How different are marine microbial natural products compared to their terrestrial counterparts?†
Tanja M.
Voser
 ab,
Max D.
Campbell
ab,
Max D.
Campbell
 ac and
Anthony R.
Carroll
ac and
Anthony R.
Carroll
 *ab
*ab
aSchool of Environment and Science, Griffith University, Gold Coast, Australia. E-mail: tanja.schlaepfer@griffithuni.edu.au
bGriffith Institute for Drug Discovery, Griffith University, Brisbane, Australia. E-mail: A.Carroll@griffith.edu.au
cAustralian Rivers Institute-Coasts and Estuaries, Griffith University, Nathan, Australia. E-mail: max.campbell@griffith.edu.au
First published on 15th October 2021
Abstract
Covering: 1877 to 2020
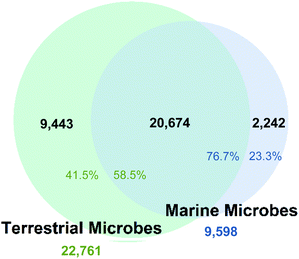
A key challenge in natural products research is the selection of biodiversity to yield novel chemistry. Recently, marine microorganisms have become a preferred source. But how novel are marine microorganism natural products compared to those reported from terrestrial microbes? Cluster analysis of chemical fingerprints and molecular scaffold analysis of 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817 compounds reported from marine and terrestrial microorganisms, and marine macro-organisms showed that 76.7% of the compounds isolated from marine microorganisms are closely related to compounds isolated from terrestrial microorganisms. Only 14.3% of marine microorganism natural products are unique when marine macro-organism natural products are also considered. Studies targeting marine specific and understudied microbial phyla result in a higher likelihood of finding marine specific compounds, whereas the depth and geographic location of microorganism collection have little influence. We recommend marine targeted strain isolation, incorporating early use of genomic sequencing to guide strain selection, innovation in culture media and cultivation techniques and the application of cheminformatics tools to focus on unique natural product diversity, rather than the dereplication of known compounds.
817 compounds reported from marine and terrestrial microorganisms, and marine macro-organisms showed that 76.7% of the compounds isolated from marine microorganisms are closely related to compounds isolated from terrestrial microorganisms. Only 14.3% of marine microorganism natural products are unique when marine macro-organism natural products are also considered. Studies targeting marine specific and understudied microbial phyla result in a higher likelihood of finding marine specific compounds, whereas the depth and geographic location of microorganism collection have little influence. We recommend marine targeted strain isolation, incorporating early use of genomic sequencing to guide strain selection, innovation in culture media and cultivation techniques and the application of cheminformatics tools to focus on unique natural product diversity, rather than the dereplication of known compounds.
1. Introduction
The search for novel, bioactive molecules is a major aim in modern drug discovery. Although it is possible to generate extensive libraries of synthetic compounds through automated processes, biodiversity is still a unique and exceptionally diverse source of novel bioactive molecules. Natural products (NPs) are encoded to interact with proteins associated with a myriad of diseases or proteins present in infectious agents such as bacteria, fungi or viruses because they are biosynthesised by enzymatically catalysed reactions involving structurally related proteins.1 Advances in technology have allowed us to venture into extreme environments, such as the depths of the ocean in the search for novel chemistry. Marine environments have proven to be an excellent source for unique and novel NPs since they harbour organisms that occur nowhere else on earth. Key features of the chemical diversity reported from marine organisms overlap with that of approved drugs making marine NPs an important source for drug discovery.2 Marine drugs are becoming more prevalent; to date 14 marine NPs or their derivatives are registered drugs, and another 23 are currently in clinical trials.3Research suggests that many compounds isolated from marine invertebrates are produced by symbiotic microorganisms.4–6 In recent years this often touted claim has resulted in a major shift from investigations that target marine macro-organisms to those that target marine microorganisms.7,8 However, some of the NPs found in marine invertebrates are produced by obligate symbionts that are nearly impossible to grow without their marine invertebrate host. Furthermore, in certain cases secondary metabolites are made through biosynthetic pathways shared between host and symbiont.9 This is due to their long evolutionary connection, to the point where some microbes have lost the ability to survive on their own, making it difficult for researchers to access these NPs.10
Marine fungi (mainly Ascomycota) are currently the most studied marine microorganism phyla, and over the last five years they have represented the most prolific source of new NPs from the marine environment.11 However, a closer examination of these compounds shows that 70% of these fungal MNPs isolated post 2015 are structurally similar to the fungal compounds isolated before 2015.8
This suggests that despite intensive isolation efforts, the discovery of novel marine fungal NPs is relatively low. Further investigation of the NPs isolated from marine fungi showed that there is no clear distinction between marine fungal NPs obtained from the different habitat niches.8 This clearly suggests that the identity of the species producing the compounds is more important than the location the species has been collected from. Likewise, Hernandez et al. (2021) have highlighted the importance of targeting more unique understudied bacterial phyla in order to fully access the microbial dark matter.12
This initial analysis of marine fungal chemical diversity highlighted a potential problem with the changed focus in marine NPs research. The redundancy in fungal marine NP structures may reflect a broader problem since a cursory analysis of the literature suggests that many of the NPs reported from marine microorganisms appear to be remarkably similar to those produced by terrestrial microorganisms. Marine microbes sourced using current methodology might therefore have a relatively low likelihood of producing novel compounds, with most of their chemical diversity being accessible from cheaper terrestrial sourced microorganisms. One could therefore hypothesise that the predominance in the use of standard isolation techniques and culture media (previously developed to specifically isolate and grow terrestrial soil microorganism), will lead to the growth of “terrestrial-like” microbes that produce “terrestrial-like” compounds.
In this highlight review we investigate in detail the structural similarity between NPs produced by marine and terrestrial microorganisms. The aim of this study was to identify limitations in current marine microorganism NP discovery efforts and help guide research in the marine environment that is likely to be the most promising for discovering unique and novel NPs.
2. Methods
This study is based on the meta-analysis of two large NP databases; the MarinLit database, which covers compounds isolated from the marine environment between 1956 and 2020 (ref. 11) and the NPAtlas database, which contains compounds isolated from microorganisms and published between 1877 and 2020.13We combined the two datasets and assigned each compound to the biome it was reported from (marine or terrestrial). This resulted in the merged MarinLit and NPAtlas dataset, containing a total of 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817 compounds. Of these compounds 22
817 compounds. Of these compounds 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 761 are from terrestrial microorganisms, 9598 are from marine microorganisms and 23
761 are from terrestrial microorganisms, 9598 are from marine microorganisms and 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 458 are from marine invertebrates, marine algae, and seagrass (for ease of reading, we will refer to this group as marine macro-organisms).
458 are from marine invertebrates, marine algae, and seagrass (for ease of reading, we will refer to this group as marine macro-organisms).
The combined dataset was cleaned up prior to further analysis. Salts and halogens (chlorine and bromine) were removed from all structures and in silico deglycosylation was performed to remove terminal ring and linear sugars, using a recently published cheminformatics tool.14
Using the ‘rcdk’ package in R,15 the isomeric smile codes of the 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817 NPs were used to generate the CDK extended molecular fingerprint for each compound.16 This path-based fingerprint considers the path of a given length (depth = 6) and takes ring features and atomic properties into account. This hashed fingerprint is captured by 1024 bits, where each bit is assigned 1 or 0 depending on the presence or absence of particular structural features in the compound.17 This fingerprint was chosen due to its ability to represent a large variety of chemical properties, thus, ensuring that different structural groups were adequately distinguished. As an alternative, the analysis was also performed using the PubChem fingerprint, but this made minimal difference to the outcome of the analysis.
817 NPs were used to generate the CDK extended molecular fingerprint for each compound.16 This path-based fingerprint considers the path of a given length (depth = 6) and takes ring features and atomic properties into account. This hashed fingerprint is captured by 1024 bits, where each bit is assigned 1 or 0 depending on the presence or absence of particular structural features in the compound.17 This fingerprint was chosen due to its ability to represent a large variety of chemical properties, thus, ensuring that different structural groups were adequately distinguished. As an alternative, the analysis was also performed using the PubChem fingerprint, but this made minimal difference to the outcome of the analysis.
Traditional k-means cluster analysis is not appropriate for binary data, therefore, a two-step cluster analysis was performed.18,19 First, the 1024 fingerprint dimensions were reduced using the logistic principal components analysis (PCA) described by Landgraf and Lee.19 This had the added benefit of avoiding overweighting of correlated dimensions, while also removing those dimensions that did not distinguish chemical differences. This resulted in a reduction of the dimensions from 1024 to 85 principal components while still explaining 95.5% of the variation in the fingerprints. Second, a k-means cluster analysis was performed using the 85 principal components identified by the logistic PCA. A clustering level that explained 85% of the within sums of squares was selected based on visual inspection of the traditional “knee plot”. This resulted in the full dataset being classified into 5000 clusters. Using a clustering level that explained >85% sum of squares (and thus more clusters) did not change the overall results.
As a second approach, Bemis and Murcko scaffolds20 were used to analyse the overlap of the scaffold diversity. Murcko frameworks are generated by removing all side chains from a molecule leaving behind only the ring systems and the linkers that connect the rings. These scaffolds are a useful way to investigate the main ring system of a compound, as this is often at the core of what defines the uniqueness or novelty of a molecule.21
Once molecules were assigned to clusters and scaffolds, their overlap/nesting amongst taxonomic and biome groupings was evaluated using common similarity metrics (Simpson and Jaccard similarity). Only a subset of structures is shown in the review but compound numbers for NPs mentioned but not included are italicized. All structures discussed in the text and more detailed additional graphics are available in the ESI† document.
3. Results and discussion
As sea water is relatively rich in halides, marine organisms often incorporate these elements into their metabolites through enzymatically catalysed reactions using enzymes such as vanadium bromoperoxidase.22,23 The high proportion of marine NPs that contain bromine and to a lesser extent chlorine has therefore become a distinguishing feature of marine NPs.24 However, analysis of our extensive data set shows that this does not translate to the microbial NPs. While 10.1% of all the marine invertebrate NPs, and 36.8% of the marine plants NPs contain bromine, only 1.8% of the marine microbial NPs contain bromine, which is only slightly higher than the 0.7% found in terrestrial microbial NPs. In comparison chlorinated NPs are present in 4.5% of the marine invertebrate and 15.6% of the marine plant NPs, while 7.0% of the marine microbial NPs and 5.1% of the terrestrial microbial NPs contain chlorine. We infer from this analysis of microbial NPs that bromination is rare in cultured microbes, while chlorination may occur under laboratory culture conditions, but is independent of microorganism biome source. Since bromine and chlorine content in the marine microbial NPs (1.8%, 7.0%) was similar to terrestrial microbial NPs (0.7%, 5.1%), it was deemed safe to remove the halogens from the structures prior to similarity analysis. This approach was chosen because we wanted to specifically analyse the core frameworks present in NPs since these are linked directly to their biosynthesis. Whereas halogenation and glycosylation often occur after the core structure is biosynthesised.A molecular fingerprint consists of binary bits that are turned on or off depending on if a compound contains a specific chemical feature, therefore giving a holistic view of the structure that allows for assessing molecular similarity.17 Cluster analysis of the NP fingerprints grouped structurally similar NPs into clusters that were able to be assigned as either marine only, terrestrial only or of mixed origin. This paper mainly focuses on the results of the fingerprint cluster analysis, while intermittently referring to the scaffold analysis. Both analyses however accorded similar results.
3.1 Structure comparison of marine versus terrestrial microbial NPs
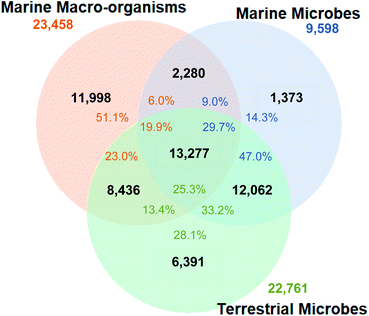
The fingerprint cluster analysis showed that 76.7% of all the NPs isolated from marine microbes are nested amongst NPs produced by terrestrial microbes (Fig. 1).Therefore, only 23.3% of the NPs isolated from microbes collected from the marine environment can be considered different from NPs derived from terrestrial microbes. The number of fingerprint clusters that define this structural diversity was even lower with only 12.8% of the clusters being unique to marine microbes to date. Further analysis of fingerprint clusters distinguished by NP taxonomic source showed that marine fungal NPs are nested with terrestrial fungal NPs at a higher relative proportion (74.6%) than marine bacterial NPs with their terrestrial counterparts (68.1%) (Fig. S3†). This indicates that marine and terrestrial fungi are more likely to share common biosynthetic gene clusters or marine sourced fungi are more likely to be terrestrial “wash ins”. Additionally, clustering across the two kingdoms led to the increased overall nesting (76.7%), which resulted from both kingdoms being prolific producers of several common NP structure classes such as cyclic peptides, terpenes, and polyketides.
Murcko scaffold cluster analysis indicated that 44.3% of marine microbe derived NPs scaffolds were also found in terrestrial microbe derived NPs. Translating this result into the total number of individual marine microbe derived NPs possessing scaffolds present in terrestrial microbes increases the nesting to 76.0%. Therefore, 76.0% of all the NPs reported from marine microbes possess the same scaffolds as NPs derived from terrestrial microbes, (an identical result to that obtained from the fingerprint cluster analysis). In comparison, when the clustered Murcko scaffolds of the total microbial NPs, were grouped by taxonomic hierarchy only a 6% overlap (Jaccard Similarity) between fungi and bacteria derived Murcko scaffolds was observed. This translated to 12.9% of the bacterial NPs, and 10.2% of fungal NPs sharing common scaffolds across the biome.
Many NPs reported from microbes, especially Cyanobacteria, are cyclic peptides or polyketide macrolides, and modifications of amino acids to generate thiazoles, oxazoles and other moieties are common, leading to unusual scaffolds. However, some of these “unique” Murcko scaffolds simply reflect variation in the amino acid substitution patterns, with incorporation of prolines into the cyclic peptide backbone or substitution of non-cyclic side chain amino acids with aromatic amino acids leading to unique scaffolds of otherwise common oligomeric cyclic peptides. These small differences, likely produced nonribosomally or by post-translational modifications, can make closely related scaffolds appear unique.25 Clustering the scaffolds in a similarity chart in DataWarrior26 confirmed these suspicions, showing clear scaffold hubs of several closely related cyclic peptides (Fig. S34†). This indicates that the overlap of scaffolds likely underestimates the clustering of NPs and suggests that some of the NPs currently found in marine only scaffold clusters are truly nested within terrestrial NP clusters.
3.2 A phylum level investigation of the marine/terrestrial chemical diversity overlap
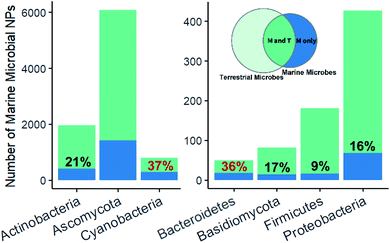
Most of the dataset contains NPs reported from seven microorganism phyla, with the largest representation from Ascomycota, followed by Actinobacteria, Basidiomycota, Cyanobacteria, Proteobacteria, Firmicutes and Bacteroidetes while the remaining NPs are reported from all other phyla (Table 1). These numbers alone highlight a skewed diversity, considering that chemical diversity correlates with species diversity.12| Phylum | Total number of NPs | Percentage of these NPs found in the marine biome |
|---|---|---|
a The dataset contains 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 359 microbial NPs, with 9598 reported from marine and 22 359 microbial NPs, with 9598 reported from marine and 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 761 from terrestrial microbes 243 of these microbial NPs have been reported in biota from both biomes. 761 from terrestrial microbes 243 of these microbial NPs have been reported in biota from both biomes.
|
||
| Ascomycota | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 455 455 |
39.4% |
| Actinobacteria | 7380 | 26.8% |
| Basidiomycota | 3766 | 2.2% |
| Cyanobacteria | 2017 | 39.3% |
| Proteobacteria | 1923 | 22.2% |
| Firmicutes | 604 | 30.0% |
| Bacteroidetes | 151 | 33.1% |
| Remaining microbial phyla | 115 | 11.3% |
Fingerprint cluster analysis of the marine microbe derived NPs versus terrestrial microbe derived NPs at a phylum level showed that the nesting occurred at different percentages across the different phyla (Fig. 2). Ascomycota and Actinobacteria, representing most of the dataset (41%), had 23% and 21% of their NPs within uniquely marine clusters, respectively. By comparison, Cyanobacteria had a substantially higher proportion (37%) of uniquely marine NPs. Cyanobacteria represent a unique group of microorganism, since they often grow as multicellular filaments or blooms in the environment, which in turn allows them to be collected en masse in a similar way to a macro-organism collections.27 This wild harvesting of Cyanobacteria is in stark contrast to the collection of a single bacteria or fungal strain in the wild, followed by laboratory isolation and culturing prior to an investigation of their NP chemistry. Cyanobacteria therefore already grow in optimum growth conditions potentially forming complex host-symbiont relationships, which leads to the production of unique secondary metabolites.
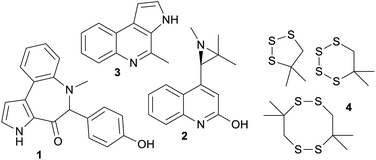
Marine bacteroidetes, also showed a higher proportion of uniquely marine compounds (36%). These uniquely marine bacteroidetes NPs are mainly from two genera; Mooreia a uniquely marine species associated with a sponge and Cytophaga (Fig. 3). Mooreia are prolific producers of unique alkaloids such as marinoazepinone A (1), marinoaziridine A (2) and marinoquinoline A (3), some of them showing nM and highly selective bioactivity against the protozoan parasite Plasmodium falciparum that causes malaria.28–30Cytophaga spp. produce a group of cyclic polysulfides 4 that are uniquely marine.31Basidiomycota and Proteobacteria both showed slightly below average uniqueness, (17% and 16% respectively). Closer inspection of the biota sources for the NPs in the marine only clusters associated with Proteobacteria revealed that 41.2% of the NPs were from strains isolated from marine macro-organisms, 33.8% isolated from sea water or sediment and 5.9% isolated from hydrothermal vents. Lastly, NPs isolated from marine Firmicutes were almost exclusively nested amongst fingerprint clusters associated with terrestrial NPs (91%). Although Firmicutes are widely distributed in marine environments and show some promising antibacterial properties, it is surprising that less than 200 NPs have been isolated from marine sourced strains of Firmicutes to date.32,33
3.3 Investigation of microbial NPs that are cosmopolitan
The significant overlap between current sourced marine and terrestrial microbial NP diversity is problematic from a marine biodiscovery perspective, because it has been estimated that the cost of doing marine-based research is an order of magnitude more expensive than equivalent land-based studies.34 Current collection, isolation and culturing strategies for marine microorganisms are clearly failing if the specific goal of marine based biodiscovery is to isolate uniquely “marine” chemistry since the majority of outcomes to date are the isolation of slightly modified terrestrial NPs. These NPs can be considered ‘me too’ compounds using the vernacular of drug development35 and there is every likelihood that simply collecting soil from a local terrestrial environment will result in the isolation of comparable chemical diversity.The 30 largest shared clusters, containing between 40–67 NPs, are associated with either subgroups of polyketides (linear, polyaromatic 5, 6 and macrocyclic 7), peptide 8, or to a lesser extent alkaloid structure classes 9–11 (Fig. 4). Comparatively, it is not surprising that NPs isolated from species in the same genera, but collected from either terrestrial or marine biomes, produce structurally similar compounds. This is especially the case if one considers that the same isolation and culturing techniques are often used irrespective of the ecological and physicochemical conditions of the natural environment where the microorganism resides. It is well known that in order to tap into uncultured microbial dark matter, a reputed source of unique chemistry, it is necessary to adjust isolation and culturing conditions to mimic the natural habitat.36 This requires the judicious use of tailored conditions, including low nutrient media, extended incubation times and natural macro-organism and/or microorganism interactions.36 However, most studies leading to the production of NPs in this dataset have used easily accessible isolation and culture methods well suited to the growth of cosmopolitan and fast growing species, with the outcome being the predominant production of ‘me too’ NPs.
To minimise the chances of this outcome, it is imperative that diverse microbial collection libraries that capture the true diversity of the marine microbial community and include understudied microbial biota are created.12 To this end bioinformatics platforms should be used early on in the isolate selection process to target unique biota or biosynthetic gene clusters within biota.37,38 Cheminformatics platforms such as GNPS should be used to prioritise unique singletons or clusters rather than prioritizing molecules that cluster with known structure classes.39 The recent developments of NMR based dereplication methods more tailored to identify uniqueness such as the DOSY NMR molecular weight prediction tool should also be considered.40
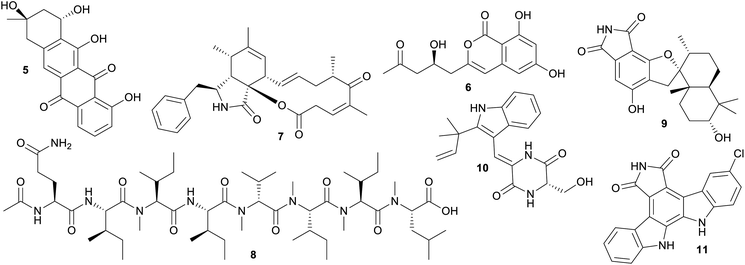 | ||
| Fig. 4 Selection of microbial NPs that are found in clusters consisting of NPs produced by marine and terrestrial microorganisms. | ||
3.4 Investigation of microbial NPs that are uniquely marine
Further analysis of the NPs that were in marine only fingerprint clusters showed that certain genera produce a higher proportion of NPs that were not related to NPs found in terrestrial microbes. This highlights the importance of the uniqueness and diversity of strain selection when considering marine microbial NP discovery. Assessing if these genera and their constituent species are exclusively present in marine habitats or are more cosmopolitan can be challenging, because there is often no clear distinction made in the literature about the habitat preference for specific microorganism species. However, a search of the World Register of Marine Species (WoRMS)41 for each of the genera and a subset of their species, showed that the majority of microbe species were reported from both marine and terrestrial environments, while only a few have been reported to be uniquely marine.The top five genera that produced the largest number of uniquely marine compounds were Aspergillus, Streptomyces, Penicillium, Moorea and Micromonospora. However, for Aspergillus, Streptomyces and Penicillium this is just a reflection of the disproportionately large number of studies that focus on these cosmopolitan genera with only a small number of species potentially being uniquely marine. By comparison, the top five genera in terms of the proportion of NPs that are uniquely marine are Cytophaga, Pseudopestalotiopsis, Chrysosporium, Pseudonocardia and Polyporales. The top five genera that have both highest number of uniquely marine and the highest proportion of uniquely marine NPs are Oceania (a uniquely marine Cyanobacteria), Spiromastix (found in the deep sea and terrestrial environments), Actinoalloteichus, Micromonospora, Epicoccum (all reportedly present in both terrestrial and marine environments), and Pseudopestalotiopsis (an endophyte reported in both terrestrial and marine environments) (see ESI† for more details).
A closer inspection of NPs associated with marine only fingerprint clusters, highlighted important discoveries that come from targeting marine microbes (Fig. 5). Specifically targeting uniquely marine species, such as the cyanobacterium genus Oceania led to the isolation of several bioactive NPs, such as the cyclic peptides odobromoamide (12) and the kohamamides A–C (13–15), both exhibiting cytotoxicity against cancer cell lines,42,43 and irijimasides A–E (16–20), a group of macrolides showing promise as therapeutic agents against bone-loss diseases.44 Extensive studies of actinobacteria from the genus Micromonospora collected from the South China Sea led to the discovery of uniquely marine NPs such as the microsporanates A–F (21–26),45 pyrazolofluostatins A–C (27–29)46 and sungeidines A–H (30–37),47 with the Micromonospora strains collected at varying depths −1565 m, −30 m and from mangrove sediments respectively. Nesteretal A (38), possessing a novel carbon scaffold was isolated from a moderately halophilic coral-derived actinomycete Nesterenkonia halobia,48 while a recently discovered species collected at a depth of 3865 m, Marinactinospora thermotolerans, produced new marine alkaloids, marinacarbolines A–D (39–42).49 These NPs highlight that chemically unique molecules can be found in the marine environment, however, it is essential that great care is taken in choosing unique marine genera with a focus on the appropriate cultivation methods to suit these understudied genera.
Although very few of the NPs isolated from Firmicutes sourced from marine habitats were found in uniquely marine fingerprint clusters, two deep-sea collections produced some uniquely marine NPs. These were the subtipyrrolines A–C (43–45), which possess an unprecedented pyrrole–pyrrole–dihydropyridine tricyclic ring system,50 and the new diterpene indole alkaloid halomide (46).51 Both were isolated from non-marine specific Bacillus species (B. subtilis and B. amyloliquefaciens), but were collected at great depth, 2476 m and 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 m respectively.50,51
000 m respectively.50,51
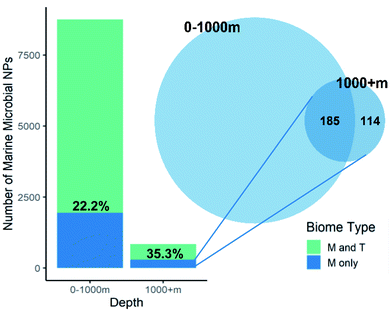
One marine ecosystem that one would consider to be unique is the deep-ocean and it should therefore be a prolific source of uniquely marine chemical diversity. Calculation of the percentage of marine microbes specific NPs versus total microbial NPs found at a depth of over 1000 m (35.3%) is higher than NPs reported from species collected at shallower depths (22.2%, Fig. 6). However, once the overlap with marine specific NPs found at lesser depth was considered, only 13.5% of the deep-sea microbial NPs were unique. What this means is that most of the NPs unique to the marine environment that have been isolated from deep-water microbes (62%) were also found in shallow water marine microbes. It is therefore questionable whether the cost of deep-sea collections is worth the reward. Surprisingly, none of the deep-sea microbe investigations have used high pressure or extreme temperatures (either ambient (4 °C) or near hydrothermal vents (100 °C)) during strain isolation and culturing and therefore do not use conditions that mimic the natural environments of the microbes.
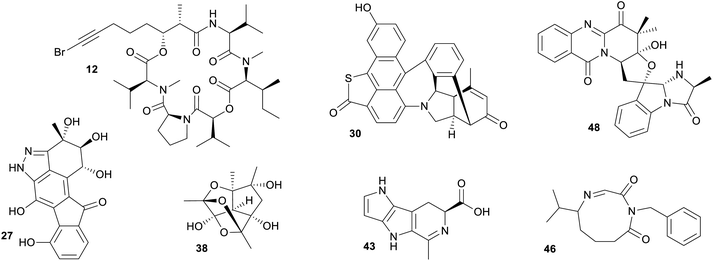 | ||
| Fig. 5 Selection of marine microbial NPs that were found in uniquely marine clusters and do not nest among terrestrial NPs. | ||
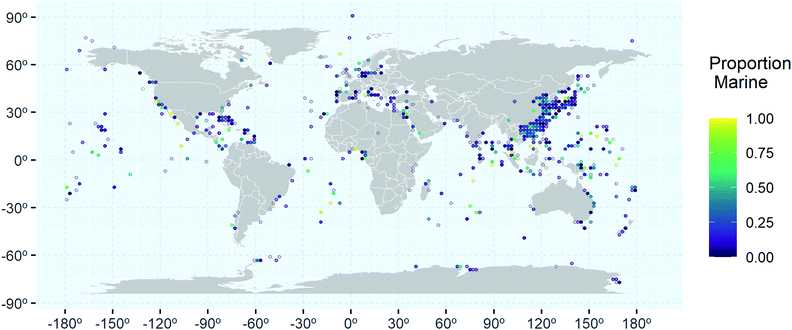
Analysis of the geographical distribution of the collection locations associated with marine microbial NPs shows that the hotspots of relative high percentage of marine only clusters (yellow and light green) are scattered throughout the world's oceans (Fig. 7). This suggests that distance from larger landmasses does not affect uniqueness of NPs found in the marine environment. However, further analysis of the uniquely marine hotspots showed that certain research groups have achieved a high success rate in finding uniquely marine NPs. Research groups led by Gerwick (Oregon, USA),52–54 Moore (Hawaii)55,56 and Tan (Singapore),57,58 have all isolated marine specific cyanobacterial NPs. A focus on genome mining has resulted in the Chinese group successfully isolating new NPs from Actinobacteria. Totopotensamide C (47) isolated from a Streptomyces sp.59 and the sungeidines (30–37) isolated from a Micromonospora,47 both collected around Malaysia are examples of their success. Groups led by Proksch (Germany) and/or Lin (China) have successfully isolated uniquely marine NPs from deep-sea sediments from the South Atlantic Ocean.60–62 Interestingly, a major sampling effort has occurred in the North Pacific Ocean, especially around the Chinese and Japanese Coastlines, with some locations yielding up to 400 new compounds (Fig. 7). Marine Ascomycota have been studied extensively in this area but most of these studies have yielded ‘me too’ compounds. Studies that resulted in the discovery of unique chemistry like the scedapins A–E (48–52), from the South China Sea,63 mainly came from either deep-sea collections,64 endophytic fungi65 or applying special culturing techniques such as OSMAC.66
An investigation of the culture media used to cultivate microorganisms that produced marine specific NPs revealed that most studies used potato dextrose or rice in seawater or sea salts, but few studies used marine specific agar. Although many of the papers use an OSMAC approach or manipulation of culturing conditions, we were surprised how few studies used more specific techniques such as iChip,67 co-culturing68 or other advanced technologies like genome mining,69 even though these techniques have proven to be effective at generating unique chemistry. The complexity in understanding the drivers of structural uniqueness in NPs obtained from marine environments is clearly challenging.
3.5 Chemical overlap between microbial and marine macro-organism NPs
Several studies have demonstrated that some NPs reported from sponges, ascidians and bryozoans are produced by symbiotic microorganisms that live in or on the animal. Considering this, a reasonably large overlap in fingerprint clusters between the NPs derived from marine microorganisms and macro-organisms was expected. However, our analysis did not support this, since marine macro-organism NPs were present in clusters associated with NPs solely isolated from terrestrial microorganism to a much higher extent (27.5%) than with NPs isolated solely from marine microorganisms (7.2%) with an additional 25.1% shared with cluster fingerprints common to both marine and terrestrial microorganisms. However, some of these shared clusters are carbon skeletons that are highly decorated with halogens in the NPs isolated from macro-organism but not within the NPs isolated from microorganism. As described earlier, the high proportion of halogenated NPs isolated from marine macro-organisms especially within the Rhodophyta, Bryozoa, Chordata and Porifera is a feature that distinguishes their chemistry from that derived from terrestrial biota. Therefore, these shared carbon skeletons are likely to reflect shared capabilities to biosynthesise carbon frameworks within both marine macro-organisms and marine and terrestrial microorganisms, but many of the cultured microorganisms do not have the ability to enzymatically halogenate these skeletons. If one considers these halogenated NPs within the shared fingerprint clusters are different to a non-halogenated NP within the same cluster then these clusters should be split. Structures 53,70versus54,71 and 55,72versus56,73 represent cases where non-halogenated microbial NPs cluster with halogenated marine macro-organism NPs (Fig. 8).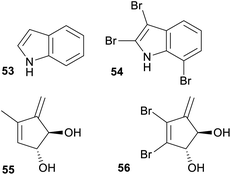 | ||
| Fig. 8 Example of where non-halogenated microbial NPs cluster with halogenated marine macro-organism NPs. | ||
When these clusters were split based on halogenation patterns, the overlap between NPs isolated from marine macro-organisms and microorganisms was reduced by 10%, resulting in a total overlap of 48.9% of marine macro-organism NPs with all microbial NPs (Fig. 9). Some uniquely macro-organism derived compounds include pyridoacridine alkaloids, isolated from several phyla of marine invertebrates,74 and exemplified by the cystodytins A–C (57–59),75 the bromopyrrole imidazole alkaloids such as oroidin (60)76,77 and the agelamadins A and B (61–62),78 and the cytotoxic macrocyclic alkaloids haliclamines A and B (63–64).79 These compounds highlight the unique and biologically interesting chemistry that macro-organisms have the potential to produce.
Interestingly, the proportion of NPs isolated from marine macro-organism that are present in clusters associated with terrestrial microorganism NPs remained much higher (23.0%, 5395 NPs) than the ones associated with marine microbial NPs (6.0%, 1407 NPs), while 4668 marine macro-organism NPs (19.9%) shared clusters containing NPs from both terrestrial and marine microorganisms. To some extent the higher representation of macro-organism NPs shared with terrestrial microorganism chemistry is unsurprising since the initial impetus to study marine microbial NPs resulted from the observation that sponge derived compounds such as the mycalamide A (65) and onnamide A (66) are remarkably similar to pederin (67) isolated from a terrestrial bacteria endosymbiont of a beetle.80
From a marine microorganism perspective, 38.7% of NPs reported from marine microbes cluster with marine macro-organism NPs but only 9% of these NPs are associated with clusters that are uniquely marine. The inclusion of marine macro-organism NPs into the similarity analysis resulted in a further reduction in total unique marine microbial NP diversity, with only 1373 marine microbe NPs (14.3%) considered to be different to either terrestrial microorganism and/or marine macro-organism NPs.
An explanation for the smaller clustering of marine macro-organism NPs with marine microbial NPs is that more NPs have been isolated from terrestrial microbial sources than from marine microbes. The study of marine microbial NPs is still somewhat in its infancy, and it is possible that the overlap between NPs reported from marine microbes and macro-organisms has not, to date, been investigated thoroughly. Additionally, some marine microbes NP studies that have yielded known NPs might simply not have been published because of the need to report “new” chemistry. Nevertheless, the large overlap of marine macro-organism NPs and terrestrial microbial NPs indicates that selection of the next marine microbe to investigate needs to be done cautiously, to not waste time and effort isolating structurally similar ‘me too’ NPs.
Our analysis shows that some of this higher overlap between terrestrial microbial and marine macro-organism NPs is associated with terpenes and steroids isolated mainly from terrestrial Basidiomycota (only 2.2% of all Basidiomycota NPs are marine). A total of 1201 NPs isolated from Basidiomycota were found at the intersection between the three groups (marine microorganism/terrestrial microorganism/marine macro-organism), and another 914 were found at the marine macro-organisms/terrestrial microorganism intersection. This indicated a large overlap of Basidiomycota NPs with NPs produced by marine macro-organisms.
For each of the 18 marine macro-organism phyla the overlap with terrestrial microbial NPs was substantially higher than that between marine macro-organisms and marine microbes, with some phyla having no exclusive overlap with marine microbial NPs (Fig. S9†). Investigating the structure classes associated with the overlap between marine macro-organism and Actinobacteria shows that it is driven by alkaloids, macrolides, and peptides, whereas the overlap between marine macro-organism and Ascomycota is driven by terpenes, polyketides, steroids, macrolides and peptides. This is consistent for marine and terrestrial microbes.
Interestingly, nearly all NPs reported from Arthropoda and Bacillariophyta show overlap with terrestrial microbial NPs, which arises from larger numbers of lipids found in Arthropoda and steroids in Bacillariophyta. Other phyla, such as Rhodophyta, Bryozoa, Porifera and Chordata show a large proportion (over 55%) of marine only NPs.
One of the marine bacteria species that stood out as having an almost complete overlap of its NPs with marine invertebrate NPs is “Candidatus Entotheonella factor”. This symbiont of the marine sponge Theonella swinhoei is a prolific producer of a range of NPs such as cyclotheonamide A (68), keramamide D (69) and onnamide A (66).81 These compounds were originally isolated from the sponge, but through genomic and biosynthetic studies are now identified as bacterial metabolites.81 Further studies on the biosynthetic gene clusters present in other Entotheonella species revealed them to be “super producers” of NPs, hence a promising source of new marine chemistry.82 Another microbial NP that did not cluster with any terrestrial NPs, is pateamine A (70), a cytotoxin initially isolated from the marine sponge Mycale hentscheli.83 The biosynthetic gene cluster producing pateamine A was recently identified to belong to Kiritimatiellaeota, a bacteria phylum previously not known to produce secondary metabolites.84,85 Further investigation of M. hentscheli revealed an extensive bacterial consortium responsible for the production of the sponge's chemistry, with many of them previously not known to produce NPs.85
Halichondrin B (71), isolated from the marine sponge Halichondria okadai is suspected to be produced by a symbiotic microorganism.86 The macrocyclic lactone pharmacophore of halichondrin B inspired the creation of eribulin mesylate (72), trademarked as Halaven® that is currently used to treat metastatic breast cancer.87 Even though halichondrin B is strongly suspected to be produced by a microbial symbiont, its microorganism source is still to be identified. In both the fingerprint clustering and the scaffold analysis, halichondrin B only clustered with other marine invertebrates' compounds. These examples show that marine invertebrates and their understudied endosymbionts, if approached with the right techniques, are prolific sources of new marine NPs. On the other hand, it also shows that time and immense effort must be put into the collection, isolation, and cultivation of these endosymbiotic microorganisms, to isolate NPs that are likely similar to NPs more easily accessible from marine invertebrates. Therefore, there needs to be good justification for the study of marine microorganisms over their marine macro-organism hosts. These justifications may include investigations to resolve compound supply issues, to isolate novel genes associated with biosynthesis for combinatorial biochemistry, or to unravel configurational assignments in complex molecular structures for example.
4. Future perspectives
Our findings indicate that there is indeed a substantial overlap of the chemistry produced by marine microorganisms compared to their terrestrial counterparts. This does not detract from the fact that the marine environment is a major source of NPs with novel chemistry. However, research focus needs to shift more towards uniquely marine microorganisms, but a continued focus on marine macro-organisms, particularly those that harbour obligate symbionts that potentially produce many unique compounds is advisable. It would also be worth considering co-cultures with marine microbes that are known to harbour genes associated with halo peroxidases, to promote halogenation.Fenical, a pioneer in marine bacteria research, reemphasised in his recent editorial the importance of using unique and often simple culturing techniques in order to grow marine bacteria that are seemingly impossible to culture.88 The three main points he noted are; (1) increasing incubation times from weeks to months, (2) using extremely low nutrient agar, hence mimicking the natural concentrations of the ocean and (3) the simple but important notion of using seawater.88 Incubating for long periods such as months might not seem very efficient; however, if the results are novel strains producing novel compounds, we would strongly suggest it is worth it.
Advances in 16S rRNA gene sequencing coupled with the analysis of biosynthetic gene clusters of unculturable bacteria can result in the identification of novel chemistry in microbial dark matter, which in the past was simply not accessible.89 Using surrogate host bacteria to express these biosynthetic gene clusters, as has been reported for the aeronamides and polytheonamides produced from deep-rock biosphere DNA, highlights significant opportunities for future discovery of new secondary metabolites.90
Genome studies on uncultured bacteria can also help identify marine bacteria that are closely related to a terrestrial relative. For example, the genome of the marine bacteria “Candidatus Desulfopertinax cowenii” modA32 shows a surprising similarity to its terrestrial relative, despite being collected from the deep subseafloor.91 In this case, it might be more cost effective to explore its terrestrial relative. In other cases such as the Entotheonella taxon, which has been identified as a rich source of biosynthetic gene clusters that promise new chemistry, it is likely to be worth pursuing culturing and biosynthetic studies.82 Furthermore, metagenomics studies that allow for improvement of cultivation techniques, by adapting to the ecology of the uncultured bacteria, brightens future outlooks.36
Common genera such as Streptomyces and Penicillium might still be worth investigating, however, only in combination with advanced dereplication tools, such as SMART92 that allow for the identification of unique chemical structures and avoids the isolation of ‘me too’ compounds. Reinventing GNPS39 as a tool to mine for unique chemistry rather than known compound clusters or DOSY NMR methods that can allow new structures to be identified from a mixture of NPs should be considered.40 Utilisation of chemical ontology combined with structure-based classification systems such as that used in ChEBI,93 which combines expert knowledge with datasets, can also aid researchers to interrogate large cheminformatics data to identify bioactivity and important structural features.94
A comparison between marine and terrestrial microorganism genomes and/or biosynthetic gene cluster analysis, analogous to the approach taken in this highlight, could further provide a complimentary perspective to direct future marine NP discovery.
5. Conclusion
This study shows that the current direction of marine microbial NP discovery is problematic. A 14.3% overall uniqueness in marine microorganism NP diversity is low both in comparison with terrestrial microbial and marine macro-organism NP diversity. Furthermore, this unique marine microbial NP diversity includes a subset of NPs obtained from Cyanobacteria, a phylum where wild harvesting is generally used to access its NP diversity and thus does not generally rely on laboratory strain isolation and culturing. In comparison all other marine microorganisms need to be isolated from marine samples and then laboratory cultured. This has led to a low success rate in the discovery of unique marine microorganism NP diversity pointing to a need for innovation in strain selection, emphasising the importance of careful selection of understudied genera, and marine specific cultivation techniques.We hope that this snapshot of chemical diversity sparks further discussion and ideas that will help guide future collection and culturing efforts for the discovery of new and unique marine NPs.
6. Author contributions
Contributions to the preparation of this manuscript are as follows; TV: conceptualization, data curation, formal analysis, investigation, methodology, resources, validation, visualization, writing – original draft, writing – review & editing, MC: investigation, formal analysis, methodology, writing – review & editing, AC: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, visualization, writing – review & editing.7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgement
We gratefully acknowledge the support of the Griffith University eResearch Services Team and the use of the High Performance Computing Cluster ‘‘Gowonda” for cluster analysis calculations. We thank Rebekka Pentti for providing us with the underwater photo for the graphical abstract. This work was funded by an Australian Postgraduate Award (APA) provided by the Australian Commonwealth Government.9. Notes and references
- R. A. Copeland, Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists, John Wiley & Sons, 2013 Search PubMed.
- J. W. Blunt, A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2018, 35, 8–53 RSC.
- K. Jekielek, H. Le, A. Wu, D. Newman, K. Glaser and A. Mayer, FASEB J., 2021, 35 DOI:10.1096/fasebj.2021.35.s1.00209.
- T. L. Simmons, R. C. Coates, B. R. Clark, N. Engene, D. Gonzalez, E. Esquenazi, P. C. Dorrestein and W. H. Gerwick, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4587–4594 CrossRef CAS.
- G. M. König, S. Kehraus, S. F. Seibert, A. Abdel-Lateff and D. Müller, Chembiochem, 2006, 7, 229–238 CrossRef PubMed.
- U. Hentschel, J. Piel, S. M. Degnan and M. W. Taylor, Nat. Rev. Microbiol., 2012, 10, 641–654 CrossRef CAS.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2019, 36, 122–173 RSC.
- A. R. Carroll, B. R. Copp, R. A. Davis, R. A. Keyzers and M. R. Prinsep, Nat. Prod. Rep., 2021, 38, 362–413 RSC.
- E. W. Schmidt, Invertebr. Biol., 2015, 134, 88–102 CrossRef PubMed.
- P. A. O’Brien, N. S. Webster, D. J. Miller and D. G. Bourne, mBio, 2019, 10, e02241-18 CrossRef.
- J. W. Blunt and M. H. Munro, MarinLit, accessed Feb 20, 2021, http://pubs.rsc.org/marinlit/ Search PubMed.
- A. Hernandez, L. T. Nguyen, R. Dhakal and B. T. Murphy, Nat. Prod. Rep., 2021, 38, 292–300 RSC.
- J. A. Van Santen, G. Jacob, A. L. Singh, V. Aniebok, M. J. Balunas, D. Bunsko, F. C. Neto, L. Castaño-Espriu, C. Chang, T. N. Clark, J. L. Cleary Little, D. A. Delgadillo, P. C. Dorrestein, K. R. Duncan, J. M. Egan, M. M. Galey, F. P. J. Haeckl, A. Hua, A. H. Hughes, D. Iskakova, A. Khadilkar, J. H. Lee, S. Lee, N. Legrow, D. Y. Liu, J. M. Macho, C. S. McCaughey, M. H. Medema, R. P. Neupane, T. J. O'Donnell, J. S. Paula, L. M. Sanchez, A. F. Shaikh, S. Soldatou, B. R. Terlouw, T. A. Tran, M. Valentine, J. J. J. Van Der Hooft, D. A. Vo, M. Wang, D. Wilson, K. E. Zink and R. G. Linington, ACS Cent. Sci., 2019, 5, 1824–1833 CrossRef CAS.
- J. Schaub, A. Zielesny, C. Steinbeck and M. Sorokina, J. Cheminf., 2020, 12, 1–20 Search PubMed.
- R. Guha, J. Stat. Softw., 2007, 18, 1–60 CrossRef.
- E. L. Willighagen, J. W. Mayfield, J. Alvarsson, A. Berg, L. Carlsson, N. Jeliazkova, S. Kuhn, T. Pluskal, M. Rojas-Chertó and O. Spjuth, J. Cheminf., 2017, 9, 1–19 Search PubMed.
- A. Cereto-Massague, M. J. Ojeda, C. Valls, M. Mulero, S. Garcia-Vallve and G. Pujadas, Methods, 2015, 71, 58–63 CrossRef CAS.
- M. Mitsuhiro and H. Yadohisa, Comput. Stat., 2015, 30, 463–475 CrossRef.
- A. J. Landgraf and Y. Lee, J. Multivar. Anal., 2020, 180, 104668 CrossRef.
- G. W. Bemis and M. A. Murcko, J. Med. Chem., 1996, 39, 2887–2893 CrossRef CAS PubMed.
- Y. Hu, D. Stumpfe and J. Bajorath, J. Med. Chem., 2016, 59, 4062–4076 CrossRef CAS.
- A. Butler and J. Walker, Chem. Rev., 1993, 93, 1937–1944 CrossRef CAS.
- J. N. Carter-Franklin and A. Butler, J. Am. Chem. Soc., 2004, 126, 15060–15066 CrossRef CAS PubMed.
- G. W. Gribble, Environ. Sci. Pollut. Res., 2000, 7, 37–49 CrossRef CAS PubMed.
- J. C. Kehr, D. Gatte Picchi and E. Dittmann, Beilstein J. Org. Chem., 2011, 7, 1622–1635 CrossRef CAS.
- T. Sander, J. Freyss, M. Von Korff and C. Rufener, J. Chem. Inf. Model., 2015, 55, 460–473 CrossRef CAS.
- A. Herrero, J. Stavans and E. Flores, FEMS Microbiol. Rev., 2016, 40, 831–854 CrossRef CAS.
- A. C. C. Aguiar, M. Panciera, E. F. Simao Dos Santos, M. K. Singh, M. L. Garcia, G. E. de Souza, M. Nakabashi, J. L. Costa, C. R. S. Garcia, G. Oliva, C. R. D. Correia and R. V. C. Guido, J. Med. Chem., 2018, 61, 5547–5568 CrossRef CAS.
- E. J. Choi, S. J. Nam, L. Paul, D. Beatty, C. A. Kauffman, P. R. Jensen and W. Fenical, Chem. Biol., 2015, 22, 1270–1279 CrossRef CAS.
- Y. Sangnoi, O. Sakulkeo, S. Yuenyongsawad, A. Kanjana-opas, K. Ingkaninan, A. Plubrukarn and K. Suwanborirux, Mar. Drugs, 2008, 6, 578–586 CrossRef CAS.
- P. Sobik, J. Grunenberg, K. Böröczky, H. Laatsch, I. Wagner-Döbler and S. Schulz, J. Org. Chem., 2007, 72, 3776–3782 CrossRef CAS PubMed.
- M. A. da Silva, A. Cavalett, A. Spinner, D. C. Rosa, R. B. Jasper, M. C. Quecine, M. L. Bonatelli, A. Pizzirani-Kleiner, G. Corção and A. O. Lima, Springerplus, 2013, 2, 1–10 CrossRef.
- V. K. Kizhakkekalam and K. Chakraborty, Arch. Microbiol., 2020, 202, 905–920 CrossRef CAS.
- G. M. Cragg and D. J. Newman, Pharm. Biol., 2001, 39, 8–17 CAS.
- J. K. Aronson and A. R. Green, Br. J. Clin. Pharmacol., 2020, 86, 2114–2122 CrossRef CAS.
- J. Overmann, B. Abt and J. Sikorski, Annu. Rev. Microbiol., 2017, 71, 711–730 CrossRef CAS.
- J. K. B. Cahn and J. Piel, Angew. Chem., Int. Ed., 2021, 60, 18412–18428 CrossRef CAS.
- M. A. Schorn, M. M. Alanjary, K. Aguinaldo, A. Korobeynikov, S. Podell, N. Patin, T. Lincecum, P. R. Jensen, N. Ziemert and B. S. Moore, Microbiol., 2016, 162, 2075–2086 CrossRef CAS.
- A. T. Aron, E. C. Gentry, K. L. McPhail, L.-F. Nothias, M. Nothias-Esposito, A. Bouslimani, D. Petras, J. M. Gauglitz, N. Sikora, F. Vargas, J. J. J. van der Hooft, M. Ernst, K. B. Kang, C. M. Aceves, A. M. Caraballo-Rodríguez, I. Koester, K. C. Weldon, S. Bertrand, C. Roullier, K. Sun, R. M. Tehan, C. A. Boya P, M. H. Christian, M. Gutiérrez, A. M. Ulloa, J. A. Tejeda Mora, R. Mojica-Flores, J. Lakey-Beitia, V. Vásquez-Chaves, Y. Zhang, A. I. Calderón, N. Tayler, R. A. Keyzers, F. Tugizimana, N. Ndlovu, A. A. Aksenov, A. K. Jarmusch, R. Schmid, A. W. Truman, N. Bandeira, M. Wang and P. C. Dorrestein, Nat. Protoc., 2020, 15, 1954–1991 CrossRef CAS PubMed.
- G. Kleks, D. C. Holland, J. Porter and A. R. Carroll, Chem. Sci., 2021, 12, 10930–10943 RSC.
- W. E. Board, World Register of Marine Species (WoRMS), 2021, accessed Aug 2021, https://www.marinespecies.org Search PubMed.
- K. Sueyoshi, T. Kudo, A. Yamano, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, Bull. Chem. Soc. Jpn., 2017, 90, 436–440 CrossRef CAS.
- A. Iwasaki, I. Shiota, S. Sumimoto, T. Matsubara, T. Sato and K. Suenaga, J. Nat. Prod., 2017, 80, 1948–1952 CrossRef CAS.
- A. Yamano, N. Natsume, M. Yamada, S. Sumimoto, A. Iwasaki, K. Suenaga and T. Teruya, J. Nat. Prod., 2020, 83, 1585–1591 CrossRef CAS.
- C. Gui, S. Zhang, X. Zhu, W. Ding, H. Huang, Y. C. Gu, Y. Duan and J. Ju, J. Nat. Prod., 2017, 80, 1594–1603 CrossRef CAS.
- W. Zhang, C. Yang, C. Huang, L. Zhang, H. Zhang, Q. Zhang, C. S. Yuan, Y. Zhu and C. Zhang, Org. Lett., 2017, 19, 592–595 CrossRef CAS.
- Z. J. Low, G. L. Ma, H. T. Tran, Y. Zou, J. Xiong, L. Pang, S. Nuryyeva, H. Ye, J. F. Hu, K. N. Houk and Z. X. Liang, J. Am. Chem. Soc., 2020, 142, 1673–1679 CrossRef CAS.
- C. L. Xie, R. Chen, S. Yang, J. M. Xia, G. Y. Zhang, C. H. Chen, Y. Zhang and X. W. Yang, Org. Lett., 2019, 21, 8174–8177 CrossRef CAS.
- H. Huang, Y. Yao, Z. He, T. Yang, J. Ma, X. Tian, Y. Li, C. Huang, X. Chen, W. Li, S. Zhang, C. Zhang and J. Ju, J. Nat. Prod., 2011, 74, 2122–2127 CrossRef CAS.
- L. Qin, W. Yi, X.-Y. Lian, N. Wang and Z. Zhang, Tetrahedron, 2020, 76, 131516 CrossRef CAS.
- C. Gao, X. Yi, L. Yu, L. Pan, X. Yang, M. Qin and R. Huang, Chem. Nat. Compd., 2016, 52, 368–369 CrossRef CAS.
- L. M. Nogle, R. T. Williamson and W. H. Gerwick, J. Nat. Prod., 2001, 64, 716–719 CrossRef CAS.
- L. M. Nogle and W. H. Gerwick, Org. Lett., 2002, 4, 1095–1098 CrossRef CAS.
- R. T. Williamson, I. P. Singh and W. H. Gerwick, Tetrahedron, 2004, 60, 7025–7033 CrossRef CAS.
- R. E. Moore, A. J. Blackman, C. E. Cheuk, J. S. Mynderse, G. K. Matsumoto, J. Clardy, R. W. Woodard and J. C. Craig, J. Org. Chem., 1984, 49, 2484–2489 CrossRef CAS.
- J. S. Mynderse and R. E. Moore, J. Org. Chem., 1978, 43, 4359–4363 CrossRef CAS.
- A. Tripathi, J. Puddick, M. R. Prinsep, P. P. Lee and L. T. Tan, Phytochemistry, 2010, 71, 307–311 CrossRef CAS.
- D. K. Gupta, P. Kaur, S. T. Leong, L. T. Tan, M. R. Prinsep and J. J. Chu, Mar. Drugs, 2014, 12, 115–127 CrossRef PubMed.
- R. Chen, Q. Zhang, B. Tan, L. Zheng, H. Li, Y. Zhu and C. Zhang, Org. Lett., 2017, 19, 5697–5700 CrossRef CAS PubMed.
- S. Niu, D. Liu, Z. Shao, P. Proksch and W. Lin, RSC Adv., 2017, 7, 33580–33590 RSC.
- S. Niu, D. Liu, Z. Shao, P. Proksch and W. Lin, Tetrahedron, 2018, 74, 7310–7325 CrossRef CAS.
- S. Niu, L. Si, D. Liu, A. Zhou, Z. Zhang, Z. Shao, S. Wang, L. Zhang, D. Zhou and W. Lin, Eur. J. Med. Chem., 2016, 108, 229–244 CrossRef CAS.
- L.-H. Huang, M.-Y. Xu, H.-J. Li, J.-Q. Li, Y.-X. Chen, W.-Z. Ma, Y.-P. Li, J. Xu, D.-P. Yang and W.-J. Lan, Org. Lett., 2017, 19, 4888–4891 CrossRef CAS.
- S. Niu, J. M. Xia, Z. Li, L. H. Yang, Z. W. Yi, C. L. Xie, G. Peng, Z. H. Luo, Z. Shao and X. W. Yang, J. Nat. Prod., 2019, 82, 2307–2331 CrossRef CAS.
- Y. Xu, C. Wang, H. Liu, G. Zhu, P. Fu, L. Wang and W. Zhu, Mar. Drugs, 2018, 16, 363 CrossRef CAS.
- H. H. Kang, H. B. Zhang, M. J. Zhong, L. Y. Ma, D. S. Liu, W. Z. Liu and H. Ren, Mar. Drugs, 2018, 16, 449 CrossRef CAS PubMed.
- B. Berdy, A. L. Spoering, L. L. Ling and S. S. Epstein, Nat. Protoc., 2017, 12, 2232–2242 CrossRef.
- D. Arora, P. Gupta, S. Jaglan, C. Roullier, O. Grovel and S. Bertrand, Biotechnol. Adv., 2020, 40, 107521 CrossRef PubMed.
- M. H. Medema, T. de Rond and B. S. Moore, Nat. Rev. Genet., 2021, 1–19 Search PubMed.
- M. Oimomi, M. Hamada and T. Hara, J. Antibiot., 1974, 27, 987–988 CrossRef CAS PubMed.
- M. R. Brennan and K. L. Ericson, Tetrahedron Lett., 1978, 19, 1637–1640 CrossRef.
- M. Weidler, J. Rether, T. Anke, G. Erkel and O. Sterner, J. Antibiot., 2001, 54, 679–681 CrossRef CAS PubMed.
- R. Kazlauskas, P. Murphy and R. Wells, Aust. J. Chem., 1982, 35, 219–220 CrossRef CAS.
- S. R. Ibrahim and G. A. Mohamed, Bull. Fac. Pharm. Cairo Univ., 2016, 54, 107–135 Search PubMed.
- J. Kobayashi, J. Cheng, M. R. Walchli, H. Nakamura, Y. Hirata, T. Sasaki and Y. Ohizumi, J. Org. Chem., 1988, 53, 1800–1804 CrossRef CAS.
- S. Forenza, L. Minale, R. Riccio and E. Fattorusso, Chem. Commun., 1971, 1129–1130 RSC.
- E. Garcia, L. Benjamin and R. I. Fryer, Chem. Commun., 1973, 78–79 RSC.
- T. Kusama, N. Tanaka, K. Sakai, T. Gonoi, J. Fromont, Y. Kashiwada and J. i. Kobayashi, Org. Lett., 2014, 16, 3916–3918 CrossRef CAS.
- N. Fusetani, K. Yasumuro, S. Matsunaga and H. Hirota, Tetrahedron Lett., 1989, 30, 6891–6894 CrossRef CAS.
- J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14002–14007 CrossRef CAS.
- M. C. Wilson, T. Mori, C. Ruckert, A. R. Uria, M. J. Helf, K. Takada, C. Gernert, U. A. Steffens, N. Heycke, S. Schmitt, C. Rinke, E. J. Helfrich, A. O. Brachmann, C. Gurgui, T. Wakimoto, M. Kracht, M. Crusemann, U. Hentschel, I. Abe, S. Matsunaga, J. Kalinowski, H. Takeyama and J. Piel, Nature, 2014, 506, 58–62 CrossRef CAS PubMed.
- T. Mori, J. K. B. Cahn, M. C. Wilson, R. A. Meoded, V. Wiebach, A. F. C. Martinez, E. J. N. Helfrich, A. Albersmeier, D. Wibberg, S. Datwyler, R. Keren, A. Lavy, C. Ruckert, M. Ilan, J. Kalinowski, S. Matsunaga, H. Takeyama and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 1718–1723 CrossRef CAS PubMed.
- P. T. Northcote, J. W. Blunt and M. H. G. Munro, Tetrahedron Lett., 1991, 32, 6411–6414 CrossRef CAS.
- M. A. Storey, S. K. Andreassend, J. Bracegirdle, A. Brown, R. A. Keyzers, D. F. Ackerley, P. T. Northcote and J. G. Owen, mBio, 2020, 11, e02997-19 CrossRef PubMed.
- M. Rust, E. J. N. Helfrich, M. F. Freeman, P. Nanudorn, C. M. Field, C. Ruckert, T. Kundig, M. J. Page, V. L. Webb, J. Kalinowski, S. Sunagawa and J. Piel, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 9508–9518 CrossRef CAS.
- Y. Hirata and D. Uemura, Pure Appl. Chem., 1986, 58, 701–710 CAS.
- M. J. Towle, K. A. Salvato, J. Budrow, B. F. Wels, G. Kuznetsov, K. K. Aalfs, S. Welsh, W. Zheng, B. M. Seletsky and M. H. Palme, Cancer Res., 2001, 61, 1013–1021 CAS.
- W. Fenical, J. Antibiot., 2020, 73, 481–487 CrossRef CAS.
- T. Zamkovaya, J. S. Foster, V. de Crecy-Lagard and A. Conesa, ISME J., 2021, 15, 228–244 CrossRef.
- A. Bhushan, P. J. Egli, E. E. Peters, M. F. Freeman and J. Piel, Nat. Chem., 2019, 11, 931–939 CrossRef CAS PubMed.
- S. P. Jungbluth, T. Glavina Del Rio, S. G. Tringe, R. Stepanauskas and M. S. Rappe, PeerJ, 2017, 5, e3134 CrossRef PubMed.
- C. Zhang, Y. Idelbayev, N. Roberts, Y. Tao, Y. Nannapaneni, B. M. Duggan, J. Min, E. C. Lin, E. C. Gerwick and G. W. Cottrell, Sci. Rep., 2017, 7, 1–17 CrossRef.
- J. Hastings, G. Owen, A. Dekker, M. Ennis, N. Kale, V. Muthukrishnan, S. Turner, N. Swainston, P. Mendes and C. Steinbeck, Nucleic Acids Res., 2016, 44, D1214–D1219 CrossRef CAS PubMed.
- J. Hastings, D. Magka, C. Batchelor, L. Duan, R. Stevens, M. Ennis and C. Steinbeck, J. Cheminf., 2012, 4, 1–20 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1np00051a |
| This journal is © The Royal Society of Chemistry 2022 |