Open Access Article
Open Access ArticleCryo-EM reconstructions of BMV-derived virus-like particles reveal assembly defects in the icosahedral lattice structure
Milosz
Ruszkowski
 a,
Aleksander
Strugala
a,
Paulina
Indyka
bc,
Guillaume
Tresset
d,
Marek
Figlerowicz
a and
Anna
Urbanowicz
a,
Aleksander
Strugala
a,
Paulina
Indyka
bc,
Guillaume
Tresset
d,
Marek
Figlerowicz
a and
Anna
Urbanowicz
 *a
*a
aInstitute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, 61-704 Poznan, Poland. E-mail: aniau@ibch.poznan.pl
bJagiellonian University, Solaris National Synchrotron Radiation Centre, Czerwone Maki 98, 30-392 Cracow, Poland
cJagiellonian University, Malopolska Centre of Biotechnology (MCB), 30-387 Cracow, Poland
dUniversité Paris-Saclay, CNRS, Laboratoire de Physique des Solides, 91405 Orsay, France
First published on 14th February 2022
Abstract
The increasing interest in virus-like particles (VLPs) has been reflected by the growing number of studies on their assembly and application. However, the formation of complete VLPs is a complex phenomenon, making it difficult to rationally design VLPs with desired features de novo. In this paper, we describe VLPs assembled in vitro from the recombinant capsid protein of brome mosaic virus (BMV). The analysis of VLPs was performed by Cryo-EM reconstructions and allowed us to visualize a few classes of VLPs, giving insight into the VLP self-assembly process. Apart from the mature icosahedral VLP practically identical with native virions, we describe putative VLP intermediates displaying non-icosahedral arrangements of capsomers, proposed to occur before the final disorder–order transition stage of icosahedral VLP assembly. Some of the described VLP classes show a lack of protein shell continuity, possibly resulting from too strong interaction with the cargo (in this case tRNA) with the capsid protein. We believe that our results are a useful prerequisite for the rational design of VLPs in the future and lead the way to the effective production of modified VLPs.
Introduction
Icosahedral plant viruses and virus-like particles (VLPs) composed of plant virus capsid proteins (CPs), have been proposed to serve as nanocontainers, biosensors, nanoreactors, and drug or gene delivery vehicles due to their nanoscale dimensions, simple yet rigid structures, self-assembly capability and empty interior.1,2 The increasing interest in VLP application has been reflected by the growing number of studies on their assembly and disassembly processes as well as on their cellular internalization, cargo capacity, or modifications of their external surface in order to direct them to target cells.3–7 Recently, VLPs composed of the native capsid proteins from plant viruses belonging to the bromovirus genus, i.e. brome mosaic virus (BMV) and cowpea chlorotic mottle virus (CCMV), have been shown to easily internalize fluorophores into breast tumor cells.8,9 Furthermore, BMV-derived VLPs loaded with small interfering RNA (siRNA) against green fluorescence protein (GFP) coding sequence efficiently silenced constitutive expression of gfp in breast tumor cells and showed no cytotoxicity.9 These results further underline the attractive potential of plant virus VLPs in therapy and diagnostic imaging.The formation of the complete VLP is a complex phenomenon, and the state-of-art is insufficient to rationally design VLPs with desired features. The kinetics of virion growth in the presence of RNA is usually faster than that of empty capsid assembly, indicating that the assembly pathways of empty and RNA-filled VLPs might be different.5,7,10,11 The assembly has been proposed to proceed either through the nucleation and growth or the en masse mechanism.12 The first pathway assumes the formation of a single nucleus followed by sequential binding of subunits to its edges, thus leading to non-symmetrical shell intermediates. In the second pathway, subunits bind to each other forming multiple nuclei, and the shell keeps growing in a disordered manner. In both scenarios, when enough subunits are incorporated, they undergo cooperative rearrangements to assemble into an ordered icosahedral capsid.5 The two pathways have been experimentally evidenced by time-resolved small-angle X-ray scattering (TR-SAXS) and by interferometric scattering microscopy for CCMV and MS2 bacteriophage capsids.7,13 Recently, thanks to the development of high-resolution experimental and computational techniques in the field of physical virology, the capsid assembly has been described in terms of conformational dynamics superseding previous static descriptions of viral properties.11 Accordingly, before the icosahedral protein shell self-assembly is accomplished, its non-symmetrical intermediate forms have to overcome multiple energy bariers.5,7,14 These barriers result from the line tension at the rim of the growing capsid and from the mechanical stress induced by the arrangement of pentameric and hexameric capsomers, which eventually have to comply with the final icosahedral symmetry.15,16 The application of small-angle X-ray scattering (SAXS), cryoelectron microscopy (Cryo-EM) and Monte Carlo simulations has revealed a disorder–order transition occurring during CCMV assembly in the presence of genomic RNA.5 Such a transition has also been seen during the assembly of empty capsids of hepatitis B virus (HBV).17 In the intermediate CCMV shells with disordered or misassembled structures, the distribution of pentamers does not satisfy the icosahedral restrictions, which impose twelve pentameric capsomers for the closure of the shell. As a consequence, the resulting elastic energy becomes prohibitively high. This leads to the dissociation of energetically-unfavorable capsomers and their subsequent reassembly (e.g., hexameric capsomer into pentameric one) in order to relax the mechanical stress and to allow the formation of perfect icosahedral shells. Thus, the phase diagram of viral shells, assembled either through the nucleation-growth or en masse pathways, is shaped by the elastic energy compared with the subunit–subunit and subunit–core energies and the chemical potential of free subunits. The elastic energy also depends on the mechanical properties of the shell, namely the stretching and bending rigidities and on the spontaneous curvature. As a rule of thumb, simulations showed that low elastic energies lead to irregular closed shapes with low symmetry, whereas high elastic energies yield incomplete and disordered structures.5
