The expanding CRISPR toolbox for natural product discovery and engineering in filamentous fungi†
Clara
Woodcraft
 ,
Yit-Heng
Chooi
,
Yit-Heng
Chooi
 * and
Indra
Roux‡
* and
Indra
Roux‡
 *
*
School of Molecular Sciences, The University of Western Australia, Perth, WA 6009, Australia. E-mail: yitheng.chooi@uwa.edu.au; Indra.roux@uwa.edu.au
First published on 7th October 2022
Abstract
Covering: up to May 2022
Fungal genetics has transformed natural product research by enabling the elucidation of cryptic metabolites and biosynthetic steps. The enhanced capability to add, subtract, modulate, and rewrite genes via CRISPR/Cas technologies has opened up avenues for the manipulation of biosynthetic gene clusters across diverse filamentous fungi. This review discusses the innovative and diverse strategies for fungal natural product discovery and engineering made possible by CRISPR/Cas-based tools. We also provide a guide into multiple angles of CRISPR/Cas experiment design, and discuss current gaps in genetic tool development for filamentous fungi and the promising opportunities for natural product research.
1 Introduction
The exponential increase in microbial genomic information has provided a window into their vastly underexplored specialised metabolism. However, experimental exploration is still needed to translate genomic information into new chemical compounds and unveil their mechanisms of biosynthesis. The specialised metabolism of filamentous fungi is particularly prolific, occupying a unique chemical space and displaying distinctive biosynthetic logics.1 Fungal secondary metabolites (SMs) are highly enriched in antibacterial, antifungal and antitumor activities2 and have found various applications in pharmaceutical and agricultural industries.3 The genes encoding the synthesis of each SM are often found grouped together in the genome as biosynthetic gene clusters (BGCs), which can span up to 50 kb. Each BGC is composed of 2–20 genes and can be classified by the type of primary metabolite substrate accepted by the core biosynthetic enzyme. These include polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), and terpene synthases (TPSs).4 Fungal PKSs and NRPSs are large enzymes with multiple catalytic domains that often operate iteratively to form the SM scaffold.5,6 The rest of the genes in a cluster usually encode tailoring enzymes, transporters, transcription factors or genes that provide resistance to toxic metabolic products.Ongoing fungal genome sequencing initiatives and bioinformatic efforts have revealed a wide diversity of BGCs, many of which have yet to be chemically characterised.1,4 Genetic engineering techniques play a key role in utilizing this data for the study and engineering of BGCs.4 Many BGCs are held under tight transcriptional regulation and as a result, their encoded compounds often remain undetectable under laboratory culturing conditions.4 As the conditions in which a SM is actively produced are often unknown, most targeted activation strategies rely on the genetic manipulation of BGCs. On the other hand, dissecting the genetic basis of actively produced secondary metabolites can be pursued by genetic inactivation of candidate biosynthetic genes. Additionally, the interrogation of biosynthetic genes in their native host can shed light on the biological functions of SMs. As many fungal species remain genetically intractable or hard to cultivate, heterologous expression is a useful alternative for exploring cryptic specialised metabolism. Some filamentous fungi have been engineered as hosts for BGC expression, or even for large-scale industrial production of SMs.7 Therefore, the genetic transfer of large BGCs to heterologous hosts and the optimisation of their expression are also relevant for SM research. Overall, the genetic toolbox available for filamentous fungi has widely impacted the pace of BGC elucidation and engineering. Therefore, breakthrough technologies like Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated proteins (Cas) offer unprecedented avenues for fungal SM research.
The development and general applications of CRISPR/Cas systems in filamentous fungi have been covered elsewhere.8–12 In this review, we highlight the innovative approaches by which CRISPR/Cas tools can be used for SM genome mining and engineering. Firstly, we will provide a bird's-eye view of the principles of CRISPR/Cas experimental design, execution and analysis used across BGC manipulation reports. Importantly, we will classify the different CRISPR/Cas genome editing strategies in relation to the DNA repair pathways they rely on in the native fungus. We will also cover emerging tools in filamentous fungi for CRISPR/Cas based effector recruitment. We will then discuss diverse applications and case studies of CRISPR/Cas gene editing for SM research across filamentous ascomycetes. Recent applications using CRISPR/Cas-based transcriptional control and chromatin editing will also be addressed. Finally, we present our vision on areas for optimisation and the potential for expansion of the CRISPR/Cas toolbox for BGC manipulation in fungi.
2 A practical guide to CRISPR/Cas tools for filamentous fungi
2.1 Principles of CRISPR/Cas systems
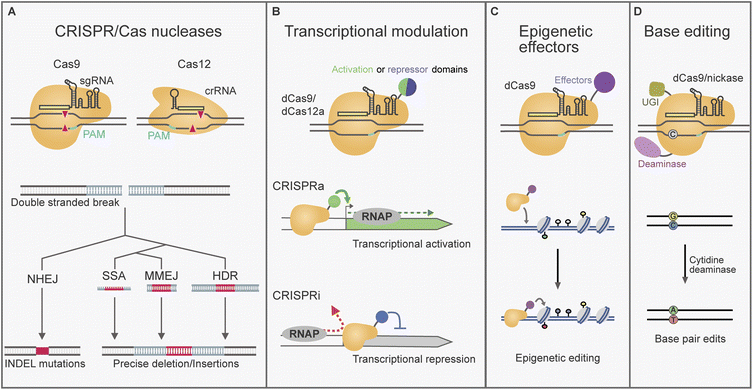
To better understand how CRISPR/Cas-gene editing can be a game-changer, it is relevant to place it in the context of previous gene manipulation strategies used for fungi. A traditional approach for the introduction of specific genetic modifications in fungal genomes is gene targeting via homologous recombination (HR).13 By flanking a selection marker gene with custom homology arms, the donor DNA is incorporated through the native homology-directed repair (HDR) pathway. However, the efficiency of HDR in filamentous fungi is often limited by the competing dominant non-homologous end-joining (NHEJ) pathway of DNA repair.14 Impairing NHEJ by mutating genes from its machinery (e.g. ΔKU, Δlig4/D) boosted the specificity of gene targeting.13 However, this strategy remained unsuccessful in several fungus species in which the efficiency was limiting. As a result, genetic engineering was held back in many filamentous fungi species until the advent of CRISPR/Cas tools.15,16 Additionally, as CRISPR/Cas gene editing can often bypass the requirement for selection markers it is well suited for sequential or parallel modifications. CRISPR/Cas gene editing also enabled efficient editing in wild type strains with an active NHEJ.17 Due to these factors, CRISPR/Cas gene editing has been established across a rapidly growing number of filamentous fungi.CRISPR/Cas are bacterial and archaeal adaptive immune response systems that have provided the foundation for powerful gene editing tools.18 Amongst the diverse variety of naturally evolved CRISPR/Cas systems, the type II systems Cas9 and Cas12a have been adapted for gene editing in filamentous fungi.11,12 The important components shared by these systems are an RNA-guided Cas nuclease that can be directed to a DNA target specified by base-pairing complementarity with a CRISPR RNA (crRNA).19 Each crRNA is composed of a scaffold region and a ∼20 nt spacer sequence that can be programmed to define the DNA target. Target selection is dependent on the presence of a short (2–6 bp) protospacer adjacent motif (PAM) required directly up or downstream of the target locus. Multiple spacer sequences can be incorporated in arrays of pre-crRNAs for multi-target editing. However, arrays require processing to yield individual crRNAs. In native bacterial Cas9 systems an endogenous housekeeping RNAse enzyme is responsible for this process, whilst Cas12a possesses inbuilt RNAse activity.20,21 Additionally, for Cas9 systems the crRNA requires the presence of a trans-activating CRISPR RNA (tracrRNA) for recognition and binding by Cas9. When used for gene editing Cas9 crRNA and tracrRNA are often expressed as a single synthetic chimera known as a single guide RNA (sgRNA).22 In this review we will use the generic term guide RNA (gRNA) when referring to either Cas9 sgRNA or Cas12a crRNA.
2.2 CRISPR/Cas nucleases and interacting DNA repair pathways
CRISPR/Cas nucleases can be directed to cleave a specific DNA target, leaving behind a double stranded DNA (dsDNA) break that activates the DNA repair response (Fig. 1A). The DNA repair pathway taken determines the nature of any edits introduced.23,24 Repair by the NHEJ machinery is error-prone and can lead to the introduction of deletion/insertion (INDEL) mutations (Fig. 1A). This can be leveraged to impair or “knockout” gene function by the random introduction of frameshift and early stop codon mutations. Single guide NHEJ-based methods for fungal BGC gene knockout have reported loss of function deletions ranging between 1–283 bp, as well as single base pair insertions.25–27 In such cases gRNA targets are typically selected in the 5′ region of a gene of interest. Alternatively, complete genes or BGCs have been deleted with NHEJ by targeting with two gRNAs flanking the desired region (examples span 0.4–8 kb).28–31 Additionally, NHEJ can be exploited to integrate heterologous DNA by ligation into the cleavage site.32,33CRISPR/Cas dsDNA breaks can also be used to increase the efficiency of gene targeting through the HDR pathway. This form of repair can be exploited for the precise introduction of sequences delivered on a tailored DNA repair template (Fig. 1A). Templates with dsDNA homology regions of 100 bp–3 kb have been reported to provide high efficiency for CRISPR/Cas HDR approaches in filamentous fungi.34–36 Improvements to the specificity of HDR have been reported by using circular donor DNA, which recombines by double crossover, instead of linear donor DNA.36–38
CRISPR/Cas nucleases can also enable editing by pathways that use short homologies such as microhomology mediated end joining (MMEJ) and single strand annealing (SSA).23,24 MMEJ and SSA are pathways recently characterised in yeast and human cell lines, where the homology required for action was reported as < 25 bp, and > 20 nt, respectively.23 Microhomology-based repair has been effectively exploited in filamentous fungi for CRISPR/Cas gene editing using homologies 30–50 bp/nt in various species.31,39–42 These pathways simplify the construction of the donor DNA, as the microhomology can be introduced in PCR primers or directly using single stranded DNA oligonucleotides.
Unlike traditional gene targeting, which is often heavily reliant on the use of markers, CRISPR/Cas based methods have allowed efficient editing with marker-free DNA repair templates. In filamentous fungi marker-free approaches have been used for insertions and deletions ranging from genes to full BGCs (see Sections 3.1 and 3.2).35,39,42–44
2.3 Modified CRISPR/Cas effectors
Engineered Cas nucleases have allowed access to a broader suite of tools. Cas9 possesses two independent nuclease domains that can be mutated to eliminate DNA cleavage while retaining RNA-guided DNA binding. Cas9 nickase was developed by single nuclease domain deactivation, resulting in a mutant capable of cleaving only one of the DNA strands.22,45 In addition to nickases, completely nuclease-deactivated variants (dCas) have been developed.22 Cas9 nickase and dCas proteins have been used as the foundation of base editing technologies, where they can be fused to a deaminase enzyme responsible for nucleotide conversions (Fig. 1D).46,47 In filamentous fungi dCas9 cytosine base editors, which convert cytidine (C) to thymine (T), have been used in Aspergillus niger and Myceliophthora thermophila (see Section 3.3).48,49CRISPR/Cas-based tools have also been built for transcriptional modulation. These tools are generally built with dCas proteins, including dCas9 and dCas12a, which can be programmed to deliver effector proteins to specific regulatory regions (Fig. 1C).22,50–53 In CRISPR-based activation (CRISPRa), dCas is used to localise transcription activation domains to promoter regions for transcription upregulation. On the other hand, CRISPR interference (CRISPRi) involves the inhibition of expression either by the recruitment of co-repressor domains, or by the binding of a dCas protein at sites where it is thought to obstruct polymerase function. Lastly, targeted epigenome editing can be achieved by fusing Cas to epigenetic effectors, although usually with more modest transcriptional changes than CRISPRa/i.54 CRISPR transcriptional and epigenetic regulators have been used for BGC manipulation in some Aspergillus and Penicillum species (See section 3.4).55–58 These tools allow access to changes in transcription circumventing the need for hardwired gene editing.
2.4 Expression and delivery of CRISPR/Cas components in filamentous fungi
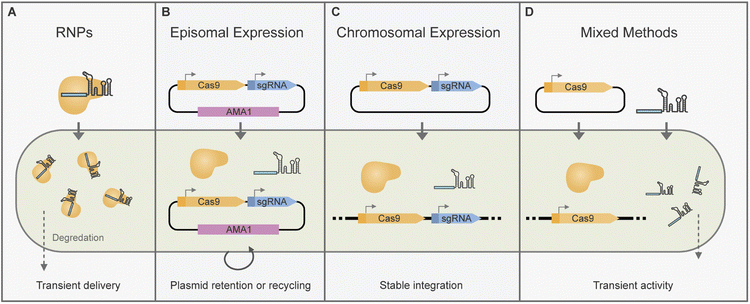
CRISPR/Cas-based gene editing or transcriptional modulation requires the delivery of CRISPR/Cas components to the cell nucleus. There are multiple ways of attaining this which we will cover in more detail in the following sections (Fig. 2).Importantly, some considerations need to be taken for the correct expression of gRNAs in vivo. As they comprise short, precise sequences, additions to the transcript such as end processing from RNA polymerase II (RNAPII) promoters can be detrimental. RNA polymerase III (RNAPIII) promoters are well suited for gRNA expression as they produce unmodified short RNAs. However, there are a limited number of characterised RNAPIII promoters for gRNA expression in fungi. Examples include several promoters from U6 and tRNA orthologues that have been used successfully for gRNA delivery in some fungi.62 As an alternative, RNAPII promoters – responsible for mRNA transcription–have been used together with RNA processing to release individual gRNAs. In filamentous fungi, ribozymes34,63 and tRNA splicing mediated gRNA release39 have been used for Cas9 gRNA release. As Cas12a has its own RNAse activity, it can process multiple individual crRNAs from RNAPII promoters such as PgpdA.55 Additionally, these strategies for processing gRNAs from a single RNAPIII transcript can be used to allow the multiplexed delivery of gRNAs.29,39
Currently, there are several CRISPR/Cas toolkits for filamentous fungi available in plasmid repositories such as Addgene.34,55,58,64,65 Although setting up a CRISPR/Cas system for the first time in a fungal species can require optimisation, once established the only component that requires tailoring in further experiments is the gRNA spacer(s). Several bioinformatics tools offer support to design gRNAs with an increased likelihood of success based on predicted on target activity and the risk of off-target effects. Popular tools for gRNA design across filamentous fungi BGC manipulation reports include sgRNA Scorer 2.0,66 WU-CRISPR,67 CHOP CHOP,68 and EuPaGDT.69 The last two also offer off-target analysis against filamentous fungi genomes. Once the 20 nt spacers are selected, they can be synthetised as short oligonucleotides which depending on the expression strategy are cloned as annealed oligonucleotides55 or as part of PCR fragments.39,70
2.4.2 Chromosomal expression of CRISPR/Cas components
Some of the early implementations of CRISPR/Cas editing in filamentous fungi relied on the random integration of cas9 and sgRNA with constitutive expression cassettes (Fig. 2C).32 This approach was adapted for BGC editing in Aspergillus oryzae,25 and Acremonium chrysogenum.71 However, for gene editing applications transient CRISPR/Cas component expression can be beneficial as it minimises the risk of off-target effects or impacts to host fitness. Inducible tetON promoters have been used to control chromosomal cas9 expression for BGC editing in an Aspergillus fumigatus strain.63 Transient CRISPR/Cas9 activity has also been achieved by mixed delivery of in vitro transcribed sgRNA(s) into fungal strains expressing cas9 chromosomally (Fig. 2D). Following early reports in Trichoderma reesei72 this approach has been adopted for BGC editing in Nodulisporium sp.(no. 65-17-2-1), Pestalotiopsis fici, and Fusarium fujikuroi.59,73,74CRISPR/Cas and gRNA expression cassettes are typically introduced to the fungi by polyethylene glycol (PEG)-mediated protoplast transformation, however in some cases agrobacterial delivery techniques have also been used.27,43,75 Transformant strains are often selected for integration of CRISPR/Cas components, and then further screened for the desired edits.25,71,76 Alternatively, selection can be placed on markers incorporated into repair templates74,77 or in co-transformed DNA fragments.73 A unique approach by Weber et al. allowed selection for CRISPR/Cas activity by use of a split marker system. A non-functional marker is introduced as part of a gRNA expression construct which upon gRNA and Cas expression, is targeted and cleaved allowing marker reconstitution.63
To avoid genome integration altogether Wang et al. developed a suicide vector in which Cas9 components were encoded on an extended linear repair template that included a marker flanked by homology to the target site.78 Upon CRISPR/Cas cleavage the marker can recombine at the cleaved target site, leaving the CRISPR/Cas expression set to be degraded. This approach has been adapted for BGC engineering in Chaetomium globosum.79
One of the main advantages of using AMA1 systems for gene editing is that they can be eliminated after growth on non-selective media.37 Due to this AMA1 vectors are uniquely placed to allow transient CRISPR/Cas expression, marker-free edits and marker recycling (Fig. 2B). For marker-free editing and plasmid recycling selection pressure may be initially placed on AMA1-plasmid uptake followed by re-culturing on nonselective media for plasmid loss.30,85,89 Alternatively, for transient plasmid uptake selection pressure may be placed on the integration of a marker from a repair template instead of for AMA1 upkeep.36,37,41
The ease of AMA1-plasmid loss may vary across different fungal species. Nielsen et al. reported the loss of AMA1 plasmids from the colony periphery after one round of growth on non-selective media, while some strains and workflows may require more than two rounds of non-selective growth to effectively lose all AMA1 plasmids.35,37,85,89 Counterselection can be leveraged to force the loss of AMA1 plasmids, including 5′FOA toxicity for the pyrG marker41 or the use of a conditionally lethal gene such as Aoace2.38 To further simplify plasmid curation, truncated AMA1 variants with low retention in conidia have also been developed for CRISPR/Cas expression.38,92 These vector sets have been widely used for the sequential integration of BGC genes for heterologous expression in As. oryzae.70,93
Selection pressure for editing events by RNPs is commonly applied by incorporating markers in DNA repair templates. Alternatively, co-transformation with marker amplicons or marker-bearing plasmids can be used to select transformation events.97,99 Marker-free edits have also been attained by RNP delivery. An approach by Pohl et al. relied on in vivo recombination of an AMA1 plasmid to reconstitute a marker, which could later be removed by growth on non-selective media.35
3 Applications of CRISPR tools in biosynthetic gene cluster engineering
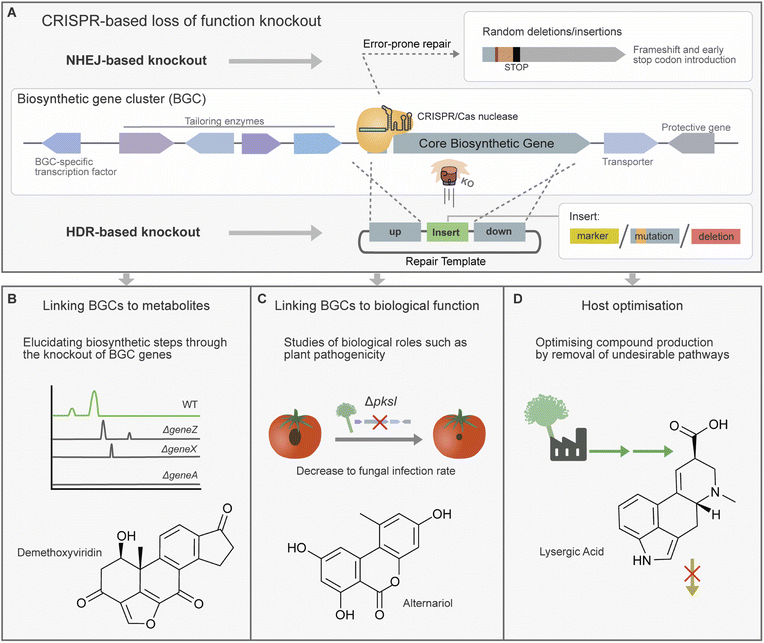
3.1. Loss of function with CRISPR/Cas gene knockout
CRISPR/Cas-based gene knockouts are a powerful tool for loss of function studies and the manipulation of biosynthetic pathways within their host organism (Fig. 3). This can include applications from the general identification of the genetic origin of a given metabolite to analysing the function of individual genes and domains. Additionally, CRISPR/Cas knockouts offer a powerful approach for studying the biological roles played by metabolites and can enable the creation of strains tailored for specific metabolic outputs.Marker-free edits using AMA1-based CRISPR/Cas toolkits have also proved effective for the knockout of core biosynthetic genes. Futyma et al. confirmed the biosynthetic origins of homopyrones in As. homomorphus by marker-free deletion of a putative PKS gene. This was attained by using two gRNA targets at the beginning and end of the gene and a repair template containing 30 nt microhomology regions.42
CRISPR/Cas-based gene knockout strains have also been used to analyse individual biosynthetic steps within a secondary metabolite pathway. For example, a strain collection harbouring CRISPR/Cas9 knockouts of 15 genes from the demethoxyviridin BGC was constructed and analysed for the accumulation of pathway intermediates (Fig. 3B). These strains contributed to the proposal of a complete biosynthetic pathway for this cluster, as well as the structural elucidation of novel chemical intermediates.73 In a similar vein, the study of piperazine-containing alkaloids the brasiliamides was carried out in the native host Penicillium brasilianum. Firstly, brasiliamide production was activated through the deletion of a histone deacetylase. Following this the identification of the core NRPS and investigation of tailoring enzymes was aided by the production of BGC gene knockout mutants.86
Characterising enzyme functions and intermediates in vivo in the native fungus is particularly valuable when studying novel canonical biocatalytic activities. To this end, a CRISPR/Cas knockout strain was used by Dan et al. to further validate the native substrate of a proposed bifunctional enantioselective Diels-Alderase/reductase as part of their investigation of the biosynthesis pathway of prenylated indole alkaloids malbranchaemide and paraherquamides.83 Examination of accumulated pathway intermediates in the ΔphaE P. simplicissimum strain helped verify an unexpected zwitterionic intermediate as the native substrate for the Diels-Alderase/reductase PhaE and complemented the in vitro characterisation of its homologue MalC. A similar approach was pursued to characterise a putative spirocyclase from the same pathway.84 CRISPR/Cas9-based gene knockout also provided in vivo support for the rare formation of a non-heme enzyme heterodimer required for the biosynthesis of the terpenoids talaromyolides in their host Talaromyces purpureogenus.102 Furthermore, CRISPR/Cas has facilitated testing of short deletions to investigate putative condensation domains within β-lactam precursor NRPS enzymes in Penicillium rubens.103
Studying secondary metabolites by gene knockout in the native host is also a robust complement to heterologous expression when the metabolic products are suspected to be artifacts of the heterologous host machinery. For example, CRISPR/Cas knockouts of core and accessory biosynthetic genes of the thermolides BGC in Thermomyces dupontii clarified key biosynthetic steps that had previously been incorrectly identified by heterologous expression. The mutant strains derived from this study also shed light on the roles that these compounds play in development and stress response in the native fungus.26
The portability of AMA1-based CRISPR delivery has made it a useful tool for investigating the biological role of SMs across various species. For example, AMA1-based CRISPR knockout was used in studies of melanin and its role in heat tolerance in prominent food spoilage fungal species,89 as well as the role of melanin in radiation tolerance in a chernobyl nuclear powerplant isolate of Pa. variotii.88 In addition, AMA1 vector-based Cas9 knockout supported the study of the biological function of NRPS siderophore enzymes in iron acquisition in Al. alternata.30
Microbes often utilise SMs as effectors for virulence or for mediating communication with other microbes. CRISPR/Cas knockout has been used to create strains for studying the roles SMs play in these often-complex inter-species interactions. The knockout of a transcription factor in Purpureocillium lilacinum revealed its role in controlling the expression of the Leucinostatins BGC and revealed that expression was linked to antagonism against Phytophthora infestans, a microbe responsible for potato blight.77 In another study CRISPR/Cas9-based knockout was used to verify the biosynthetic origins of toxic virulence factor alternariol and derivative alternariol monomethyl ether in Al. alternata (Fig. 3C).29 The resulting deletion strains displayed a reduction in plant virulence. Another study in Sclerotinia sclerotiorum confirmed a putative melanin PKS by CRISPR/Cas-based knockout, and in Leptosphaeria maculans a putative cytochrome P450 was linked to abscisic acid production. However, in both of these studies examination of resulting knockout strains showed no obvious links between these metabolites and virulence.104,105
Additionally, knockouts can be used to study BGC regulation. In P. decumbens CRISPR/Cas9 knockouts aided in the identification of the calbistrin BGC and shed light on the importance of a putative transport pump for calbistrin export.28 In another fungus, Pes. fici, transcription factor knockouts were used in the study of regulatory mechanisms for the dihydroxynaphthalene melanin BGC.74
CRISPR/Cas knockouts have also been used to study the interplay between enzymes from the pathways that provide precursors and the production of specific secondary metabolites. For example, the interrogation of three HMG-CoA reductase genes from the mevalonate pathway of Mu. circinelloides, allowed their individual impact on terpenoid and ergosterol biosynthesis to be analysed.98
Knockouts can also be used to inactivate competing pathways to increase the production of specific compounds of interest. This approach was used by a study aiming to increase the production of the Aspergillus terreus polyketide FR901512, a tetralin statin with potent cholesterol-lowering activity. The SREBP gene involved in the regulation of sterol metabolism was knocked out with CRISPR/Cas9 after deletion attempts by homologous recombination failed. These deletions resulted in increased production of FR901512, apparently due to increased levels of polyketide precursors such as acetyl-CoA and malonyl-CoA from decreased sterol production.15 In a similar vein, the production of the β-lactam antibiotic cephalosporin C in Ac. chrysogenum was increased by knocking out the sorbicillinoid gene cluster which was thought to be competing for substrates.75
CRISPR/Cas knockouts have also been used to rewire secondary metabolite biosynthetic pathways to divert resources towards specific products of interest (Fig. 3D). A study by Shi et al. engineered the gibberellic acid metabolic pathway in Fu. fujikuroi to improve the accumulation of specific gibberellic acids by knockout of a P450 gene responsible for catalysing a branching reaction path.59 In another case, Me. brunneum produces lysergic acid which is a desirable lead compound, however, exists largely as a transient intermediate. Accumulation of lysergic acid was achieved by knockout of downstream enzyme lpsB, halting the biosynthetic pathway.97
Manipulating fungal morphology and development is another strategy to increase the yields of some compounds. CRISPR/Cas was used to delete a membrane protein gene involved in arthrospore formation in an industrial strain of Ac. chrysogenum used for the production of cephalosporin C. The mutant strain possessed increased arthrospore formation and triplicated compound titres, in agreement with the previously observed correlation between morphology and production of cephalosporin.109
3.2 Gain of function with CRISPR/Cas Gene knock-in
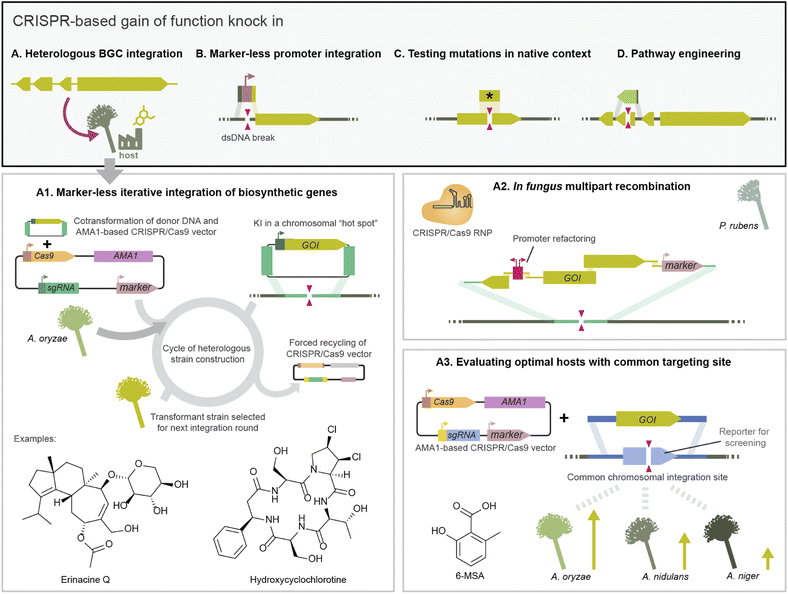
CRISPR/Cas gene editing can also be used to replace the sequence within a gene, enabling the study of mutations in the native context of a gene (Fig. 4C). For example, Weber et al. eliminated a single nucleotide polymorphism by CRISPR/Cas9 integration using template DNA with the corresponding functional sequence. For seamless integration within a coding sequence, they selected clones which had integrated in parallel a marker in a different locus.63 In other work, gene replacement was used to engineer the production of pneumocandin B0 in the industrial fungus Glarea lozoyensis by replacing a promiscuous proline hydroxylase with a heterologous variant which was more regiospecific, leading to a purer product.43 Finally, CRISPR/Cas integration has been used for genetic complementation to validate mutant phenotypes affecting development and pathogenicity. For example, in-cis complementation of the PKS15 gene in a ΔPKS15 strain of the entomopathogenic fungus Be. bassiana restored insect virulence and conidia morphology to wild type levels.87
CRISPR/Cas dsDNA breaks can also boost the efficiency of targeted integration of multi-kilobase DNA fragments, to the extent that marker-less integration of large biosynthetic genes becomes feasible. Pathway elucidation projects often opt for stepwise gene integration to obtain strains with truncated BGC variants which produce pathway intermediates. BGC integration via traditional HR often relied on recycling a marker encoded in the integrated donor DNA.116 As an alternative, Katayama et al. developed a CRISPR/Cas9 toolkit delivered on half-AMA1 vectors harbouring the inducible counter selection marker Aoace2 for forced recycling, facilitating subsequent marker-less integration events in the host As. oryzae (Fig. 4A).38 This toolkit was used to integrate sesquiterpene synthases (STs) from basidiomycetes into As. oryzae for heterologous expression in a locus previously identified in high production As. oryzae strains.76 They obtained 80–90% integration efficiency as evaluated by compound production, higher than HR alone, which facilitated screening 30 ST genes by heterologous expression. This platform was expanded to iteratively or in parallel target additional high expression loci in As. oryzae, and used to integrate ten genes in two transformation rounds to reconstitute the diterpene erancine BGC (Fig. 4A).70 CRISPR/Cas9 also facilitated integrating genes to produce UDP-xylose, which is needed for a xylose-specific glycosylation in the biosynthesis of erinacine Q.
An advantage of using CRISPR/Cas-integration for heterologous expression is that once the integration loci are selected, the gRNA(s) and homology arms can be reused in subsequent projects, only tailoring the donor DNA to incorporate the gene of interest. Effectively, the CRISPR/Cas9 platform for As. oryzae was repurposed to integrate other pathways to shed light on their biosynthetic mechanisms, for example the BGC encoding the indole terpenes lolitrems.117 Although terpene encoding genes are short and thus simpler to integrate, CRISPR/Cas9-KI in As. oryzae also proved effective for the stepwise integration of a larger NRPS BGC responsible for cyclochlorotine biosynthesis (Fig. 4A).93 Throughout these reports of BGC integration70,93,117 the transformation efficiency obtained was up to 100% as defined as the proportion of transformant colonies producing the expected compound from the total, which is not informative of integration specificity.
CRISPR/Cas9 has also enabled one-step integration of a large DNA construct split into multiple fragments containing homology regions to recombine in vivo in fungi. Following this strategy, Pohl et al. recombined up to 8 DNA fragments from the calbistrin BGC in the host P. rubens performing a dsDNA break with CRISPR/Cas9 RNP transformation.44 They also performed simultaneous BGC integration and promoter refactoring in P. rubens using multiple template fragments (Fig. 4A).44
CRISPR/Cas knock-in has also been used to optimise the heterologous production of BGCs. For example, to test different hosts for optimal production the DIVERSIFY platform uses the same sgRNA and homology arms to integrate a gene across multiple Aspergilli. This is achieved by previously integrating a common target site in all strains, in this case a gene for blue/white screening flanked by an exogenous homology sequence (Fig. 4A).82,118 In another work, CRISPR/Cas-integration was used to optimise an As. oryzae host for fungal terpenoid production by integrating additional copies of genes from the mevalonate pathway increasing the precursor pool.119 Beyond Aspergillus and Penicillium, it is relevant to develop heterologous hosts from other taxonomical classes. For example, gain of function and deletions via CRISPR/Cas9 RNP were used to engineer the ergot alkaloid pathway in Me. brunneum to produce lysergic acid and dihydrolysergic acid. The authors report increased metabolite yields and secretion compared to hosts from other lineages.97
3.3 Introducing nucleotide substitutions with CRISPR/Cas base editors
Cytidine to thymine (C-to-T) CRISPR base editing has been performed in As. niger to introduce an early stop codon in a PKS responsible for pigment production.48 Base editing has potential applications in protein engineering and multiplexing mutant combinations, and hence it shows promise as a tool that could be further developed for precision BGC engineering in fungi.1203.4 Modulating the expression of BGCs with CRISPR/dCas-effectors
Strategies for CRISPR-based activation of BGCs can be grouped into two main approaches. Some BGCs contain a cluster-specific transcription factor, which when targeted may allow the upregulation of the entire cluster. This was leveraged by Mozsik et al. for the activation of macrophorin production.58 This approach was also used by Schuller et al. who demonstrated transcriptional upregulation of the monodictyphenone cluster and an unknown NRPS.57 However, this targeting criterion is constrained by the fact that many clusters do not contain a transcription factor. Targeting individual biosynthetic genes within a cluster is another valuable approach. Roux et al. targeted the core NRPS-like gene and a cytochrome P450, allowing stepwise investigation of the gene cluster and leading to the discovery of the final product, dehydromicroperfuranone.55 In the same vein, Li et al. observed transcriptional upregulation from p300-based activation of the core biosynthetic gene in the brevianamide F cluster and increased compound production when targeting the fumonisin B2 cluster.56 Additionally, CRISPRa has been used to increase the expression of an episomally encoded biosynthetic gene micA for microperfuranone production, which shows that CRISPRa could be valuable in the context of heterologous expression.55
CRISPRa efficiency is strongly tied to gRNA target locations. In general, the optimum window for CRISPRa targeting is upstream of the transcription start site (TSS). However, targeting too close to or downstream of the TSS is associated with repressive effects.121 In filamentous fungi TSS data is not always available therefore the screening of multiple targets may be necessary to identify functional sites relative to start codon location. Moszik et al. screened a pool of 20 guide sites tiling the entire range of a promoter and found that only 2/20 resulted in sufficient transcription factor activation for compound production.58 While Li et al. screened roughly four sites in a region 100–600 bp upstream from the start codon of each promoter, finding significant transcriptional activation from 30–50% of their gRNAs.56 Analysis of characterised promoters in As. nidulans by Roux et al. revealed that many genes have short 5′ untranslated regions (UTRs) (median 100 bp) Using this data, Roux et al. tested a generic targeting criterion focused on a region 140–330 bp upstream of the start codon, as well as a tailored criterion 119–303 bp upstream of the predicted TSS. Interestingly, when targeted with arrays of four Cas12a crRNAs both regions resulted in successful activation despite the generic criterion falling within a predicted 5′ UTR region for the particular gene targeted.
A different approach employed by Schuller et al. used RNA sequencing and nucleosome positioning data to prioritise guide targets. When targeting a nucleosome-free region in a bi-promoter two guides tested resulted in increases to gene expression of between 3.5 – 4000-fold. However, at a second promoter no significant activation was observed from individual guides targeting either a nucleosome-free region or a region near the predicted TSS. Despite this, combinations of two to four of these guides allowed significant transcriptional activation.57 This suggests that for some promoters targeting with arrays of multiple guides may be necessary for strong activation. In line with this, a synergistic activation effect was observed from the four-gRNA per gene strategy used by Roux et al. and no individual gRNA was observed to significantly support activation.55 Overall, CRISPRa represents a potentially scalable strategy for BGC activation, but more evaluation is needed to refine targeting strategies in filamentous fungi.
3.5 Cloning and editing of fungal BGCs in plasmids with CRISPR/Cas
Genomic DNA capture is a technique that has been used to clone large fungal BGCs into vectors for heterologous expression. While it has previously relied on random shearing of genomic DNA,122in vitro CRISPR/Cas digestion can be used to selectively release a DNA fragment containing a BGC of interest for cloning. Recently, in vitro CRISPR/Cas digestion of the gDNA from Neosartorya fischeri coupled with transformation associated recombination in yeast (TAR cloning) has been used to capture an 18-kb BGC into fungal expression vectors. However, the authors report that more optimisation is needed to capture larger clusters.123 Manipulating large plasmids containing BGCs can be troublesome with classic molecular biology techniques. As an alternative, in vivo CRISPR/Cas cleavage in yeast coupled with TAR cloning was used to eliminate tailoring enzyme genes from a shuttle AMA1-based plasmid containing the 27 kb elsinochrome BGC from Parastagonospora nodorum.1244. Perspectives
4.1 Avoiding pitfalls and refining the toolbox
To continue optimising the CRISPR/Cas toolbox for filamentous fungi it is important to know its current limitations. In general, precision and accuracy are key aspects to validate in CRISPR tool development. The risk of unintended CRISPR/Cas off-target and on-target effects remains actively investigated across biological systems.24,125 However, this area remains comparatively understudied in fungal BGC manipulation, with just a few examples of empirical genome-wide off-target analysis. For example, using whole genome sequencing, Pohl et al. compared a P. rubens parental strain with a CRISPR mutant strain after four BGCs were inactivated using Cas9 RNPs.44 Although this revealed multiple mutations, the involvement of CRISPR off-targets was ruled out by comparing mutated loci with in silico predicted off-targets. It is understandable that genome wide monitoring of off-targets is not accessible in all projects but characterising possible pitfalls of CRISPR/Cas tools across fungi remains valuable.125On the other hand, undesired on-target effects such as genome rearrangements, large deletions and undesired integration have been observed when using CRISPR/Cas nucleases in human cell lines.126 In fungal BGC engineering some unexpected on-target outcomes have been reported such as integration of multiple copies of template DNA in tandem38 or the integration of AMA1-vector fragments.28,89 Errors have also been observed when co-integrating multiple fragments harbouring identical genetic parts such as the same promoter.44 These reflect the importance of reporting unexpected outcomes of CRISPR/Cas tools in filamentous fungi, as they can guide us to further refine our editing strategies.
Establishing CRISPR/Cas tools in underexplored fungal taxa is valuable for natural product discovery as there is BGC novelty at the fungal species level. However, filamentous fungi display a wide diversity of growth temperatures, which could limit the application of CRISPR/Cas tools that are temperature sensitive. Progress has been made in this area by using mutant Cas proteins, or by branching out to temperature tolerant Cas variants. For example, a thermophilic CRISPR/Cas system, GeoCas9, was adopted for BGC interrogation at 45 °C in Th. dupontii.26 On the other hand, the mutant dLbCas12aD156R was proven effective for CRISPRa in As. nidulans at 25 °C, a limiting temperature for WT LbCas12a which performs optimally at 37 °C.55
4.2 More tools on the way? Prospects and opportunities
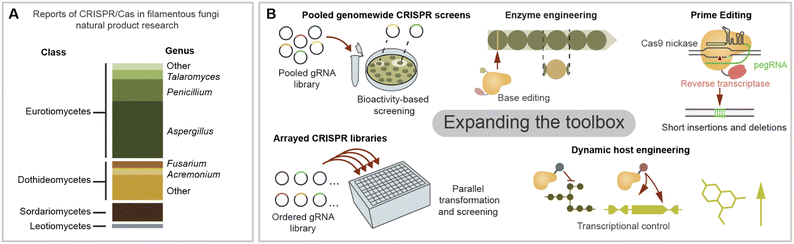
As next-generation CRISPR/Cas tools emerge, several bottlenecks in natural product discovery and engineering can be re-examined in a new light. CRISPR/Cas-based technologies are often first developed in more popular eukaryotic model systems such as human cell lines or yeast. Adapting them to filamentous fungi is increasingly feasible thanks to the current knowledge on CRISPR/Cas expression strategies and system validation across diverse fungal species. In the last few years, a suite of gene editing tools free of dsDNA cleavage has been developed including base editors, transposases/recombinases, and prime editors.127 CRISPR prime editing makes use of a Cas protein linked to a retrotranscriptase which uses an engineered gRNA as template for targeted integrations (Fig. 5B).128 Prime editing has been used to integrate sequences <100 bp, including short landing sites for integration via site-specific recombinases such as Cre/loxp. Site specific recombination has recently been used for the integration of a large BGC in filamentous fungi,114 and CRISPR/Cas could facilitate the integration of recombinase landing sites in fungi in which HR remains limiting. Additionally, CRISPR prime editing stands as a promising tool for protein and promoter engineering.Beyond new CRISPR/Cas functions, we can also take inspiration from the experimental strategies used in other organisms to boost BGC research in filamentous fungi. One of the biggest bottlenecks in fungal genome mining is the gap between BGC identification and the characterisation of chemical phenotype. As genome sequences continue accumulating in public databases, it is crucial to scale up the functional genetic interrogation of promising BGC candidates. One strength of CRISPR-based technologies is their potential scalability for high-throughput knockout-, knockdown- and overexpression screening. In several eukaryotic organisms it is now possible to interrogate a whole genome or a large subset of genes using a gRNA library.129 In a pooled CRISPR screen, a mixed gRNA library is used in bulk for a single transformation generating diverse transformants, each harbouring a different gRNA, which are assayed for the desired phenotype. The identity of gRNAs responsible for the phenotype is later identified by PCR amplification and sequencing (Fig. 5B).130 In contrast, individual gRNA libraries can be used to create ordered strain collections which are tested in parallel (Fig. 5B).129 For example, recently a small arrayed library of As. niger was developed using CRISPR/Cas integration for morphology engineering, which is known to impact the yield of some compounds.131 CRISPR/Cas protocols optimised for microtiter plates and robotics could enable the construction of larger arrayed libraries by scaled down transformation reactions and increases to throughput.132 Although pooled assays could be simpler to scale up, sorting chemical phenotypes can be troublesome. This could be circumvented making use of microfluidics to sort large libraries and biosensors or bioactivities to screen for small molecules production.133 For example, recently ∼200 engineered As. oryzae strains were screened to produce heterologous terpenoids with anti-inflammatory proprieties by automated robotics in a biofoundry set up.119
When considering future applications of CRISPR for fungal BGC engineering, we can also take inspiration from initiatives in bacterial BGCs.134 For example, developing chimeric or mutant core enzymes capable of synthetising new-to-nature natural products has been performed successfully for bacterial NRPS by swapping domains in vivo aided with CRISPR/Cas9.135 On the other hand, in vitro CRISPR/Cas9 cleavage was used to swap an acyltransferase domain within a bacterial PKS encoded in a >100 kb vector.136 Another area which is widely developed in actinomycete BGC manipulation is the use of CRISPR/Cas tools for BGC cloning in vitro or by TAR cloning, and we refer the reader to some relevant examples,137–140 in which similar strategies could be adopted for fungal BGC engineering. In the area of microbial compound production, CRISPR/Cas transcriptional circuits have gathered interest as a tool for the dynamic control of pathways for improved yields (Fig. 5B).141
Finally, we consider the CRISPR/Cas toolbox has great potential to be expanded to underexploited biosynthetically relevant fungal taxa. The literature in this review centred on fungi from the pezizomycotina taxon, we noticed that more than half of CRISPR/Cas reports in natural product research were on Eurotiomycetes fungi, mainly from the Aspergillus and Penicillium genera (Fig. 5A). Interestingly, applications in Sordariomycetes are comparatively underexplored considering their prominence in natural product research.142 Nevertheless, there are auspicious examples of engineering BGCs in unconventional fungus, including those from the Dothiodeomycetes and Leotiomycetes classes (Fig. 5A). There is also potential to expand the toolbox to other biosynthetically relevant fungal taxa not covered in their review, such as Basidiomycetes143 or anaerobic fungi.144 Considering the multiple intersections of filamentous fungi diversity (nucleus number, sexual cycle, growth temperature, lichenised lifestyle, etc.), leveraging the potential of currently available tools will also require awareness of the uniqueness of their physiology. Expanding the CRISPR/Cas toolbox will then not only allow us to uncover the cryptic biosynthetic space of diverse fungi but also shed light on their underexplored biology.
5. Conflicts of interest
There are no conflicts to declare.6. Acknowledgements
This work was supported by the Australian Research Council (DP210102180 and FT160100233). C.W. is the recipient of a UWA PhD Scholarship.7. Notes and references
- M. T. Robey, L. K. Caesar, M. T. Drott, N. P. Keller and N. L. Kelleher, Proc. Natl. Acad. Sci., 2021, 118, e2020230118 CrossRef CAS PubMed.
- F. Peláez, Handbook of Industrial Mycology, CRC Press, 2004, pp. 68–111 Search PubMed.
- A. Schueffler and T. Anke, Nat. Prod. Rep., 2014, 31, 1425–1448 RSC.
- N. P. Keller, Nat. Rev. Microbiol., 2019, 17, 167–180 CrossRef CAS.
- Y. H. Chooi and Y. Tang, J. Org. Chem., 2012, 77, 9933–9953 CrossRef CAS.
- A. A. Brakhage, Nat. Rev. Microbiol., 20122012, 11111, 21–32 Search PubMed.
- A. Madhavan, K. B. Arun, R. Sindhu, A. Alphonsa Jose, A. Pugazhendhi, P. Binod, R. Sirohi, R. Reshmy and M. Kumar Awasthi, Bioresour. Technol., 2022, 344, 126209 CrossRef CAS PubMed.
- R. Song, Q. Zhai, L. Sun, E. Huang, Y. Zhang, Y. Zhu, Q. Guo, Y. Tian, B. Zhao and H. Lu, Appl. Microbiol. Biotechnol., 2019, 103, 6919–6932 CrossRef CAS.
- M. Schuster and R. Kahmann, Fungal Genet. Biol., 2019, 130, 43–53 CrossRef CAS.
- M. Ullah, L. Xia, S. Xie and S. Sun, Biotechnol. Appl. Biochem., 2020, 67, 835–851 CrossRef CAS.
- J. P. Ouedraogo and A. Tsang, Fungal Biol. Rev., 2020, 34, 189–201 CrossRef.
- A. M. Rozhkova and V. Y. Kislitsin, Biochem, 2021, 86, S120–S139 CAS.
- S. Krappmann, Fungal Biol. Rev., 2007, 21, 25–29 CrossRef.
- U. Kück and B. Hoff, Appl. Microbiol. Biotechnol., 2010, 86, 51–62 CrossRef.
- H. Itoh, A. Miura, M. Matsui, T. Arazoe, K. Nishida, T. Kumagai, M. Arita, K. Tamano, M. Machida and T. Shibata, Appl. Microbiol. Biotechnol., 2018, 102, 1393–1405 CrossRef CAS PubMed.
- H. Deng, R. Gao, X. Liao and Y. Cai, J. Biotechnol., 2017, 259, 228–234 CrossRef CAS PubMed.
- S. E. Clemmensen, K. J. K. Kromphardt and R. J. N. Frandsen, Fungal Genet. Biol., 2022, 160, 103689 CrossRef CAS.
- J. A. Doudna and E. Charpentier, Science, 2014, 346(6213), 1258096-1–1258096-9 CrossRef PubMed.
- K. S. Makarova, Y. I. Wolf, J. Iranzo, S. A. Shmakov, O. S. Alkhnbashi, S. J. J. Brouns, E. Charpentier, D. Cheng, D. H. Haft, P. Horvath, S. Moineau, F. J. M. Mojica, D. Scott, S. A. Shah, V. Siksnys, M. P. Terns, Č. Venclovas, M. F. White, A. F. Yakunin, W. Yan, F. Zhang, R. A. Garrett, R. Backofen, J. van der Oost, R. Barrangou and E. V. Koonin, Nat. Rev. Microbiol., 2019, 18, 67–83 CrossRef PubMed.
- E. Deltcheva, K. Chylinski, C. M. Sharma, K. Gonzales, Y. Chao, Z. A. Pirzada, M. R. Eckert, J. Vogel and E. Charpentier, Nature, 2011, 471, 602–607 CrossRef CAS.
- I. Fonfara, H. Richter, M. BratoviÄ, A. Le Rhun and E. Charpentier, Nature, 2016, 532, 517–521 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- C. D. Yeh, C. D. Richardson and J. E. Corn, Nat. Cell Biol., 2019, 21, 1468–1478 CrossRef CAS.
- T. S. Nambiar, L. Baudrier, P. Billon and A. Ciccia, Mol. Cell, 2022, 82, 348–388 CrossRef CAS.
- T. Katayama, Y. Tanaka, T. Okabe, H. Nakamura, W. Fujii, K. Kitamoto, J. ichi Maruyama, J. ichi Maruyama and J. ichi Maruyama, Biotechnol. Lett., 2016, 38, 637–642 CrossRef CAS PubMed.
- W. P. Huang, Y. J. Du, Y. Yang, J. N. He, Q. Lei, X. Y. Yang, K. Q. Zhang and X. M. Niu, Appl. Environ. Microbiol., 2020, 86(20), 1–18 CrossRef.
- A. S. Urquhart, C. E. Elliott, W. Zeng and A. Idnurm, PLoS One, 2021, 16, e0252333 CrossRef CAS.
- S. Grijseels, C. Pohl, J. C. Nielsen, Z. Wasil, Y. Nygård, J. C. Nielsen, J. C. Frisvad, K. F. Nielsen, M. Workman, T. O. Larsen, A. J. M. Driessen and R. J. N. Frandsen, Fungal Biol. Biotechnol., 2018, 5, 18 CrossRef.
- M. Wenderoth, F. Garganese, M. Schmidt-Heydt, S. T. Soukup, A. Ippolito, S. M. Sanzani and R. Fischer, Mol. Microbiol., 2019, 112, 131–146 CrossRef CAS PubMed.
- B. Voß, F. Kirschhöfer, G. Brenner-Weiß and R. Fischer, Sci. Rep., 2020, 10, 3587 CrossRef PubMed.
- M. Ferrara, M. Haidukowski, A. F. Logrieco, J. F. Leslie and G. Mulè, Sci. Rep., 2019, 9, 19836 CrossRef CAS PubMed.
- K. K. Fuller, S. Chen, J. J. Loros and J. C. Dunlap, Eukaryot. Cell, 2015, 14, 1073–1080 CrossRef CAS.
- Y. M. Zheng, F. L. Lin, H. Gao, G. Zou, J. W. Zhang, G. Q. Wang, G. D. Chen, Z. H. Zhou, X. S. Yao and D. Hu, Sci. Rep., 20172017, 717, 9250 Search PubMed.
- C. S. Nødvig, J. B. Nielsen, M. E. Kogle and U. H. Mortensen, PLoS One, 2015, 10, e0133085 CrossRef PubMed.
- C. Pohl, J. A. K. W. Kiel, A. J. M. Driessen, R. A. L. Bovenberg and Y. Nygård, ACS Synth. Biol., 2016, 5, 754–764 CrossRef CAS.
- W. Liu, C. An, X. Shu, X. Meng, Y. Yao, J. Zhang, F. Chen, H. Xiang, S. Yang, X. Gao and S. S. Gao, ACS Synth. Biol., 2020, 9, 2087–2095 CrossRef CAS PubMed.
- M. L. Nielsen, T. Isbrandt, K. B. Rasmussen, U. Thrane, J. B. Hoof, T. O. Larsen and U. H. Mortensen, PLoS One, 2017, 12, e0169712 CrossRef.
- T. Katayama, H. Nakamura, Y. Zhang, A. Pascal, W. Fujii and J. I. Maruyama, Appl. Environ. Microbiol., 2019, 85(3), 1–16 CrossRef.
- C. S. Nødvig, J. B. Hoof, M. E. Kogle, Z. D. Jarczynska, J. Lehmbeck, D. K. Klitgaard and U. H. Mortensen, Fungal Genet. Biol., 2018, 115, 78–89 CrossRef.
- S. Valente, E. Piombo, V. Schroeckh, G. R. Meloni, T. Heinekamp, A. A. Brakhage and D. Spadaro, Front. Microbiol., 2021, 12, 660871 CrossRef PubMed.
- H. Dong, J. Zheng, D. Yu, B. Wang and L. Pan, J. Microbiol. Methods, 2019, 163, 105655 CrossRef CAS PubMed.
- M. E. Futyma, Y. Guo, C. Hoeck, J. B. Hoof, C. H. Gotfredsen, U. H. Mortensen and T. O. Larsen, Appl. Microbiol. Biotechnol., 2021, 5113–5121 CrossRef CAS.
- T. Y. Wei, Y. J. Wu, Q. P. Xie, J. W. Tang, Z. T. Yu, S. B. Yang and S. X. Chen, ACS Synth. Biol., 2020, 9, 1968–1977 CrossRef CAS PubMed.
- C. Pohl, F. Polli, T. Schütze, A. Viggiano, L. Mózsik, S. Jung, M. de Vries, R. A. L. Bovenberg, V. Meyer and A. J. M. Driessen, Sci. Rep., 2020, 10, 7630 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. Lin, R. Barretto, N. Habib, P. D. Hsu, X. Wu, W. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2015, 533, 420–424 CrossRef.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS.
- L. Huang, H. Dong, J. Zheng, B. Wang and L. Pan, Microbiol. Res., 2019, 223–225, 44–50 CrossRef CAS PubMed.
- C. Zhang, N. Li, L. Rao, J. Li, Q. Liu and C. Tian, Microbiol. Spectr., 2022, e0232121 CrossRef.
- B. Zetsche, J. S. Gootenberg, O. O. Abudayyeh, I. M. Slaymaker, K. S. Makarova, P. Essletzbichler, S. E. Volz, J. Joung, J. Van Der Oost, A. Regev, E. V. Koonin and F. Zhang, Cell, 2015, 163, 759–771 CrossRef CAS PubMed.
- A. A. Dominguez, W. A. Lim and L. S. Qi, Nat. Rev. Mol. Cell Biol., 2016, 17, 5–15 CrossRef CAS PubMed.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173 CrossRef CAS.
- Y. E. Tak, B. P. Kleinstiver, J. K. Nuñez, J. Y. Hsu, J. E. Horng, J. Gong, J. S. Weissman and J. K. Joung, Nat. Methods, 2017, 14, 1163–1166 CrossRef CAS PubMed.
- M. Nakamura, Y. Gao, A. A. Dominguez and L. S. Qi, Nat. Cell Biol., 2021, 23, 11–22 CrossRef CAS PubMed.
- I. Roux, C. Woodcraft, J. Hu, R. Wolters, C. L. M. Gilchrist and Y. H. Chooi, ACS Synth. Biol., 2020, 9, 1843–1854 CrossRef CAS PubMed.
- X. Li, L. Huang, L. Pan, B. Wang and L. Pan, Microbiol. Res., 2021, 245, 126694 CrossRef CAS.
- A. Schüller, L. Wolansky, H. Berger, L. Studt, A. Gacek-matthews, M. Sulyok and J. Strauss, Appl. Microbiol. Biotechnol., 2020, 104, 9801–9822 CrossRef.
- L. Mózsik, M. Hoekzema, N. A. W. de Kok, R. A. L. Bovenberg, Y. Nygård and A. J. M. Driessen, Sci. Rep., 2021, 11, 1118 CrossRef.
- T. Q. Shi, J. Gao, W. J. Wang, K. F. Wang, G. Q. Xu, H. Huang and X. J. Ji, ACS Synth. Biol., 2019, 8, 445–454 CrossRef CAS.
- Q. Wang, P. A. Cobine and J. J. Coleman, Fungal Genet. Biol., 2018, 117, 21–29 CrossRef CAS.
- S. Krappmann, Med. Mycol., 2017, 55, 16–23 CrossRef CAS.
- L. Song, J. P. Ouedraogo, M. Kolbusz, T. T. M. Nguyen and A. Tsang, PLoS One, 2018, 13, e0202868 CrossRef.
- J. Weber, V. Valiante, C. S. Nødvig, D. J. Mattern, R. A. Slotkowski, U. H. Mortensen and A. A. Brakhage, ACS Synth. Biol., 2017, 6, 62–68 CrossRef CAS PubMed.
- L. Mózsik, C. Pohl, V. Meyer, R. A. L. Bovenberg, Y. Nygård and A. J. M. Driessen, ACS Synth. Biol., 2021, 10, 2850–2861 CrossRef.
- L. Song, J. P. Ouedraogo, M. Kolbusz, T. T. M. Nguyen and A. Tsang, PLoS One, 2018, 13, e0202868 CrossRef PubMed.
- R. Chari, N. C. Yeo, A. Chavez and G. M. Church, ACS Synth. Biol., 2017, 6, 902 CrossRef CAS.
- K. Hiranniramol, Y. Chen, Y. Chen, W. Liu, W. Liu and X. Wang, Bioinformatics, 2020, 36, 2684 CrossRef CAS PubMed.
- K. Labun, T. G. Montague, M. Krause, Y. N. Torres Cleuren, H. Tjeldnes and E. Valen, Nucleic Acids Res., 2019, 47, W171–W174 CrossRef CAS.
- D. Peng and R. Tarleton, Microb. Genomics, 2015, 1, e000033 Search PubMed.
- C. Liu, A. Minami, T. Ozaki, J. Wu, H. Kawagishi, J. I. Maruyama and H. Oikawa, J. Am. Chem. Soc., 2019, 141, 15519–15523 CrossRef CAS PubMed.
- G. Chen and J. Chu, Appl. Biochem. Biotechnol., 2019, 188, 1134–1144 CrossRef CAS.
- R. Liu, L. Chen, Y. Jiang, Z. Zhou and G. Zou, Cell Discov., 2015, 1, 1–11 Search PubMed.
- G. Q. Wang, G. D. Chen, S. Y. Qin, D. Hu, T. Awakawa, S. Y. Li, J. M. Lv, C. X. Wang, X. S. Yao, I. Abe and H. Gao, Nat. Commun., 2018, 9, 1–13 CrossRef.
- P. Zhang, S. Zhou, G. Wang, Z. An, X. Liu, K. Li and W. B. Yin, Mol. Microbiol., 2019, 112, 649–666 CrossRef CAS.
- C. Chen, J. Liu, C. Duan, Y. Pan and G. Liu, Fungal Genet. Biol., 2020, 134, 103279 CrossRef CAS PubMed.
- S. Nagamine, C. Liu, J. Nishishita, T. Kozaki, K. Sogahata, Y. Sato, A. Minami, T. Ozaki, C. Schmidt-Dannert, J. ichi Maruyama and H. Oikawa, Appl. Environ. Microbiol., 2019, 85, e00409–e00419 CrossRef CAS PubMed.
- Y. Jiao, Y. Li, Y. Li, H. Cao, Z. Mao, J. Ling, Y. Yang and B. Xie, Appl. Microbiol. Biotechnol., 2019, 103, 6187–6194 CrossRef CAS.
- Y. Wang, D. Wei, X. Zhu, J. Pan, P. Zhang, L. Huo and X. Zhu, Sci. Rep., 20162016, 616, 31145 Search PubMed.
- B. Xiang, X. Hao, Q. Xie, G. Shen, Y. Liu and X. Zhu, J. Fungi, 2021, 7, 750 CrossRef CAS.
- D. Gems, I. L. Johnstone and A. J. Clutterbuck, Gene, 1991, 98, 61–67 CrossRef CAS.
- S. Theobald, T. C. Vesth, J. K. mmer Rendsvig, K. F. Nielsen, R. Riley, L. M. de Abreu, A. Salamov, J. C. Frisvad, T. O. Larsen, M. R. Andersen and J. B. Hoof, Manuscr. Rev., 2018, 17957 CAS.
- Z. D. Jarczynska, J. K. H. Rendsvig, N. Pagels, V. R. Viana, C. S. Nødvig, F. H. Kirchner, T. Strucko, M. L. Nielsen and U. H. Mortensen, ACS Synth. Biol., 2021, 10, 579–588 CrossRef CAS.
- Q. Dan, S. A. Newmister, K. R. Klas, A. E. Fraley, T. J. McAfoos, A. D. Somoza, J. D. Sunderhaus, Y. Ye, V. V. Shende, F. Yu, J. N. Sanders, W. C. Brown, L. Zhao, R. S. Paton, K. N. Houk, J. L. Smith, D. H. Sherman and R. M. Williams, Nat. Chem., 2019, 11, 972–980 CrossRef CAS.
- A. E. Fraley, K. Caddell Haatveit, Y. Ye, S. P. Kelly, S. A. Newmister, F. Yu, R. M. Williams, J. L. Smith, K. N. Houk and D. H. Sherman, J. Am. Chem. Soc., 2020, 142, 2244–2252 CrossRef CAS.
- M. Wenderoth, C. Pinecker, B. Voß and R. Fischer, Fungal Genet. Biol., 2017, 101, 55–60 CrossRef CAS.
- B. Yuan, D. Liu, X. Guan, Y. Yan, J. Zhang, Y. Zhang, D. Yang, M. Ma and W. Lin, Appl. Microbiol. Biotechnol., 2020, 104, 6149–6159 CrossRef CAS.
- S. Udompaisarn, W. Toopaang, U. Sae-Ueng, C. Srisuksam, N. Wichienchote, R. Wasuwan, N. A. S. Nahar, M. Tanticharoen and A. Amnuaykanjanasin, Sci. Rep., 2020, 10, 12630 CrossRef PubMed.
- S. Lim, S. Bijlani, A. Blachowicz, Y. M. Chiang, M. S. Lee, T. Torok, K. Venkateswaran and C. C. C. Wang, Fungal Genet. Biol., 2021, 152, 103567 CrossRef CAS.
- S. J. Seekles, P. P. P. Teunisse, M. Punt, T. van den Brule, J. Dijksterhuis, J. Houbraken, H. A. B. Wösten and A. F. J. Ram, Fungal Biol. Biotechnol., 2021, 8, 4 CrossRef CAS.
- T. T. Dao, K. M. J. de Mattos-Shipley, I. M. Prosser, K. Williams, M. K. Zacharova, C. M. Lazarus, C. L. Willis and A. M. Bailey, Front. Fungal Biol., 2021, 2, 632542 CrossRef.
- J. Kuivanen, Y. M. J. Wang and P. Richard, Microb. Cell Fact., 2016, 15, 210 CrossRef.
- P. Sarkari, H. Marx, M. L. Blumhoff, D. Mattanovich, M. Sauer and M. G. Steiger, Bioresour. Technol., 2017, 245, 1327–1333 CrossRef CAS PubMed.
- Y. Jiang, T. Ozaki, C. Liu, Y. Igarashi, Y. Ye, S. Tang, T. Ye, J. I. Maruyama, A. Minami and H. Oikawa, Org. Lett., 2021, 23, 2616–2620 CrossRef.
- L. Yu, M. Xiao, Z. Zhu, Y. Wang, Z. Zhou, P. Wang and G. Zou, Synth. Syst. Biotechnol., 2022, 7, 664–670 CrossRef PubMed.
- C. A. Vakulskas and M. A. Behlke, Nucleic Acid Ther., 2019, 29, 167–174 CrossRef CAS PubMed.
- Q. Al Abdallah, W. Ge and J. R. Fortwendel, mSphere, 2017, 2(6), 1–14 CrossRef.
- K. A. Davis, J. K. Sampson and D. G. Panaccione, Appl. Environ. Microbiol., 2020, 86(19), 1–14 CrossRef PubMed.
- G. Nagy, A. G. Vaz, C. Szebenyi, M. Takó, E. J. Tóth, Á. Csernetics, O. Bencsik, A. Szekeres, M. Homa, F. Ayaydin, L. Galgóczy, C. Vágvölgyi and T. Papp, Fungal Genet. Biol., 2019, 129, 30–39 CrossRef CAS PubMed.
- S. Florea, J. Jaromczyk and C. L. Schardl, Toxins, 2021, 13, 153 CrossRef CAS.
- G. D. Chen, D. Hu, M. J. Huang, J. Tang, X. X. Wang, J. Zou, J. Xie, W. G. Zhang, L. D. Guo, X. S. Yao, I. Abe and H. Gao, Chem. Commun., 2020, 56, 4607–4610 RSC.
- S. W. Yuan, S. H. Chen, H. Guo, L. T. Chen, H. J. Shen, L. Liu and Z. Z. Gao, Org. Lett., 2022, 24, 3069–3074 CrossRef CAS PubMed.
- X. Li, T. Awakawa, T. Mori, M. Ling, D. Hu, B. Wu and I. Abe, J. Am. Chem. Soc., 2021, 143, 21425–21432 CrossRef CAS PubMed.
- R. Iacovelli, L. Mózsik, R. A. L. Bovenberg and A. J. M. Driessen, Microbiologyopen, 2021, 10, e1145 CrossRef CAS PubMed.
- J. Li, Y. Zhang, Y. Zhang, P. L. Yu, H. Pan and J. A. Rollins, MBio, 2018, 9(3), 1–19 CrossRef.
- R. Darma, A. Lutz, C. E. Elliott and A. Idnurm, Fungal Genet. Biol., 2019, 130, 62–71 CrossRef CAS PubMed.
- N. Liu, Y. S. Hung, S. S. Gao, L. Hang, Y. Zou, Y. H. Chooi and Y. Tang, Org. Lett., 2017, 19, 3560–3563 CrossRef CAS.
- Y. Zhu, J. Wang, P. Mou, Y. Yan, M. Chen and Y. Tang, Org. Biomol. Chem., 2021, 19, 1985–1990 RSC.
- I. C. Okorafor, M. Chen and Y. Tang, ACS Synth. Biol., 2021, 10, 2159–2166 CrossRef CAS PubMed.
- Y. Xu, L. Liu, Z. Chen, X. Tian and J. Chu, J. Biotechnol., 2022, 347, 26–39 CrossRef CAS PubMed.
- J. C. Frisvad, L. L. H. Møller, T. O. Larsen, R. Kumar and J. Arnau, Appl. Microbiol. Biotechnol., 2018, 102, 9481–9515 CrossRef CAS.
- X. Zheng, P. Zheng, K. Zhang, T. C. Cairns, V. Meyer, J. Sun and Y. Ma, ACS Synth. Biol., 2018, 8, 1568–1574 CrossRef PubMed.
- H. Deng, W. Liang, T. P. Fan, X. Zheng and Y. Cai, Int. J. Biol. Macromol., 2020, 165, 796–803 CrossRef CAS PubMed.
- G. Evdokias, C. Semper, M. Mora-Ochomogo, M. Di Falco, T. T. M. Nguyen, A. Savchenko, A. Tsang and I. Benoit-Gelber, J. Fungi, 2021, 7, 374 CrossRef CAS PubMed.
- I. Roux and Y. H. Chooi, ACS Synth. Biol., 2022, 11, 1186–1195 CrossRef CAS PubMed.
- X. Wei, T. Matsuyama, H. Sato, D. Yan, P. M. Chan, K. Miyamoto, M. Uchiyama and Y. Matsuda, J. Am. Chem. Soc., 2021, 143, 17708–17715 CrossRef CAS PubMed.
- M. T. Nielsen, J. B. Nielsen, D. C. Anyaogu, D. K. Holm, K. F. Nielsen, T. O. Larsen and U. H. Mortensen, PLoS One, 2013, 8, e72871 CrossRef CAS PubMed.
- Y. Jiang, T. Ozaki, M. Harada, T. Miyasaka, H. Sato, K. Miyamoto, J. Kanazawa, C. Liu, J. ichi Maruyama, M. Adachi, A. Nakazaki, T. Nishikawa, M. Uchiyama, A. Minami and H. Oikawa, Angew. Chem., Int. Ed., 2020, 59, 17996–18002 CrossRef CAS PubMed.
- Y. Guo, F. J. Contesini, X. Wang, S. Ghidinelli, D. S. Tornby, T. E. Andersen, U. H. Mortensen and T. O. Larsen, Org. Lett., 2022, 24, 804–808 CrossRef CAS.
- Y. Yuan, S. Cheng, G. Bian, P. Yan, Z. Ma, W. Dai, R. Chen, S. Fu, H. Huang, H. Chi, Y. Cai, Z. Deng and T. Liu, Nat. Catal., 2022, 5, 277–287 CrossRef CAS.
- Y. Wang, Y. Liu, P. Zheng, J. Sun and M. Wang, Trends Biotechnol., 2021, 39, 165–180 CrossRef CAS.
- M. K. Jensen, FEMS Yeast Res., 2018, 18, 39 CrossRef.
- J. W. Bok, R. Ye, K. D. Clevenger, D. Mead, M. Wagner, A. Krerowicz, J. C. Albright, A. W. Goering, P. M. Thomas, N. L. Kelleher, N. P. Keller and C. C. Wu, BMC Genomics, 2015, 16, 343 CrossRef.
- X. Xu, J. Feng, P. Zhang, J. Fan and W. B. Yin, J. Microbiol. Biotechnol., 2021, 31, 8–15 CrossRef CAS PubMed.
- J. Hu, F. Sarrami, H. Li, G. Zhang, K. A. Stubbs, E. Lacey, S. G. Stewart, A. Karton, A. M. Piggott and Y. H. Chooi, Chem. Sci., 2019, 10, 1457–1465 RSC.
- D. Kim, K. Luk, S. A. Wolfe and J. S. Kim, Annu. Rev. Biochem., 2019, 88, 191–220 CrossRef CAS PubMed.
- J. Boutin, D. Cappellen, J. Rosier, S. Amintas, S. Dabernat, A. Bedel and F. Moreau-Gaudry, Cris. J., 2022, 5, 19–30 CrossRef CAS PubMed.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Nat. Biotechnol., 20202020, 387(38), 824–844 Search PubMed.
- A. V. Anzalone, X. D. Gao, C. J. Podracky, A. T. Nelson, L. W. Koblan, A. Raguram, J. M. Levy, J. A. M. Mercer and D. R. Liu, Nat. Biotechnol., 2021, 40, 731–740 CrossRef PubMed.
- C. Bock, P. Datlinger, F. Chardon, M. A. Coelho, M. B. Dong, K. A. Lawson, T. Lu, L. Maroc, T. M. Norman, B. Song, G. Stanley, S. Chen, M. Garnett, W. Li, J. Moffat, L. S. Qi, R. S. Shapiro, J. Shendure, J. S. Weissman and X. Zhuang, Nat. Rev. Methods Prim., 2022, 21, 8 CrossRef.
- J. G. Doench, Nat. Rev. Genet., 2017, 19, 67–80 CrossRef PubMed.
- T. C. Cairns, X. Zheng, C. Feurstein, P. Zheng, J. Sun and V. Meyer, Front. Bioeng. Biotechnol., 2022, 9, 1335 Search PubMed.
- J. Kuivanen, V. Korja, S. Holmström and P. Richard, Fungal Biol. Biotechnol., 2019, 6, 3 CrossRef PubMed.
- E. K. Bowman, J. M. Wagner, S. F. Yuan, M. Deaner, C. M. Palmer, S. D'Oelsnitz, L. Cordova, X. Li, F. F. Craig and H. S. Alper, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2106818118 CrossRef CAS.
- C. Beck, J. F. G. Garzón and T. Weber, Biotechnol. Bioprocess Eng., 2020, 25, 886–894 CrossRef CAS.
- W. L. Thong, Y. Zhang, Y. Zhuo, K. J. Robins, J. K. Fyans, A. J. Herbert, B. J. C. Law and J. Micklefield, Nat. Commun., 2021, 12, 6872 CrossRef CAS.
- K. Kudo, T. Hashimoto, J. Hashimoto, I. Kozone, N. Kagaya, R. Ueoka, T. Nishimura, M. Komatsu, H. Suenaga, H. Ikeda and K. Shin-ya, Nat. Commun., 2020, 11, 2–4 CrossRef.
- S. H. Kim, W. Lu, M. K. Ahmadi, D. Montiel, M. A. Ternei and S. F. Brady, ACS Synth. Biol., 2019, 8, 109–118 CrossRef CAS.
- H. S. Kang, Z. Charlop-Powers and S. F. Brady, ACS Synth. Biol., 2016, 5, 1002–1010 CrossRef CAS.
- M. Liang, L. Liu, F. Xu, X. Zeng, R. Wang, J. Yang, W. Wang, L. Karthik, J. Liu, Z. Yang, G. Zhu, S. Wang, L. Bai, Y. Tong, X. Liu, M. Wu, L. X. Zhang and G. Y. Tan, Nucleic Acids Res., 2022, 50, 3581–3592 CrossRef CAS PubMed.
- B. Enghiad, C. Huang, F. Guo, G. Jiang, B. Wang, S. K. Tabatabaei, T. A. Martin and H. Zhao, Nat. Commun., 20212021, 121(12), 1171 Search PubMed.
- J. Fontana, W. E. Voje, J. G. Zalatan and J. M. Carothers, J. Ind. Microbiol. Biotechnol., 2018, 45, 481–490 CrossRef CAS.
- Y. F. Li, K. J. S. Tsai, C. J. B. Harvey, J. J. Li, B. E. Ary, E. E. Berlew, B. L. Boehman, D. M. Findley, A. G. Friant, C. A. Gardner, M. P. Gould, J. H. Ha, B. K. Lilley, E. L. McKinstry, S. Nawal, R. C. Parry, K. W. Rothchild, S. D. Silbert, M. D. Tentilucci, A. M. Thurston, R. B. Wai, Y. Yoon, R. S. Aiyar, M. H. Medema, M. E. Hillenmeyer and L. K. Charkoudian, Fungal Genet. Biol., 2016, 89, 18 CrossRef CAS PubMed.
- F. Alberti, S. Kaleem and J. A. Weaver, Biol. Open, 2020, 9, 056010 Search PubMed.
- C. L. Swift, K. B. Louie, B. P. Bowen, H. M. Olson, S. O. Purvine, A. Salamov, S. J. Mondo, K. V. Solomon, A. T. Wright, T. R. Northen, I. V. Grigoriev, N. P. Keller and M. A. O'Malley, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2019855118 CrossRef CAS.
Footnotes |
| † This paper was published as a part of a special issue on “Engineering the Biosynthesis of Fungal Natural Products”. |
| ‡ Present address: The Medical Research Council Toxicology Unit, University of Cambridge, Cambridge CB2 1QR, UK. Email: ir340@cam.ac.uk |
| This journal is © The Royal Society of Chemistry 2023 |