Open Access Article
Open Access ArticleMolecular mechanisms of the antibacterial activity of polyimide fibers in a skin-wound model with Gram-positive and Gram-negative bacterial infection in vivo
Xia
Yang†
 *a,
Wei
Ma†
a,
Hua
Lin†
b,
Shengxiang
Ao
a,
Haoru
Liu
a,
Hao
Zhang
a,
Wanqi
Tang
a,
Hongyan
Xiao
a,
Fangjie
Wang
a,
Junyu
Zhu
a,
Daoyan
Liu
a,
Shujun
Lin
c,
Ying
Zhang
d,
Zhongfu
Zhou
e,
Changbin
Chen
f and
Huaping
Liang
*a
*a,
Wei
Ma†
a,
Hua
Lin†
b,
Shengxiang
Ao
a,
Haoru
Liu
a,
Hao
Zhang
a,
Wanqi
Tang
a,
Hongyan
Xiao
a,
Fangjie
Wang
a,
Junyu
Zhu
a,
Daoyan
Liu
a,
Shujun
Lin
c,
Ying
Zhang
d,
Zhongfu
Zhou
e,
Changbin
Chen
f and
Huaping
Liang
*a
aDepartment of Wound Infection and Drug, State Key Laboratory of Trauma, Burn and Combined Injury, Daping Hospital, Army Medical University (Third Military Medical University), Chongqing 400042, P. R. China. E-mail: oceanyx@126.com; 13638356728@163.com
bFaculty of Materials and Energy, Southwest University, Chongqing 400715, P. R. China
cChangchun HiPolyking Co. Ltd., No. 666B, Super Street, Jilin 132000, P. R.China
dShanghai Kington Technology Limited, 8 Jinian Road, Shanghai 200433, P. R. China
eSchool of Materials Science & Engineering, Shanghai University, 99 Shangda Road, Shanghai 200444, P. R. China
fThe Center for Microbes, Development, and Health, Key Laboratory of Molecular Virology and Immunology, Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai 200031, P. R. China
First published on 2nd June 2022
Abstract
Recently, the need for antibacterial dressings has amplified because of the increase of traumatic injuries. However, there is still a lack of ideal, natural antibacterial dressings that show an efficient antibacterial property with no toxicity. Polyimide (PI) used as an implantable and flexible material has been recently reported as a mixture of particles showing more desirable antibacterial properties. However, we have identified a novel type of natural polyimide (PI) fiber that revealed antibacterial properties by itself for the first time. The PI fiber material is mainly composed of C, N, and O, and contains a small amount of Ca and Cl; the characteristic peaks of polyimide appear at 1774 cm−1, 1713 cm−1, 1370 cm−1, 1087 cm−1, and 722 cm−1. PI fibers displayed significant antibacterial activities against Escherichia coli (as a Gram-negative bacteria model) and methicillin-resistant Staphylococcus aureus (MRSA, as a Gram-positive bacteria model) according to the time-kill kinetics in vitro, and PI fibers damaged both bacterial cell walls directly. PI fibers efficiently ameliorated a local infection in vivo, inhibited the bacterial burden, decreased infiltrating macrophages, and accelerated wound healing in an E. coli- or MRSA-infected wound model. In conclusion, PI fibers used in the present study may act as potent antibacterial dressings protecting from MRSA or E. coli infections and as promising candidates for antimicrobial materials for trauma and surgical applications.
Introduction
Trauma is the leading major cause of death worldwide among those below 45 years of age.1 The characteristic of major trauma is complicated by the systematic inflammatory response to major trauma and the adjacent extensive soft tissue injuries. Surgical treatment for those patients may lead to approximately 24% of all surgical site infections in Germany, and approximately 160![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–300
000–300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 surgical site infections per year in America.2 Some bacterial species have major infective traits and can lead to fatal diseases; some evoke serious antibiotic resistance; and some develop into super-bacteria. These problems have become a global health issue according to the World Health Organization (WHO)3,4 and provoked an urgent need to discover effective discriminatory and antibacterial strategies or drugs or dressings.
000 surgical site infections per year in America.2 Some bacterial species have major infective traits and can lead to fatal diseases; some evoke serious antibiotic resistance; and some develop into super-bacteria. These problems have become a global health issue according to the World Health Organization (WHO)3,4 and provoked an urgent need to discover effective discriminatory and antibacterial strategies or drugs or dressings.
One of the promising strategies that may reduce the risk of infection is applying a certain type of antibacterial dressing. Thus far, to improve the antimicrobial dressing properties, antimicrobial materials comprising essential antibiotics (e.g., tetracycline,5 ciprofloxacin,6 gentamicin7 and sulfadiazine8), nanoparticles (NPs, e.g., silver nanoparticles9), and natural products (e.g., honey,10 essential oils11 and chitosans12) have been investigated.13 For example, biocompatible UV-photocrosslinkable methacrylated gelatin-coated gentamicin sulfate inhibited the growth of MRSA and P. aeruginosa.14 Surgical dressings containing antibiotics could be available to treat wound infections; however, the application can trigger bacterial resistance and lead to a more serious infection.15 This increases morbidity and mortality and leads to a high economic burden of caring for patients infected with multidrug-resistant bacteria.16 Silver nanoparticles display bactericidal activity against a large number of bacteria (such as E. coli ATCC 25922, Enterococcus faecalis NCTC 775, Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus ATCC 25923, and Streptococcus mutans ATCC 25175);17 unfortunately, silver nanoparticles still stimulate the potency of microbial resistance18 and easily enter the human body through various biological barriers such as skin and tissues, and can cause serious toxicity to the human cells.19,20 Natural honey,21 essential oils,22 and chitosans23 and its modified derivatives such as adenine-modified chitosan (AC) hydrogels displayed excellent antibacterial activities against Gram-negative bacteria, Gram-positive bacteria, fungi, and drug-resistant bacteria.24
Nevertheless, natural biofilms are still found to enhance bacterial tolerance in the presence of chitosans.25 Honey consists of many active compounds such as flavonoids, organic acids, and enzymes that may inhibit bacterial growth;25 unfortunately, it is not easily modifiable. Interestingly, other relatively antimicrobial agents such as natural phenolic used as an antibacterial ingredient in the hydrogel dressing26 and lysozyme loaded in polyurethane foam dressings27 showed high antimicrobial activity. However, the introduction of organic substances and enzymes into the content of human dressings needs to consider the adjuvants and immunogenic characteristics, respectively. These materials have shown promising antibacterial activities following their application after a wound surgery and provided the potential abilities to reduce wound infections; however, there is limited clinical evidence among patients with wounds associated with trauma or surgery. Thus, there is a clear need for more novel natural dressings that enable the efficient inhibition of wound infection at the site of injury, which presents advantages of low cost, high antimicrobial activity, nontoxicity, and absence of microbial resistance.