The native BMV capsids result from the assembly of 180 identical proteins with T = 3 symmetry that forms the icosahedral shell with a diameter of 28 nm.18 The N-terminal region of the capsid protein is highly basic and positively charged, which allows for the binding of four viral RNA molecules.19 Genomic RNA1 (3.2 kb) and RNA2 (2.8 kb) are each packaged separately into individual virions, whereas genomic RNA3 (2.1 kb) and the subgenomic RNA4 (0.8 kb) are co-packaged into the third virion.20 The three virions have indistinguishable capsid structures and almost identical overall mass densities.21,22 The recent Cryo-EM studies of native BMV virions have indicated that the genomic RNA (regardless of its type) forms an inner layer close to the capsid shell. In contrast, the interior of the virion is nearly empty. Interestingly, the BMV RNA is not associated with any particular structure but forms an ensemble of secondary or tertiary structures interacting with the protein shell.22
VLPs can assemble not only from native but also from recombinant CP (rCP), obtained via heterologous expression.23 Such an approach offers beneficial possibilities of genetic engineering of the rCP sequence to produce VLPs with desired features, such as amino acid side chains prone to chemical modifications, additional epitopes, or altered VLP stability.3,23 On the contrary, the genetic modification of the viruses produced in the native host is severely restricted due to the negative influence of the mutations on the virus life cycle and fitness.24,25 In this paper, we present the high-resolution Cryo-EM structures of VLPs obtained by assembly of the BMV rCP produced in E. coli. The VLP assembly described here was performed in the absence and in the presence of yeast tRNA. Previously, we showed that the latter can substitute the tRNA-like structure (TLS) of BMV genomic RNA, and can be efficiently incorporated into the capsid shell.23 We compared the structures elaborated by us with the structure of the native BMV virion.21,22 We also analyzed nearly 1.5 million of Cryo-EM particles to visualize a few—possibly intermediate—classes of VLPs, giving insight into the VLP self-assembly process. These putative intermediates present non-icosahedral arrangements of capsomers proposed to occur before the final disorder–order transition stage of icosahedral VLP assembly. In fact, we observed the non-symmetrical VLPs with forbidden pentamer–pentamer contacts, which may immediately precede the transitions to the icosahedral symmetry. Finally, we show that the formation of both non-symmetrical and icosahedral BMV-derived VLPs is possible even in the absence of RNA. This indicates the capability of VLP adjustment to the size of encapsidated cargo and is an important step towards using BMV-derived VLPs as a shuttle, e.g., in drug delivery.
Results and discussion
Icosahedral reconstructions of BMV-derived VLPs reveal a very conserved structure
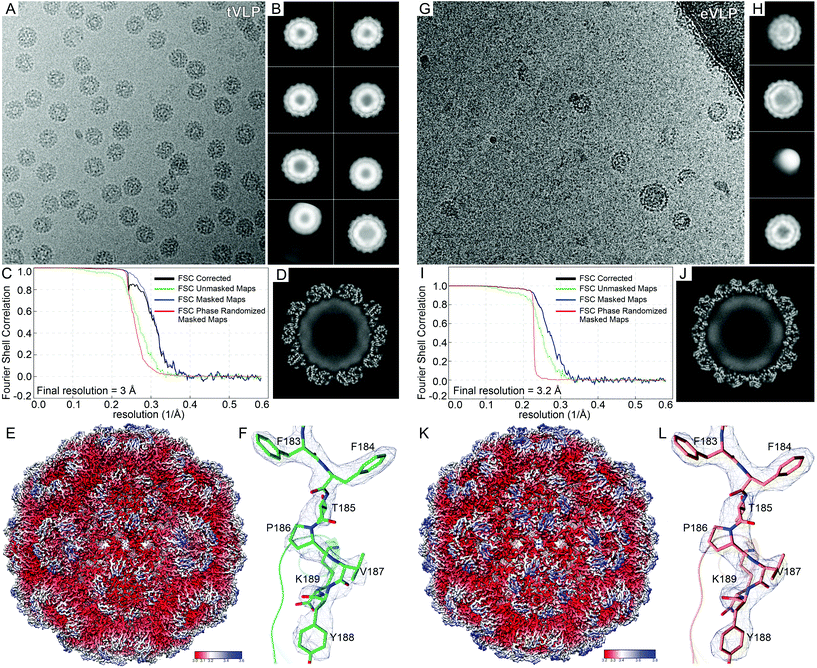
To date, twenty-four EM maps showing BMV reconstructions have been deposited in the Electron Microscopy Data Bank (EMDB) (accession no. EMD-21260, EMD-21261, and EMD-21279 – EMD-21300). However, only one atomic model is available in the Protein Data Bank (PDB ID 6VOC). The later model has been built based on the icosahedral, 3.1 Å EM map resulting from BMV isolated from N. benthamiana plants. Here we report another two atomic models of BMV-derived VLPs constituted in the presence of tRNA, named tVLP (3.0 Å map resolution, Fig. 1A–F), and empty VLP (eVLP) (3.2 Å map resolution, Fig. 1G–L). Quality of the EM maps allowed us to trace most of the protein chain unambiguously, including individual side chains (Fig. 1F and L). The atomic models contain residues 41/42–189 in the subunits forming pentameric capsomers (chain A), whereas N-termini of the subunits in hexameric capsomers (chains B and C) extend until residues 25/26. We also observed a layer of Cryo-EM density inside the VLPs that was impossible to interpret unambiguously. This density appears in both tVLP and eVLP (Fig. 1D and J, respectively) but is stronger in tVLP. Therefore it most likely corresponds to disordered N-terminal regions, possibly mixed with one or more tRNA molecules.The sequence of the presented VLPs contains three substitutions (R26G, L35I, R189K) with respect to the sequence in the 6VOC structure. We compared the structures to identify features that differ between VLPs assembled (i) in vivo, (ii) in vitro in the presence of tRNA, and (iii) in vitro in the absence of polyelectrolytes. Surprisingly, all three structures are remarkably similar despite different sample preparation protocols and sequences. The similarity is best reflected by pairwise structural comparison of individual chains (A–C, Fig. 2A) in the three structures. The root-mean-square deviations (RMSDs) between superposed Cα atoms in 6VOC and tVLP or eVLP are ∼0.4 Å (Table 1). The differences between tVLP and eVLP are even smaller, as the RMSD values are less than 0.25 Å. It is also important to note that those minor differences in single chains do not accumulate in the icosahedral assembly, i.e., no variations are observed even on opposite poles of the VLP. The diameter inside the VLP is approximately 190 Å (Fig. 2B), which makes the available “cargo” volume (inside the VLP) nearly 3.6 × 106 Å3. The outer VLP diameter is 280 Å, like that of native virions.
