Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluorescent styrene maleic acid copolymers to facilitate membrane protein studies in lipid nanodiscs†
George M.
Neville
 ,
Karen J.
Edler
,
Karen J.
Edler
 and
Gareth J.
Price‡
and
Gareth J.
Price‡
 *
*
Department of Chemistry, University of Bath, Bath, BA2 7AY, UK
First published on 17th March 2022
Abstract
Fluorescently-labelled variants of poly(styrene-co-maleic acid), SMA, have been synthesised by RAFT copolymerisation. We show that low ratios of vinyl fluorophores, analogous to styrene, can be successfully incorporated during polymerisation without detriment to nanodisc formation upon interaction with lipids. These novel copolymers are capable of encapuslating lipids and the model membrane protein, gramicidin, and hence have the potential to be applied in fluorescence-based biological studies. To demonstrate this, energy transfer is used to probe polymer–protein interactions in nanodiscs. The copolymers may also be used to monitor nanodisc self assembly by exploiting aggregation-caused-quenching (ACQ).
Essential to the processes that mediate life, membrane proteins (MPs) are of keen interest across biological and medical research. Thought to account for over 30% of the human proteome,1 MPs are now the target for an estimated 50% of all pharmaceuticals.2,3 However, components of cell membranes raise challenges during the purification, stabilisation and analysis of MPs in solution.4–6 Despite advances in this area, such as the development of cryogenic electron microscopy, the percentage of MP structures in the protein structural database remains notoriously low, at less than 2%.5,7 This disparity between usefulness and understanding has been attributed to the heavy dependence of MP structure, and hence activity, on the native membrane environment. Without the surrounding lipids, MPs become liable to denaturation. This is further exacerbated by a reliance on harsh detergents for extraction, where soft membrane interactions become disrupted.4,6–8 Hence, the study of MPs has previously been restricted to those known to be extractable in a folded, functional state.
Copolymers comprising styrene and maleic acid (Fig. 1a) promise to replace detergent methodologies for MP study. They can directly capture MPs from cells whilst retaining native lipid interactions present in the immediate bilayer.3,9 This is achieved through the self-assembly of a SMALP (styrene maleic acid lipid particle) nanodisc (Fig. 1b). The amphiphilic copolymer annulus can both stabilise hydrophobic interactions between the phospholipid bilayer and MPs, as well as provide sufficient hydrophilicity to disperse the SMALPs in aqueous media. Reported to both better conserve MP dynamics and improve spectral quality,3,9 SMA has been rapidly adopted by biologists and chemists alike. While most studies have been performed using commercially-available materials, a range of structural and compositional variants have become available using controlled radical methods, such as RAFT.10–12 The copolymer features necessary for nanodisc formation are now sufficiently well understood that novel materials can be feasibly designed and optimised for particular conditions or applications.
Fluorescence is widely used in biological assays and has been employed in the study of MPs.13–15 Hence, it is of interest to synthesise fluorescently-labelled SMA to enable such studies, either in conjunction with existing protein tags, or without need for complex protein modification. Fluorescence correlation spectroscopy was recently used to monitor coupling between fluorescent ligands and SMALP encapsulated G-coupled protein receptors,16 key targets for drug discovery.2 Similar experiments could be envisaged using fluorescently-labelled SMA.
Such a polymer could also potentially be used to monitor nanodisc self-assembly. Previously, fluorescent SMA variants have been prepared by cysteamine modification of commercial anhydride precursors of SMA, allowing attachment of biologically-relevant reagents, such as biotin or other fluorophores.17 Observation of Förster resonance energy transfer (FRET) in such copolymers demonstrated that nanodiscs contain multiple copolymer chains.18 However, these strategies have exclusively relied on post-polymerisation side-chain extension, which places restrictions on reaction control.19 Moreover, commercial SMA is inherently polydisperse (Table S2†), and while useful for biological research, precise control over copolymer structure is needed if the mechanistic implications of modification are to be investigated.
SMALP formation is governed by an intricate balance between hydrophilic and hydrophobic interactions, steric packing, electrostatics and surface tension,10,12,20 so it is likely that this would be at least somewhat perturbed by the introduction of large hydrophobic side chains. Indeed, it has been shown that extending side chains by even a single carbon atom, from methyl to ethyl, supresses detergency behaviour, lowers pH tolerance through loss of inter-chain repulsion, and generally increases nanodisc radii.19,21 By instead incorporating fluorophores directly into the feed of RAFT polymerisations, these challenges may be alleviated, and more effective polymers rationally designed.
In this work, aromatic fluorophores, either 9-vinyl anthracene (9-VA) or 1-pyrenemethyl acrylate (PY)22 (Fig. 1a) were directly copolymerised at low levels (0.01% wt feed) with styrene (Sty) and maleic anhydride (MAnh) by RAFT,23 before hydrolysis,24 to yield fluorescently-labelled SMA (SMA-VA0.01 & SMA-Py0.01, respectively). Full synthetic details and data, including isolated yields, are in the ESI (Table S2†). These moieties were chosen as to have minimal impact on both RAFT polymerisations as well as eventual nanodisc formation, and at 0.01% wt incorporation, there will be less than one fluorescent unit per chain. To both assess the effects of fluorophore incorporation and explore higher levels of functionalisation, a copolymer with a higher 9-VA content (0.1% wt), SMA-VA0.1, was also prepared.
All copolymers synthesised had a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 styrene
1 styrene![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) maleic anhydride ratio (Fig. S2†) with comparable molecular weights (Mn = 3–5 kDa) (Table S2†). As expected, all those prepared by RAFT copolymerisation had a lower PDI (1.2–1.3) compared with the commercial free-radical SMA 2000 (1.8). Unlike commercial SMA, RAFT-made SMA with this composition (2
maleic anhydride ratio (Fig. S2†) with comparable molecular weights (Mn = 3–5 kDa) (Table S2†). As expected, all those prepared by RAFT copolymerisation had a lower PDI (1.2–1.3) compared with the commercial free-radical SMA 2000 (1.8). Unlike commercial SMA, RAFT-made SMA with this composition (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) is a diblock copolymer comprising an alternating Sty-MA block with the excess Sty forming a homoblock ‘tail’ (Fig. 1a).25,26 This is confirmed through 13C NMR (Fig. S3†).27,28 Given the low levels and similar chemical nature of the fluorophores used, it can be assumed that all RAFT-made copolymers here, given their similar degrees of polymerisation, also present comparable monomer sequences (Table S2†).
1) is a diblock copolymer comprising an alternating Sty-MA block with the excess Sty forming a homoblock ‘tail’ (Fig. 1a).25,26 This is confirmed through 13C NMR (Fig. S3†).27,28 Given the low levels and similar chemical nature of the fluorophores used, it can be assumed that all RAFT-made copolymers here, given their similar degrees of polymerisation, also present comparable monomer sequences (Table S2†).
Upon incubation, all copolymers caused turbid lipid vesicle suspensions to clear, indicating the spontaneous stabilisation of lipid nanoparticles less than 100 nm in diameter, as confirmed by dynamic light scattering (DLS) (Fig. 2a) and small angle X-ray scattering (SAXS) (Fig. S6†). Formation of these structures occurred under the usual conditions for SMALP preparation (25 °C, pH = 8.0), and were able to incorporate the model MP, gramicidin, using 1,2-dimystoyl-sn-glycero-3-phosphocholine (DMPC) as the lipid.29 SAXS data for these structures fit parameters using a core shell bicelle model (Fig. S5†),30,31 confirming typical nanodisc morphologies (Table S4†). Although all of our novel copolymers formed nanodiscs that were significantly larger than with the commercial variant, the diameters were comparable with those from SMA R, a non-fluorescent, similarly prepared, RAFT-made variant. Using RAFT-made SMA typically results in larger diameter SMALPs as previously reported,10,26 and hence these results show that low levels of fluorophores do not significantly influence nanodisc size. However, it is notable that, SMA-VA0.1, containing the highest fluorophore content, formed the largest and most disperse nanodiscs, with or without gramicidin. This may be a result of the increased hydrophobicity of the copolymer as noted elsewhere.12
A key difference between SMA 2000 and RAFT-made SMA concerns copolymer aggregation in solution. SMA 2000 has a random structure prepared by starved-feed free radical copolymerisation. Hence, at this concentration it does not form higher-order aggregates in solution. The small structures in Fig. 2b are predominantly isolated single chains whose diameters can be well predicted from a Gaussian coil model (∼0.6 nm).
By contrast, the block architecture of the RAFT copolymers leads to the formation of aggregates in aqueous suspensions of either the polymer alone (Fig. 2b) or when lipid is added to form nanodiscs (Table S4†). A more detailed structural investigation of these aggregates is ongoing, but as others have suggested,32–34 we believe them to be assembled from multiple chains, and that the reduction of size and polydispersity upon heating (Fig. 2b) represents the partitioning of hydrophobic groups into a Sty core, protected from solvent by a Sty-MA shell. Given that these aggregates are present at concentrations relevant for nanodisc formation, it is likely they must first dissociate prior to interaction with lipids, and hence are of interest in discerning the origins of behavioural differences. Again, SMA-VA0.1 is seen to form the largest and most polydisperse aggregates at operational temperature, further indicating a deviation from typical SMA behaviour.35
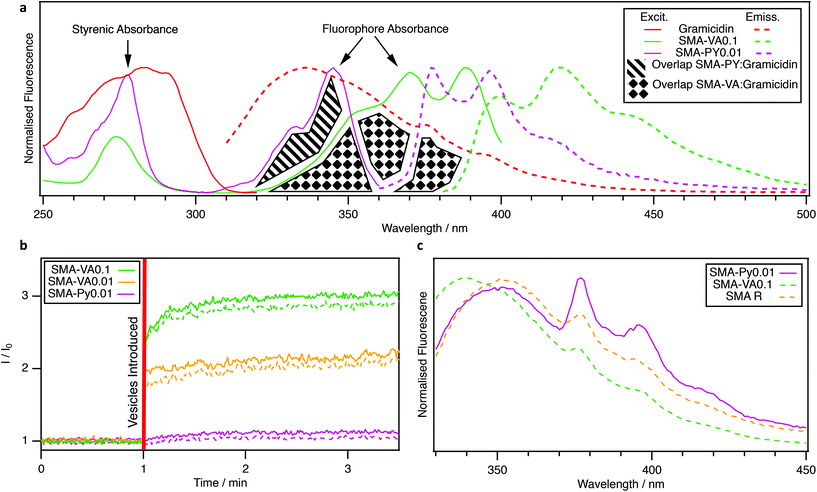
The excitation and emission spectra of the fluorescently-labelled copolymers are presented in Fig. 3a. Although the low ratios of incorporation, and similarity of the fluorophores to styrene, made their presence challenging to detect by NMR or IR spectroscopies (Fig. S1 and 2†), it was possible to observe fluorophore emission by exciting at wavelengths where only styrene absorbs (Fig. S7a and b†), indicative of close proximity. Moreover, the extensive purification undertaken during hydrolysis gives confidence that the 9-VA and PY units were successfully incorporated into copolymer chains. As expected, SMA R, synthesised in the absence of fluorophores, had no fluorescence emission at these wavelengths (Fig. S7c†).
As spectra have been normalised, the relative absorbance arising from fluorophore and styrene units are proportional to their concentration ratio within the chain. As the styrene content between SMA-VA0.1 and SMA-PY0.01 are similar, the lower ratio of styrene to fluorophore absorbance for SMA-VA0.1 (Fig. 3a), is indicative of a higher fluorophore loading.
It is important to note that these spectra were recorded at very low copolymer concentrations (∼6 × 10−4% wt), far below that used for nanodisc formation (1.65% wt) and below that at which aggregates form. At higher concentrations (1.65% wt), suspensions of polymer-only aggregates presented significantly lower fluorescent emission which increased upon interaction with lipids (Fig. 3b and Fig. S8†). Anthracene and pyrene both undergo aggregation-caused-quenching (ACQ),36,37 through pi-conjugated stacking interactions driven by hydrophobicity in polar solvents. This ground state interaction quenches fluorescence and can be enhanced by incorporation into hydrophobic polymers.38 Although solvatochromic effects were also considered, we prefer ACQ as an explanation of the results here. ACQ, whilst typically a challenge to the design of polymers for biomedical application,39 here presents an opportunity to use fluorescently-labelled RAFT-SMA to monitor the transition from aggregated polymer to nanodisc annulus over time (Fig. 3b), informing the discussion of how aggregates might mediate nanodisc formation.
The potential to study interactions occurring between the copolymer annulus and MPs was also explored using SMALPs encapsulating gramicidin. Gramicidin contains fluorescent tryptophan residues, the emission of which overlaps with the absorbance of the fluorophores (Fig. 3a). Hence, FRET may be induced within nanodiscs over distances less than about 10 nm. To evaluate this, suspensions of gramicidin-encapsulated SMALPs were excited at 295 nm, a wavelength where excitation of pyrene and the polymers is negligible. The resulting emission was normalised so that the intensity ratios between peaks could be compared.40 FRET was evaluated for all copolymers (Fig. 3c) but was only found to occur when using SMA-PY0.01. Comparing the peak intensities at 340 and 375 nm, gramicidin typically has an emission ratio around 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 respectively, also observed when the non-fluorescent copolymer, SMA R was used. In SMA-Py0.01 SMALPs this ratio was close to 1
1 respectively, also observed when the non-fluorescent copolymer, SMA R was used. In SMA-Py0.01 SMALPs this ratio was close to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, as gramicidin emission is lost to that of PY (Fig. 3c). FRET was not observed using the 9-VA containing copolymers. However, the nanodiscs formed by these copolymers were much larger (Fig. 2a) and hence were likely too wide (>10 nm radius) to produce the required proximity between the copolymer annulus and gramicidin. Nonetheless, these 9-VA-containing copolymers may be useful in facilitating studies at alternate ranges of wavelength. In future, it could also be possible to use this technique to precisely probe interactions between encapsulated MPs with specific locations in the copolymer chain, for example, by using a fluorescently-labelled initiator and/or RAFT agent.
1, as gramicidin emission is lost to that of PY (Fig. 3c). FRET was not observed using the 9-VA containing copolymers. However, the nanodiscs formed by these copolymers were much larger (Fig. 2a) and hence were likely too wide (>10 nm radius) to produce the required proximity between the copolymer annulus and gramicidin. Nonetheless, these 9-VA-containing copolymers may be useful in facilitating studies at alternate ranges of wavelength. In future, it could also be possible to use this technique to precisely probe interactions between encapsulated MPs with specific locations in the copolymer chain, for example, by using a fluorescently-labelled initiator and/or RAFT agent.
Conclusions
RAFT copolymerisation has been successfully employed to produce fluorescently-labelled SMA copolymers which are capable of forming lipid nanodiscs. Aromatic fluorophores were incorporated in the reaction feed up to 0.1% wt with little effect upon copolymer behaviour. However, the overall increased hydrophobicity of these copolymers did appear to induce the formation of larger, more disperse, aggregates and nanodiscs. Hence, increasing the fluorophore content beyond this should first include evaluation of possible implications to nanodisc self-assembly. Able to encapsulate the model MP, gramicidin, the potential usefulness of these novel copolymers as research tools has been illustrated. To our knowledge, this is the first report of FRET between a SMALP-encapsulated MP and the copolymer annulus, providing a new avenue for study into the extent of polymer–protein interactions in these systems. Although this work has featured a single pH and salt concentration, the method is potentially applicable across a wide range of conditions, including with other polymer–lipid combinations. With development, there is the opportunity to exploit these novel capabilities to expand the range of possible experiments and produce new insights into SMALP nanodisc formation and the interactions of MPs within these environments.Author contributions
GN: investigation, writing – original draft, writing–review & editing; KJE: conceptualization, funding acquisition, supervision, writing–review & editing; GJP: conceptualization, methodology, supervision, writing–review & editing.Conflicts of interest
There are no conflicts to declare.Data availability
Data supporting the results in this paper are available through the University of Bath Research Data Archive. https://doi.org/10.15125/BATH-01123.Acknowledgements
This work was supported by the Engineering and Physical Sciences Research Council EP/L016354/1.Notes and references
- R. M. Bill, P. J. Henderson, S. Iwata, E. R. Kunji, H. Michel, R. Neutze, S. Newstead, B. Poolman, C. G. Tate and H. Vogel, Nat. Biotechnol., 2011, 29, 335–340 CrossRef CAS PubMed.
- G.-M. Hu, T.-L. Mai and C.-M. Chen, Sci. Rep., 2017, 7, 15495 CrossRef PubMed.
- T. J. Knowles, R. Finka, C. Smith, Y.-P. Lin, T. Dafforn and M. Overduin, J. Am. Chem. Soc., 2009, 131, 7484–7485 CrossRef CAS PubMed.
- A. M. Seddon, P. Curnow and P. J. Booth, Biochim. Biophys. Acta, Biomembr., 2004, 1666, 105–117 CrossRef CAS PubMed.
- Z. Stroud, S. C. L. Hall and T. R. Dafforn, Methods, 2018, 147, 106–117 CrossRef CAS PubMed.
- S. C. Lee, T. J. Knowles, V. L. Postis, M. Jamshad, R. A. Parslow, Y. P. Lin, A. Goldman, P. Sridhar, M. Overduin, S. P. Muench and T. R. Dafforn, Nat. Protoc., 2016, 11, 1149–1162 CrossRef CAS PubMed.
- J. M. Dörr, S. Scheidelaar, M. C. Koorengevel, J. J. Dominguez, M. Schäfer, C. A. van Walree and J. A. Killian, Eur. Biophys. J., 2016, 45, 3–21 CrossRef PubMed.
- D. E. Otzen, Biophys. J., 2002, 83, 2219–2230 CrossRef CAS PubMed.
- H. E. Autzen, D. Julius and Y. Cheng, Curr. Opin. Struct. Biol., 2019, 58, 259–268 CrossRef CAS PubMed.
- S. C. L. Hall, C. Tognoloni, G. J. Price, B. Klumperman, K. J. Edler, T. R. Dafforn and T. Arnold, Biomacromolecules, 2018, 19, 761–772 CrossRef CAS PubMed.
- S. C. L. Hall, L. A. Clifton, C. Tognoloni, K. A. Morrison, T. J. Knowles, C. J. Kinane, T. R. Dafforn, K. J. Edler and T. Arnold, J. Colloid Interface Sci., 2020, 574, 272–284 CrossRef CAS PubMed.
- A. F. Craig, E. E. Clark, I. D. Sahu, R. Zhang, N. D. Frantz, M. S. Al-Abdul-Wahid, C. Dabney-Smith, D. Konkolewicz and G. A. Lorigan, Biochim. Biophys. Acta, 2016, 1858, 2931–2939 CrossRef CAS PubMed.
- J.-M. Swiecicki, J. T. Santana and B. Imperiali, Cell Chem. Biol., 2020, 27, 245–251 CrossRef CAS PubMed.
- H. Raghuraman, S. Chatterjee and A. Das, Front. Mol. Biosci., 2019, 6 Search PubMed.
- M. S. Rana, X. Wang and A. Banerjee, Biochemistry, 2018, 57, 6741–6751 CrossRef CAS PubMed.
- R. L. Grime, J. Goulding, R. Uddin, L. A. Stoddart, S. J. Hill, D. R. Poyner, S. J. Briddon and M. Wheatley, Nanoscale, 2020, 12, 11518–11525 RSC.
- S. Lindhoud, V. Carvalho, J. W. Pronk and M.-E. Aubin-Tam, Biomacromolecules, 2016, 17, 1516–1522 CrossRef CAS PubMed.
- V. Schmidt and J. N. Sturgis, Biochim. Biophys. Acta, Biomembr., 2018, 1860, 777–783 CrossRef CAS PubMed.
- M. Esmaili, C. Acevedo-Morantes, H. Wille and M. Overduin, Biochim. Biophys. Acta, Biomembr., 2020, 1862, 183360 CrossRef CAS PubMed.
- S. Scheidelaar, M. C. Koorengevel, J. D. Pardo, J. D. Meeldijk, E. Breukink and J. A. Killian, Biophys. J., 2015, 108, 279–290 CrossRef CAS PubMed.
- K. M. Burridge, B. D. Harding, I. D. Sahu, M. M. Kearns, R. B. Stowe, M. T. Dolan, R. E. Edelmann, C. Dabney-Smith, R. C. Page, D. Konkolewicz and G. A. Lorigan, Biomacromolecules, 2020, 21, 1274–1284 CrossRef CAS PubMed.
- X. Lou, R. Daussin, S. Cuenot, A.-S. Duwez, C. Pagnoulle, C. Detrembleur, C. Bailly and R. Jérôme, Chem. Mater., 2004, 16, 4005–4011 CrossRef CAS.
- S. Harrisson and K. L. Wooley, Chem. Commun., 2005, 3259–3261, 10.1039/B504313A.
- S. C. L. Hall, C. Tognoloni, J. Charlton, É. C. Bragginton, A. J. Rothnie, P. Sridhar, M. Wheatley, T. J. Knowles, T. Arnold, K. J. Edler and T. R. Dafforn, Nanoscale, 2018, 10, 10609–10619 RSC.
- R. D. Cunningham, A. H. Kopf, B. O. W. Elenbaas, B. B. P. Staal, R. Pfukwa, J. A. Killian and B. Klumperman, Biomacromolecules, 2020, 21, 3287–3300 CrossRef CAS PubMed.
- A. A. A. Smith, H. E. Autzen, T. Laursen, V. Wu, M. Yen, A. Hall, S. D. Hansen, Y. Cheng and T. Xu, Biomacromolecules, 2017, 18, 3706–3713 CrossRef CAS PubMed.
- P. F. Barron, D. J. T. Hill, J. H. O'Donnell and P. W. O'Sullivan, Macromolecules, 1984, 17, 1967–1972 CrossRef CAS.
- J. Luo, D. Cheng, M. Li, M. Xin, W. Sun and W. Xiao, Adv. Polym. Technol., 2020, 3695234, DOI:10.1155/2020/3695234.
- S. S. Rawat, D. A. Kelkar and A. Chattopadhyay, Biophys. J., 2004, 87, 831–843 CrossRef CAS PubMed.
- M. Jamshad, V. Grimard, I. Idini, T. J. Knowles, M. R. Dowle, N. Schofield, P. Sridhar, Y. Lin, R. Finka, M. Wheatley, O. R. T. Thomas, R. E. Palmer, M. Overduin, C. Govaerts, J.-M. Ruysschaert, K. J. Edler and T. R. Dafforn, Nano Res., 2015, 8, 774–789 CrossRef CAS PubMed.
- M. B. Smith, D. J. McGillivray, J. Genzer, M. Lösche and P. K. Kilpatrick, Soft Matter, 2010, 6, 862–865 RSC.
- N. G. Brady, S. Qian and B. D. Bruce, Eur. Polym. J., 2019, 111, 178–184 CrossRef CAS.
- P. S. Orekhov, M. E. Bozdaganyan, N. Voskoboynikova, A. Y. Mulkidjanian, H.-J. Steinhoff and K. V. Shaitan, Langmuir, 2019, 35, 3748–3758 CrossRef CAS PubMed.
- J. Claracq, S. F. C. R. Santos, J. Duhamel, C. Dumousseaux and J.-M. Corpart, Langmuir, 2002, 18, 3829–3835 CrossRef CAS.
- M. D. Phan, O. I. Korotych, N. G. Brady, M. M. Davis, S. K. Satija, J. F. Ankner and B. D. Bruce, Langmuir, 2020, 36, 3970–3980 CrossRef CAS PubMed.
- Y. Shen, H. Liu, S. Zhang, Y. Gao, B. Li, Y. Yan, Y. Hu, L. Zhao and B. Yang, J. Mater. Chem. C, 2017, 5, 10061–10067 RSC.
- X. Feng, Z. Xu, Z. Hu, C. Qi, D. Luo, X. Zhao, Z. Mu, C. Redshaw, J. W. Y. Lam, D. Ma and B. Z. Tang, J. Mater. Chem. C, 2019, 7, 2283–2290 RSC.
- J. Royakkers, A. Minotto, D. G. Congrave, W. Zeng, A. Hassan, A. Leventis, F. Cacialli and H. Bronstein, Chem. Mater., 2020, 32, 10140–10145 CrossRef CAS.
- X. Zhang, K. Wang, M. Liu, X. Zhang, L. Tao, Y. Chen and Y. Wei, Nanoscale, 2015, 7, 11486–11508 RSC.
- M. A. Soto-Arriaza, C. Olivares-Ortega and E. A. Lissi, J. Colloid Interface Sci., 2012, 385, 48–57 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Copolymer synthesis, copolymer characterisation, nanodisc formation and scattering, fluorescence spectra. See https://doi.org/10.1039/d1nr07230g |
| ‡ Present address: Department of Chemistry, Khalifa University, PO Box 127788, Abu Dhabi, UAE. E-mail: gareth.price@ku.ac.ae. |
| This journal is © The Royal Society of Chemistry 2022 |