PI is widely used in engineering polymers.28,29 The number of PI monomer precursors is enormous. Thousands of derivative products can be used.30 In many cases, the structure–property relationship of PI is not fully predictable and illustrated. Due to high-temperature tolerance, low-temperature resistance, chemical and radiation tolerance, excellent mechanical capacity, flexibility, excellent dielectric properties, and PI-based shape memory polymers (SMPs), it has been successfully applied in microelectronics, military armors, radomes, aerospace areas, solar-to-electrochemical energy storage, photocatalysis, electrocatalysis, porous materials, and drug delivery.31,32 However, the natural form of PI fibers alone has not been reported as an antibacterial dressing yet.
In the initial stage of the infectious process of the skin, Gram-positive bacteria such as Staphylococcus aureus (S. aureus) and Streptococcus pyogenes (S. pyogenes) are the dominant organisms concerned, while Gram-negative bacteria such as E. coli and Pseudomonas aeruginosa (P. aeruginosa) are only found in later stages of the process, such as chronic wounds.33 In addition, methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci are two major multiple resistant organisms involved in wound infections.34 In the present research, MRSA and E. coli were applied to access the antibacterial function of PI.
Among hundreds of PI fibers, a novel type of natural PI fiber was first identified in the present research for antibacterial properties. The chemical component and structure, characteristic peaks, morphology, antibacterial activity, and the mechanism of antibacterial activity in vitro of PI were systematically evaluated to assess their potential as a natural functional antibacterial wound dressing; then, a wound infection model to perform the potential mechanisms in vivo was drafted. The scientific significance of this work is to develop novel natural PI fibers that have both inherent antibacterial and bioactive functionalities as wound dressings to accelerate the healing of contaminated or infected wounds in traumatic injuries. Our data revealed that the application of PI fibers to traumatic injuries could efficiently inhibit the growth of bacteria, relieve inflammatory response and improve wound healing efficiency.
Material and methods
Characterization
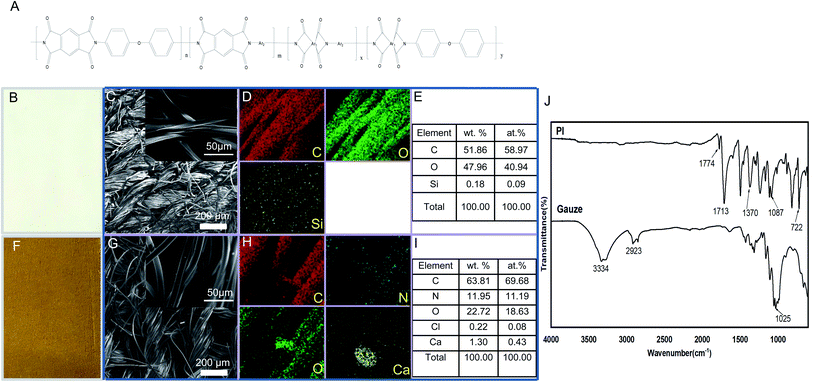
PI fibers used in the present research work were mainly prepared by Changchun HiPolyking Co. Ltd., China. Simply, PI fibers used in the present research were obtained by a two-step method that involves reacting dianhydride and diamine monomers in the wet-spinning process. Pyromellitic dianhydride and 3,4′-oxydianiline were crosslinked with 0.5 wt% N,N-dimethylacetamide; the resulting fibers were treated by annealing, rapidly cooled with water and then stretched to the final forms. The main structure of PI fibers is shown as Fig. 1 A and identified by FESEM, EDS, and FTIR. Field-emission scanning electron microscopic (FESEM) images35 and energy-dispersive X-ray spectroscopy (EDS) mappings were obtained using a JEOL-7800F field emission scanning electron microscope equipped with an energy-dispersive X-ray spectrometer operating at 10 kV.36 Fourier transform infrared (FTIR) spectra were recorded using a Nicolet-5700 FT-IR spectrometer (Thermo Electron Corporation, USA),37 in a test range of 4000–500 cm−1 and a resolution of 4 cm−1.Antibacterial activity of PI on E. coli and MRSA ATCC43300 in vitro
The time-kill kinetics of PI against E. coli and MRSA were measured following a previously described method.38E. coli (BL21 DE3) and MRSA ATCC43300 strains stored in our own lab were cultured in a LB medium at 37 °C overnight with shaking at 200 rpm to reach the logarithmic phase. The growth of E. coli and MRSA were analyzed by measuring the absorbance at OD600. The different size of PI (1, 4 cm2) co-cultured with E. coli or MRSA inoculum containing 5 × 105 CFUs was shaked and incubated at 37 °C. When PI and bacteria were cultured at each designated time point, the OD600 values were measured using Biotek Synergy 2, and then the CFUs per well were calculated. At this point, the CFU values were compared simultaneously with PI and gauze to assess the antibacterial activity. In another independent experiment, the morphological properties or CFU of bacteria (E. coli or MRSA) co-cultured for 24 h were also measured to assess the antibacterial property of PI.Determination of effects of PI on the integrity of bacterial cell membrane in vitro
Bacteria viability tests for PI and gauze were evaluated. A Live/Dead®Baclight bacterial viability kit (#L7012, Life Technologies, Carlsbad, CA, USA) was used to assess the live/dead bacteria.39 The same area of PI and gauze was incubated with MRSA or E. coli (1 × 106 CFU mL−1) separately in a 5 mL LB medium with shaking at 37 °C to study the possible effects of the bacterial colony formation. At indicated incubation time periods (12 and 24 h), the same volume of cultured bacteria was stained according to the manufacturer's instruction. According to the instruction, bacteria with intact cell membranes stained with SYTO 9 exhibit fluorescent green; however, bacteria with damaged membranes stained with propidium iodide show fluorescent red. Then, relative fluorescence units (RFU) were determined using a broad spectrum reader (Thermo Scientific, Varioskan Flash) equipped with a filter (excitation: 480 nm; emission: 500 nm) for SYTO 9 stain and another filter (excitation: 490 nm, emission: 635 nm) for propidium iodide stain.40Animal model
All experimental animal protocols were strictly carried out following the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines and approved by the Institutional Animal Care and Use Committee of Third Military Medical University (No. AMUWEC20201546).Aged male rats or mice were maintained in a temperature-controlled facility under a standard light/darkness cycle and randomly assigned to either gauze or PI-treated groups on the day of surgery, and body weights were recorded throughout the experiments. Rats were used to analyze the above-described in vivo antibacterial model because of their larger body surface area than that of mice and favorable HE and immunofluorescence staining. For wounding, rats were anesthetized with inhaled isoflurane, and then, the hairs in the thoracodorsal region were removed with hair removal cream and aseptically prepared for surgery. Sterile scissors were used to create 2 symmetrical 1 × 1 cm2 wounds in the thoracodorsal region, one on each side of the midline.41 Ten microliters of the lethal dose of MRSA or E. coli bacterial suspension (MRSA 1 × 108 CFU cm−2; E. coli 2 × 108 CFU cm−2, n = 5–8 animals, >4 independent experiments) were applied to both sides of the incision sites and allowed to absorb for 20 minutes. One wound was sutured with sterile gauze (control), and another was sutured with sterile PI. The rats were monitored and vitals were recorded daily including wounds, body weights, and signs of pain and illness on postoperative days 0, 1, 2, and 4.