 | ||
| Fig. 2 Structure of VLP. (A) Comparison of the native BMV (PDB ID 6VOC, gray), tVLP (green) and eVLP (orange). (B) Dimensions of the tVLP EM maps, color-coded according to the distance from the center (bottom-right). | ||
| Chains | RMSD |
|---|---|
| 6VOC-A/tVLP-A | 0.374 |
| 6VOC-B/tVLP-B | 0.410 |
| 6VOC-C/tVLP-C | 0.376 |
| 6VOC-A/eVLP-A | 0.378 |
| 6VOC-B/eVLP-B | 0.391 |
| 6VOC-C/eVLP-C | 0.397 |
| eVLP-A/tVLP-A | 0.225 |
| eVLP-B/tVLP-B | 0.241 |
| eVLP-C/tVLP-C | 0.231 |
Malformed VLPs group into several non-icosahedral classes, giving insights into the capsid assembly
As described in the previous section, the icosahedral protein shell of BMV-derived VLP, regardless of whether it is empty or not, consists of 12 non-adjacent pentamers and 20 hexamers.21,22 The intermediate-assembly VLP forms may contain an altered number of capsomers, and we were able to reconstruct a few types of such forms in the course of our Cryo-EM studies. The 2D classification of the total pool of 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 759 particles picked from 10
759 particles picked from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 935 micrographs of eVLP preparation revealed that apart from the icosahedral eVLPs described in the previous chapter (7.3% of total particles), 6.9% of total particles represented non-icosahedral eVLPs, hereafter referred to as malformed eVLPs (meVLPs). These included two meVLP types: 2.7% of meVLPs type 1 and 3.95% of meVLPs type 2 (the remaining 0.25% was not applicable in the reconstruction). The remaining 85.8% of particles were recognized as smaller parts of the protein shell, too heterogeneous to be used for 3-D reconstruction (Table 2). Heterogeneity of VLPs assembled in vitro from the native bromoviral CP has also been observed by others.7,10
935 micrographs of eVLP preparation revealed that apart from the icosahedral eVLPs described in the previous chapter (7.3% of total particles), 6.9% of total particles represented non-icosahedral eVLPs, hereafter referred to as malformed eVLPs (meVLPs). These included two meVLP types: 2.7% of meVLPs type 1 and 3.95% of meVLPs type 2 (the remaining 0.25% was not applicable in the reconstruction). The remaining 85.8% of particles were recognized as smaller parts of the protein shell, too heterogeneous to be used for 3-D reconstruction (Table 2). Heterogeneity of VLPs assembled in vitro from the native bromoviral CP has also been observed by others.7,10
| eVLP (%) | tVLP (%) | |
|---|---|---|
| Icosahedral | 7.3 | 2 |
| Non-icosahedral | 6.9 | 89 |
| Parts of shell | 85.8 | 9 |
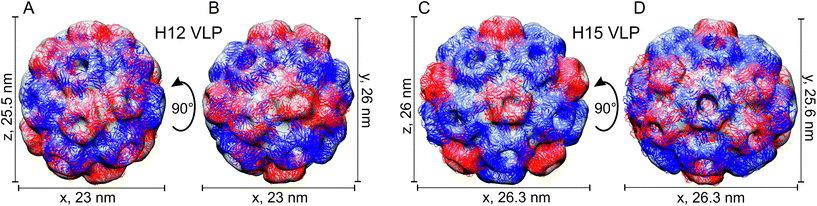
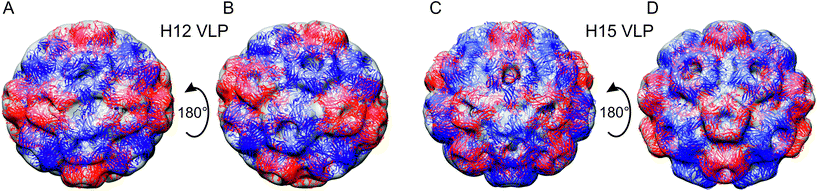
The asymmetric 3D reconstruction of meVLP type 1 revealed an elliptic structure consisting of 132 rCP subunits arranged in a closed non-icosahedral shell containing 12 pentameric and 12 hexameric capsomers (Fig. 3A and B). We called it H12 according to a nomenclature proposed by Bond et al., as per the amount of hexameric capsomers.4 The H12 VLP has a pair of pentamers surrounded by six hexamers forming a cup at both opposing polar ends, which connect by a central belt of eight pentamers. The H12 measures 23 × 26 × 25.5 nm (Fig. 3A and B). The asymmetric 3D reconstruction of meVLP type 2, named H15, resulted in the non-icosahedral VLP consisting a total of 150 rCP subunits forming 12 pentamers and 15 hexamers (Fig. 3C and D). In H15, the capsomers are arranged as follows: a cup at both poles is formed by one pentamer surrounded by five hexamers and both cups are divided by the central belt made of five pairs of pentamers, each pair being separated by one hexamer. The H15 measures 26.3 × 25.6 × 26 nm (Fig. 3C and D). A non-icosahedral BMV-derived VLP, similar to the observed H15 shell, has recently been described by Bond et al. but was assembled from the native BMV CP and a 52-nt long single-stranded DNA (ssDNA) oligonucleotide.4 This ssDNA-containing H15 VLP showed an identical capsomer arrangement but had slightly different dimensions, 27.1 × 25.9 nm. The second type of BMV-derived VLP loaded with the same ssDNA oligonucleotide described by Bond et al. and called H8 is composed of 12 pentamers and 8 hexamers.4 This suggests that during the BMV capsid assembly, other VLP types can be formed, apart from the mature icosahedral protein shell. All of them contain 12 pentamers and fewer than 20 hexamers, and they have been proposed to show pseudo-T = 2 symmetry.4,10 Similar to Bond et al., we observed the H15 shell, which appears stable in eVLP or with ssDNA oligonucleotides inside it. However, we detected H12 shells but not H8 shells as Bond et al. did.4 Most probably, H12 is stable enough in the absence of the polyelectrolyte, while the stabilization of the H8 shell requires a polyanion similar to ssDNA that can interact with CP through electrostatic interactions. The stabilization of H8 by the CP–ssDNA electrostatic interactions might compensate for the excess of elastic energy and curvature resulting from the high ratio of pentamers to hexamers in this type of shell. On the other hand, the lack of H12 shell with ssDNA oligonucleotides indicates that the cargo polyelectrolyte length or structure defines the type of formed pseudo-T = 2 VLPs. One can speculate that the formation of other VLP types with the number of hexamers different than already described by Bond et al. and by us could be possible with the application of ssDNA or another polyelectrolyte with a different length. Interestingly, neither we nor Bond et al. did observe the T = 1 shells. This suggests that such shells composed only of pentamers are not formed by the full-length BMV CP but only when the 35 N-terminal residues are truncated.4,26 We also did not observe multiplets (a few protein shells joined together), which confirms that such VLP species are formed only in the presence of a long polyelectrolyte.4,10,22
Analyzing the topography of both meVLP types, we observed that in each type, one side of the protein shell seems to be better ordered and complete than the opposite side (Fig. 4). Such ordering polarization suggests that the assembly proceeds according to the nucleation and growth pathway. This scenario has also been inferred from computational simulations of the empty CCMV virion assembly published by Panahandeh et al.5 Accordingly, after the first pentamer is formed, the consecutive capsomers assemble in adjacent positions. Subsequently, the growing shell can undergo a few steps of relaxation and ordering that involve the dissociation of misplaced pentamers and their transformation into hexamers. The observed disorder at one side of both meVLP types might result from the disorder–order transitions within the capsomers in the newly formed parts of the protein shell. It is hard to tell whether the observed meVLPs are intermediate forms capable of transformation into icosahedral eVLP or are stable final forms. Bond et al. have suggested that the pseudo-T = 2 shells are metastable due to the excess of elastic energy and curvature resulting from the higher ratio of pentamers to hexamers compared to the mature T = 3 shell.4 Accordingly, such metastable shells tend to dissociate into fragments, which initiate new nucleation and growth events, leading to icosahedral protein shells. Taking this into account, the presence of disordered regions at the surface of meVLPs might reflect either the ordering/relaxation of the capsomers or their fragmentation. The total pool of the 2D classified objects in the analyzed eVLP sample included 85.8% of particles smaller than meVLPs which might represent either growing or dissociating shells.
It has been pointed out that the course of the assembly process strongly depends on the CP concentration and proceeds a few times slower in the absence of RNA or other polyelectrolyte.5 It is likely that dissociating and growing capsids coexist, but the formation of VLPs slows down as a result of the depletion of available CP subunits in the assembly milieu.
The second dataset in our studies shows BMV-derived VLPs formed in the presence of tRNA (tVLPs).23 During the 2D classification of tVLP, we also observed non-icosahedral tVLP. The total pool of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419
419![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 349 particles picked in 15
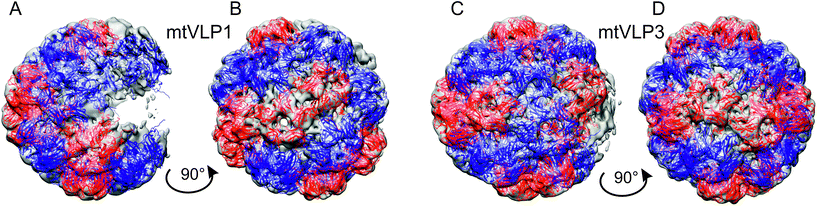
349 particles picked in 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 micrographs contained 2% of icosahedral tVLPs described in the previous section, 89% of non-icosahedral tVLPs with a size range between 22 nm and 25 nm, and 9% of smaller species too heterogeneous for further analysis (Table 2). 3D classification of non-icosahedral tVLPs revealed that only 25% of them were applicable in the asymmetric reconstruction. The 3D asymmetric reconstruction let us divide the non-icosahedral tVLPs into four types. Unlike the pseudo-T = 2 symmetrical meVLPs, all non-icosahedral tVLPs were missing some parts of the protein shell, and we called them malformed tVLPs (mtVLPs) (Fig. 5 and 6). mtVLPs type 1 (mtVLP1) were represented by 4.5% of the particles and were composed of 7 pentamers and 9 hexamers, mtVLPs type 2 (mtVLP2), represented by 7% of the particles were assembled from 9 pentamers and 10 hexamers, mtVLPs type 3 (mtVLP3), 7.5% of the particles, contained 10 pentamers and 10 hexamers, and finally, mtVLPs type 4 (mtVLP4), represented by 5.6% of the particles, were composed of 11 pentamers and 10 hexamers. The observed size range of tVLPs (23 nm to 25 nm) is in line with our earlier observations relying on DLS and low-resolution Cryo-EM measurements of the size range of assembled tVLPs.23 Interestingly, the distribution of pentamers in mtVLP1 and mtVLP3 resembles H12 eVLP with some capsomers missing. Compared to H12, mtVLP1 lacks one pentamer and two hexamers in the cup at one polar end, one hexamer in the cup at the opposite polar end, and four pentamers from the belt joining the two cups. mtVLP3, compared to H12, lacks one hexamer in each of the two cups at polar ends and two adjacent pentamers from the belt joining the two cups (Fig. 5). The arrangement of capsomers in mtVLP2 and mtVLP4 is not similar to any of the above-described protein shells (Fig. 6).
861 micrographs contained 2% of icosahedral tVLPs described in the previous section, 89% of non-icosahedral tVLPs with a size range between 22 nm and 25 nm, and 9% of smaller species too heterogeneous for further analysis (Table 2). 3D classification of non-icosahedral tVLPs revealed that only 25% of them were applicable in the asymmetric reconstruction. The 3D asymmetric reconstruction let us divide the non-icosahedral tVLPs into four types. Unlike the pseudo-T = 2 symmetrical meVLPs, all non-icosahedral tVLPs were missing some parts of the protein shell, and we called them malformed tVLPs (mtVLPs) (Fig. 5 and 6). mtVLPs type 1 (mtVLP1) were represented by 4.5% of the particles and were composed of 7 pentamers and 9 hexamers, mtVLPs type 2 (mtVLP2), represented by 7% of the particles were assembled from 9 pentamers and 10 hexamers, mtVLPs type 3 (mtVLP3), 7.5% of the particles, contained 10 pentamers and 10 hexamers, and finally, mtVLPs type 4 (mtVLP4), represented by 5.6% of the particles, were composed of 11 pentamers and 10 hexamers. The observed size range of tVLPs (23 nm to 25 nm) is in line with our earlier observations relying on DLS and low-resolution Cryo-EM measurements of the size range of assembled tVLPs.23 Interestingly, the distribution of pentamers in mtVLP1 and mtVLP3 resembles H12 eVLP with some capsomers missing. Compared to H12, mtVLP1 lacks one pentamer and two hexamers in the cup at one polar end, one hexamer in the cup at the opposite polar end, and four pentamers from the belt joining the two cups. mtVLP3, compared to H12, lacks one hexamer in each of the two cups at polar ends and two adjacent pentamers from the belt joining the two cups (Fig. 5). The arrangement of capsomers in mtVLP2 and mtVLP4 is not similar to any of the above-described protein shells (Fig. 6).
