Open Access Article
Open Access ArticleSynthesis and functionalization of casein nanoparticles with aptamers for triple-negative breast cancer targeting†
Chiara
Spanu
a,
Simona
Camorani
b,
Silvia
Tortorella
 ab,
Lisa
Agnello
b,
Mirko
Maturi
ab,
Lisa
Agnello
b,
Mirko
Maturi
 a,
Mauro
Comes Franchini
a,
Mauro
Comes Franchini
 a,
Laura
Cerchia
a,
Laura
Cerchia
 *b and
Erica
Locatelli
*b and
Erica
Locatelli
 *a
*a
aDepartment of Industrial Chemistry “Toso Montanari”, University of Bologna, Viale Risorgimento 4, 40136, Bologna, Italy. E-mail: erica.locatelli2@unibo.it
bInstitute of Experimental Endocrinology and Oncology “Gaetano Salvatore”, CNR, Via S. Pansini 5, 80131 Naples, Italy. E-mail: cerchia@unina.it
First published on 26th October 2022
Abstract
This work shows the synthesis of a drug delivery system made of casein nanoparticles able to host hydrophobic molecules and be functionalized with aptamers targeting the epidermal growth factor receptor. In vitro cell viability and uptake analyses, performed on triple-negative breast cancer cells, confirmed the safety profile and the target selectivity.
The most studied types of nano-carriers for nano-medical applications in drug delivery systems have been bio-degradable polymers that could either be natural or synthetic but, in general, with the latter being more easily processed for the development of tailored systems with a wide range of properties.1–3 On the other hand, natural polymers have great potential for their intrinsic properties, such as bio-degradability, bio-compatibility, and non-toxicity.4 Food proteins, a natural class of macromolecules, represent one of the most interesting nanocarriers investigated so far.5,6 Thanks to their low immunogenicity, amphiphilic character, and easy modification of exposed functional groups, they could be easily employed for addressing the issues of nano-systems for medical applications.7 Several systems based on food proteins such as albumin, gelatin, collagen, zein, or casein have been studied and synthesized to deliver drugs, nutrients, and probiotic organisms.8–10 Casein is the most abundant protein in milk, along with β-lactoglobulin. Its isoelectric point is at pH 4.6, whereas its alkaline forms such as sodium caseinate (NaCas) are completely soluble in water.11 What makes casein unique and interesting for pharmaceutical applications in drug delivery systems is its natural properties of being a transporter of essential micronutrients, such as calcium and phosphate,12,13 thanks to its ability to self-assemble into a micellar structure with a hydrophobic core and a hydrophilic surface. This self-assembling ability makes casein an excellent material for encapsulating hydrophobic substances and protect them from oxidation,7 UV light14 or temperature.15,16 Only few and recent works aimed at the preparation of drug delivery systems based on casein nanomicelles.17–19 Overall, these studies highlight that casein is perfectly able to encapsulate hydrophobic drugs in nanomicelles with improved stability, solubility, and bioavailability, but often requires complex and invasive techniques such as microfluidization and spray drying for their manufacture. More importantly, no studies have yet extended the drug delivery system towards chemical targeting.20 Aptamers are short oligonucleotide sequences of RNA or DNA, which fold into unique three-dimensional structures and bind targets with high affinity and specificity. They represent a new emergent class of components that find their application in chemical targeting.21 Cancer treatments have been the most investigated area for the application of highly specialized aptamer-based drug delivery systems.22 Like antibodies, aptamers are highly selective to cancer cell-surface receptors, but offer great advantages over antibodies for targeted cancer therapy such as cost- and time-effective discovery, easy synthesis and modification, and very high thermal stability. Furthermore, they have a small size for effective tumor penetration and possess no or low immunogenicity.23 Among the various cancer types, many studies were dedicated to investigate the possibility of active targeting of triple-negative breast cancer (TNBC). TNBC is one of the most aggressive subtypes of breast cancer that still lacks an efficacious therapy, due to the lack of receptors for progesteron, estrogen, or HER2/neu.24 The CL4 nuclease-resistant 2′-fluoro-pyrimidine (2′-F-Py) containing RNA aptamers is an ideal candidate for TNBC-active targeting.25 It binds to domain IV in the epidermal growth factor receptor (EGFR) extracellular region and its binding to EGFR-positive cancer cell lines correlates with the expression's level of the receptor on the cell surface, while it does not bind to EGFR-negative non tumor fibroblasts.26 Because of its high affinity to the surface of cells belonging to different tumor types, including TNBC, CL4 has been extensively used as a component of novel nano-formulations for cancer therapy.21,27 EGFR is frequently overexpressed in both primary and metastatic TNBC.28 We previously demonstrated the exquisite selectivity of CL4 for EGFR-positive TNBC cells both in vitro and in preclinical mouse models.27,29 The present research explores for the first time the functionalization of the hydrophilic surface of casein nanoparticles (CNPs) with aptamers to address the selectivity of the system towards TNBC cells, thus ensuring active targeting and high internalization of lipophilic drugs into the cell compartments, while limiting the toxicity of the nanocarrier itself. CNPs encapsulating 5,5-difluoro-5H-4λ5-dipyrrolo[1,2-c:2′,1′-f][1,3,2]diazaborinin-4-ylium-5-uide (BODIPY) as a model lipophilic compound (BODIPY@CNPs) were synthesized using mild conditions achieving a controlled size and polydispersity index (PDI). The model drug delivery system functionalized with CL4-aptamers was thus analyzed in vitro using MDA-MB-231 and BT-549 cells in order to evaluate the effectiveness of the targeting, for the ability of our casein-aptamer system to deliver lipophilic compounds, such as BODIPY, inside the tumor cells, thus opening the way to further and deeper studies.
Casein solubility is strongly dependent on the pH, temperature, and ionic strength and it is affected also by the presence of calcium or phosphate ions.30 Near the isoelectric point (pI 4.6), casein is completely insoluble, while moving away from it the solubility increases. This occurs because around pH 5 the functional groups present on the protein have a neutral overall charge, causing aggregation and precipitation of the protein. Moving towards higher or lower pH, the external functional groups become negatively/positively charged.31 Consequently, not only the repulsion between protein chains increases but also the dispersion is improved. In this work, a synthetic procedure to dissolve casein was adopted. Following the addition of sodium hydroxide until pH 8 to an aqueous suspension of casein, a significant increase in the solubility was recorded as a result of the deprotonation of acidic moieties in casein. Furthermore, soluble sodium caseinate (NaCas) obtained may be in equilibrium with its aggregated form, nano-micelles. The fabrication of casein nanoparticles starts using lyophilized NaCas, eventually a lipophilic compound and an aqueous solution of calcium chloride (1 M). After less than one hour at 30 °C, the mixture was emulsified with a trip probe sonicator and the final CNPs are obtained. Indeed, the addition of calcium chloride solution causes a rearrangement of the NaCas towards a more packed state as a result of the electrostatic interactions established between Ca2+ ions and phosphoserine residues present in the hydrophilic side of the protein.19,32,33 Two different types of CNPs were synthesized and characterized: CNPs and BODIPY@CNPs. The BODIPY dye was chosen and added to allow better traceability and quantification of nanoparticles during in vitro release tests. Furthermore, BODIPY is a lipophilic compound and we report an easy methodology to entrap it efficiently. Finally, after purification of the obtained CNPs, the conjugation of the amino-terminated CL4 aptamer for each sample was performed. Due to the activation of casein's carboxylic acid via EDC/NHS chemistry and the thermal activation of the aptamers, amide bonds between the aptamer's amine group and casein's carboxylic acid may form. The same synthetic procedure was used to conjugate CNPs with CL4-scrambled to be used as a negative control (Fig. 1 and Fig. S1, ESI†). All the details of the materials, experimental procedures and set-ups are available in the ESI.† All samples were purified and then characterized by dynamic light scattering (DLS) at a constant pH of 6.5–7.0 in water. As shown in Table 1, the nanoparticles display a reproducible hydrodynamic diameter that does not exceed over 200 nm, even after the aptamer's conjugation. Comparing the empty nanomicelles to BODIPY@CNPs_CL4 or BODIPY@CNPs_SCR, there is an increase in size, which may be due to the dye encapsulation (more details in Fig. S2 and S3, ESI†). Furthermore, a low polydispersity index (PDI), which reveals a homogeneous size distribution and negative ζ-potential values, due to negatively charged groups on the protein's surface, was achieved. Together these results provide significant evidence of the homogeneity of the particles in terms of size, surface charge and suitability of the particles for in vivo experiments. Encapsulation efficiency (EE%) and loading capacity (LC%) were determined via UV-VIS analysis (more details are in the ESI† and Fig. S4). EE% expresses the percentage of dye that is effectively trapped in the CNPs, while LC% expresses the mass percentage of the dye relative to the total nanosystem mass (Table 1). We should underline that the observed values are derived from a two-step reaction: nanoparticle synthesis and aptamer conjugation. For this reason, the results obtained had to be considered more than satisfactory and in line with the features required to perform in vitro cellular tests. ATR-FTIR analysis was performed on freeze-dried samples to explore the chemical features of the different preparations (Fig. 2a). The FTIR spectra of all systems show two characteristic bands for amide at 1635 cm−1 corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of amide I and at 1513 cm−1 corresponding to the N–H bending vibration of amide II. These signals are related to the protein's peptide bond and their maintenance during later stages suggests that casein was not damaged. Furthermore, at around 3247 cm−1 and 2949 cm−1 are shown the stretching vibrations of −OH/−NH and of aliphatic C–H, respectively. To see the distribution and morphology of particles, the SEM images were acquired. As shown in Fig. 2b, c and Fig. S4, ESI,† both CNPs and BODIPY@CNPs exhibit a homogeneous distribution and size with a globular shape. These images additionally support the presence of nano-vectors perfectly suitable for drug delivery application.
O stretching vibration of amide I and at 1513 cm−1 corresponding to the N–H bending vibration of amide II. These signals are related to the protein's peptide bond and their maintenance during later stages suggests that casein was not damaged. Furthermore, at around 3247 cm−1 and 2949 cm−1 are shown the stretching vibrations of −OH/−NH and of aliphatic C–H, respectively. To see the distribution and morphology of particles, the SEM images were acquired. As shown in Fig. 2b, c and Fig. S4, ESI,† both CNPs and BODIPY@CNPs exhibit a homogeneous distribution and size with a globular shape. These images additionally support the presence of nano-vectors perfectly suitable for drug delivery application.
 | ||
| Fig. 1 Schematic representation of the development of casein aptamer-based nanoparticles with encapsulated BODIPY: BODIPY@CNPs_CL4 and BODIPY@CNPs_SCR. | ||
| Samples | Size (nm) | PDI | ζ-Pot (mV) | Dry matter (mg mL−1) | [BODIPY] (mM) | BODIPY EE% | BODIPY LC% |
|---|---|---|---|---|---|---|---|
| CNPs@CL4 | 156 ± 5 | 0.197 | −13.3 | 17.0 | — | — | — |
| CNPs@SCR | 167 ± 3 | 0.212 | −13.8 | 16.0 | — | — | — |
| BODIPY@CNPs_CL4 | 194 ± 9 | 0.244 | −12.1 | 16.0 | 0.163 | 43.6 | 0.25 |
| BODIPY@CNPs_SCR | 189 ± 8 | 0.237 | −13.1 | 15.5 | 0.201 | 53.6 | 0.32 |
 | ||
| Fig. 2 Characterization of BODIPY nanoparticles: the ATR-FTIR analysis (a) and FEG-STEM images of CNPs (b) and BODIPY@CNPs (c). | ||
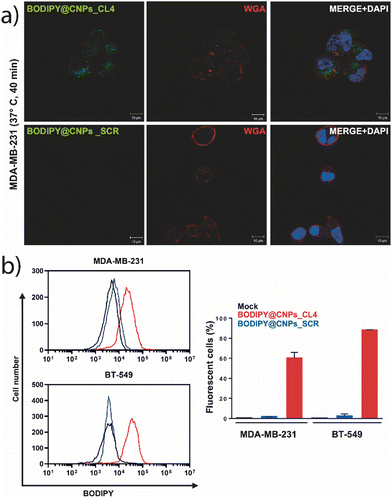
To assess the CL4 aptamer-mediated delivery of nanoparticles to cancer cells, the human EGFR-positive MDA-MB-231 cell line was chosen because it represents an established model for the mesenchymal stem-like TNBC and well recapitulates the high malignant and invasive phenotype of this poorly differentiated subtype.34 Importantly, we have already demonstrated that CL4 not only binds to EGFR on the cell surface, but also is rapidly internalized into MDA-MB-231 cells both in a nude form29 and conjugated to polymeric nanoparticles,27,35 or antibodies.36 The specificity of the cellular uptake of the nanovectors was assessed in EGFR-depleted MDA-MB-231 cells (MDA-MB-231 EGFR-KO), which represent the more appropriate negative control to validate the targeting specificity of an aptamer against its cell surface receptor.37 Thus, cultured MDA-MB-231 and MDA-MB-231 EGFR-KO cells were treated with BODIPY@CNPs_CL4 or BODIPY@CNPs_SCR for 40 min at 37 °C and then washed to remove unbound nanoparticles. Cell nuclei and membranes were stained with DAPI and WGA-647, respectively. Confocal microscopy revealed a clear nanoparticle-associated fluorescence signal within the cytoplasm of MDA-MB-231 cells, but not MDA-MB-231 EGFR-KO, treated with the CL4-equipped CNPs (Fig. 3a and Fig. S5a, ESI†). No signal was observed in both cell lines upon treatment with the SCR-nanoparticles, indicating that CNP internalization is specifically driven by the CL4 aptamer into EGFR-positive TNBC cells. Furthermore, the flow cytometry assays revealed that according to the efficient recognition of EGFR-positive BT-549 TNBC cells by CL4,27,29,35 the aptamer induces a strong uptake of CNPs into these cells (Fig. 3b). As a final test, the safety profile of the nanocarrier conjugated with the aptamer was checked: no cytotoxicity of unloaded CNPs conjugated to either CL4 or SCR aptamers was observed at 0.1 mg ml−1 and 0.2 mg ml−1 (Fig. S5b, ESI†).
Conclusions
In conclusion, an easy and versatile method for casein-based nanocarrier preparation and drug loading has been developed. Casein showed promising peculiarity in terms of safety and high lipophilic compound payloads as well as the possibility to surface functionalization: indeed, CL4-aptamer was conjugated onto the external surface of the nanocarrier, thus increasing the targeting ability and internalization in EGFR-positive triple-negative breast cancer MDA-MB-231 and BT-549 cells.Author contributions
Conceptualization: EL and LC. Data curation: LA, MM, and ST. Formal analysis: CS and SC. Funding acquisition: MCF and LC. Investigation: CS and LA. Methodology: CS and MM. Supervision: EL and ST. Writing – original draft: CS and LA. Writing – review and editing: EL, MCF, and LC.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The University of Bologna is gratefully acknowledged. This research was partially funded by Fondazione AIRC per la Ricerca sul Cancro (IG 23052) to LC.Notes and references
- S. Tortorella, V. Vetri Buratti, M. Maturi, L. Sambri, M. Comes Franchini and E. Locatelli, Int. J. Nanomed., 2020, 15, 9909–9937 CrossRef CAS.
- A. Kumari, S. K. Yadav and S. C. Yadav, Colloids Surf., B, 2010, 75, 1–18 CrossRef CAS PubMed.
- S. Tortorella, M. Maturi, F. Dapporto, C. Spanu, L. Sambri, M. Comes Franchini, M. Chiariello and E. Locatelli, Cellulose, 2020, 27, 8503–8511 CrossRef.
- H. Chen, H. Wooten, L. Thompson and K. Pan, in Biopolymer Nanostructures for Food Encapsulation Purposes, Elsevier, 2019, pp. 39–68 Search PubMed.
- A. O. Elzoghby, W. M. Samy and N. A. Elgindy, J. Controlled Release, 2012, 161, 38–49 CrossRef CAS PubMed.
- E. Assadpour and S. Mahdi Jafari, Crit. Rev. Food Sci. Nutr., 2019, 59, 3129–3151 CrossRef CAS PubMed.
- M. Haham, S. Ish-Shalom, M. Nodelman, I. Duek, E. Segal, M. Kustanovich and Y. D. Livney, Food Funct., 2012, 3, 737 RSC.
- Y. Qin, L. Xu, Y.-Y. Guan and H. Liu, ACS Appl. Bio Mater., 2020, 3, 3196–3202 CrossRef CAS PubMed.
- D. Lickorish, J. A. M. Ramshaw, J. A. Werkmeister, V. Glattauer and C. R. Howlett, J. Biomed. Mater. Res., 2004, 68A, 19–27 CrossRef CAS PubMed.
- T. K. Głąb and J. Boratyński, Top. Curr. Chem., 2017, 375, 71 CrossRef PubMed.
- Y. Liu and R. Guo, Biophys. Chem., 2008, 136, 67–73 CrossRef CAS PubMed.
- P. F. Fox and A. Brodkorb, Int. Dairy J., 2008, 18, 677–684 CrossRef CAS.
- F. Song, L.-M. Zhang, J.-F. Shi and N.-N. Li, Colloids Surf., B, 2010, 79, 142–148 CrossRef CAS PubMed.
- A. O. Elzoghby, W. S. Abo El-Fotoh and N. A. Elgindy, J. Controlled Release, 2011, 153, 206–216 CrossRef CAS PubMed.
- Y. Zhang, S. He, Y. Ma, W. Xu and H. Tang, RSC Adv., 2015, 5, 77595–77600 RSC.
- A. Matalanis, O. G. Jones and D. J. McClements, Food Hydrocolloids, 2011, 25, 1865–1880 CrossRef CAS.
- B.-S. Chu, S. Ichikawa, S. Kanafusa and M. Nakajima, J. Am. Oil Chem. Soc., 2007, 84, 1053–1062 CrossRef CAS.
- A. O. Elzoghby, M. W. Helmy, W. M. Samy and N. A. Elgindy, Pharm. Res., 2013, 30, 2654–2663 CrossRef CAS PubMed.
- K. C. Barick, A. Tripathi, B. Dutta, S. B. Shelar and P. A. Hassan, J. Pharm. Sci., 2021, 110, 2114–2120 CrossRef CAS PubMed.
- J. M. Morachis, E. A. Mahmoud and A. Almutairi, Pharmacol. Rev., 2012, 64, 505–519 CrossRef CAS PubMed.
- S. Shigdar, L. Agnello, M. Fedele, S. Camorani and L. Cerchia, Pharmaceutics, 2021, 14, 28 CrossRef PubMed.
- F. He, N. Wen, D. Xiao, J. Yan, H. Xiong, S. Cai, Z. Liu and Y. Liu, Curr. Med. Chem., 2020, 27, 2189–2219 CrossRef CAS PubMed.
- S. Camorani, E. Crescenzi, M. Fedele and L. Cerchia, Biochim. Biophys. Acta, Rev. Cancer, 2018, 1869, 263–277 CrossRef CAS PubMed.
- S. Camorani, M. Fedele, A. Zannetti and L. Cerchia, Pharmaceuticals, 2018, 11, 123 CrossRef CAS PubMed.
- C. L. Esposito, D. Passaro, I. Longobardo, G. Condorelli, P. Marotta, A. Affuso, V. de Franciscis and L. Cerchia, PLoS One, 2011, 6, e24071 CrossRef CAS PubMed.
- S. Camorani, E. Crescenzi, D. Colecchia, A. Carpentieri, A. Amoresano, M. Fedele, M. Chiariello and L. Cerchia, Oncotarget, 2015, 6, 37570–37587 CrossRef PubMed.
- L. Agnello, S. Tortorella, A. D’Argenio, C. Carbone, S. Camorani, E. Locatelli, L. Auletta, D. Sorrentino, M. Fedele, A. Zannetti, M. C. Franchini and L. Cerchia, J. Exp. Clin. Cancer Res., 2021, 40, 239 CrossRef CAS PubMed.
- R. Costa, A. N. Shah, C. A. Santa-Maria, M. R. Cruz, D. Mahalingam, B. A. Carneiro, Y. K. Chae, M. Cristofanilli, W. J. Gradishar and F. J. Giles, Cancer Treat. Rev., 2017, 53, 111–119 CrossRef CAS PubMed.
- S. Camorani, E. Crescenzi, M. Gramanzini, M. Fedele, A. Zannetti and L. Cerchia, Sci. Rep., 2017, 7, 46659 CrossRef CAS PubMed.
- E. D. Strange, D. L. Van Hekken and V. H. Holsinger, J. Dairy Sci., 1994, 77, 1216–1222 CrossRef CAS.
- A. E. Post, B. Arnold, J. Weiss and J. Hinrichs, J. Dairy Sci., 2012, 95, 1603–1616 CrossRef CAS PubMed.
- D. J. McMahon and B. S. Oommen, Advanced Dairy Chemistry, Springer US, Boston, MA, 2013, pp. 185–209 Search PubMed.
- F. Rehan, N. Ahemad and M. Gupta, Colloids Surf., B, 2019, 179, 280–292 CrossRef CAS PubMed.
- S. Camorani, B. S. Hill, F. Collina, S. Gargiulo, M. Napolitano, M. Cantile, M. Di Bonito, G. Botti, M. Fedele, A. Zannetti and L. Cerchia, Theranostics, 2018, 8, 5178–5199 CrossRef CAS PubMed.
- L. E. Ibarra, S. Camorani, L. Agnello, E. Pedone, L. Pirone, C. A. Chesta, R. E. Palacios, M. Fedele and L. Cerchia, Pharmaceutics, 2022, 14, 626 CrossRef CAS PubMed.
- M. Passariello, S. Camorani, C. Vetrei, L. Cerchia and C. De Lorenzo, Cancers, 2019, 11, 1268 CrossRef CAS PubMed.
- M. McKeague, V. Calzada, L. Cerchia, M. DeRosa, J. M. Heemstra, N. Janjic, P. E. Johnson, L. Kraus, J. Limson, G. Mayer, M. Nilsen-Hamilton, D. Porciani, T. K. Sharma, B. Suess, J. A. Tanner and S. Shigdar, Aptamers, 2022, 10–18 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2nj03367d |
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2022 |