RNA sequencing
Mice were used for the in vivo antibacterial model described following RNA sequence analysis because of the existing mice database. Granulation tissue under the wound from each treated mouse was collected. One milliliter TRIzol LS Reagent (Invitrogen, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 296
296![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010) was added to each sample to extract RNA, and each RNA sample was sent to Beijing Genomics institution for RNA-seq analysis (Beijing, China). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, Gene Set Enrichment Analysis (GSEA), and Gene Ontology (GO) functional analyses were carried out to analyze genes regulated by PI treatment.42 The P-value < 0.05 was applied to nonsignificant cut-off data.
010) was added to each sample to extract RNA, and each RNA sample was sent to Beijing Genomics institution for RNA-seq analysis (Beijing, China). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, Gene Set Enrichment Analysis (GSEA), and Gene Ontology (GO) functional analyses were carried out to analyze genes regulated by PI treatment.42 The P-value < 0.05 was applied to nonsignificant cut-off data.
HE staining and immunofluorescence staining
Muscular tissue samples of each treated rat group were fixed with paraformaldehyde and processed for paraffin embedding. Each section was deparaffinized in xylene to prepare for H&E histological analysis, rehydrated in a decreasing alcohol gradient, stained with hematoxylin for 5 min, washed with water, stained with eosin for 3 min, dehydrated in a fresh absolute alcohol series until eosin is completely removed, soaked in absolute xylene, and finally, mounted.43The H&E results were assessed at 200× magnification using a microscope (BX51, Olympus) and an accompanying image-capture system. The rehydrate sections were also blocked with 5% bovine serum and stained at 4 °C overnight in a humid chamber with primary antibodies, including rabbit anti-E. coli antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; ab137967 from Abcam, Cambridge, UK), mouse anti-MRSA antibodies (1
100; ab137967 from Abcam, Cambridge, UK), mouse anti-MRSA antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100; ab62742 from Abcam), rabbit anti-CD3 antibodies (1
100; ab62742 from Abcam), rabbit anti-CD3 antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, ab16669 from Abcam), rabbit anti-CD68 antibodies (1
50, ab16669 from Abcam), rabbit anti-CD68 antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, ab125212 from Abcam), or rabbit anti-CD15 antibodies (1
100, ab125212 from Abcam), or rabbit anti-CD15 antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, ab15377 from Abcam) and no primary antibody was used as a negative control. The following day, those sections were incubated with a secondary antibody for immunofluorescence studies conjugated with Alexa Fluor 488 (Thermo Fisher) in 1% BSA for 1 h. Nuclear stain DAPI (Thermo Fisher) was used for staining at room temperature for 10 min. The slides were washed at least three times with PBS in each step. Coverslips were mounted on slides using an antifade kit (Beyotime) and then examined using an Olympus SpinSR10 confocal fluorescence microscope.
100, ab15377 from Abcam) and no primary antibody was used as a negative control. The following day, those sections were incubated with a secondary antibody for immunofluorescence studies conjugated with Alexa Fluor 488 (Thermo Fisher) in 1% BSA for 1 h. Nuclear stain DAPI (Thermo Fisher) was used for staining at room temperature for 10 min. The slides were washed at least three times with PBS in each step. Coverslips were mounted on slides using an antifade kit (Beyotime) and then examined using an Olympus SpinSR10 confocal fluorescence microscope.
Western blotting analysis
Muscular tissue samples from each treated mouse group were collected after 48 h following treatment, weighed, and then lysed using a RIPA Buffer (CST, 9806) containing a Protease/Phosphatase Inhibitor Cocktail (CST, 5872). Cleared lysates were used for routine immunoblot analysis and ELISA analysis of indicated proteins. Anti-CXCL1 antibody (ABclonal, A5802) was used.Enzyme-linked immunosorbent assay
The serum, supernatant of muscular cells, and muscular cell samples from each group of mice treated for 48 h were separated to determine the concentration of MCP/CCL2, TNF alpha, and IL-6. The protocols were in accordance with the instruction provided by the manufacturers. MCP-1/CCL2 Mouse Uncoated ELISA Kit with Plates (ThermoFisher Scientific, #88-7391-86), TNF alpha Mouse Uncoated ELISA Kit with Plates (ThermoFisher Scientific, #88-7324-76), and IL-6 alpha Mouse Uncoated ELISA Kit with Plates (ThermoFisher Scientific, #88-7064-76) were used for ELISA analysis.Statistical analysis
All the quantitative data were analyzed using GraphPad Prism 7 and showed variance as mean ± SEM. Statistical analysis was carried out with unpaired Student's t-test with data from gauze and PI groups. A p-value less than 0.05 was considered statistically significant.Results
Characterization and appearance of PI
PI synthesis routes include polycondensation or addition mechanisms. This spinning method was considered promising for the commercial application of PI fibers because of its low toxic solvents during the process.44 Some published comprehensive works focused on PI chemistry, synthesis, and characterization.45 In the present research work, the main structure of PI fiber is shown as Fig. 1A; and the appearance of gauze fibers, used as control, and PI fibers is observed in Fig. 1B and F. The surface morphology, FESEM images, EDS mapping, and element distribution diagram of samples gauze and PI are shown in Fig. 1B–I. It can be seen from the figure that the morphology of the samples is fabric-shaped, which is made by winding a bunch of wires and then weaving them. Compared to Fig. 1C and G, it can be found that the yarn bundles of the gauze sample are irregular, and the surface of the braid is somewhat disordered and collapsed, which is caused by the flat band of a single gauze fiber. In the PI sample, each filament is columnar, regular, elastic, and smooth, with a diameter of about 10 μm. The EDS results indicate that the main components of gauze are C and O, and a very small amount of Si is detected. The PI is mainly composed of C, N, and O, and contains a small amount of Ca and Cl. The weight percentage and the atom percentage of each element composed of gauze and PI are exhibited in Fig. 1E and I, respectively.
Fig. 1J shows the FTIR spectra of gauze and PI. It can be seen from the figure that the sample of gauze mainly has the following distinct groups: the broad absorption peak with a wavenumber of 3334 cm−1 corresponds to O–H, the wavenumber of 2923 cm−1 belongs to the C–H stretching vibration peak, and the wavenumber of 1025 cm−1 belongs to the C–O stretching vibration peak. The PI spectra show that the characteristic peaks of PI appeared at 1774, 1713, 1370, 1087, and 722 cm−1, respectively. Among them, 1774 cm−1 corresponds to the asymmetric stretching vibration peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and 1713 cm−1 is the symmetric stretching vibration peak of C
O, and 1713 cm−1 is the symmetric stretching vibration peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O; 1370 cm−1 is the stretching vibration peak of C–N, 1087 cm−1 is the stretching vibration peak of C–N–C, and 722 cm−1 is the bending vibration peak of C–N–C.46 In addition, another similar PI fiber that did not exhibit any antibacterial activity was also investigated to identify the main characteristics (data not show); the results indicated that the PI fiber used in the present research exhibited the specific composition, morphology, and characteristic of a broad absorption peak.
O; 1370 cm−1 is the stretching vibration peak of C–N, 1087 cm−1 is the stretching vibration peak of C–N–C, and 722 cm−1 is the bending vibration peak of C–N–C.46 In addition, another similar PI fiber that did not exhibit any antibacterial activity was also investigated to identify the main characteristics (data not show); the results indicated that the PI fiber used in the present research exhibited the specific composition, morphology, and characteristic of a broad absorption peak.
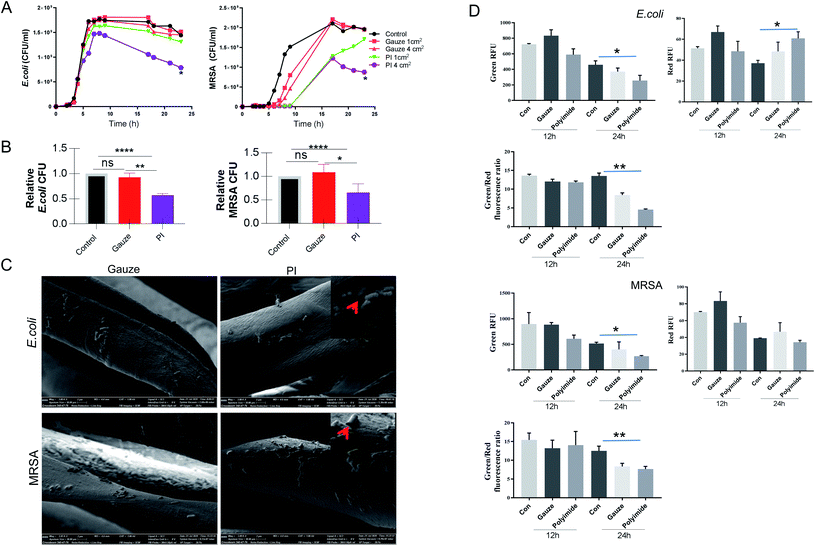
Antibacterial activity in vitro
The antibacterial activity of PI and gauze was evaluated by determining the effects against E. coli and MRSA according to the time-kill kinetics (from 0 to 24 h). The different areas of PI and gauze measuring 1 cm2 or 4 cm2 were used in this experiment. The time-kill kinetics results (Fig. 2A) showed that gauze (control) has no antibacterial activity whatsoever using either 1 cm2 or 4 cm2 pieces, while PI exhibited a relatively higher antibacterial activity against both Gram-negative and Gram-positive bacteria models, especially when using the 4 cm2 strip. The antibacterial activity of PI compared to gauze against E. coli exhibited no significant changes in the lag phase, logarithmic phase, and stationary phase; however, PI covering a large area showed significant antibacterial activities at the decline phase. The antibacterial activity of 4 cm2 PI against MRSA was nearly 2-fold higher than that of the gauze during all the bacterial growth phases. The amount of E. coli and MRSA co-cultured with PI and gauze for 24 h was reduced by 45% and 35%, respectively (Fig. 2B). Those results indicated that PI exhibits antibacterial property against Gram-negative and Gram-positive bacteria.More than 10 PI materials were applied to determine their antibacterial properties at the beginning of this research (data not show); however, only the specific material, shown in Fig. 1F, exhibited antibacterial abilities in vitro. This special PI fiber was then explored more in depth. Natural PI fibers alone showed antibacterial properties in the present research. Various PIs combined with other antibacterial agents or with a modified structure were also investigated as a possible antibacterial dressing. The TiO2-patterned PI films were successfully shown to degrade amoxicillin, atrazine, and 4-chlorophenol.47 PI/NH2-UiO-66 composites modified by photocatalytic degradation of sulfonamides showed antibacterial activity to eliminate water pollutants compared to PI and NH2-UiO-66 alone.27 PI with a modified surface by 15 wt% tantalum oxide or surface-silver-metalized showed superior antibacterial activities.48,49 A plant polyphenol (tannic acid) metal complex introduced on the surface of an electrospun PI nanofibrous membrane displayed antibacterial activities.50 It was also reported that a commercial biomedical PI film modified by topography and chemistry exhibited significantly decreased bacterial (P. aeruginosa) adhesion and antibacterial properties.40 2-Methacryloyloxyethyl phosphorylcholine-modified hyperbranched polyimide exhibited improved biocompatibility and reduced the number of bacterial adhesion.51 This research of using PI combined with other reagents evokes promising further applications as antibacterial dressings; however, more attention needs to be paid to the effectiveness and safety of this compound. The biocompatibility of PI is considered to be related to the chemical, physical, and manufacturing elements.52
PI damages the bacterial cell wall
The morphological properties of PI co-cultured with E. coli and MRSA for 24 h were observed using a SEM, as shown in Fig. 2C. The major change of PI when compared to gauze interaction with bacteria showed that PI attached more E. coli and MRSA. In addition, the morphology of E. coli and MRSA co-cultured with PI exhibited more holes or collapse versus gauze co-cultured for 24 h, leading to the consideration that PI destroyed Gram-negative and Gram-positive bacterial integrity walls. Then, under the same condition, LIVE/DEAD staining was performed to verify the viability of bacteria co-cultured for 12 and 24 h. According to the manufacturer's instruction, SYTO 9 could stain the bacteria with intact cell membranes with fluorescent green color, whereas propidium iodide could stain bacteria with damaged membranes with fluorescent red color.53 The value of Green RFU (excitation/emission: 480/500 nm) suggests the number of live bacteria; the value of Red RFU (excitation/emission: 490/635 nm) suggests the number of dead bacteria. In Fig. 2D, E. coli co-cultured with PI exhibit less Green RFU for 12 and 24 h and more Red RFU for 24 h compared with gauze. The total number of live MRSA co-cultured with PI for 24 h was decreased to almost 50% of non-fiber control; but both the value of Red RFU were nearly the same. Meanwhile, the value of Green/Red from the co-culture with PI showed the lowest number; these results indicated that PI could damage bacterial cell walls while gauze could not. The literature data indicate that cell membrane damage, loss of membrane integrity, inhibition of DNA repair and protein synthesis, inhibition of virulence factors and biofilm production, suppression of adhesion to the host, induction of oxidative stress, and inhibition of metabolic pathways of bacteria contribute to the mechanism of antibacterial property.54 The chlorinated polypropylene-grafted methacrylamide exhibited disruption and lysis of the bacterial cellular structures as its antibacterial mechanism.55 We hypothesised that this special PI fiber showed antibacterial properties due to its special structures. PI fibers first provide a potent antibacterial property and safety potential as a natural wound dressing for Gram-negative and Gram-positive bacterial infections, which is achieved by the disruption and lysis of the bacterial cellular structures.In vivo bacterial killing in MRSA and E. coli wound infection by PI fibers
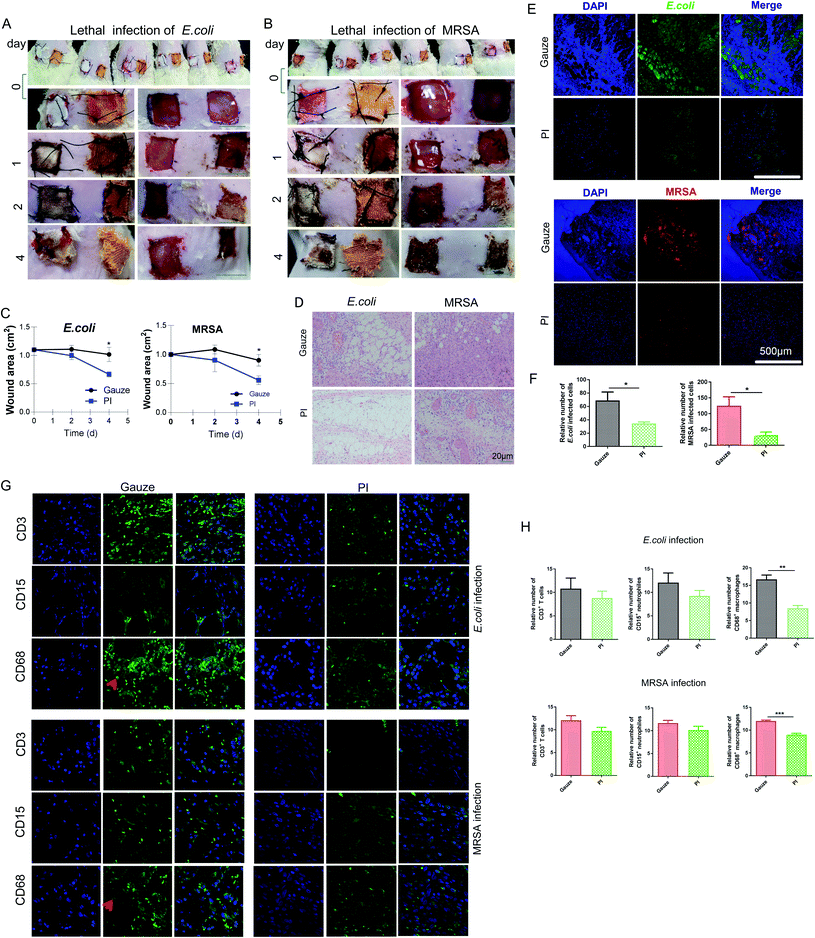
To evaluate the antibacterial efficiency of PI fibers in combating skin wound infection, a MRSA- and E. coli-infected wound model of Wistar rats or mice was established. The data of mice didn't show Fig. 3A and B illustrates the antibacterial process. Two skin-wound infection models were established on the back of the rat subjects because of their larger body surface area compared to mice. On day 0, the same-size-wound bacterial infection model on both sides of the thoracodorsal region was prepared and then sutured with gauze (white color fiber) and PI fiber (yellow color fiber) on each side, separately. On day 1, wounds treated with control gauze showed heavier exudate levels than that of the PI treatment after the same lethal dose of MRSA and E. coli infection; gauze enriched more excreta, making them more durable; however, PI fibers could help to absorb wound exudate (the third row of Fig. 3A and B). On day 2, all wounds treated with PI fibers disappeared from the exudate and promoted scab formation in the E. coli- or MRSA-infected model (the fourth row of Fig. 3A and B). On day 4, wounds treated with gauze after infection by E. coli or MRSA were festered and had purulent expulsion. Inversely, wounds treated with PI fibers were beginning to heal (the fifth row of Fig. 3A and B) and established scar tissue. We then detected the wound area treated with gauze and PI fibers on indicated days. The wound area treated with PI fibers decreased compared with gauze after E. coli and MRSA infection for 0, 1, 2, and 4 days (Fig. 3C). The granulation tissue was also stained with H&E to further investigate the histopathological structure during wound healing between the two groups. H&E staining showed the accumulation of many inflammatory infiltrated cells in the granulation tissue within the gauze-treated control group. Inversely, inflammatory changes in the PI fiber-treated group was less after E. coli and MRSA infection for 2 days (Fig. 3D). Bacterial quantification was then assessed by immunofluorescence using MRSA- and E. coli-specific antibodies and positive cells calculated in the indicated area for graphical presentation and statistical analysis. From Fig. 3E and F, there were considerably increasing numbers of E. coli (green)- and MRSA (red)-infiltrated wound sutures in the gauze-treated group compared to the PI fiber-treated group. Consistently, the numbers of E. coli and MRSA in those two groups were 68.88 ± 12.68% vs. 34.5 ± 2.306% and 124.8 ± 28.29% vs. 32.5 ± 9.47% (p < 0.05), respectively.At the early stage of inflammation, neutrophils were recruited to the infected sites, which engulfed bacteria, and then decontaminated the wound by releasing proteases and antimicrobial peptides and producing reactive oxygen intermediates. Monocytes/macrophages were also recruited, which phagocytized the apoptotic neutrophils and other debris, and also secreted cytokines and multiple growth factors. T lymphocytes secreted cytokines, which promoted the cytolytic activity.56 In addition, we performed immunohistochemical staining to detect the expression levels of CD3+ T cells, CD15+ neutrophils, and CD68+ macrophages in tissue sections from each group and found that the expression of the CD68 macrophage in the gauze-treated group was significantly higher than that in the PI fiber-treated group (Fig. 3G and H). In conclusion, the number of infiltrated T cells and neutrophils in the tissue treated by PI fibers decreased slightly; significantly fewer macrophages and bacteria infiltrated into the region of wound areas under the same condition after E. coli and MRSA infection for 2 days. In contrast, gauze-treated groups showed more infiltration cells and bacteria into the wound area, exhibiting infection and inflammatory responses. In addition, neutrophils, mononuclear macrophages, and T lymphocytes play important roles in eliminating bacteria in the wounds during inflammatory reactions and infections.57 Macrophages can also secrete proinflammatory factors such as TNF-α and IL1β, which undergo LPS stimulation and amplify inflammation to provide antibacterial effects; however, larger or smaller chitosans could inhibit or increase this function.58 It has been reported in the literature that stimulating NO production and TNF-α release from CS-GMA/Arg-PEUU hybrid hydrogels is useful for antibacterial wound dressing biomaterials;59 however, the production of inflammatory chemokines/cytokines such as IL1β and TNF-α initiates and amplifies inflammation response.60 In the present study, PI fibers relieve the inflammatory response.
Antibacterial effects of PI on immune response in vivo
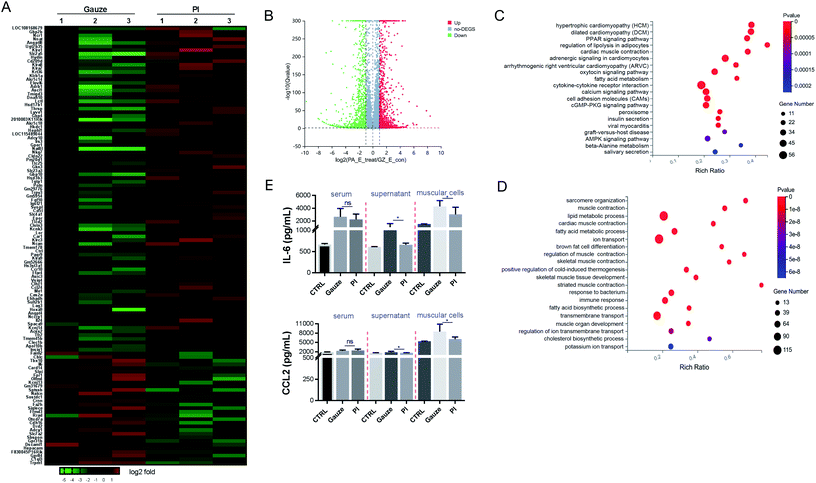
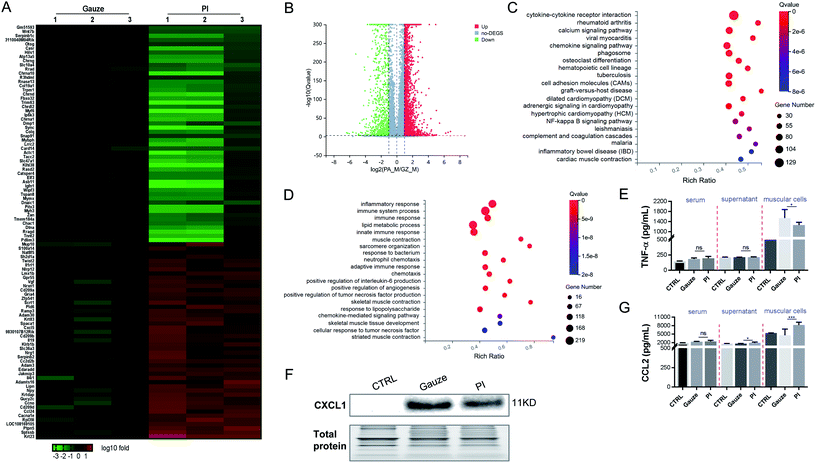
To determine the mechanism of immune response aroused by the antibacterial activity of PI in vivo, we performed an RNAseq analysis of the differentially expressed genes (DEGs) from the granulation tissues in response to PI and gauze treatment for 2 days. We analyzed the transcriptional changes in DEG values calculated by fragments per kilobase million (FPKM) values; Student's t-test was used, and DEGs with at least a 2-fold statistically significant (P < 0.05) difference in expression levels were selected.We identified 4134 genes for E. coli infection, which were DEGs in response to the treatment with PI compared to gauze. Next, heatmap and volcano plot analyses were used to visualize some of these DEGs, which are shown in Fig. 4A and B. KEGG pathway enrichment and GO functional analyses were used to determine biological processes affected by PI. As shown in Fig. 4C, the upregulated genes in gauze-treated granulation tissues compared to PI-treated tissues were significantly enriched in cytokine–cytokine receptor interaction, calcium signaling pathway, cell adhesion molecules, regulation of lipolysis in adipocytes, and the PPAR signaling pathway according to the KEGG pathway. Next, a GO pathway enrichment analysis in the same model was conducted to explore DEGs' most significantly enriched pathways (Fig. 4D). The most enriched DEGs in gauze-treated tissues vs. PI have significantly upregulated the pathways related to ion transport, lipid metabolic process, transmembrane transport, immune response, fatty acid metabolic process, response to the bacterium, cholesterol biosynthetic process, etc. It was reported that chemokines CCL2 and IL-6 were elevated in the culture supernatant after macrophages were exposed to Gram-negative bacteria.61 As expected, PI treatment significantly reduced the CCL2 and IL-6 chemokines due to the lethal dose of E. coli in the supernatant of muscular cells and the muscle cells compared to gauze treatment according to the ELISA analysis (Fig. 4E). These data suggested that PI treatment during E. coli infection significantly relieved cytokine, chemokine, and immune responses.
It was found that 3610 DEGs were upregulated and downregulated from gauze-treated granulation tissues compared to PI treatment during MRSA infection. Heatmap and volcano plot analyses showed some of these DEGs (Fig. 5A and B). The top 20 pathways were shown by KEGG pathway enrichment (Fig. 5C). The most significantly elevated pathways in gauze-treated granulation tissues were cytokine–cytokine receptor interaction, calcium signaling pathway, phagosome, cell adhesion molecules, and the NF-κB signaling pathway. The GO pathway enrichment analysis indicated that inflammatory response, immune system process, immune response, lipid metabolic process, innate immune response, response to the bacterium, neutrophil chemotaxis, adaptive immune response, positive regulation of interleukin-6 production, response to lipopolysaccharide, and chemokine-mediated signaling pathway were increased in gauze-treated granulation tissues compared to PI treatment. These data suggested the inhibition of the inflammatory response, and innate and adaptive immune response by PI treatment during MRSA infection. The chemokine CXCL1 and the cytokine TNF were elevated in the culture supernatant after macrophages were exposed to Gram-positive bacteria.61 We also found that PI treatment significantly reduced the TNF-α and CXCL1 levels when infected by a lethal dose of MRSA bacteria in the muscular cells compared to gauze treatment according to the ELISA (Fig. 5E) and western blot analysis (Fig. 5F), respectively. Surprisingly, in this same lethal dose of Gram-positive bacterial infection, PI treatment elevated the CCL2 expression levels in the supernatant of muscular cells and the muscle cells compared to gauze treatment (Fig. 5G); however, PI treatment reduced the CCL2 expression level after exposure to lethal doses of Gram-negative bacteria (Fig. 4E). The report demonstrated that equine MSCs secrete CCL2 that increases the antimicrobial activity of equine keratinocytes by stimulating the expression of antimicrobial peptides during MRSA infection.62 It has been suggested that PI elevated some antimicrobial elements to act against MRSA infection.
Conclusions
In summary, we have identified a natural antibacterial PI fiber for the effective application of Gram-positive and Gram-negative bacteria antibiosis. Therefore, this natural PI fiber provides a protecting antibiosis over wounds and avoids further external trauma infection. The PI fibers exhibited special surface morphology, chemical composition, and characteristic groups. The time-kill kinetics results indicated that the bigger area of PI showed a relatively high antibacterial activity, especially at the decline phase against both E. coli and MRSA; however, gauze (control) showed no antibacterial activity. PI fiber directly damaged both E. coli and MRSA bacterial cell walls. From the SEM image, it has been observed PI fibers attached more E. coli and MRSA, and the morphology of E. coli and MRSA co-cultured with PI for 24 h exhibited more holes or cell-membrane collapse vs. gauze co-cultured. The rats in vivo infected wound model demonstrated that PI fibers showed strong antibacterial characteristics against E. coli and MRSA, helped absorb wound exudate, accelerated wound closure, and decreased the number of bacteria and inflammatory infiltrating cells in granulation tissues. PI fiber reduced the infiltrated macrophages and downregulated cytokine–cytokine receptor interaction and immune response against E. coli and MRSA infection. The data suggested that the novel type of natural PI fiber provides extraordinary performance in the use of covering a bacterially infected wound, especially for MRSA (multiple resistant bacteria), and as a hopeful therapeutic for the next natural generation of antibacterial materials.Author contributions
Conceptualization, Xia Yang and Huaping Liang. Data curation, Wei Ma and Xia Yang. Formal analysis, Wei Ma, Hua Lin, and Haoru Liu. Funding acquisition, Xia Yang and Huaping Liang. Investigation and Project administration, Wei Ma, Shengxiang Ao, Hao Zhang, Wanqi Tang, Hongyan Xiao, Fangjie Wang, and Junyu Zhu. Methodology, Xia Yang and Huaping Liang. Resources, Shujun Lin, Ying Zhang and Zhongfu Zhou. Supervision, Daoyan Liu, Changbin Chen, Zhongfu Zhou and Huaping Liang. Validation, Xia Yang, Wei Ma, and Shengxiang Ao. Writing, Xia Yang and Wei Ma.Conflicts of interest
The authors declare that they have no competing interests.Abbreviations
| PI | Polyimide |
| FESEM | Field-emission scanning electron microscopy |
| EDS | Energy-dispersive X-ray spectroscopy |
| FTIR | Fourier transform infrared |
| MRSA | Methicillin-resistant Staphylococcus aureus |
| E. coli | Escherichia coli |
| WHO | World Health Organization |
| NPs | Nanoparticles |
| SMPs | Shape memory polymers |
| RFU | Relative fluorescence unit |
| KEGG | Kyoto encyclopedia of genes and genomes |
| GSEA | Gene set enrichment analysis |
| GO | Gene ontology |
| DEGs | Differentially expressed genes |
| FPKM | Fragments per kilobase million |
Acknowledgements
This work was supported by special Funds of China (21QNPY028) and the 2020 Chongqing Talent Program (CQ2020TP09). We would like to thank my friend Alex Cain for polishing the grammatical presentation of the manuscript.References
- F. Mas-Celis, J. Olea-Lopez and J. A. Parroquin-Maldonado, Arch. Med. Res., 2021, 52, 808–816 CrossRef CAS PubMed.
- A. Brunauer, R. D. Verboket, D. M. Kainz, F. von Stetten and S. M. Fruh, Biosensors, 2021, 11(3), 74 CrossRef CAS PubMed.
- J. Zhang, Y. P. Chen, K. P. Miller, M. S. Ganewatta, M. Bam, Y. Yan, M. Nagarkatti, A. W. Decho and C. Tang, J. Am. Chem. Soc., 2014, 136, 4873–4876 CrossRef CAS PubMed.
- K. M. O'Connell, J. T. Hodgkinson, H. F. Sore, M. Welch, G. P. Salmond and D. R. Spring, Angew. Chem., Int. Ed., 2013, 52, 10706–10733 CrossRef PubMed.
- A. Marino, A. Nostro, N. Mandras, J. Roana, G. Ginestra, N. Miceli, M. F. Taviano, F. Gelmini, G. Beretta and V. Tullio, BMC Complementary Med. Ther., 2020, 20, 89 CrossRef PubMed.
- M. P. Chrysouli, C. N. Banti, N. Kourkoumelis, E. E. Moushi, A. J. Tasiopoulos, A. Douvalis, C. Papachristodoulou, A. G. Hatzidimitriou, T. Bakas and S. K. Hadjikakou, Dalton Trans., 2020, 49, 11522–11535 RSC.
- F. Heno, Z. Azoulay, B. Khalfin, H. A. Craddock, E. Silberstein, J. Moran-Gilad and H. Rapaport, Aesthetic Plast. Surg., 2021, 45, 2980–2989 CrossRef PubMed.
- M. S. E. von Silva-Tarouca, G. Wolf and R. S. Mueller, Vet. Dermatol., 2019, 30, 145 CrossRef PubMed.
- J. Mussin, V. Robles-Botero, R. Casanas-Pimentel, F. Rojas, L. Angiolella, E. San Martin-Martinez and G. Giusiano, Sci. Rep., 2021, 11, 14566 CrossRef CAS PubMed.
- M. Ratajczak, D. Kaminska, E. Matuszewska, E. Holderna-Kedzia, J. Rogacki and J. Matysiak, Molecules, 2021, 26(13), 4007 CrossRef CAS PubMed.
- L. Federico, M. Filippo and T. Bruno, Nat. Prod. Res., 2021, 35, 5452–5458 CrossRef CAS PubMed.
- D. Yan, Y. Li, Y. Liu, N. Li, X. Zhang and C. Yan, Molecules, 2021, 26(23), 7136 CrossRef CAS PubMed.
- D. Simoes, S. P. Miguel, M. P. Ribeiro, P. Coutinho, A. G. Mendonca and I. J. Correia, Eur. J. Pharm. Biopharm., 2018, 127, 130–141 CrossRef CAS PubMed.
- J. Zhou, D. Yao, Z. Qian, S. Hou, L. Li, A. T. A. Jenkins and Y. Fan, Biomaterials, 2018, 161, 11–23 CrossRef CAS PubMed.
- B. N. Green, C. D. Johnson, J. T. Egan, M. Rosenthal, E. A. Griffith and M. W. Evans, J. Chiropr Med., 2012, 11, 64–76 CrossRef PubMed.
- M. A. A. Majumder, S. Rahman, D. Cohall, A. Bharatha, K. Singh, M. Haque and M. Gittens-St Hilaire, Infect. Drug Resist., 2020, 13, 4713–4738 CrossRef CAS PubMed.
- J. F. B. Bruniera, L. Gabriel-Silva, R. S. Goulart, Y. T. C. Silva-Sousa, M. G. Lara, A. Pitondo-Silva and C. E. S. Miranda, Braz. Dent. J., 2020, 31, 485–492 CrossRef PubMed.
- B. P. Backx, M. S. Dos Santos, O. A. L. Dos Santos and S. A. Filho, Curr. Pharm. Biotechnol., 2021, 22, 762–772 CAS.
- H. Bahadar, F. Maqbool, K. Niaz and M. Abdollahi, Iran. Biomed. J., 2016, 20, 1–11 Search PubMed.
- C. Liao, Y. Li and S. C. Tjong, Int. J. Mol. Sci., 2019, 20(2), 449 CrossRef PubMed.
- V. C. Nolan, J. Harrison and J. A. G. Cox, Antibiotics, 2019, 8(4), 251 CrossRef CAS PubMed.
- K. Winska, W. Maczka, J. Lyczko, M. Grabarczyk, A. Czubaszek and A. Szumny, Molecules, 2019, 24(11), 2130 CrossRef CAS PubMed.
- W. Al-Gethami and N. Al-Qasmi, Polymers, 2021, 13(20), 3557 CrossRef PubMed.
- P. Deng, L. Yao, J. Chen, Z. Tang and J. Zhou, Carbohydr. Polym., 2022, 276, 118718 CrossRef CAS PubMed.
- S. Abusrewil, J. L. Brown, C. Delaney, M. C. Butcher, M. Tiba, J. A. Scott, G. Ramage and W. McLean, Antibiotics, 2021, 10(11), 1317 CrossRef CAS PubMed.
- W. Ning Chen, J. Wound Care, 2021, 30, 330 CrossRef PubMed.
- J. Wang, M. Yuan, C. Li, B. Zhang, J. Zhu, X. Hao, H. Lu and Y. Ma, J. Colloid Interface Sci., 2022, 612, 536–549 CrossRef CAS PubMed.
- M. Hatami, Ultrason. Sonochem., 2018, 44, 261–271 CrossRef CAS PubMed.
- M. Hasegawa, Polymers, 2017, 9(10), 520 CrossRef PubMed.
- C. N. Hoang, Y. H. Dang, C. T. Pham and D. Hoang, ACS Omega, 2020, 5, 7044–7050 CrossRef CAS PubMed.
- Y. Zhang, Z. Huang, B. Ruan, X. Zhang, T. Jiang, N. Ma and F. C. Tsai, Macromol. Rapid Commun., 2020, 41, e2000402 CrossRef PubMed.
- I. Gouzman, E. Grossman, R. Verker, N. Atar, A. Bolker and N. Eliaz, Adv. Mater., 2019, 31, e1807738 CrossRef PubMed.
- A. F. Cardona and S. E. Wilson, Clin. Infect. Dis., 2015, 61(suppl. 2), S69–S78 CrossRef CAS PubMed.
- G. R. Rudramurthy, M. K. Swamy, U. R. Sinniah and A. Ghasemzadeh, Molecules, 2016, 21(7), 836 CrossRef PubMed.
- I. G. Subramani, V. Perumal, S. C. B. Gopinath, N. M. Mohamed, M. Ovinis and L. L. Sze, Sci. Rep., 2021, 11, 20825 CrossRef CAS PubMed.
- A. Mehdinia, M. Ramezani and A. Jabbari, Food Chem., 2017, 237, 1112–1117 CrossRef CAS PubMed.
- B. S. Huang, G. H. Lai, T. I. Yang, M. H. Tsai and Y. C. Chou, Polymers, 2020, 12(1), 91 CrossRef CAS PubMed.
- S. K. Sy, L. Zhuang and H. Derendorf, Expert Opin. Drug Metab. Toxicol., 2016, 12, 93–114 CrossRef CAS PubMed.
- J. Robertson, C. McGoverin, F. Vanholsbeeck and S. Swift, Front. Microbiol., 2019, 10, 801 CrossRef PubMed.
- E. A. Cuello, L. E. Mulko, C. A. Barbero, D. F. Acevedo and E. I. Yslas, Colloids Surf. B Biointerfaces, 2020, 188, 110801 CrossRef CAS PubMed.
- K. M. Guthrie, A. Agarwal, D. S. Tackes, K. W. Johnson, N. L. Abbott, C. J. Murphy, C. J. Czuprynski, P. R. Kierski, M. J. Schurr and J. F. McAnulty, Ann. Surg., 2012, 256, 371–377 CrossRef PubMed.
- T. Shi and G. Gao, Biosci. Rep., 2022, 42(2), BSR20212523 CrossRef CAS PubMed.
- Y. He, J. Leng, K. Li, K. Xu, C. Lin, Z. Yuan, R. Zhang, D. Wang, B. Tao, T. J. Huang and K. Cai, Biomaterials, 2021, 278, 121164 CrossRef CAS PubMed.
- M. Zhang, H. Niu and D. Wu, Macromol. Rapid Commun., 2018, 39, e1800141 CrossRef PubMed.
- M. Y. Khan, I. Khan, M. Zeama and A. Khan, Chem. Asian J., 2021, 16, 1979–1987 CrossRef CAS PubMed.
- X. Liu, J. Ma, P. Jiang, J. Shen, R. Wang, Y. Wang and G. Tu, ACS Appl. Mater. Interfaces, 2020, 12, 45332–45341 CrossRef CAS PubMed.
- S. Ramasundaram, M. G. Seid, W. Lee, C. U. Kim, E. J. Kim, S. W. Hong and K. J. Choi, J. Hazard. Mater., 2017, 340, 300–308 CrossRef CAS PubMed.
- E. Han, Y. Wang, X. Chen, G. Shang, W. Yu, H. Niu, S. Qi, D. Wu and R. Jin, ACS Appl. Mater. Interfaces, 2013, 5, 4293–4301 CrossRef CAS PubMed.
- S. Asadullah, S. Mei, K. Yang, X. Hu, F. Wang, B. Yu, Z. Wu and J. Wei, J. Mech. Behav. Biomed. Mater., 2021, 124, 104800 CrossRef CAS PubMed.
- W. Ma, Y. Ding, M. Zhang, S. Gao, Y. Li, C. Huang and G. Fu, J. Hazard. Mater., 2020, 384, 121476 CrossRef CAS PubMed.
- Q. Li, J. Li, G. Liao and Z. Xu, J. Mater. Sci. Mater. Med., 2018, 29, 126 CrossRef PubMed.
- C. P. Constantin, M. Aflori, R. F. Damian and R. D. Rusu, Materials, 2019, 12(19), 3166 CrossRef CAS PubMed.
- R. Gao, X. Liao, X. Zhao, D. Liu and T. Ding, Compr. Rev. Food Sci. Food Saf., 2021, 20, 2146–2175 CrossRef CAS PubMed.
- S. Kang, F. Kong, X. Liang, M. Li, N. Yang, X. Cao, M. Yang, D. Tao, X. Yue and Y. Zheng, J. Agric. Food Chem., 2019, 67, 12322–12332 CrossRef CAS PubMed.
- Y. Ma, N. Wisuthiphaet, H. Bolt, N. Nitin, Q. Zhao, D. Wang, B. Pourdeyhimi, P. Grondin and G. Sun, ACS Biomater. Sci. Eng., 2021, 7, 2329–2336 CrossRef CAS PubMed.
- J. R. Davidson, Vet. Clin. N. Am. - Small Anim. Pract., 2015, 45, 537–564 CrossRef PubMed.
- L. C. Yang, S. W. Hu, M. Yan, J. J. Yang, S. H. Tsou and Y. Y. Lin, J. Periodontol., 2015, 86, 310–318 CrossRef PubMed.
- S. H. Chang, Y. Y. Lin, G. J. Wu, C. H. Huang and G. J. Tsai, Int. J. Biol. Macromol., 2019, 131, 167–175 CrossRef CAS PubMed.
- M. Yin, S. Wan, X. Ren and C. C. Chu, ACS Appl. Mater. Interfaces, 2021, 13, 14688–14699 CrossRef CAS PubMed.
- S. R. Holdsworth and P. Y. Gan, Clin. J. Am. Soc. Nephrol., 2015, 10, 2243–2254 CrossRef CAS PubMed.
- R. A. Gottschalk, M. G. Dorrington, B. Dutta, K. S. Krauss, A. J. Martins, S. Uderhardt, W. Chan, J. S. Tsang, P. Torabi-Parizi, I. D. Fraser and R. N. Germain, Elife, 2019, 8, e46836 CrossRef PubMed.
- C. Marx, S. Gardner, R. M. Harman, B. Wagner and G. R. Van de Walle, Stem Cells Transl. Med., 2021, 10, 1666–1679 CrossRef CAS PubMed.
Footnote |
| † Xia Yang, Wei Ma, and Hua Lin have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |