Open Access Article
Open Access ArticleThe chemical ecology of the fungus-farming termite symbiosis†
Suzanne
Schmidt
 a,
Sara
Kildgaard
a,
Sara
Kildgaard
 a,
Huijuan
Guo
a,
Huijuan
Guo
 b,
Christine
Beemelmanns
b,
Christine
Beemelmanns
 b and
Michael
Poulsen
b and
Michael
Poulsen
 *a
*a
aSection for Ecology and Evolution, Department of Biology, University of Copenhagen, Universitetsparken 15, 2100 Copenhagen, Denmark. E-mail: mpoulsen@bio.ku.dk
bLeibniz Institute for Natural Product Research and Infection Biology e.V., Hans-Knöll-Institute (HKI), Beutenbergstraße 11a, 07745 Jena, Germany
First published on 19th August 2021
Abstract
Covering: September 1972 to December 2020
Explorations of complex symbioses have often elucidated a plethora of previously undescribed chemical compounds that may serve ecological functions in signalling, communication or defence. A case in point is the subfamily of termites that cultivate a fungus as their primary food source and maintain complex bacterial communities, from which a series of novel compound discoveries have been made. Here, we summarise the origins and types of 375 compounds that have been discovered from the symbiosis over the past four decades and discuss the potential for synergistic actions between compounds within the complex chemical mixtures in which they exist. We go on to highlight how vastly underexplored the diversity and geographic distribution of the symbiosis is, which leaves ample potential for natural product discovery of compounds of both ecological and medical importance.
1 Introduction
Natural products represent structurally and functionally diverse molecules that exhibit a plethora of functional roles in signalling, communication, or defence in the natural context within which they are produced.1,2 Hosts may produce their own defensive compounds, while symbiont-derived natural products might serve as vital mediators between hosts and their antagonists by conferring protection directly through biological activity3 or indirectly by stimulating host immune systems to improve protection during infection.4 Despite increasing scientific interest, the precise functions of secreted natural products, their possible targets and modes of action, and synergies between compounds within complex chemical mixtures remain mostly unknown. In contrast, ecology and genome-mining driven exploration of complex host-symbiont associations using state-of-the-art analytical dereplication tools has proven to be a successful strategy for the discovery of novel chemical scaffolds and bioactivities.5–8 Amongst the many symbiosis-related model systems, fungus-farming insects have been intensively studied, both from a chemical ecology and pharmacological perspective. As both the fungus-growing termite and ant symbioses rely on the successful protection of a fungal cultivar, their primary food source, microbial symbionts are likely to act as defensive partners and thus as prolific natural product sources for novel chemistry.2,6–8Fungus-farming termites (Macrotermitinae, Termitidae: Blattodea) engage in a symbiosis with a fungal cultivar (genus Termitomyces; Agaricales: Lyophyllaceae) that they have co-evolved with since the origin of fungiculture 30 mya.9–12 In addition, termite guts and fungus combs harbour diverse and co-adapted microbiomes that play roles in plant biomass decomposition and potentially prophylaxis.13–16 Symbiont complementarity ensures near-complete plant biomass decomposition with contribution from the termite-nurtured fungal garden and gut microbiomes.17,18 Consequently, the termites provide important ecological services as major decomposers of dead plant material, and translocation of water and nutrients that sustain vegetation growth in the vicinity of termite mounds.19,20
Foraging on decaying plant biomass should make the termites vulnerable to antagonists or competitors of their fungal crop entering colonies with the plant substrate.21 Remarkably, the symbiosis does not appear to suffer from specialised diseases despite maintaining fungal cultivars in monoculture within colonies that can become decades old.11,15 This suggests the presence of very effective defences, which extend to the use of natural products produced by the insect host22–24 and fungal and bacterial symbionts.25,26 However, despite receiving substantial – and increasing – attention, we lack a comprehensive overview of the natural products that have been identified in the fungus-growing termite symbiosis and reflection on their putative functions.
To do this, we screened the literature for natural products from fungus-farming termites and identified more than 60 sources that collectively report 375 natural products discovered from members of the termite symbiosis. For each compound, we subsequently identified known or putative activities reported in the literature. This overview allows us to discuss the identity of natural product classes and their activities, in addition to their origins. Lastly, by quantifying the vast under-sampling of the diversity and geographic distribution of the symbiosis, we outline opportunities for future studies on natural products of ecological and pharmacological importance.
2 The fungus-farming termite symbiosis
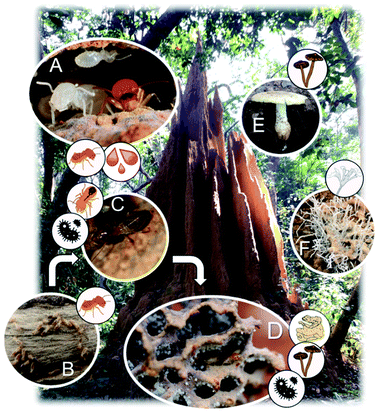
Fungus-farming in termites originated once ∼30MYA in the African rainforest in the subfamily Macrotermitinae.10 This monophyletic subfamily includes ∼404 described termite species in 13 genera that all cultivate specialised basidiomycete fungi in the genus Termitomyces.11,27 To date, 49 Termitomyces species have been described, all of which depend on association with termites. Co-phylogenetic patterns between the termites and Termitomyces indicate host-symbiont co-diversification,10,11 and over the course of time, the association has dispersed to inhabit most of sub-Saharan Africa and large parts of Southeast Asia.10 Most fungus-farming termites acquire their fungal mutualist horizontally as mushrooms growing from termite colonies produce basidiospores that are dispersed to the environment and collected by workers of new nests to establish incipient fungus garden.28 However, some degree of specialisation exists despite horizontal transmission, so that termite species preferentially associate with specific fungal species.11,29Termitomyces is maintained within termite mounds in structures called fungus combs (gardens), within which the termites manure the fungus as a monoculture on decaying plant biomass that older workers bring to the nest. This biomass is passed to younger workers that ingest it along with asexual Termitomyces spores from mature parts of the fungus comb, and this process ensures efficient mixing and deposition of Termitomyces with the plant substrate as ‘fresh’ comb (Fig. 1).28Termitomyces proliferates while decomposing the plant substrate and after near-complete degradation of all plant biomass, comb biomass is consumed by older termites.14,28,29 The two gut passages (inoculation and final consumption) place the termite gut central in the symbiosis, and guts harbour diverse and distinct bacterial communities, containing 100s of bacterial lineages, predominantly in the phyla Firmicutes, Bacteroidetes, and Proteobacteria.13,30,31 Dominant members are distinct from those of other termites – and the ancestral cockroaches – implying that community shifts in compositions and functions have been associated with the transition from a plant to a fungal biomass-based diet of the termite host.30,31 Across termite species within the fungus-growing termite sub-family, conserved gut community compositions suggest specificities in association for reasons that remain unresolved.13 Gut contents are deposited during the inoculation of fungus combs, leading to an abundance of gut-inhabiting lineages within gardens; however, these bacterial communities fluctuate more in composition than those observed in guts.13
Maintaining a dense monoculture fungus15,32 within a colony in an environment that is optimised for fungal growth with high humidity,10 constant temperature,33 and continuous addition of plant biomass should make fungal gardens prone to infections. However, despite being in close contact with substrates containing potentially competing fungi,21 fungus combs appear free from actively growing competitors or antagonists.15 Only if colonies are compromised, e.g., due to the removal or death of workers, will combs rapidly get infested and overgrown by generalist (e.g., Trichoderma34) and specialist (Pseudoxylaria35) ascomycete fungi. A series of defences helps secure that fungus gardens are disease-free, including avoidance of antagonists by the termites,34 burial of unwanted fungi,36 and utilisation of antimicrobial compounds of termite, Termitomyces, and bacterial origins.23,24,37
3 Natural products reported from the termite symbiosis
Our extensive literature survey from 1972 to 2020 revealed that 375 natural products have been identified in the fungus-farming termite symbiosis. Subsequent literature screening of hundreds of scientific papers allowed us to generate a searchable database of all compounds (ESI, Table 1†) that includes molecular formulas, mass, compound name, natural product class, the producing organism and termite host (when known), geographic origin of discovery, method of identification, and known bioactivities or lack thereof. In addition to citing the articles that elucidated the presence of the compounds from the fungus-growing termite symbiosis, we also include references associated to bioactivities, modes of action, and synergies between compounds (ESI Table 1†).Below we summarise the discoveries of natural products from the symbiosis. While early reports focused on the termite host and particularly glandular defensive secretions, research from the late 1990s and onwards shifted towards analyses of symbiotic partners (Fig. 2A). Current reports include natural products from 14 termite and 11 Termitomyces species, five bacterial genera, and two studies on compound mixtures from the complex gut and comb environments (Fig. 2A). The natural products present are structurally diverse, often derived either from the phenylpropanoid pathway, or of polyketide, terpene or fatty acid origin (Fig. 2B). Unsurprisingly, their bioactivities also range in targets (antifungal and antibacterial) and levels (ESI Table 1† and Fig. 2B). We structure the sections below by organism, first discussing compounds of termite origin, thereafter Termitomyces, bacteria and Pseudoxylaria, and lastly symbiont communities.
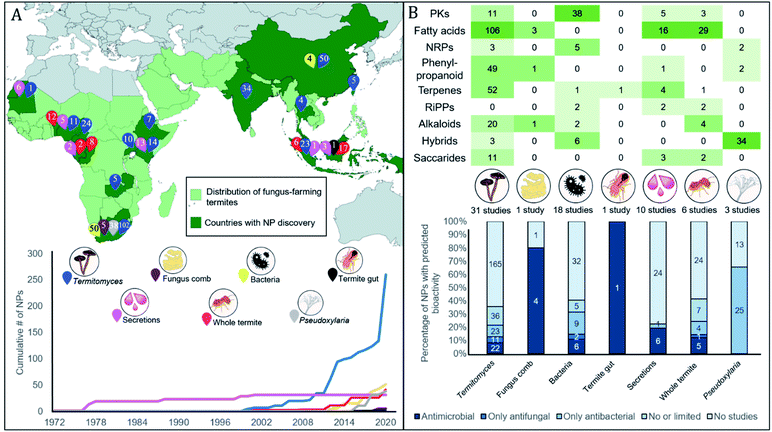 | ||
| Fig. 2 Thirty-five years of natural product discovery from the fungus-farming termite symbiosis. (A) Map showing in light green the distribution of the fungus-farming termites (Macrotermitinae) and in dark green countries and areas from which discoveries of natural products have been made. A total of 375 compounds have been discovered from the symbiosis, from across the symbiotic partners, as indicated with numbered pins of different colours. Below the map, we plot the timeline for the cumulative discovery of fungus-farming termite-associated natural products compounds by symbiosis source. (B) The top section provides a heatmap of compounds discovered across biosynthetic classes by symbiont origin. Two S-containing compounds from Termitomyces are not shown. The bottom section provides the percentage of natural products for each source where antimicrobial activities have been reported (MIC less than 128 μg μl−1 or ZOI more than 10 mm). The number of compounds that each bar represents is indicated within the stacked bar categories. For full details, see ESI Table 1.† | ||
3.1 The fungus-farming termite host
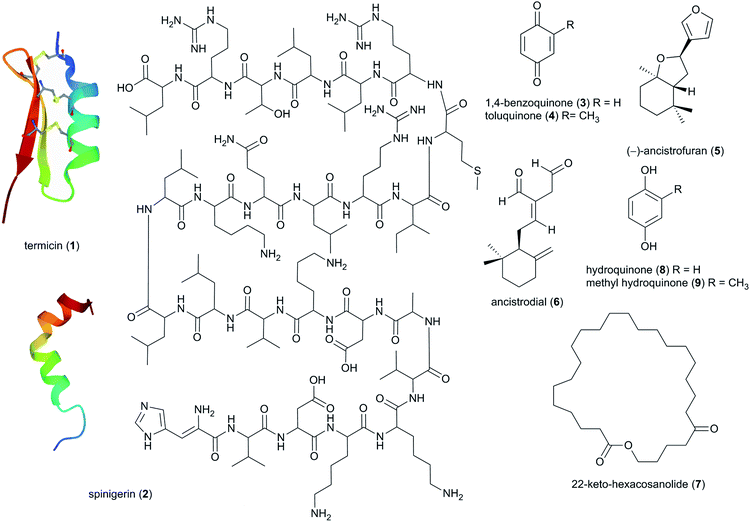
The insect host was the first source of termite-associated natural products to be exploited already in the 1970s, focusing on soldier oral and salivary gland secretions that serve in the chemical defence against e.g., ants.38 Since then, 73 natural products have been discovered, primarily being steroids and lipids/hydrocarbons, with 13 being tied to potential defensive or antimicrobial activities (ESI Table 1†).3.2 Termitomyces
In the late 90s, the first reports of natural products from Termitomyces appeared, and now include work on 11 species from fourteen countries (Fig. 2A and B). By December 2020, 31 studies have identified 257 natural products from Termitomyces, of which at least 53 have antimicrobial properties (ESI Table 1† and Fig. 2B). Studies on Termitomyces mushrooms and cultures have revealed their prominent nutritional value, and several Termitomyces species are consumed also for medicinal purposes by indigenous communities.48 Some of the major bioactive compounds found in Termitomyces could have potential antioxidant, anti-tumour and antimicrobial effects.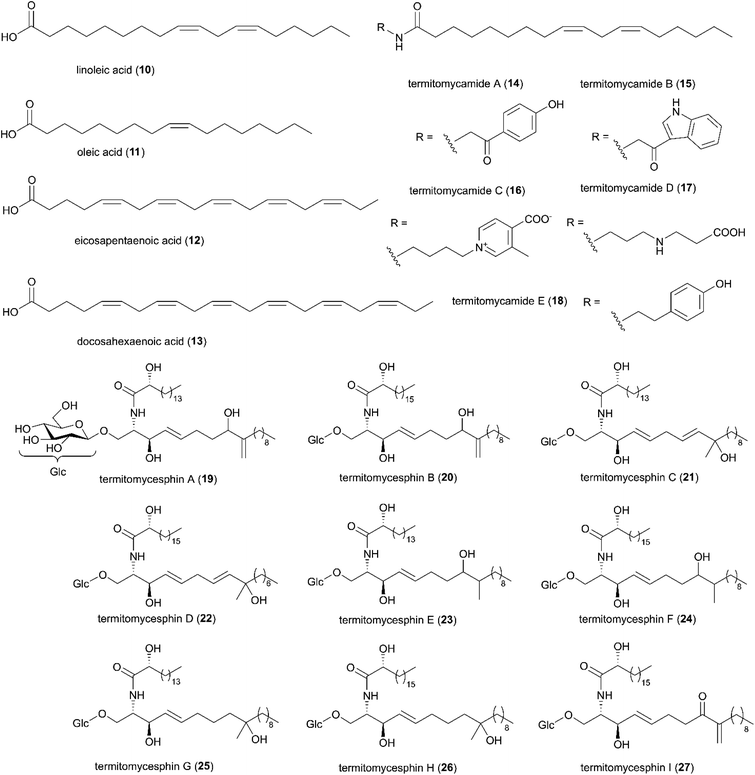 | ||
| Fig. 4 A selection of fatty acids identified in Termitomyces spp. Compound 19–27 were redrawn from original articles.58–61 | ||
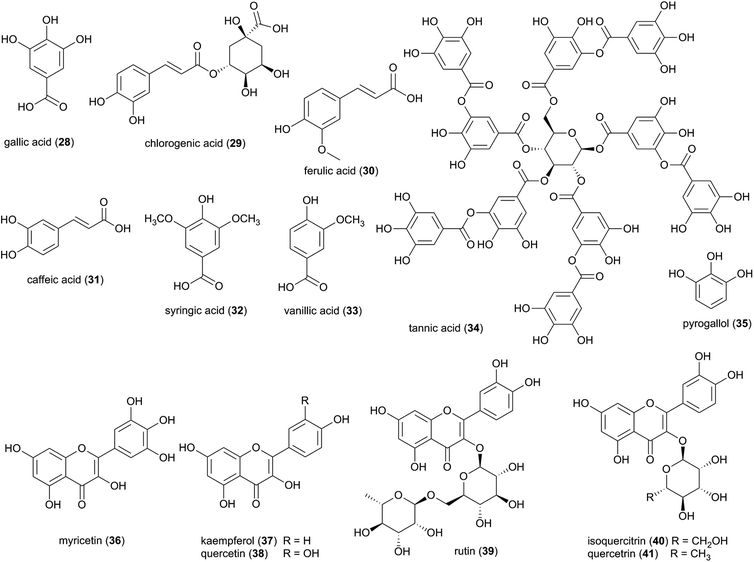
Characterized flavonoids include myricetin (36), kaempferol (37), quercetin (38) rutin (39), and isoquercetrin (40), which vary in their degree of antibacterial and antifungal activities (see ESI Table 1†)73 but have been proposed to exhibit complementary and synergistic activities (in vitro and in vivo) through various mode of actions and thus may be important in fungus garden defence. Specifically, synergistic activities of flavonoids in combination treatments have been observed for e.g., rutin, quercitrin (41), and quercetin, which revealed stronger inhibitory effects on Bacillus cereus and Salmonella enterica Serotype Enteritidis compared to treatment with either flavonoid alone.74 Notably, rutin (39) does not exhibit any antibacterial activity alone, but significantly enhances antibacterial effects of other flavonoids, including quercetin, quercitrin, kaempferol and myricetin.74 Causality for the complementary activities remains obscure, although modes of action for the various flavonoids allows for speculation. Rutin, might for example enhance the activity of other hydrophilic antibiotics by disrupting the bacterial cell walls of multi-drug-resistant Staphylococcus aureus (MRSA)75,76 and allowing its effective penetration. Despite not showing any effect alone, rutin have been shown to improves the antibiotic effect of quercetin,76 Quercetin (38) has been linked to multiple modes of action, including inhibition of DNA gyrase,77 inhibition of efflux pumps,78 membrane disruption79 and cell envelope synthesis.80 It has also both potentiating and synergistic antibacterial actions in combination with a broad range of antibiotics with different mode of actions, such as penicillins (ampicillin, amoxicillin and methicillin),76,81,82 cephalosporins (ceftriaxone, cefixime, and cephradine),76,83 tetracycline81 and aminoglycosides (tobramycin, gentamycin, amikacin).83 It is thus conceivable that combinations of multiple compounds play a significant role in termite nest defence through contributions to the antimicrobial activity observed in Termitomyces.24,84
Phenolic compounds are found widely throughout the plant kingdom, and it is possible that the phenolic compounds identified in Termitomyces could have been synthesized and absorbed from plants. However, the Shikimate pathway has been correlated to the presence of phenolic compounds in fungi85,86 and a recent study showed that despite flavonoids being well-known plant metabolites, all genes and protein sequences associated with the biosynthesis of flavonoids can be found in some fungi.87 Future genome analyses and molecular biological studies will thus be needed to confirm the putative fungal origin of these metabolites.
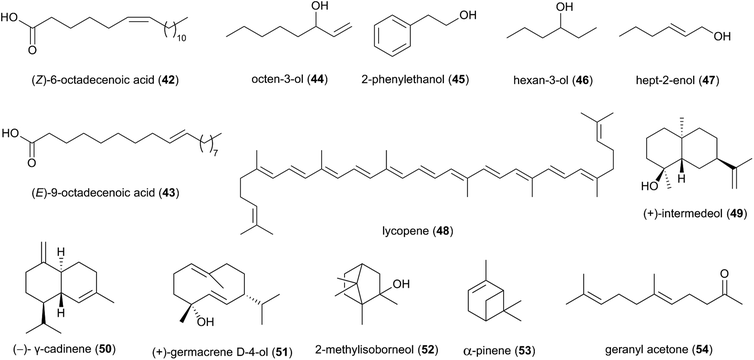
Studies based on head-space gas-chromatograph mass spectrometry (GC-MS) have allowed for the first insights into the volatilome of Termitomyces sp. and revealed several odorous compounds including typical C8 components (mushroom odours), such as octen-3-ol (44), 2-phenyl-ethanol (45), hexan-3-ol (46) and hept-2-enol (47) (Fig. 6), as well as benzaldehyde (almond odour), benzyl alcohol (sweet-spicy odour), phenylethanol (rose odour), and monoterpenes.16,89,90
More than 50 terpenes and their functional derivatives (terpenoids) have been reported from Termitomyces (ESI Table 1†). Both compound classes are found in most living organisms and contribute to flavour, scent and colour in plants and fungi, and often exhibit insecticidal and/or antimicrobial activities.91 Just to give an example: in 2016, the terpenoid lycopene (48), a carotenoid hydrocarbon, was detected in T. heimii and T. microcarpus63 and it exhibits strong antimicrobial activity (Fig. 6).
Terpenes and terpenoids are derived from the conversion of geranyl or farnesyl pyrophosphate catalysed by dedicated terpene synthases and/or terpene cyclases.92 A first bioinformatics analysis of the draft genome sequence of Termitomyces sp. J132 revealed more than 20 putative type I terpene cyclase genes, three of which were functionally characterized as (+)-intermedeol (49) synthase, (−)-γ-cadinene (50) synthase and (+)-germacrene D-4-ol (51) synthase.93 The cyclization reactions were surveyed for all three enzymes by incubation with isotopomers of FPP, yielding valuable insights into the stereochemical course of the ring closing mechanism, reprotonation steps and hydride shifts. A follow-up study in early 2021 identified 13 additional terpenoids from Termitomyces, including 2-methylisoborneol (52), α-pinene (53) and geranylacetone (54),16 which have been linked to antifungal and antibacterial activities.94–96 α-Pinene has also been observed to modulate the sensibility of antibiotic-resistant bacterial strains by preventing the efflux of multiple antibacterial agents.95,97
3.3 Bacterial symbionts
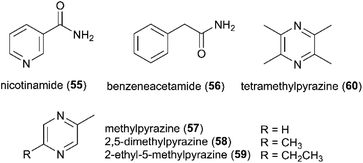
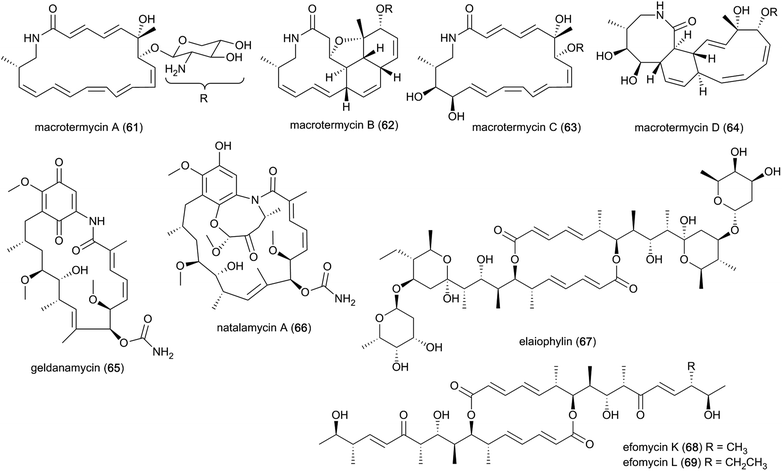
The past decade has seen an increased focus on chemical analyses of termite-associated bacteria that have been isolated from the termite cuticle, termite gut or fungus comb. Most studies have been on termite species from South Africa (15 studies/50 compounds) and China (3 studies/4 compounds)105–107 (Fig. 2A). Termite guts and combs harbour diverse bacterial communities,13,15,108 however, chemical studies and novel natural products have only been reported from Bacillus, Streptomyces, Amycolatopsis and Actinomadura, of which three belong to the phylum Actinobacteria. Of the 54 natural products discovered from termite-associated bacteria, 17 have been reported to exhibit strong antimicrobial activities (ESI Table 1† and Fig. 2B).Ecological-driven co-cultivation studies of the M. natalensis-associated Amycolatopsis sp. with Pseudoxylaria sp. X802, which is often found dormant within fungus combs,35 lead to the isolation of four glycosylated macrolactams (macrotermycin A–D; 61–64). Macrotermycin A and C are 20-membered glycosylated polyene macrolactams, which are transformed by an intramolecular cycloaddition sequence leading to macrotermycin B and D. While polyene macrotermycin A and C exhibited strong antimicrobial activity, including selective inhibition of Pseudoxylaria, the cyclized derivatives were inactive (Fig. 8).26
In a similar co-cultivation study, Streptomyces sp. M56 was also found to strongly inhibit Pseudoxylaria spp. (and Termitomyces) growth. This led to the isolation of the antifungal polyketide geldanamycin (65) along with the novel geldanamycin analogue, natalamycin A (66),111 which contains a rare C-5 unit on the 3-amino-5-hydroxybenzoic acid (AHBA) head group to form fused bicyclic[6.4.0]ansa macrolide. The same bacterium was also found to produce the antibacterial macrolactone elaiophylin (67), along with several analogues efomycins K and L (68–69), which are antibacterial towards Gram-positive bacteria.112 Efomycins are C2 symmetric 16-membered macrolides that are derived from two linear polyketide chains with an unsaturated enone moiety, while elaiophylins carry a hemiketal moiety and are glycosylated. However, neither of the compounds alone were found to be responsible for inhibition of Pseudoxylaria, and synergistic effects were postulated as geldanamycin has previously been reported to possess synergistic antifungal activities with e.g., triazoles, echinocandins and fluconazole.113,114 Indeed, elaiophylin enhances the activity of the co-produced antifungal macrolide rapamycin against Candida albicans, while lacking activity alone.115 Geldanamycin is a well-known potent inhibitor of the heat shock protein Hsp90,116 whereas the mode of action for elaiophylin has not yet been established, but may be associated with the alteration of membrane permeability by destabilization and formation of ion-penetrable channels.117 A study investigating the role of Hsp90 in the mTOR-signalling pathway reported that geldanamycin targets this pathway by suppressing mTOR activity, similar to the antifungal rapamycin (originally identified to target the TOR kinases in the yeast Saccharomyces cerevisiae), which induces dissociation of the mTOR-raptor complex. Both exhibit anti-cancer activities by targeting mTOR, but through different mechanisms of action.118 As TOR kinases are ubiquitously conserved in eukaryotic organisms, the same target may be responsible for the antifungal activity observed towards Candida.113 Geldanamycin and elaiophylin are often found to be co-produced, with the TetR-family regulator GdmRIII apparently having an inverse effect on the two compounds, positively regulating geldanamycin and to some extend repressing elaiophylin.119 No explanation for this possible co-production has been reported, but it has been hypothesized that reverse regulation is essential in controlling the flux of precursor metabolites for the two compounds.120 Further investigation into potential complementary activities of geldanamycin and elaiophylin against ecologically relevant fungal strains that challenge the symbiosis could help unravel their ecological importance.
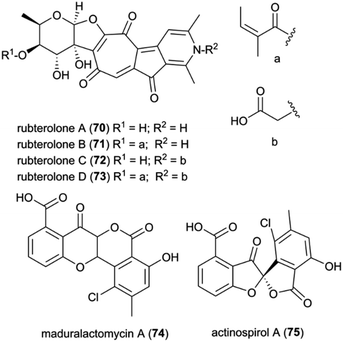
Recently, Actinomadura rubteroloni was found to produce two types of polyketide-derived, highly oxidized and rearranged types of natural products (Fig. 9). In a first study, a rare group of glycosylated tropolone-containing natural products, named rubterolones A–D (70–73), were identified and found to have anti-inflammatory activity.121 Rubterolones feature a tropolone moiety, a fused cyclopentanone ring, an O,C-condensated sugar and a highly substituted pyridine or pyridine inner salt moiety, and are structurally related to the recently identified rubrolone B122 and isarubrolones.123 Several biosynthetic studies and complementing isotope feeding experiments suggested that the tropolone and cyclopentanone containing carbon skeleton is of type II polyketide origin, but undergoes a series of complex oxidative rearrangements induced by the cluster-encoded oxygenases to yield first the later-identified pre-rubterolones, and then undergoes a spontaneous pyridine formation in the presence of amines.
While high biomass cultivation induced mostly the formation of rubterolones, growth from diluted spore cultures resulted in the dominant formation of chlorinated maduralactomycin (74) containing a rare bicyclic 4-chromanone fused with isocoumarin core structure and spirocyclic actinospirols (75).124 Intriguingly, the tested cultivation conditions likely simulate the natural ecological environment bacterial strains would face when entering the fungus comb after obligate gut passage of a termite worker. Based on comparative genome studies and HRMS2-based GNPS analyses, a putative biosynthetic mechanism was proposed that is based on a non-canonical angucycline biosynthesis and extensive oxidative modifications that ultimately result in the formation of the unique halogenated tetracyclic polyketides and after additional oxidative rearrangements in the formation of actinospirols. Bioactivity studies of maduralactomycin A revealed antibacterial activity towards both Vancomycin-Resistant Enterococcus faecalis and Mycobacterium vaccae.124
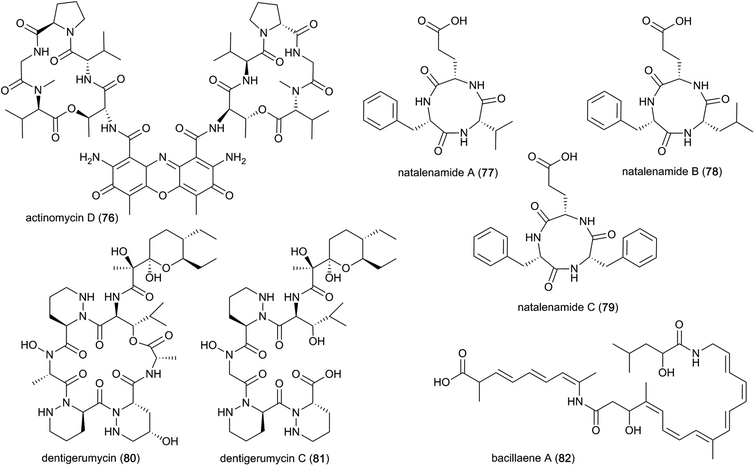
A well-known compound that has been discovered multiple times from termite-associated Streptomyces is the chromopeptide antibiotic actinomycin D (76). Actinomycin D belongs to a family of bicyclic chromopeptide lactones sharing the chromophoric phenoxazinone dicarboxylic acid attached to two cyclic pentapeptide lactones of non-ribosomal origin containing L-Thr, D-Val, L-Pro, Sar and L-MeVal. It appears to have broad-spectrum antibacterial activity and potent activity against several ecologically-relevant fungi, including Pseudoxylaria (Fig. 10).107,125 Actinomycin D may have this effect due to mode(s) of action that include DNA-dependent inhibition of RNA synthesis, and potentially targeting the fungal plasma membrane through a membrane splitting mechanism.126
Interestingly, Actinomycins are also produced by Streptomyces isolated from fungus-farming ants, where they may be involved in fungistatic activity against Escovopsis spp. antagonists of the mutualistic fungus of the ants.127 Another study showed strong synergistic effects of mixtures of antifungals against E. weberi.128 However, strong antibacterial activities were also reported towards ant bacterial symbionts, suggesting that they may play roles in competition between bacteria.128 Actinomycin D has also been observed to be involved in synergistic effects with the antimicrobial agents amphotericin B and colistin,129,130 both of which disrupt the cell membrane, thereby facilitating actinomycin D access.
Lastly, a study directed at identifying novel pharmacologically relevant natural products elucidated three new cyclic tripeptides, containing L-Phe, L-Val/L-Leu and L-Glu, named natalenamides A–C (77–79).131 These compounds are presumably of non-ribosomal origin, and were obtained from Actinomadura sp. RB99 using an LC/MS/UV-based dereplication approach.131 Natalenamide A and B showed weak cytotoxicity against cancer cell lines HepG2 (liver) and HeLa (cervical), and A549 (lung), respectively. Natalenamide C (79) significantly inhibits the production of 3-isobutyl-1-methylxanthine induced melanin.
In a subsequent study, dentigerumycins (80–81) were isolated from Streptomyces sp. M41 from the South African fungus-growing termite species Macrotermes natalensis, based on its unique metabolomic profile from principal component analysis (PCA) of 41 Actinobacteria isolates.136 Dentigerumycin is a 19-membered macrocyclic hexapeptide containing three piperazic acids, ester-forming Ala, N-OH-Ala, β-OH-Leu, and a pyran-bearing polyketide acyl chain, whereas dentigerumycin C is the linear form lacking the ester-forming alanine residue (Fig. 10).136–138 The identified derivatives are structurally related to the Pseudonocardia-derived antifungal dentigerumycin that inhibits the specialized mycopathogen Escovopsis spp. that invade and consume the ants' fungal cultivar.137
3.4 Pseudoxylaria stowaway fungus
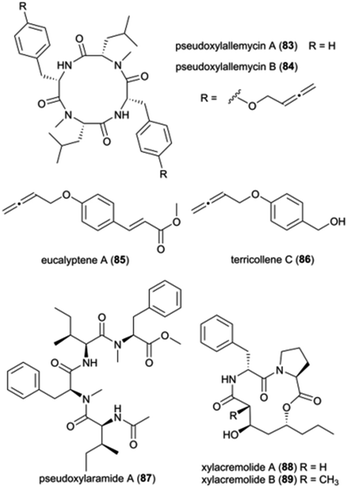
Pseudoxylaria spp. (Ascomycota: Xylariaceae) are frequently found on deteriorating comb material and is presently considered a stowaway fungus, waiting as a substrate specialist and opportunistic weed until conditions are favourable for outcompeting Termitomyces.21,35 Members of the Xylariaceae are ecologically important as saprotrophs, but they are also well-known for their biosynthetic capabilities to produce structurally diverse metabolites with a broad spectrum of biological activities.139 While the exact mechanisms of the interaction and role of Pseudoxylaria in the comb remains unknown, given the competitive nature of the association,35 it is conceivable that termite-associated Pseudoxylaria produce biologically active small molecules. Indeed, crude Pseudoxylaria extracts can be both antifungal and antibacterial, and a recent study discovered six novel compounds. Using an MS-based imaging and a dereplication strategy, Guo et al. discovered six new antimicrobial cyclic peptides, named pseudoxylallemycins A–F (83–84), and eucalyptene A (85) and terricollene C (86) from Pseudoxylaria sp. X802 (Fig. 11).140 Pseudoxylallemycins contain symmetric or asymmetric tetracyclic consisting L-Phe, NMe-L-Leu, and a very rare allenyl modification on tyrosine. More recently, high-resolution tandem mass spectrometry (HRMS2) based approaches led to the identification of chemical features unique to Pseudoxylaria sp. X187: four linear non-ribosomally synthesized peptides (NRPs) and two cyclic NRPS-polyketide synthase (PKS) derived natural products, pseudoxylaramides (87) and xylacremolides (88–89) (Fig. 11).141 Pseudoxylaramides are linear tetrapeptides containing L-Phe and L-Ile with methylation in the C-terminal and acylation in the N-terminal, while xylacremolides are cyclic polyketide-dipeptide hybrids composed of L-Pro and L-Phe and 3,5-dihydroxy octanoic acid (DHOA) (Fig. 11). The pseudoxylaramides share features with other antimicrobial fungal compounds,142 and the xylacremolides share some similarity to trapoxin, an antitumor cyclic tetrapeptide.143 None of the compounds identified in Pseudoxylaria showed activity towards the tested strains; thus, the bioactivity of these NRPS and NRPS–PKS hybrids still needs to be resolved. However, their structural similarities to other peptides point to ecologically-relevant roles as antimicrobials.3.5 Analysis of symbiont communities
The identification and interactions of chemical compounds in the complex communities of organisms in termite guts and fungus combs hold particular promise for insights into ecologically-relevant functions. To date, only two studies have investigated the mixtures of natural products from the complex gut and fungus comb symbiont communities, one of which focused on putative defensive roles in Macrotermes and Odontotermes nests from South Africa15 and one on termite guts.144 The latter involved GC-MS analysis of dissected gut of Macrotermes gilvus workers, which led to the identification of the terpenoid β-sitosterol (90) (Fig. 12), which possesses selective antifungal and antibacterial activities.145,146 As β-sitosterol is one of the most abundant occurring phytosterols, its identification in the gut most likely stem from the plant material the termites gathered.The chemical analysis of fungus combs led to the identification of five small molecules, of which stearamide (91), azelaic acid (92), indole-3-carboxaldehyde (93) and 4-hydroxybenzaldehyde (94) have reported antifungal and antibacterial activities (Fig. 12; ESI Table 1†). Indole-3-carboxaldehyde (93) was previously identified from the bacterium Janthinobacterium lividum on the skin of the red-backed salamander (Plethodon cinereus), where it appears to be involved in the chemical defence against the fungal pathogen Batrachochytrium dendrobatidis causing chytridiomycosis in amphibians.147 However, none of the identified compounds were verified to be responsible for the observed antifungal activity of chemical extracts of fungus comb against an ecological-relevant competitor of Termitomyces (Trichoderma) and the entomopathogen B. bassiana.15
Although comb metabolites may exhibit weak activity, it is conceivable that synergistic or additive antimicrobial effects exist. Synergistic interactions between multiple small molecules present in extracts of Termitomyces, fungus combs or Actinomadura, have been shown in other work to improve the antibiotic effect of multidrug resistant bacteria, like MRSA.148 These include phenolic acids of Termitomyces origin (see above) and the small molecule 4-hydroxybenzaldehyde (94) that was identified in antifungal fungus comb extracts.15 This compound appears to improve the sensitivity of Acinetobacter baumannii to amphenicol.149 Similarly, the two alkaloids, indole-3-carboxaldehyde and banegasine (95) enhance the efficacy of multiple antifungal agents against a diversity of fungal species.150–153 Instead of relying on one single metabolite, the combination of diverse molecules with synergistic or potentiating effects is likely essential for defence of the symbiosis, mirroring strategies employed by humans to overcome resistance evolution in target pathogens.154
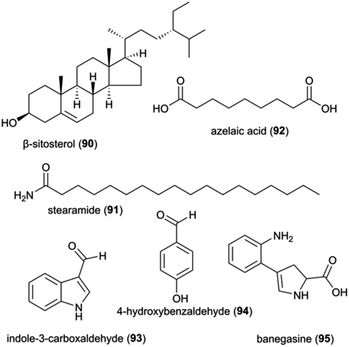 | ||
| Fig. 12 A selection of natural product and small molecules identified from analyses on complex communities. | ||
4 Prospects for defence potential and novel natural product discovery
The fungus-farming termite symbiosis is extraordinarily successful in hindering the entry and spread of antagonists,15,21 likely through a multi-layer defence that integrates behaviours with sophisticated use of antimicrobial compounds. The roles of the vast majority of identified natural products from the symbiosis have yet to be established, including for general bioactivity and in ecologically-relevant contexts (ESI Table 1†). Current reports rarely provide evidence for importance in defence, leaving promising insights to come from elucidation of their concentrations, how and when compounds are applied, and their targets. Efforts to bridge objectives and methods of natural product discovery and elucidation of roles in defence are warranted to shed light on the implications of natural product use in defence in the environments within which they act.The natural product discovery potential from the symbiosis remains extraordinary, with only a small fraction of the geographical distribution and host and symbiont diversity, having been explored (Fig. 13A). Efforts so far have remained on the conspicuous termite genus Macrotermes (80% of studies), which accounts for only a fifth of the known termite species.27 Similarly, only a fourth of the described Termitomyces species, and a handful of gut and comb bacteria and antagonists of the symbiosis, have been explored (Fig. 13B). Among the bacteria, Actinobacteria dominate the focus,35,125,155 for good reason given their prolific potential,125,155,156 including as defensive symbionts in fungiculture.35,157 Recent genome analyses support that we are far from elucidating their full chemical potential: more than 100 biosynthetic gene clusters present within 16 actinobacterial genomes encode putative novel chemistry.155 It is thus undeniable that further investigation carries promising potential for novel discoveries. This potential extends to so-far unexplored organisms that inhabit termite mounds as guests, such as rove beetles and scuttle flies and their gut symbionts,158–160 and other conceivably beneficial symbionts (e.g., yeasts161). Understanding these multi-species interactions, and the compounds involved, remains a major challenge and an outstanding opportunity for future work on the symbiosis. To this end, comparison of the chemical repertoire that the farming termite symbiosis with other fungus-farming symbioses holds great potential to shed light on unique and convergently evolved utilisation of natural products in fungus farming.
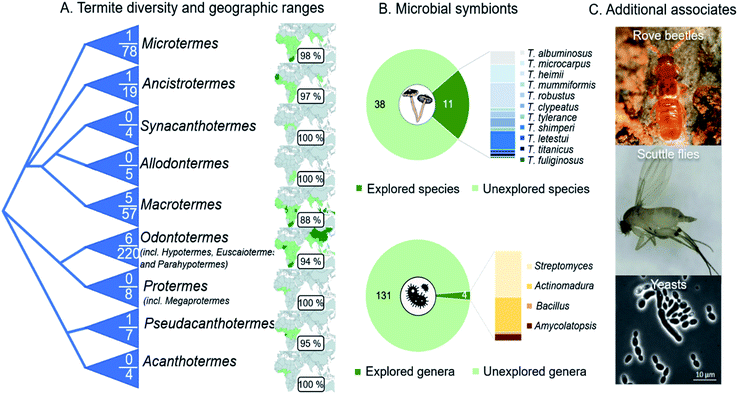 | ||
Fig. 13 Outstanding opportunities for natural product discovery from the fungus-farming termite symbiosis. (A) A schematic phylogeny of the Macrotermitinae (based on ref. 11), indicating the number of termite species out of the total number of known species that have been explored for natural products. The geographic ranges of termite genera and the percentage of countries from which no work has been reported on natural product from any members of the symbiosis. (B) Eleven of the 49 described Termitomyces species have been explored for natural products, and the distribution of compounds discovered illustrated by species in the bar plot. Only four of the estimated 102–149 genus-level phylotypes (average 131![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14) have been cultured and explored for natural product discoveries, not including metabolomic analyses of complex microbial communities in guts and fungus combs. (C) Natural product discoveries have largely overlooked guest species, including beetles (Tom Murray) and flies,158–160 and additional symbionts (e.g., yeast species161). 14) have been cultured and explored for natural product discoveries, not including metabolomic analyses of complex microbial communities in guts and fungus combs. (C) Natural product discoveries have largely overlooked guest species, including beetles (Tom Murray) and flies,158–160 and additional symbionts (e.g., yeast species161). | ||
Bioactive natural products from the fungus-farming symbiosis occupy a unique chemical space and their bacterial and fungal symbiont represents an extraordinary discovery and studying potential from both ecological and chemical perspectives. Previous investigations have elucidated a large number of novel compounds and theorised their functions. Insights from reported activities show their potential to be used by the symbiosis in multi-target combination defences. In recent years, the importance of synergies between natural products has gained significant attention, including in the development of new methods and optimisation of approaches to identify mixture constituents, characterise the nature of compound interactions and unravel synergistic or potentiating modes of action. The continued studies on this topic are of the utmost importance and we are still only at the beginning of investigating the chemical potential of the fungus-farming termite symbiosis. If given proper attention, the symbiosis holds great promise for the future of natural product drug discovery.
5 Author contributions
SS: conceptualization, data curation, methodology, formal analysis, validation, visualization, writing – original draft, writing – review & editing. SK: conceptualization, data curation, methodology, formal analysis, visualization, writing – original draft, writing – review & editing. HG: data curation, methodology, validation, writing – review & editing. CB: conceptualization, funding acquisition, validation, visualization, writing – original draft, writing – review & editing. MP: conceptualization, funding acquisition, project administration, resources, supervision, validation, visualization, writing – voriginal draft, writing – review & editing.6 Conflicts of interest
There are no conflicts to declare.7 Acknowledgements
We thank members of the Social and Symbiotic Evolution group at the University of Copenhagen for comments on a previous version of the manuscript. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy – EXC 2051 – Project-ID 390713860 and CRC 1127 – Project-ID 239748522 to CB; and a European Research Council Consolidator Grant (ERC-CoG-771349) to MP.8 Notes and references
- J. Macheleidt, D. J. Mattern, J. Fischer, T. Netzker, J. Weber, V. Schroeckh, V. Valiante and A. A. Brakhage, Annu. Rev. Genet., 2016, 50, 371–392 CrossRef CAS PubMed.
- K. Scherlach and C. Hertweck, Annu. Rev. Microbiol., 2020, 74, 267–290 CrossRef CAS PubMed.
- J. Hrcek, A. H. McLean and H. C. Godfray, J Anim Ecol, 2016, 85, 1605–1612 CrossRef.
- T. Ohta, A. Ido, K. Kusano, C. Miura and T. Miura, PLoS One, 2014, 9, e114823 CrossRef.
- A. L. Demain, Appl. Microbiol. Biotechnol., 1999, 52, 455–463 CrossRef CAS PubMed.
- E. B. Van Arnam, C. R. Currie and J. Clardy, Chem. Soc. Rev., 2018, 47, 1638–1651 RSC.
- N. Adnani, S. R. Rajski and T. S. Bugni, Nat. Prod. Rep., 2017, 34, 784–814 RSC.
- D. Heine, N. A. Holmes, S. F. Worsley, A. C. A. Santos, T. M. Innocent, K. Scherlach, E. H. Patrick, D. W. Yu, J. C. Murrell, P. C. Vieria, J. J. Boomsma, C. Hertweck, M. I. Hutchings and B. Wilkinson, Nat. Commun., 2018, 9, 2208 CrossRef.
- T. Nobre, C. Rouland-Lefèvre and D. K. Aanen, in Biology of Termites: a Modern Synthesis, ed. D. E. Bignell, Y. Roisin and N. Lo, Springer Netherlands, Dordrecht, 2011, pp. 193–210 Search PubMed.
- D. K. Aanen and P. Eggleton, Curr. Biol., 2005, 15, 851–855 CrossRef CAS PubMed.
- D. K. Aanen, P. Eggleton, C. Rouland-Lefevre, T. Guldberg-Froslev, S. Rosendahl and J. J. Boomsma, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 14887–14892 CrossRef CAS.
- L. N. van de Peppel, M. Auxier, B. Grum-Grzhimaylo, A. Cárdenas, M. de Beer, Z. W. Lodge, D. J. Smith, M. E. Kuyper, T. Franco-Molano, T. Baroni and D. K. Aanen, 2021, DOI:10.2139/ssrn.3828201.
- S. Otani, L. H. Hansen, S. J. Sorensen and M. Poulsen, Microb. Ecol., 2016, 71, 207–220 CrossRef CAS PubMed.
- R. R. da Costa, H. Hu, H. Li and M. Poulsen, Insects, 2019, 10, 87 CrossRef PubMed.
- S. Otani, V. L. Challinor, N. B. Kreuzenbeck, S. Kildgaard, S. Krath Christensen, L. L. M. Larsen, D. K. Aanen, S. A. Rasmussen, C. Beemelmanns and M. Poulsen, Sci. Rep., 2019, 9, 8819 CrossRef PubMed.
- F. Schalk, C. Gostincar, N. B. Kreuzenbeck, B. H. Conlon, E. Sommerwerk, P. Rabe, I. Burkhardt, T. Kruger, O. Kniemeyer, A. A. Brakhage, N. Gunde-Cimerman, Z. W. de Beer, J. S. Dickschat, M. Poulsen and C. Beemelmanns, mBio, 2021, 12, e0355120 CrossRef PubMed.
- H. J. Li, S. E. Young, M. Poulsen and C. R. Currie, Annu. Rev. Entomol., 2021, 66, 297–316 CrossRef CAS PubMed.
- M. Poulsen, H. F. Hu, C. Li, Z. S. Chen, L. H. Xu, S. Otani, S. Nygaard, T. Nobre, S. Klaubauf, P. M. Schindler, F. Hauser, H. L. Pan, Z. K. Yang, A. S. M. Sonnenberg, Z. W. de Beer, Y. Zhang, M. J. Wingfield, C. J. P. Grimmelikhuijzen, R. P. de Vries, J. Korb, D. K. Aanen, J. Wang, J. J. Boomsma and G. J. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 14500–14505 CrossRef CAS.
- H. Erens, B. B. Mujinya, F. Mees, G. Baert, P. Boeckx, F. Malaisse and E. Van Ranst, Geoderma, 2015, 249, 40–50 CrossRef.
- G. W. Sileshi, M. A. Arshad, S. Konate and P. O. Y. Nkunika, J. Veg. Sci., 2010, 21, 923–937 CrossRef.
- N. Bos, L. Guimaraes, R. Palenzuela, J. Renelies-Hamilton, L. Maccario, S. K. Silue, N. A. Kone and M. Poulsen, BMC Evol. Biol., 2020, 20, 163 CrossRef PubMed.
- J. Sobotnik, A. Jirosova and R. Hanus, J. Insect Physiol., 2010, 56, 1012–1021 CrossRef CAS.
- V. Plasman, D. Daloze, J. C. Braekman, S. Connetable, A. Robert and C. Bordereau, Tetrahedron Lett., 1999, 40, 9229–9232 CrossRef CAS.
- O. Mahamat, N. André-Ledoux, T. Chrisopher, A. A. Mbifu and A. Kamanyi, Clin. Phytosci., 2018, 4, 28 CrossRef CAS.
- C. Beemelmanns, H. J. Guo, M. Rischer and M. Poulsen, Beilstein J. Org. Chem., 2016, 12, 314–327 CrossRef CAS.
- C. Beemelmanns, T. R. Ramadhar, K. H. Kim, J. L. Klassen, S. Cao, T. P. Wyche, Y. Hou, M. Poulsen, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2017, 19, 1000–1003 CrossRef CAS PubMed.
- Y. Roskov, G. Ower, T. Orrell, D. Nicolson, N. Bailly, P. M. Kirk, T. Bourgoin, R. E. DeWalt, W. Decock, E. J. P. van Nieukerken and L. Journal, Species 2000 & ITIS Catalogue of Life, 2028th March 2018. Digital resource at, Species 2000: Naturalis, Leiden, the Netherlands, 2021, ISSN 2405-8858, https://www.catalogueoflife.org Search PubMed.
- R. Sieber and R. H. Leuthold, Ins. Soc, 1981, 28, 371–382 CrossRef.
- R. R. da Costa, S. M. E. Vreeburg, J. Z. Shik, D. K. Aanen and M. Poulsen, Fungal Ecol., 2019, 38, 54–61 CrossRef.
- C. Dietrich, T. Kohler and A. Brune, Appl. Environ. Microbiol., 2014, 80, 2261–2269 CrossRef.
- S. Otani, A. Mikaelyan, T. Nobre, L. H. Hansen, N. A. Kone, S. J. Sorensen, D. K. Aanen, J. J. Boomsma, A. Brune and M. Poulsen, Mol. Ecol., 2014, 23, 4631–4644 CrossRef CAS PubMed.
- D. K. Aanen, H. H. D. Licht, A. J. M. Debets, N. A. G. Kerstes, R. F. Hoekstra and J. J. Boomsma, Science, 2009, 326, 1103–1106 CrossRef CAS.
- L. Katariya, P. B. Ramesh and R. M. Borges, Environ. Microbiol., 2018, 20, 971–979 CrossRef CAS PubMed.
- K. H. Bodawatta, M. Poulsen and N. Bos, Insects, 2019, 10, 185 CrossRef PubMed.
- A. A. Visser, P. W. Kooij, A. J. M. Debets, T. W. Kuyper and D. K. Aanen, Fungal Ecol., 2011, 4, 322–332 CrossRef.
- L. Katariya, P. B. Ramesh, A. Sharma and R. M. Borges, Insect Soc., 2018, 65, 561–569 CrossRef.
- S. Um, A. Fraimout, P. Sapountzis, D. C. Oh and M. Poulsen, Sci. Rep., 2013, 3, 3250 CrossRef.
- G. D. Prestwich, B. A. Bierl, E. D. Devilbiss and M. F. B. Chaudhury, J. Chem. Ecol., 1977, 3, 579–590 CrossRef CAS.
- M. Lamberty, D. Zachary, R. Lanot, C. Bordereau, A. Robert, J. A. Hoffmann and P. Bulet, J. Biol. Chem., 2001, 276, 4085–4092 CrossRef CAS.
- N. Hegedus and F. Marx, Fungal Biol. Rev., 2013, 26, 132–145 CrossRef.
- C. Hamilton and M. S. Bulmer, Dev. Comp. Immunol., 2012, 36, 372–377 CrossRef CAS PubMed.
- C. Hamilton, F. Lay and M. S. Bulmer, J. Insect Physiol., 2011, 57, 1259–1266 CrossRef CAS PubMed.
- C. Landon, H. Meudal, N. Boulanger, P. Bulet and F. Vovelle, Biopolymers, 2006, 81, 92–103 CrossRef CAS.
- J. Ruther, L. Podsiadlowski and M. Hilker, Chemoecology, 2001, 11, 225–229 CrossRef CAS.
- G. P. Dahal and R. E. Viola, SLAS Discov., 2018, 23, 520–531 CAS.
- R. Baker, P. H. Briner and D. A. Evans, J. Chem. Soc. Chem. Comm., 1978, 9, 410–411 RSC.
- D. H. Mahdi, J. Hubert, J. H. Renault, A. Martinez, A. Schubert, K. M. Engel, B. Koudogbo, Z. Vissiennon, V. Ahyi, K. Nieber and C. Vissiennon, Molecules, 2020, 25, 5015 CrossRef CAS PubMed.
- A. Venkatachalapathi and S. Paulsamy, Mycosphere, 2016, 7, 118–130 CrossRef.
- J. J. Kabara, D. M. Swieczkowski, J. P. Truant and A. J. Conley, Antimicrob. Agents Chemother., 1972, 2, 23–28 CrossRef CAS PubMed.
- S. N. A. Malek, G. Kanagasabapathy, V. Sabaratnam, N. Abdullah and H. Yaacob, Int. J. Food. Prop., 2012, 15, 809–814 CrossRef.
- F. Dilika, P. D. Bremner and J. J. Meyer, Fitoterapia, 2000, 71, 450–452 CrossRef CAS PubMed.
- C. J. Zheng, J. S. Yoo, T. G. Lee, H. Y. Cho, Y. H. Kim and W. G. Kim, Febs Lett., 2005, 579, 5157–5162 CrossRef CAS PubMed.
- J. B. Parsons, J. Yao, M. W. Frank, P. Jackson and C. O. Rock, J. Bacteriol., 2012, 194, 5294–5304 CrossRef CAS PubMed.
- D. L. Greenway and K. G. Dyke, J. Gen. Microbiol., 1979, 115, 233–245 CrossRef CAS PubMed.
- M. Sun, J. Dong, Y. Xia and R. Shu, Microb. Pathog., 2017, 107, 212–218 CrossRef CAS PubMed.
- M. Sun, Z. Zhou, J. Dong, J. Zhang, Y. Xia and R. Shu, Microb. Pathog., 2016, 99, 196–203 CrossRef CAS PubMed.
- J. H. Choi, K. Maeda, K. Nagai, E. Harada, M. Kawade, H. Hirai and H. Kawagishi, Org. Lett., 2010, 12, 5012–5015 CrossRef CAS.
- J. H. Choi, K. Maeda, H. Hirai, E. Harada, M. Kawade, J. H. Qi, M. Ojika and H. Kawagishi, Biosci. Biotech. Bioch., 2012, 76, 1407–1409 CrossRef CAS PubMed.
- J. H. Qi, M. Ojika and Y. Sakagami, Tetrahedron, 2000, 56, 5835–5841 CrossRef CAS.
- J. H. Qi, M. Ojika and Y. Sakagami, Bioorg. Med. Chem., 2001, 9, 2171–2177 CrossRef CAS.
- Y. Qu, K. Y. Sun, L. J. Gao, Y. Sakagami, H. Kawagishi, M. Ojika and J. H. Qi, Biosci. Biotech. Bioch., 2012, 76, 791–793 CrossRef CAS PubMed.
- P. Mitra, N. C. Mandal and K. Acharya, J. Verbraucherschutz Lebensmittelsicherh., 2016, 11, 25–31 CrossRef CAS.
- P. Mitra, N. C. Mandal and K. Acharya, Int. Food Res. J., 2016, 23, 2384–2389 CAS.
- N. G. Puttaraju, S. U. Venkateshaiah, S. M. Dharmesh, S. M. N. Urs and R. Somasundaram, J. Agr. Food. Chem., 2006, 54, 9764–9772 CrossRef CAS PubMed.
- T. Taguri, T. Tanaka and I. Kouno, Biol. Pharm. Bull., 2006, 29, 2226–2235 CrossRef CAS.
- F. M. Campos, J. A. Couto, A. R. Figueiredo, I. V. Toth, A. O. S. S. Rangel and T. A. Hogg, Int. J. Food. Microbiol., 2009, 135, 144–151 CrossRef CAS.
- M. J. Saavedra, A. Borges, C. Dias, A. Aires, R. N. Bennett, E. S. Rosa and M. Simoes, Med. Chem., 2010, 6, 174–183 CrossRef CAS PubMed.
- A. Borges, C. Ferreira, M. J. Saavedra and M. Simoes, Microb. Drug Resist., 2013, 19, 256–265 CrossRef CAS PubMed.
- V. N. Lima, C. D. M. Oliveira-Tintino, E. S. Santos, L. P. Morais, S. R. Tintino, T. S. Freitas, Y. S. Geraldo, R. L. S. Pereira, R. P. Cruz, I. R. A. Menezes and H. D. M. Coutinho, Microb. Pathogenesis, 2016, 99, 56–61 CrossRef CAS.
- T. H. Tinh, T. Nuidate, V. Vuddhakul and C. Rodkhum, Procedia Chem., 2016, 18, 162–168 CrossRef CAS.
- F. Borokini, L. Lajide, T. Olaleye, A. Boligon, M. Athayde and I. Adesina, J. Microb. Biotec. Food, 2016, 5, 416–423 CAS.
- I. Gorniak, R. Bartoszewski and J. Kroliczewski, Phytochem. Rev., 2019, 18, 241–272 CrossRef CAS.
- A. Adamczak, M. Ozarowski and T. M. Karpinski, J. Clin. Med., 2020, 9, 109 CrossRef CAS PubMed.
- H. Arima, H. Ashida and G. Danno, Biosci. Biotech. Bioch., 2002, 66, 1009–1014 CrossRef CAS PubMed.
- D. Sanver, B. S. Murray, A. Sadeghpour, M. Rappolt and A. L. Nelson, Langmuir, 2016, 32, 13234–13243 CrossRef CAS PubMed.
- M. U. Amin, M. Khurram, B. Khattak and J. Khan, BMC Complementary Altern. Med., 2015, 15, 59 CrossRef.
- B. Suriyanarayanan, K. Shanmugam and R. S. Santhosh, Rom. Biotechnol. Lett., 2013, 18, 8587–8593 CAS.
- B. Suriyanarayanan and R. S. Santhosh, J. Biomol. Struct. Dyn., 2015, 33, 1819–1834 CrossRef CAS PubMed.
- A. Biharee, A. Sharma, A. Kumar and V. Jaitak, Fitoterapia, 2020, 146, 104720 CrossRef CAS PubMed.
- L. Zhang, Y. Kong, D. Wu, H. Zhang, J. Wu, J. Chen, J. Ding, L. Hu, H. Jiang and X. Shen, Protein Sci., 2008, 17, 1971–1978 CrossRef CAS PubMed.
- A. C. Abreu, S. C. Serra, A. Borges, M. J. Saavedra, A. J. McBain, A. J. Salgado and M. Simoes, Microb. Drug Resist., 2015, 21, 600–609 CrossRef CAS PubMed.
- S. Siriwong, Y. Teethaisong, K. Thumanu, B. Dunkhunthod and G. Eumkeb, BMC Pharmacol. Toxicol., 2016, 17, 39 CrossRef PubMed.
- C. Vipin, K. Saptami, F. Fida, M. Mujeeburahiman, S. S. Rao, Athmika, A. B. Arun and P. D. Rekha, PLoS One, 2020, 15, e0241304 CrossRef CAS.
- G. Gebreyohannes, A. Nyerere, C. Bii and D. B. Sbhatu, Sci. World J., 2019, 2019, 7357048 CrossRef.
- P. L. N. de Carvalho, E. D. Silva, D. A. Chagas-Paula, J. H. H. Luiz and M. Ikegaki, Mini-Rev. Med. Chem., 2016, 16, 259–271 CrossRef.
- T. Tohge, M. Watanabe, R. Hoefgen and A. R. Fernie, Front. Plant Sci., 2013, 4, 62 Search PubMed.
- T. K. Mohanta, J. Funct. Foods, 2020, 68, 103910 CrossRef CAS.
- I. A. Eko Kuswanto, R. E. Putra and I. S. Harahap, J. Entomol., 2015, 12, 87–94 CrossRef.
- G. Yang, F. Ahmad, S. Liang, H. Fouad, M. Guo, H. A. Gaal and J. Mo, Appl. Biochem. Biotechnol., 2020, 192, 1270–1283 CrossRef CAS PubMed.
- M. Nyegue, P.-H. A. Zollo, J.-M. Bessière and s. Rapior, J. Essent. Oil-Bear. Plants, 2003, 6, 153–160 CrossRef CAS.
- B. A. S. Reyes, E. C. Dufourt, J. Ross, M. J. Warner, N. C. Tanquilut and A. B. Leung, Stud. Nat. Prod. Chem., 2018, 55, 111–143 Search PubMed.
- A. C. Guimaraes, L. M. Meireles, M. F. Lemos, M. C. C. Guimaraes, D. C. Endringer, M. Fronza and R. Scherer, Molecules, 2019, 24, 2471 CrossRef CAS PubMed.
- I. Burkhardt, N. B. Kreuzenbeck, C. Beemelmanns and J. S. Dickschat, Org. Biomol. Chem., 2019, 17, 3348–3355 RSC.
- R. Bonikowski, P. Switakowska, M. Sienkiewicz and M. Zaklos-Szyda, Molecules, 2015, 20, 11272–11296 CrossRef CAS PubMed.
- A. C. R. da Silva, P. M. Lopes, M. M. B. de Azevedo, D. C. M. Costa, C. S. Alviano and D. S. Alviano, Molecules, 2012, 17, 6305–6316 CrossRef CAS.
- M. Nakajima, T. Ogura, Y. Kusama, N. Iwabuchi, T. Imawaka, A. Araki, T. Sasaki, E. Hirose and M. Sunairi, Water Res., 1996, 30, 2508–2511 CrossRef CAS.
- J. Kovac, K. Simunovic, Z. W. Wu, A. Klancnik, F. Bucar, Q. J. Zhang and S. S. Mozina, Plos One, 2015, 10, e0122871 CrossRef PubMed.
- K. S. Rajini, P. Aparna, C. Sasikala and V. Ramana Ch, Crit. Rev. Microbiol., 2011, 37, 99–112 CrossRef CAS PubMed.
- A. Woolfson and M. Rothschild, Proc. Royal Soc. B, 1990, 242, 113–119 CrossRef CAS PubMed.
- F. B. Mortzfeld, C. Hashem, K. Vrankova, M. Winkler and F. Rudroff, Biotechnol. J., 2020, 15 Search PubMed.
- K. Schulz-Bohm, L. Martin-Sanchez and P. Garbeva, Front. Microbiol., 2017, 8, 2484 CrossRef PubMed.
- R. Muller and S. Rappert, Appl. Microbiol. Biotechnol., 2010, 85, 1315–1320 CrossRef PubMed.
- S. Rappert, K. C. Botsch, S. Nagorny, W. Francke and R. Muller, Appl. Environ. Microbiol., 2006, 72, 1437–1444 CrossRef CAS PubMed.
- R. K. V. Meer, C. A. Preston and M. Y. Choi, J. Chem. Ecol., 2010, 36, 163–170 CrossRef.
- S. F. Bi, Z. K. Guo, N. Jiang, R. H. Jiao, H. M. Ge and R. X. Tan, J. Asian Nat. Prod. Res., 2013, 15, 422–425 CrossRef CAS PubMed.
- S. F. Bi, F. Li, Y. C. Song, R. X. Tan and H. M. Ge, Nat. Prod. Commun., 2011, 6, 353–355 CrossRef CAS.
- C. Yin, L. Jin, S. Li, X. Xu and Y. Zhang, 3 Biotech, 2019, 9, 45 CrossRef PubMed.
- G. M. Mathew, Y. M. Ju, C. Y. Lai, D. C. Mathew and C. C. Huang, FEMS Microbiol. Ecol., 2012, 79, 504–517 CrossRef CAS.
- J. F. Aparicio, I. Molnar, T. Schwecke, A. Konig, S. F. Haydock, L. E. Khaw, J. Staunton and P. F. Leadlay, Gene, 1996, 169, 9–16 CrossRef CAS.
- J. K. Weng and J. P. Noel, Methods Enzymol., 2012, 515, 317–335 CAS.
- K. H. Kim, T. R. Ramadhar, C. Beemelmanns, S. G. Cao, M. Poulsen, C. R. Currie and J. Clardy, Chem. Sci., 2014, 5, 4333–4338 RSC.
- J. L. Klassen, S. R. Lee, M. Poulsen, C. Beemelmanns and K. H. Kim, Front. Microbiol., 2019, 10, 1739 CrossRef PubMed.
- S. Mahmoudi, S. Rezaie, R. D. Ghazvini, S. J. Hashemi, H. Badali, A. Foroumadi, K. Diba, A. Chowdhary, J. F. Meis and S. Khodavaisy, Mycopathologia, 2019, 184, 607–613 CrossRef CAS PubMed.
- C. Jia, J. Zhang, Y. Z. Zhuge, K. Xu, J. H. Liu, J. L. Wang, L. Li and M. P. Chu, Free Radical Res., 2019, 53, 618–628 CrossRef CAS PubMed.
- A. Q. Fang, G. K. Wong and A. L. Demain, J. Antibiot., 2000, 53, 158–162 CrossRef CAS.
- H. J. Ochel, K. Eichhorn and G. Gademann, Cell Stress Chaperones, 2001, 6, 295 CrossRef CAS.
- J. Genova and M. Dencheva-Zarkova, J. Phys. Conf., 2017, 794, 012031 CrossRef.
- G. Ohji, S. Hidayat, A. Nakashima, C. Tokunaga, N. Oshiro, K. Yoshino, K. Yokono, U. Kikkawa and K. Yonezawa, J. Biochem., 2006, 139, 129–135 CrossRef CAS PubMed.
- T. C. McLean, B. Wilkinson, M. I. Hutchings and R. Devine, Antibiotics, 2019, 8, 83 CrossRef CAS.
- M. Jiang, M. Yin, S. Wu, X. Han, K. Ji, M. Wen and T. Lu, Sci. Rep., 2017, 7, 4803 CrossRef.
- H. Guo, R. Benndorf, D. Leichnitz, J. L. Klassen, J. Vollmers, H. Görls, M. Steinacker, C. Weigel, H. M. Dahse, A. K. Kaster, Z. W. de Beer, M. Poulsen and C. Beemelmanns, Chem. -Eur. J, 2017, 23, 9338–9345 CrossRef CAS.
- Y. Yan, Y. T. Ma, J. Yang, G. P. Horsman, D. Luo, X. Ji and S. X. Huang, Org. Lett., 2016, 18, 1254–1257 CrossRef CAS.
- L. L. Li, S. F. Li, B. Y. Jiang, M. Q. Zhang, J. P. Zhang, B. B. Yang, L. Li, L. Y. Yu, H. Y. Liu, X. F. You, X. X. Hu, Z. Wang, Y. H. Li and L. Z. Wu, J. Nat. Prod., 2019, 82, 1149–1154 CrossRef CAS PubMed.
- H. Guo, J. W. Schwitalla, R. Benndorf, M. Baunach, C. Steinbeck, H. Görls, Z. W. de Beer, L. Regestein and C. Beemelmanns, Org. Lett., 2020, 22, 2634–2638 CrossRef CAS PubMed.
- R. Benndorf, H. J. Guo, E. Sommerwerk, C. Weigel, M. Garcia-Altares, K. Martin, H. F. Hu, M. Küfner, Z. W. de Beer, M. Poulsen and C. Beemelmanns, Antibiotics, 2018, 7, 83 CrossRef CAS.
- H. Zeng, P. X. Feng and C. X. Wan, Nat. Prod. Res., 2019, 33, 1751–1755 CrossRef CAS.
- C. A. Boya, H. Fernandez-Marin, L. C. Mejia, C. Spadafora, P. C. Dorrestein and M. Gutierrez, Sci. Rep., 2017, 7, 5604 CrossRef.
- I. Schoenian, M. Spiteller, M. Ghaste, R. Wirth, H. Herz and D. Spiteller, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 1955–1960 CrossRef PubMed.
- C. N. Kwan, G. Medoff, G. S. Kobayashi, D. Schlessinger and H. J. Raskas, Antimicrob. Agents Chemother., 1972, 2, 61–65 CrossRef CAS.
- K. Kudo, N. Saito, K. Otsuki, M. Kikuchi and N. Ishida, J. Antibiot., 1971, 24, 907–910 CrossRef CAS.
- S. R. Lee, D. Lee, J. S. Yu, R. Benndorf, S. Lee, D. S. Lee, J. Huh, Z. W. de Beer, Y. H. Kim, C. Beemelmanns, K. S. Kang and K. H. Kim, Molecules, 2018, 23, 3003 CrossRef.
- E. J. N. Helfrich and J. Piel, Nat. Prod. Rep., 2016, 33, 231–316 RSC.
- J. Moldenhauer, X. H. Chen, R. Borriss and J. Piel, Angew. Chem. Int. Ed., 2007, 46, 8195–8197 CrossRef CAS PubMed.
- P. S. Patel, S. Huang, S. Fisher, D. Pirnik, C. Aklonis, L. Dean, E. Meyers, P. Fernandes and F. Mayerl, J. Antibiot., 1995, 48, 997–1003 CrossRef CAS.
- P. Nonejuie, R. M. Trial, G. L. Newton, A. Lamsa, V. R. Perera, J. Aguilar, W. T. Liu, P. C. Dorrestein, J. Pogliano and K. Pogliano, J. Antibiot., 2016, 69, 353–361 CrossRef CAS.
- T. P. Wyche, A. C. Ruzzini, C. Beemelmanns, K. H. Kim, J. L. Klassen, S. G. Cao, M. Poulsen, T. S. Bugni, C. R. Currie and J. Clardy, Org. Lett., 2017, 19, 1772–1775 CrossRef CAS PubMed.
- D. C. Oh, M. Poulsen, C. R. Currie and J. Clardy, Nat. Chem. Biol., 2009, 5, 391–393 CrossRef CAS PubMed.
- D. Shin, W. S. Byun, K. Moon, Y. Kwon, M. Bae, S. Um, S. K. Lee and D. C. Oh, Front. Chem., 2018, 6, 498 CrossRef CAS.
- S. Yan, S. F. Li, W. Wu, F. Zhao, L. Bao, R. Ding, H. Gao, H. A. Wen, F. H. Song and H. W. Liu, Chem. Biodivers., 2011, 8, 1689–1700 CrossRef CAS PubMed.
- H. Guo, N. B. Kreuzenbeck, S. Otani, M. Garcia-Altares, H. M. Dahse, C. Weigel, D. K. Aanen, C. Hertweck, M. Poulsen and C. Beemelmanns, Org. Lett., 2016, 18, 3338–3341 CrossRef CAS.
- F. Schalk, S. Um, H. Guo, N. B. Kreuzenbeck, H. Gorls, Z. W. de Beer and C. Beemelmanns, Chembiochem, 2020, 21, 2991–2996 CrossRef CAS.
- X. Liang, X. H. Nong, Z. H. Huang and S. H. Qi, J. Agr. Food. Chem., 2017, 65, 5114–5121 CrossRef CAS.
- M. Kijima, M. Yoshida, K. Sugita, S. Horinouchi and T. Beppu, J. Biol. Chem., 1993, 268, 22429–22435 CrossRef CAS.
- N. Subekti, F. Fibriana, P. Widyaningrum and M. Adfa, Ukr. Biochem. J., 2017, 89, 77–82 CrossRef CAS.
- K. Ding, Y. Y. Tan, Y. Ding, Y. Fang, X. Yang, J. Fang, D. C. Xu, H. Zhang, W. Lu, M. Li, S. C. Huang, M. L. Cai, Y. Song, Y. J. Ding and S. M. Zhang, J. Cell. Biochem., 2019, 120, 5687–5694 CrossRef CAS PubMed.
- P. C. Kiprono, F. Kaberia, J. M. Keriko and J. N. Karanja, Z. Naturforsch., C: J. Biosci., 2000, 55, 485–488 CrossRef CAS.
- R. M. Brucker, R. N. Harris, C. R. Schwantes, T. N. Gallaher, D. C. Flaherty, B. A. Lam and K. P. C. Minbiole, J. Chem. Ecol., 2008, 34, 1422–1429 CrossRef CAS PubMed.
- S. H. Eom, S. K. Kang, D. S. Lee, J. I. Myeong, J. Lee, H. W. Kim, K. H. Kim, J. Y. Je, W. K. Jung and Y. M. Kim, J. Microbiol. Biotechnol., 2016, 26, 784–789 CrossRef CAS.
- B. Shin, C. Park, J. A. Imlay and W. Park, Appl. Microbiol. Biotechnol., 2018, 102, 2323–2335 CrossRef CAS.
- J. H. Kim, K. L. Chan, N. Mahoney and B. C. Campbell, Ann. Clin. Microbiol. Antimicrob., 2011, 10, 23 CrossRef CAS PubMed.
- J. H. Kim, N. Mahoney, K. L. Chan, R. J. Molyneux, G. S. May and B. C. Campbell, FEMS Microbiol. Lett., 2008, 281, 64–72 CrossRef CAS.
- M. Himejima and I. Kubo, J. Agr. Food. Chem., 1993, 41, 1776–1779 CrossRef CAS.
- C. C. Cain, D. H. Lee, R. H. Waldo, A. T. Henry, E. J. Casida, M. C. Wani, M. E. Wall, N. H. Oberlies and J. O. Falkinham, Antimicrob. Agents Chemother., 2003, 47, 2113–2117 CrossRef CAS PubMed.
- N. Singh and P. J. Yeh, J. Antibiot., 2017, 70, 1033–1042 CrossRef CAS.
- R. Murphy, R. Benndorf, Z. W. de Beer, J. Vollmers, A.-K. Kaster, C. Beemelmanns and M. Poulsen, mSphere, 2021, 6, e01233-20 CrossRef PubMed.
- Z. Y. Wang, Z. Y. Yu, J. W. Zhao, X. X. Zhuang, P. Cao, X. W. Guo, C. X. Liu and W. S. Xiang, Front. Microbiol., 2020, 11, 201 CrossRef PubMed.
- H. J. Li, J. Sosa-Calvo, H. A. Horn, M. T. Pupo, J. Clardy, C. Rabeling, T. R. Schultz and C. R. Currie, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 10720–10725 CrossRef CAS.
- R. Noknoy, S. Sunantaraporn, A. Phumee, P. Siriyasatien and S. Sanguansub, Insects, 2020, 11, 318 CrossRef PubMed.
- J. Parker, Myrmecol. News, 2016, 22, 65–108 Search PubMed.
- G. C. Liu and G. Chen, Zootaxa, 2019, 4551, 237–243 CrossRef PubMed.
- P. Zran, T. Yoro, S. Dabonné and K. N'Guessan, Int. J. Approx. Reason., 2017, 5, 46–59 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table 1: a comprehensive overview of the 375 natural products that have been discovered from termites, Termitomyces, bacteria, complex communities and Pseudoxylaria. These natural products were identified by an extensive screening of literature and is the first in-depth investigation into all natural products associated with this termite symbiosis. The table includes molecular formula, mass, compound name, natural product class, termite host (when known), producing organism, country the source material was found in, method of identification and the articles that have discovered the compound from sources related to the fungus-growing termites symbiosis. Subsequently, hundreds of articles were screened for antimicrobial activities linked to each individual natural product, or lack thereof. If possible, the presumed mode of action(s) and reported synergies are indicated as are other known bioactivities. The references associated to bioactivities, mode of action and synergies are reported in the “Reference” column indicated by (no.). An asterisk (*) Indicates that the compound was found from multiple symbiosis sources. See DOI: 10.1039/d1np00022e |
| This journal is © The Royal Society of Chemistry 2022 |