, S. L. Seyler, J. Domański, D. L. Dotson, S. Buchoux, I. M. Kenney and O. Beckstein, Proceedings of the 15th Python in Science Conference, 2016, pp. 98–105.
- N. Michaud-Agrawal, E. J. Denning, T. B. Woolf and O. Beckstein, J. Comput. Chem., 2011, 32, 2319–2327 CrossRef CAS PubMed.
- B. Lee and F. M. Richards, J. Mol. Biol., 1971, 55, 379–400 CrossRef CAS PubMed.
- S. Mitternacht, F1000Research, 2016, 5, 189 Search PubMed.
- M. Doucet, J. H. Cho, G. Aline, J. Bakker, W. Bouwman, P. Butler, K. Campbell, M. Gonzales, R. Heenan, A. Jackson, P. Juhas, S. King, P. Kienzle, J. Krzywon, A. Markvardsen, T. Nielsen, L. O'Driscoll, W. Potrzebowski, R. F. Leal, T. Richter, P. Rozycko, T. Snow and A. Washington, SasView version 4.2, 2018 Search PubMed.
- D. T. Allen and C. D. Lorenz, J. Mol. Model., 2017, 23, 1–11 CrossRef PubMed.
- D. T. Allen, Y. Saaka, M. J. Lawrence and C. D. Lorenz, J. Phys. Chem. B, 2014, 118, 13192–13201 CrossRef CAS PubMed.
- R. M. Ziolek, P. Smith, D. L. Pink, C. A. Dreiss and C. D. Lorenz, Macromolecules, 2021, 54, 3755–3768 CrossRef CAS.
- C. D. Lorenz, C.-M. Hsieh, C. A. Dreiss and M. J. Lawrence, Langmuir, 2011, 27, 546–553 CrossRef CAS PubMed.
- P. H. Elworthy and M. S. Patel, J. Pharm. Pharmacol., 1982, 34, 543–546 CrossRef CAS PubMed.
- T. Arnarson and P. H. Elworthy, J. Pharm. Pharmacol., 1982, 34, 87–89 CrossRef CAS PubMed.
- P. H. Elworthy and M. S. Patel, J. Pharm. Pharmacol., 1984, 36, 116–117 CrossRef CAS PubMed.
- P. H. Elworthy and M. S. Patel, J. Pharm. Pharmacol., 1984, 36, 565–568 CrossRef CAS PubMed.
- M. N. Alizadeh, A. Shayanfar and A. Jouyban, J. Mol. Liq., 2018, 268, 410–414 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Packmol input for building initial micelle; radial distribution functions for water around polar atoms in ibuprofen and indomethacin; contact maps of indomethacin and ibuprofen in their crystalline structures; snapshots of representative configurations showing the interactions between indomethacin and ibuprofen encapsulated within Triton X-100 micelles and between the Triton X-100 tails; plots showing the amount of water found in the core of the different micelles. See https://doi.org/10.1039/d1nr07936k |
| This journal is © The Royal Society of Chemistry 2022 |