It is important to note that we performed the assembly procedure starting with the same concentration of rCP to obtain both eVLPs and tVLPs. However, in the case of tVLPs, it resulted in many more assembled protein shells, including both icosahedral and non-icosahedral species (14.2% of eVLPs versus 91% of tVLPs) (Table 2). This corroborates the earlier published data indicating that capsid assembly in the presence of genomic RNA is more efficient than the assembly of empty shells due to the energetically favorable CP–RNA electrostatic interactions.5 It was observed, e.g., in CCMV, that RNA-filled icosahedral capsids form under physicochemical conditions in which empty capsids cannot self-assemble.5 However, a comparison of the percentage of icosahedral shells in the total pools of reconstructed eVLPs and tVLPs revealed that the efficiency of icosahedral tVLP formation is significantly lower than that of eVLP (7.3% of icosahedral eVLP versus 2% of icosahedral tVLP). Among the mtVLPs, we found only incomplete structures with holey shells, which support the nucleation and growth pathway for assembly. The number of pentamers in each mtVLP was lower than 12, and we assume they all represent earlier stages of protein shell formation compared with the above-described pseudo-T = 2 symmetrical shells. The mtVLP1 and mtVLP3, however, show an arrangement similar to H12 eVLP. The H12 VLP has a pair of pentamers surrounded by six hexamers at both opposing polar ends, which are joined by a central belt of eight pentamers. mtVLP1 and mtVLP3 show a similar arrangement of the capsomers, but some of them are missing (Fig. 5). This suggests that H12 shell formation is one of the directions of the rCP assembly, which was arrested because of the strong electrostatic interactions between rCP and tRNA. These electrostatic interactions outcompete the elastic energy and the interactions between rCPs, impeding the protein shell relaxation and its disorder–order transition, disabling the icosahedral arrangement. The negative effect of the electrostatic tRNA–rCP interactions on the proper capsid formation is also reflected by the relatively small fraction of icosahedral tVLPs (Table 2). It is worth noting that the present results are in excellent qualitative agreement with the phase diagrams established by Panahandeh and coworkers.5 In the presence of tRNA, the spontaneous radius is lowered because tRNA enforces a smaller capsid size than in the absence of a core. As a result, stronger rCP–rCP interactions are required to overcome the elastic energy barrier to icosahedral shells. This explains why more icosahedral shells were formed with eVLPs. Besides, because of the high elastic energy induced by the electrostatic tRNA–rCP interactions, the resulting tVLPs are incomplete and disordered, whereas eVLPs exhibit mostly irregular closed shapes resulting from weaker mechanical stress.
Additional consolidation of tRNA and rCP interaction might result from the highly structured and stable character of tRNA.27 During our previous studies, we observed that tVLPs were far more resistant to thermal aggregation than the eVLPs or PSS-filled VLPs (PSS, polystyrene sulfonic acid, a synthetic polymer with a negative charge).23 H8 and H15 VLPs described by Bond et al. also contained nucleic acids (52-nt ssDNA oligomer), but perhaps due to the lack of tRNA-like compact structure, it did not interfere with the VLP formation.4 The tRNA-like structure (TLS) has been described as a packaging signal in native BMV genomic molecules and in-trans complementation of 3′-terminally truncated genomic BMV RNAs devoid of TLS with tRNA rescued genomic RNA encapsidation.28 Together with our results, it can be concluded that tRNA is a strong encapsidation signal that brings rCP subunits together, but its excess in the assembly milieu causes undesirable arrangement of capsomers, leading to malformed VLPs. This is in line with the earlier observations indicating that both the CP concentration and the CP to polyelectrolyte ratio are crucial for the assembly process.5,29
Here one question arises as to how it is possible that during the BMV replication in the plant cell, the abundant tRNAs do not package into the nascent virions. BMV replicates in special structures in the endoplasmic reticulum (ER) of infected plant cells. These involve spherules formed with the contribution of BMV non-structural protein 1a, which recruits BMV non-structural protein 2a with genomic RNA to form BMV replication complex.30,31 It has also been shown that the sites of BMV RNA replication and CP synthesis overlap because the interaction between CP and the replicase protein 2a is obligatory for RNA packaging specificity. What is more, CP itself is capable of modifying ER to form vesicles.31–33 Most probably, the specific conditions inside the spherules or vesicles induced by the viral proteins in ER of the infected plant cells prevent packaging tRNA into the nascent virions.
The total efficiency of icosahedral capsid formation in our in vitro assays is surprisingly low and raises a question about the efficiency of this process in vivo. However, the native conditions of BMV assembly in plant cells have been “optimized” by the evolution of the virus and are much more complex than the in vitro system with the buffer, CP and RNA. The understanding of in planta BMV assembly is poor, but the process appears efficient, considering that BMV may constitute at least 0.1% of total plant tissue.18 The lack of empty capsids in BMV-infected plants suggests that the CP sequence evolved towards the most efficient formation of the infective virions with genomic RNA inside.20 It is also likely that the cargo type itself has a huge impact on the assembly—genomic RNA of BMV is different from tRNA used in vitro. The low efficiency of formation of here described icosahedral eVLPs and tVLPs and the low stability of eVLP suggests that the in vitro conditions are suboptimal.23 The efficiency improvement should be reached by tuning the assembly solution composition in terms of the buffer, RNA (or other cargo) concentration, its size and compactness34 and the CP concentration.5,29 In fact, genetic engineering of the CP sequence appears a promising, alternative solution. In all cases, this work brings us a step closer to a rationalized design of BMV-based nanocontainers, biosensors, nanoreactors, and drug or gene delivery vehicles.
Experimental
Protein production and purification
The rCP was produced and purified as described earlier.23 Briefly, DNA coding for BMV CP was cloned into the pMCSG48 expression plasmid and expressed in Rosetta™ 2(DE3)pLysS E. coli cells (Novagen) according to the manufacturer's instructions. The bacteria were grown at 37 °C in LB medium containing 100 mg l−1 ampicillin and 34 mg l−1 chloramphenicol. After induction (0.5–0.8 M IPTG), the bacteria were cultured for 16 h at 20 °C. The cells were harvested and suspended in lysis buffer I [25 mM Tris pH 8, 0.5 M NaCl, 0.2 mg ml−1 lysozyme (BioShop), 250 U benzonase nuclease (Novagen)] and sonicated. Cell debris was removed by centrifugation, and the supernatant was mixed with the same amount of buffer II [25 mM Tris pH 8, 0.5 M NaCl, 20 mM imidazole (BioShop)]. To purify BMV rCP, affinity chromatography was used. After protein binding with Ni-NTA resin, the column was washed with buffer II. Next, the protein was eluted from the column using 500 mM imidazole in buffer II. The eluted protein was dialyzed using buffer III (25 mM Tris pH 8, 1 M NaCl). To remove the N-terminal tag, 1 mg of TEV protease was added per 10 mg of the eluted protein. After TEV digestion, the sample was once again applied to a Ni-NTA column. The first flowthrough was collected, and the purified proteins were concentrated by ultrafiltration (Amicon Ultra-10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO, Millipore). The concentration of the protein was determined by measuring the absorbance at 280 nm (NanoPhotometer N60, Implen). The purity of the protein samples was assessed by SDS-PAGE, and the monodisperse state of the protein solutions was confirmed by dynamic light scattering (DLS) measurements using a Zetasizer μV (Malvern).
000 MWCO, Millipore). The concentration of the protein was determined by measuring the absorbance at 280 nm (NanoPhotometer N60, Implen). The purity of the protein samples was assessed by SDS-PAGE, and the monodisperse state of the protein solutions was confirmed by dynamic light scattering (DLS) measurements using a Zetasizer μV (Malvern).
VLP assembly
eVLPs and tVLPs were assembled as described earlier.23 The rCP solution (concentration of 1 mg ml−1) was subjected to a two-step dialysis with the application of SnakeSkin Dialysis Tubing with a 3500 MWCO (Thermo Fisher Scientific). Assembly of the VLP with tRNA was performed in a mass ratio 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 (tRNA
6 (tRNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CP). The first dialysis was performed in VLP assembly buffer I [50 mM NaCl (BioShop), 10 mM KCl (BioShop), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 7.2] for 16 h at 4 °C. The second dialysis was performed in VLP assembly buffer II [25 mM NaCl (BioShop), 10 mM KCl (BioShop), 25 mM NaOAc (Merck), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 4.8] for 16 h at 4 °C. Afterwards, the samples were centrifuged (10 min, 10
CP). The first dialysis was performed in VLP assembly buffer I [50 mM NaCl (BioShop), 10 mM KCl (BioShop), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 7.2] for 16 h at 4 °C. The second dialysis was performed in VLP assembly buffer II [25 mM NaCl (BioShop), 10 mM KCl (BioShop), 25 mM NaOAc (Merck), 5 mM MgCl2·6H2O (Merck), 50 mM Tris (BioShop), pH 4.8] for 16 h at 4 °C. Afterwards, the samples were centrifuged (10 min, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rcf, 4 °C), and the supernatant was collected for further examination. The presence of assembled VLPs was confirmed by DLS.
000 rcf, 4 °C), and the supernatant was collected for further examination. The presence of assembled VLPs was confirmed by DLS.
Cryo-EM data collection
Quantifoil TEM grids (300 mesh R2/1 copper) were glow-discharged on a Leica EM ACE200 (Leica Microsystems) glow discharger. A total of 3.0 μl of the sample (0.25 to 3.0 mg ml−1) was plunge-frozen in liquid ethane using FEI Vitrobot Mark IV set to 100% humidity and 4 °C (blotting parameters: blot time 4 s; wait time 10 s; drain time 0 s; blot force 0; blot total 1). The optimal sample concentrations, based on grid screening, were 1.0 and 1.5 mg ml−1, respectively for tVLP and eVLP. Cryo-EM data were collected at National Cryo-EM Centre SOLARIS (Kraków, Poland) with a Titan Krios G3i microscope (Thermofisher Scientific) operated at 300 kV equipped with a Gatan BioQuantum energy filter (slit width of 20 eV) and a Gatan K3 direct electron detector (16 e px−1 s−1 dose rate). The K3 detector was operated in a counting mode at 105![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× magnification, resulting in a calibrated physical pixel size of 0.86 Å px−1. The images were acquired at under-focus with a defocus range of −3.0 to −0.9 μm and 0.3 μm defocus step. A total of 40 movie frames accumulating to total doses of 40 e− Å−2 were collected with a total number of 17
000× magnification, resulting in a calibrated physical pixel size of 0.86 Å px−1. The images were acquired at under-focus with a defocus range of −3.0 to −0.9 μm and 0.3 μm defocus step. A total of 40 movie frames accumulating to total doses of 40 e− Å−2 were collected with a total number of 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 955 micrographs for the tVLP reconstruction and 12
955 micrographs for the tVLP reconstruction and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 569 for the eVLP reconstruction.
569 for the eVLP reconstruction.
Cryo-EM data processing
EM map reconstructions were performed in Relion 3.1 within the CCP-EM package.35,36 Contrast transfer function (CTF) was estimated using CTFFIND4.37,38 Data processing stages are presented in Fig. 1. Particles were picked based on reference images from 2D classes after manual picking of ∼800 particles. For the tVLP dataset, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 419
419![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 349 particles were picked from 15
349 particles were picked from 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 861 micrographs (after filtering based on CTF-estimated resolution of 5 Å or better). The icosahedral tVLP reconstruction was based on a single 2D class containing 28
861 micrographs (after filtering based on CTF-estimated resolution of 5 Å or better). The icosahedral tVLP reconstruction was based on a single 2D class containing 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 412 particles. The box size of 512 pixels corresponded to 440 Å. Icosahedral symmetry was imposed after the generation of the initial 3D model and was maintained during high-resolution refinement. Map regions outside CP were flattened by masking. CTF refinement and Bayesian particle polishing preceded the final refinement and post-processing. mtVLP EM maps were calculated after 2D classification (5 classes containing 498
412 particles. The box size of 512 pixels corresponded to 440 Å. Icosahedral symmetry was imposed after the generation of the initial 3D model and was maintained during high-resolution refinement. Map regions outside CP were flattened by masking. CTF refinement and Bayesian particle polishing preceded the final refinement and post-processing. mtVLP EM maps were calculated after 2D classification (5 classes containing 498![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 281 particles out of 50 classes), followed by 3D classification (4 ellipsoidal classes out of 8). eVLP EM maps were reconstructed similarly, although with fewer particles. Namely, a single 3D class with 11
281 particles out of 50 classes), followed by 3D classification (4 ellipsoidal classes out of 8). eVLP EM maps were reconstructed similarly, although with fewer particles. Namely, a single 3D class with 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 521 particles yielded the icosahedral eVLP, whereas meVLP maps were calculated for three 3D classes containing 18
521 particles yielded the icosahedral eVLP, whereas meVLP maps were calculated for three 3D classes containing 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 037 particles. Molecular models were built using Phenix.Autobuild,39 followed by manual corrections in Coot40 and real-space refinement in Phenix.Refine,41 including validation of the model geometry and model to map correlation. Unbiased final map resolutions were calculated based on “gold-standard” Fourier-shell correlation curves.
037 particles. Molecular models were built using Phenix.Autobuild,39 followed by manual corrections in Coot40 and real-space refinement in Phenix.Refine,41 including validation of the model geometry and model to map correlation. Unbiased final map resolutions were calculated based on “gold-standard” Fourier-shell correlation curves.
Data availability
Atomic models resulting from icosahedral reconstructions and corresponding EM maps were deposited in the PDB and EMDB with the following accession numbers for tVLP, 7PE1, EMD-13344; eVLP, 7PE2, EMD-13345.Conclusions
We showed that the BMV rCP overexpressed in E. coli is capable of forming icosahedral VLP both without and with a cargo. Two types of icosahedral VLPs are accompanied by a few types of meVLPs and mtVLPs with topography indicating the assembly according to the nucleation and growth pathway. The analysis of the meVLP indicates that rCP can form pseudo-T = 2 symmetrical shells of two types, which is an important feature in the context of VLP biotechnological application as nanocontainer. All meVLPs have 12 pentamers and a variable number of hexamers. The VLPs with particular dimensions and conformation of capsomers might be formed and stabilized by the cargo, as shown by Bond et al., who used ssDNA oligonucleotide for the assembly and obtained the H8 VLP type.4 However, the presented herein holey mtVLP indicate that the cargo (in this case tRNA) may also negatively influence the assembly. In the case of here described mtVLPs, strong tRNA–rCP interactions apparently induce high mechanical stress leading to a lack of protein shell continuity. Such unwanted effects could be eliminated by optimizing the tRNA content or by genetic engineering of rCP for better adjustment with the cargo. We believe that the high-resolution Cryo-EM structures presented in this paper will provide a valuable prerequisite for the rational design of such VLPs.Author contributions
Conceptualization, A. U.; data curation and formal analysis, M. R.; funding acquisition, A. U.; investigation, P. I. and A. U.; methodology, M. R., M. F., A. S. and A. U.; project administration, A. U.; resources, M. F. and A. U.; supervision, A. U.; writing—original draft preparation, M. R., G. T. and A. U.; writing—review and editing, A. S., G. T. and M. F.; validation and visualization, M. R. and A. U.; all authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Cryo-EM sample preparation and data collection took place at SOLARIS National Synchrotron Radiation Centre, at the Cryo-EM facility infrastructure. The experiment was performed thanks to the collaboration of the SOLARIS Staff. We acknowledge the MCB Structural Biology Core Facility (supported by the TEAM TECH CORE FACILITY/2017-4/6 grant from the Foundation for Polish Science) for valuable support.References
- C. Arcangeli, P. Circelli, M. Donini, A. A. Aljabali, E. Benvenuto, G. P. Lomonossoff and C. Marusic, J. Biomol. Struct. Dyn., 2014, 32, 630–647 CrossRef CAS PubMed.
- Y. Ren, S. M. Wong and L. Y. Lim, Bioconjugate Chem., 2007, 18, 836–843 CrossRef CAS PubMed.
- Z. C. Zhao, J. C. Wang, M. Zhang, N. A. Lyktey, M. F. Jarrold, S. C. Jacobson and A. Zlotnick, Nat. Commun., 2021, 12, 589 CrossRef CAS PubMed.
- K. Bond, I. B. Tsvetkova, J. C. Y. Wang, M. F. Jarrold and B. Dragnea, Small, 2020, 16, e2004475 CrossRef PubMed.
- S. Panahandeh, S. Li, L. Marichal, R. Leite Rubim, G. Tresset and R. Zandi, ACS Nano, 2020, 14, 3170–3180 CrossRef CAS PubMed.
- A. A. Mieloch, M. Krecisz, J. D. Rybka, A. Strugala, M. Krupinski, A. Urbanowicz, M. Kozak, B. Skalski, M. Figlerowicz and M. Giersig, AIP Adv., 2018, 8, 035005 CrossRef.
- M. Chevreuil, D. Law-Hine, J. Chen, S. Bressanelli, S. Combet, D. Constantin, J. Degrouard, J. Möller, M. Zeghal and G. Tresset, Nat. Commun., 2018, 9, 3071 CrossRef PubMed.
- A. Biddlecome, H. H. Habte, K. M. McGrath, S. Sambanthamoorthy, M. Wurm, M. M. Sykora, C. M. Knobler, I. C. Lorenz, M. Lasaro, K. Elbers and W. M. Gelbart, PLoS One, 2019, 14, e0215031 CrossRef CAS PubMed.
- A. Nuñez-Rivera, P. G. J. Fournier, D. L. Arellano, A. G. Rodriguez-Hernandez, R. Vazquez-Duhalt and R. D. Cadena-Nava, Beilstein J. Nanotechnol., 2020, 11, 372–382 CrossRef PubMed.
- R. F. Garmann, M. Comas-Garcia, M. S. Koay, J. J. Cornelissen, C. M. Knobler and W. M. Gelbart, J. Virol., 2014, 88, 10472–10479 CrossRef PubMed.
- R. F. Bruinsma, G. J. L. Wuite and W. H. Roos, Nat. Rev. Phys., 2021, 3, 76–91 CrossRef PubMed.
- J. D. Perlmutter and M. F. Hagan, Annu. Rev. Phys. Chem., 2015, 66, 217–239 CrossRef CAS PubMed.
- R. F. Garmann, A. M. Goldfain and V. N. Manoharan, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 22485–22490 CrossRef CAS PubMed.
- R. Zandi, B. Dragnea, A. Travesset and R. Podgornik, Phys. Rep., 2020, 847, 1–102 CrossRef CAS.
- R. Zandi, P. van der Schoot, D. Reguera, W. Kegel and H. Reiss, Biophys. J., 2006, 90, 1939–1948 CrossRef CAS PubMed.
- L. Menou and M. Castelnovo, Soft Matter, 2019, 15, 6180–6189 RSC.
- M. Chevreuil, L. Lecoq, S. Wang, L. Gargowitsch, N. Nhiri, E. Jacquet, T. Zinn, S. Fieulaine, S. Bressanelli and G. Tresset, J. Phys. Chem. B, 2020, 124, 9987–9995 CrossRef CAS PubMed.
- A. Strugala, M. Krecisz, J. D. Rybka, A. Urbanowicz, K. Szpotkowski, P. Bierwagen, M. Figlerowicz, M. Kozak, C. Böttcher and M. Giersig, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1068, 157–163 CrossRef PubMed.
- C. C. Kao, P. Ni, M. Hema, X. L. Huang and B. Dragnea, Mol. Plant Pathol., 2011, 12, 403–412 CrossRef CAS PubMed.
- A. L. N. Rao, Annu. Rev. Phytopathol., 2006, 44, 61–87 CrossRef CAS PubMed.
- C. Beren, Y. Cui, A. Chakravarty, X. Yang, A. L. N. Rao, C. M. Knobler, Z. H. Zhou and W. M. Gelbart, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10673–10680 CrossRef CAS PubMed.
- Z. Wang, C. F. Hryc, B. Bammes, P. V. Afonine, J. Jakana, D. H. Chen, X. Liu, M. L. Baker, C. Kao, S. J. Ludtke, M. F. Schmid, P. D. Adams and W. Chiu, Nat. Commun., 2014, 5, 4808 CrossRef CAS PubMed.
- A. Strugala, J. Jagielski, K. Kamel, G. Nowaczyk, M. Radom, M. Figlerowicz and A. Urbanowicz, Int. J. Mol. Sci., 2021, 22, 3098 CrossRef CAS PubMed.
- M. Alejska, N. Malinowska, A. Urbanowicz and M. Figlerowicz, Acta Biochim. Pol., 2005, 52, 833–844 CrossRef CAS PubMed.
- A. Urbanowicz, M. Alejska, P. Formanowicz, J. Blazewicz, M. Figlerowicz and J. J. Bujarski, J. Virol., 2005, 79, 5732–5742 CrossRef CAS PubMed.
- R. W. Lucas, Y. G. Kuznetsov, S. B. Larson and A. McPherson, Virology, 2001, 286, 290–303 CrossRef CAS PubMed.
- A. G. Torres, E. Batlle and L. R. de Pouplana, Trends Mol. Med., 2014, 20, 306–314 CrossRef CAS PubMed.
- Y. G. Choi, T. W. Dreher and A. L. N. Rao, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 655–660 CrossRef CAS PubMed.
- R. D. Cadena-Nava, M. Comas-Garcia, R. F. Garmann, A. L. Rao, C. M. Knobler and W. M. Gelbart, J. Virol., 2012, 86, 3318–3326 CrossRef CAS PubMed , erratum in: J. Virol., 2019, 93, e01939–e02018.
- X. Wang, W. M. Lee, T. Watanabe, M. Schwartz, M. Janda and P. Ahlquist, J. Virol., 2005, 79, 13747–13758 CrossRef CAS PubMed.
- S. Chaturvedi and A. L. N. Rao, Virology, 2014, 464, 67–75 CrossRef PubMed.
- P. Annamalai and A. L. N. Rao, J. Virol., 2006, 80, 10096–10108 CrossRef CAS PubMed.
- D. Bamunusinghe, J. K. Seo and A. L. N. Rao, J. Virol., 2011, 85, 2953–2963 CrossRef CAS PubMed.
- L. Marichal, L. Gargowitsch, R. L. Rubim, C. Sizun, K. Kra, S. Bressanelli, Y. Dong, S. Panahandeh, R. Zandi and G. Tresset, Biophys. J., 2021, 120, 3925–3936 CrossRef CAS PubMed.
- S. H. W. Scheres, J. Struct. Biol., 2012, 180, 519–530 CrossRef CAS PubMed.
- C. Wood, T. Burnley, A. Patwardhan, S. Scheres, M. Topf, A. Roseman and M. Winn, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71, 123–126 CrossRef CAS PubMed.
- T. Burnley, C. M. Palmer and M. Winn, Acta Crystallogr., Sect. D: Struct. Biol., 2017, 73, 469–477 CrossRef CAS PubMed.
- A. Rohou and N. Grigorieff, J. Struct. Biol., 2015, 192, 216–221 CrossRef PubMed.
- T. C. Terwilliger, R. W. Grosse-Kunstleve, P. V. Afonine, N. W. Moriarty, P. H. Zwart, L. W. Hung, R. J. Read and P. D. Adams, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2008, 64, 61–69 CrossRef CAS PubMed.
- P. Emsley, B. Lohkamp, W. G. Scott and K. Cowtan, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2010, 66, 486–501 CrossRef CAS PubMed.
- P. V. Afonine, B. K. Poon, R. J. Read, O. V. Sobolev, T. C. Terwilliger, A. Urzhumtsev and P. D. Adams, Acta Crystallogr., Sect. D: Struct. Biol., 2018, 74, 531–544 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |