Open Access Article
Open Access ArticleAerolysin nanopore-based identification of proteinogenic amino acids using a bipolar peptide probe†
Yaxian
Ge
a,
Mengjie
Cui
a,
Qiuqi
Zhang
b,
Ying
Wang
*a and
Dongmei
Xi
 *a
*a
aShandong Provincial Key Laboratory of Detection Technology for Tumor Markers, College of Life Science, Linyi University, Linyi 276005, P. R. China. E-mail: dongmxi@126.com; m13854869219@163.com
bThe First School of Clinical Medicine, Southern Medical University, Guangzhou 510515, P. R. China
First published on 11th August 2022
Abstract
Nanopore technology has attracted extensive attention due to its rapid, highly sensitive, and label-free performance. In this study, we aimed to identify proteinogenic amino acids using a wild-type aerolysin nanopore. Specifically, bipolar peptide probes were synthesised by linking four aspartic acid residues to the N-terminal and five arginine residues to the C-terminal of individual amino acids. With the help of the bipolar peptide carrier, 9 proteinogenic amino acids were reliably recognised based on current blockade and dwell time using an aerolysin nanopore. Furthermore, by changing the charge of the peptide probe, two of the five unrecognized amino acids above mentioned were identified. These findings promoted the application of aerolysin nanopores in proteinogenic amino acid recognition.
Introduction
Nanopores have proven to be an attractive and powerful platform in the single-molecule analysis.1–5 Over the past two decades, they have been explored for a wide range of applications including DNA sequencing,6–8 analysis of the structure of peptides,9–11 dynamics of peptide transport,12–15 conformational changes of proteins,16,17 unfolding18–20 and folding of proteins,21,22 and application of disease detection.23,24 In this rapidly evolving field, it is hoped that nanopore technology can make a breakthrough in the determination of protein primary structure. However, there are 20 amino acids that make up proteins, and nanopore technology is more challenging to identify individual amino acids than single bases.25–28 Recently, an aerolysin nanopore was used to enable size discrimination of short uniformly charged arginine peptides at a single amino acid resolution by virtue of its high spatial resolution.29 Furthermore, H. Ouldali et al. demonstrated a strategy that allowed the identification of 13 of the 20 unmodified amino acids in an aerolysin nanopore using a short polycationic carrier, and 2 amino acids upon chemical modification.30 In the absence of any modification or labelling, a single cysteine molecule produced distinctive current blockage, while asparagine or glutamine with large volumes caused unrecognized current blockage.31 A T232K/K238Q mutant aerolysin nanopore could identify unphosphorylated Tau with almost 100% accuracy by changing the charge of amino acid sites.32 All these advancements indicate that the aerolysin nanopore holds great potential for identification of proteinogenic amino acids.Polypeptide is a good carrier for amino acid recognition. The bipolar peptide probe structure was first developed by Asandei et al. (2015), and they demonstrated that the positively and negatively charged tails of the bipolar peptide produce a tug-of-war effect after entering the nanopore under the action of the electric field, which may increase its residence time in the nanopore and help to identify the target amino acid.33–35 Inspired by this, we designed a bipolar peptide probe aimed to identify proteinogenic amino acids using a wild-type aerolysin nanopore. The N-terminus of the amino acid was chemically linked to a peptide with aspartic acid residues (D) and the C-terminus of the amino acid was chemically connected to a peptide with arginine residues (R) to form bipolar peptide probes. In this case, the peptide probe had a strong negatively charged N-terminus and a strong positively charged C-terminus at neutral pH. With this probe, we demonstrated the identification of 9 amino acids using the aerolysin nanopore without any chemical or physical modifications. Our efforts will help in proteinogenic amino acid recognition by the aerolysin nanopore.
Experimental
Reagents and chemicals
1,2-Diphytanoyl-sn-glycero-3-phosphocholine was purchased from Avanti Polar Lipids, Inc. (Alabaster, AL, USA). Pro-aerolysin was kindly provided by Professor Yi-Tao Long (East China University of Science and Technology). Trypsin-EDTA, potassium chloride, and decane were purchased from Sigma-Aldrich Co., Ltd (St Louis, MO, USA). The reagents and materials used in this study were of analytical grade. All peptide samples were synthesised and purified by high-performance liquid chromatography at GL Biochem Ltd (Shanghai, China). The sequences of peptides in the assay are shown in Table S1.† All solutions were prepared using ultrapure water (18.2 MΩ cm at 25 °C) obtained using the Milli-Q Academic A10 system (EMD Millipore, Billerica, MA, USA).Nanopore electrical recording
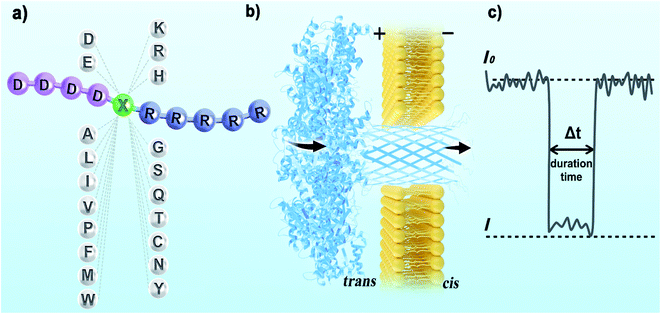
Pro-aerolysin was digested with trypsin for 6 h at room temperature. The pro-peptide sequence of pro-aerolysin was completely degraded, and then aerolysin was released.30 1,2-Diphytanoyl-sn-glycero-3-phosphocholine was dissolved in decane solution to obtain a final concentration of 50 mg L−1 and then added to the trans chamber in a Delrin bilayer cup (Warner Instruments, Hamden, CT). After forming a lipid bilayer, aerolysin was inserted into the lipid bilayer to obtain a final concentration of 1.0 μg mL−1, which partitioned the aperture into a cis chamber and a trans chamber. The electrolyte solutions in the cis and trans chambers of the lipid bilayer were buffered with tris-EDTA (10 mM tris, 1 mM EDTA, 3.0 M KCl, pH 7.5). The pore insertion was identified upon observing a well-defined leap of current values.29 Once a stable single-pore insertion was detected, the bipolar peptide was added near the newly formed nanopore in the trans chamber at a final concentration of 2 μM. The room temperature was maintained at 25 ± 2 °C for this assay, and the error represented the standard deviation of three independent experiments. In the detection process through the aerolysin nanopore, the distinct characteristics of each bipolar peptide were recorded, including current blockades, dwell times, and signal shape under the same experimental conditions.Data analysis
The current recordings were measured with a patch-clamp amplifier (Axon 200 B, AXON, USA) and converted into data using a DigiData 1440A converter (Molecular Devices, Danaher, USA). The signals were filtered at 5 kHz and acquired at a sampling rate of 100 kHz using PClamp 10.6 (Axon Instruments, Forest City, CA, USA). Finally, the data were analysed using Matlab (R2013b, MathWorks, Natick, MA, USA) software, OriginLab 9.0 (OriginLab Corporation, Northampton, MA, USA), and programs that were developed by Long's group.36,37 Typically, more than 1000 blockade events are collected in each ionic current measurement. The original ABF file data of the nanopore experiment is converted to TXT files using the Cutting software programmed by Long's group. Then the TXT files are processed using Matlab, the current signals through the nanopore are screened. The screened data are imported into OriginLab 9.0 for function fitting, and the scatter plot, blocking degree and duration time can be obtained respectively. From the current blockades corresponding to each peptide, the mean blockade degree and duration are calculated for each peptide. The mean blockade degree and duration correspond to the arithmetic mean of the blockade degree and durations, respectively. The error bar is calculated using the “Statistics” function of OriginLab 9.0.Identification of proteinogenic amino acids
The peptide powder was incubated with ultrapure water at room temperature for 30 min. Before nanopore evaluation, the peptide was dissolved in a buffer containing 3 M KCl, 10 mM Tris, and 1 mM EDTA at pH 7.5 to a final concentration of 2 μM. Unless otherwise specified, the nanopore experiments in the assay were performed at a constant voltage of +70 mV applied to the trans compartment.For the peptide probe D6XR5 (named X−), the experimental process was the same as the procedure mentioned above, except that a constant voltage of +70 mV was applied to the cis compartment.
Results and discussion
Feasibility of the electrical recognition of proteinogenic amino acids using the aerolysin nanopore
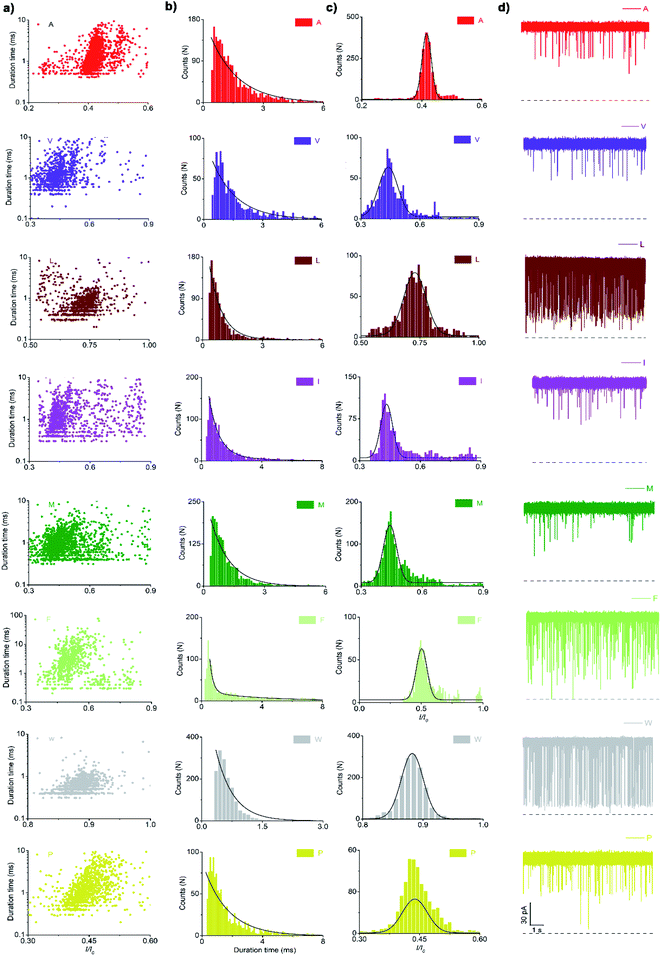
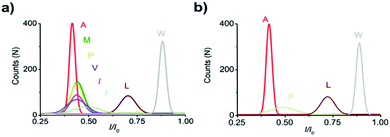
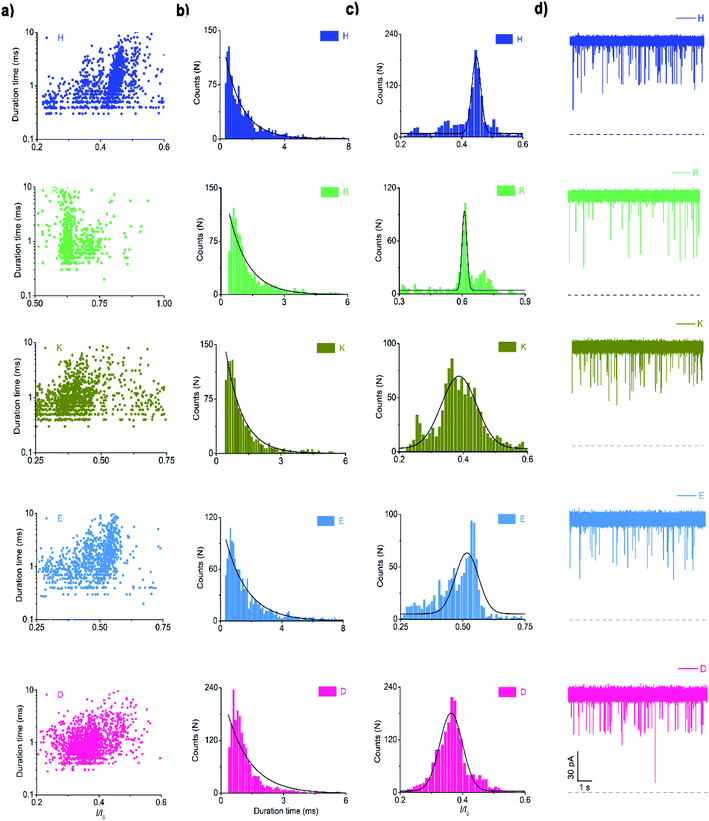
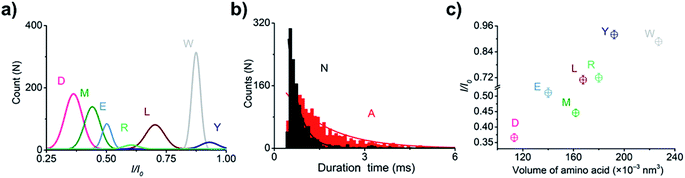
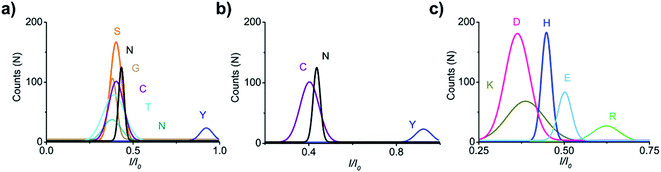
For electrical recognition of the 20 proteinogenic amino acids using an aerolysin nanopore, bipolar peptide probes were synthesised by linking D residues to the N-terminal and R residues to the C-terminal of individual amino acids (Fig. 1a).38,39 An external positive voltage (70 mV) was applied to the trans side of the lipid bilayer(Fig. 1b). When the peptide passed through the aerolysin nanopore, transient blockade of the ionic current was observed (Fig. 1c). Each characteristic current blockade corresponded to the process of an individual peptide through the nanopore, and each peptide was recognized based on the blockage current and duration time. The feasibility of the assay was first tested by three groups of peptide probes (D4XR5, D6XR8, and D8XR10) to distinguish alanine (A), cysteine (C), and serine (S). In the case of D4XR5, all three peptides (D4AR5, abbreviated as A, D4CR5, abbreviated as C, D4SR5, abbreviated as S) produced several short single-level current signals (Fig. S1†). I0 is defined as the open pore current for current amplitude analysis and I is defined as the blockage current when the analyte remained within the pore. Gaussian fitting (Fig. S1a†) revealed that A had the highest I/I0 value.40,41 The current signals produced by S were characterised by small spikes, whereas peptide C showed a jagged pattern (Fig. S1b†). In contrast, in the case of probe D6XR8 and D8XR10, there was no significant difference among the three peptides in any group (D6XR8: A6, C6, and S6; D8XR10: A8, C8, and S8) in terms of blocking degree, duration time, or signal shape (Fig. S1b†). Thus, the best identification was achieved with the bipolar probe D4XR5.With the initial success, we set out to fully evaluate the potential of this strategy for the identification of all proteinogenic amino acids. Twenty D4XR5 peptides were divided into three groups according to the polarity of the side chain of amino acid X, namely, non-polar amino acid group, polar neutral amino acid group, and polar charged amino acid group. The group of non-polar amino acids comprised alanine (A), valine (V), leucine (L), isoleucine (I), proline (P), tryptophan (W), methionine (M), and phenylalanine (F). All these peptides produced current blocks when they passed through the aerolysin pore (Fig. 2d). Using statistical analysis, the distribution of blocking current was fitted to Gaussian functions and the duration time of the peptides was fitted by exponential distributions based on the first passage process42,43 (Fig. 2a–c and 3a). Among those, the I/I0 histograms of A, L, W, and F were located at 0.42 ± 0.0145, 0.70 ± 0.0140, 0.88 ± 0.0129, and 0.50 ± 0.0121, respectively, exhibiting four well-separated peaks (Fig. 3b). The current blockades and duration of the peptides containing V, I, P, and M were too close to be easily distinguished, most likely due to their similarities in spatial structures, volumes, or molecular weights.
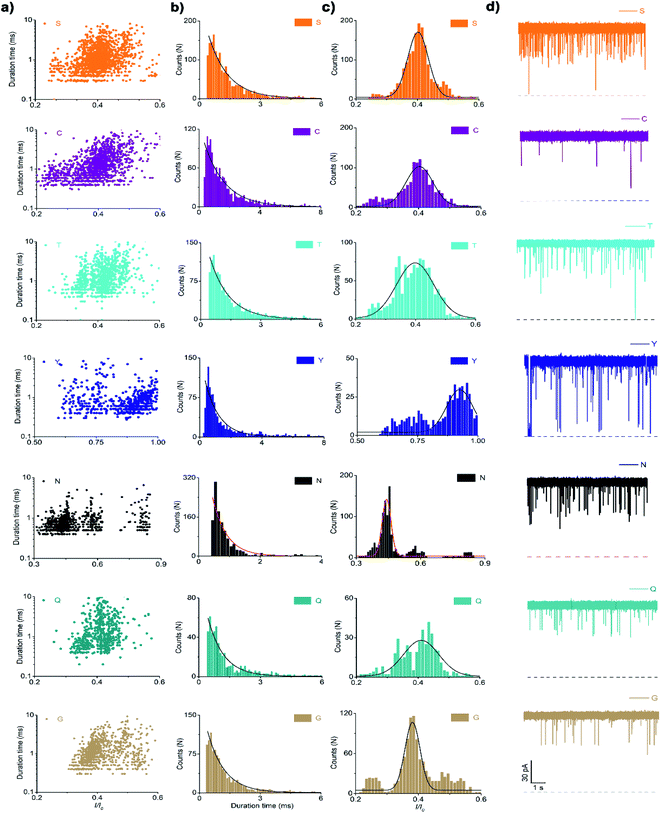
In the case of the polar uncharged amino acid group, glycine (G), serine (S), glutamine (Q), threonine (T), cysteine (C), asparagine (N), and tyrosine (Y) were analysed under the same conditions as that of non-polar amino acids (Fig. 4 and 5a). Among those, the peptides containing C, N, and Y produced blockade populations distinct from one another (Fig. 5b), with the I/I0 value of 0.39 ± 0.015, 0.44 ± 0.012, and 0.93 ± 0.048, respectively. The current amplitude and duration time distribution of G, S, Q, and T were too similar to be differentiated.
 | ||
| Fig. 5 Superimposed I/I0 histograms of G, S, Q, T, C, N, and Y (a). I/I0 histograms of C, N, and Y (b). Superimposed I/I0 histograms of K, H, R, D, and E (c). | ||
In the same way, the polar charged amino acid group contains three amino acids with positive charges, lysine (K), histidine (H), and R, and two amino acids with negative charges, D and glutamic acid (E). D4XR5 probes bearing them were identified via the nanopore assay (Fig. 6 and 5). Peptides H, R, and E could be well distinguished based on the degree of blocking, and the fitted values located in 0.45 ± 0.015 (H), 0.72 ± 0.012 (R), and 0.52 ± 0.011 (E), respectively (Fig. 5c). While the I/I0 of D and K were too similar to be differentiated, their current blockades were clearly distinguished from those of H, R and E. Overall, with the help of a bipolar peptide probe, it was feasible to recognise proteinogenic amino acids using an aerolysin nanopore electrically. Bipolar probes containing A, F, L, W, C, N, Y, H, E, and R could be identified using I/I0 and duration time, whereas the remaining 10 proteinogenic amino acids cannot be recognized.
In previous studies, the tug-of-war between positive and negative forces acting on the two oppositely charged tails of the dipolar peptide contributed to stretching the peptide and to increase the dwell time.33–35 Consistent with what was observed for α-hemolysin pores in these reports, the dwell time of the dipolar peptides bearing H or W increases when the applied voltage increases from +50 to +80 mV (Fig. S2†).
Distinction of 9 of the 20 proteinogenic amino acids with the bipolar probe D4XR5
When grouped by the polarity of the side chain of X, a few amino acids could not be distinguished by the nanopore. We further aimed to examine if amino acids could be identified when not grouped (Fig. S3†). As shown in Fig. 8, I/I0 histograms of peptides D, M, E, R, L, W, and Y exhibited well-separated peaks (Fig. 7a). The duration times of the remaining 13 peptides were combined for exponential fitting, and N and A were distinguished (Fig. 7b). It is noteworthy that W and Y showed large I/I0 (0.88 ± 0.0129 and 0.93 ± 0.0480), which may be attributed to the large side chain of an indole group or a phenol group.To probe the mechanism underlying the blockade current, we plotted the relationship of the current amplitude against the volume of amino acids (Fig. 7c). Overall, the blocking current was found to increase as the volume of amino acid increased, which is largely consistent with the findings of H. Ouldali's study used a short polycationic carrier to identify amino acids.30 However, there were notable exceptions in this study, such as A, D, T, Q, and K (Fig. S4†). We speculated that this may be related to the structures of the probes. We used the bipolar probe (D4XR5) with the target amino acid in the middle of the polypeptide chain, while H. Ouldali's group used the unipolar probe (XR7) with the target amino acid at the N-terminal. Current blockades are likely to be affected by the position of the target amino acid in the peptide and the probe sequence. The current amplitude (I/I0) in our assay ranged from 0.35 to 0.93, whereas theirs lay within the range of 0.32–0.4. The variation range of I/I0 in this study is much larger than theirs. On the other hand, in addition to the volume, the structure, charge properties of the side chain, and molecular masses of target amino acid also greatly influence the current signal. Different side chains will cause different non-covalent interactions between amino acids and nanopores, including electrostatic interactions, π-effect, and hydrophobic interactions. Previous reports of Long's group have also indicated that the steric hindrance effect is not a dominant factor for amino acid discrimination, in which the smaller cysteine produced the evident current signatures, while the larger asparagine and glutamine induced fast and undistinguishable current variations.31
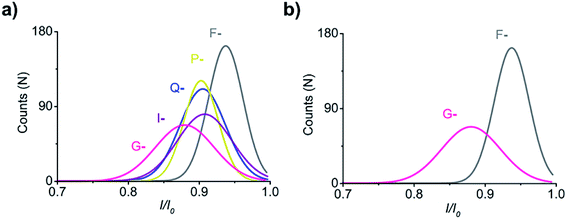
Distinction of I, F, G, P, and Q employing the negatively charged probe D6XR5
To improve the ability of our strategy to recognise the remaining 11 amino acids, I, F, G, P, and Q were randomly selected as identification objects. The charge of peptides was reversed by adding two aspartic acid residues to the N-terminus of D4XR5, and the obtained probe D6XR5 (X-) was employed to distinguish the five amino acids. The negatively charged peptide was added to the cis side of the aerolysin chamber, and a positive voltage (70 mV) was applied to drive it into the aerolysin nanopore. In this case, the I/I0 histograms of F- and G-exhibited well-separated populations (Fig. 8). Notably, compared with that of the positively charged peptides (D4XR5), the blocking degree of the negatively charged peptides (D6XR5) significantly increased (from 0.35–0.55 to 0.61–0.95). This result shows that changing the charge of the bipolar peptide probe can improve the ability of the aerolysin nanopore to distinguish amino acids, which may be attributed to the anionic selectivity of the aerolysin nanopore.44,45 Previous studies have demonstrated that there are two positively charged amino acids located at the entrance of the pore and the bottom of the lumen in wild-type aerolysin.46 This would produce strong electrostatic interactions between the peptide probe and the inner wall of the nanopore, significantly affecting the translocation of negatively charged peptides through the nanopore. As a result, the blocking degrees of negatively charged bipolar peptide probes are greatly increased, thus improving the ability of the aerolysin nanopore to distinguish different amino acids.Conclusions
In the present work, we proposed a bipolar peptide probe to enable electrical recognition of proteinogenic amino acids using an aerolysin nanopore. The bipolar peptide probe was formed by linking aspartic acid residues to the N-terminal and arginine residues to the C-terminal of individual amino acids. The advantage of this structure is that it contains amino acids with opposite charges, which is intended to improve the interaction between the probe and the inner wall of the nanopore, thus enhancing the ability to recognise amino acids. Among the 20 amino acids, our strategy permitted the reliable identification of 9 amino acids, based on distinct current blockades and duration times. In the case of the positively charged peptide probe D4XR5, amino acids D, M, E, R, L, W, and Y were effectively identified by current blockages, and N and A could be distinguished by duration times. Furthermore, the ability to distinguish peptides can be improved by employing the negatively charged probe D6XR5 (X-), as proved by the identification of G, and F. It is speculated that this is due to the strong electrostatic interactions caused by the positively charged residues at the entrances and inside the lumen during the translocation of the aerolysin nanopore. In our future study, we will include all types of peptides with a negatively charged probe. Recently, Long et al. pioneered in designing a series of mutant aerolysin that modulates charge distribution via site-directed mutagenesis.32 Inspired by this, we could design a specific biological nanosensor with a high resolution to identify amino acids in the future. We anticipate that the development of mutant aerolysin, combined with the bipolar peptide probe, would facilitate the application of aerolysin nanopores in the identification of proteinogenic amino acids.Author contributions
D. X. conceived the work. Y. G., M. C., and Q. Z. performed the experiments. D. X., Y. G., M. C., and Q. Z. analysed the data. D. X., Y. G. and Y. W. wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (21874062, 22076073), Collaborative Innovation Center in Universities of Shandong Province (Linyi University), the Natural Science Foundation of Shandong Province (ZR2017ZC0226), Key Research and Development Program of Linyi City (2021030), and the Open Project Foundation of Shandong (Linyi) Institute of Modern Agriculture, Zhejiang University (No. ZDNY-2021-FWLY02012).Notes and references
- L. Liu and H. C. Wu, Angew. Chem., Int. Ed. Engl., 2016, 55, 15216–15222 CrossRef CAS PubMed.
- D. Xi, J. Shang, E. Fan, J. You, S. Zhang and H. Wang, Anal. Chem., 2016, 88, 10540–10546 CrossRef CAS PubMed.
- B. Wei, N. Liu, J. Zhang, X. Ou, R. Duan, Z. Yang, X. Lou and F. Xia, Anal. Chem., 2015, 87, 2058–2062 CrossRef CAS PubMed.
- D. Xi, Z. Li, L. Liu, S. Ai and S. Zhang, Anal. Chem., 2018, 90, 1029–1034 CrossRef CAS PubMed.
- M. Cui, Y. Ge, X. Zhuge, X. Zhou, D. Xi and S. Zhang, Chin. J. Chem., 2021, 39, 2035–2043 CrossRef CAS.
- D. Branton, D. W. Deamer, A. Marziali, H. Bayley, S. A. Benner, T. Butler, M. Di Ventra, S. Garaj, A. Hibbs, X. Huang, S. B. Jovanovich, P. S. Krstic, S. Lindsay, X. S. Ling, C. H. Mastrangelo, A. Meller, J. S. Oliver, Y. V. Pershin, J. M. Ramsey, R. Riehn, G. V. Soni, V. Tabard-Cossa, M. Wanunu, M. Wiggin and J. A. Schloss, Nat. Biotechnol., 2008, 26, 1146–1153 CrossRef CAS PubMed.
- A. Meller and D. Branton, Electrophoresis, 2002, 23, 2583–2591 CrossRef CAS PubMed.
- I. M. Derrington, T. Z. Butler, M. D. Collins, E. Manrao, M. Pavlenok, M. Niederweis and J. H. Gundlach, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 16060–16065 CrossRef PubMed.
- E. Kennedy, Z. Dong, C. Tennant and G. Timp, Nat. Nanotechnol., 2016, 11, 968–976 CrossRef CAS PubMed.
- Q. Liu, Y. Wang, Y. Liu, H. Wang, W. Li, P. Tang, T. Weng, S. Zhou, L. Liang, J. Yuan, D. Wang and L. Wang, Nanoscale, 2020, 12, 19711–19718 RSC.
- G. M. Roozbahani, X. Chen, Y. Zhang, L. Wang and X. J. S. M. Guan, Small Methods, 2020, 4, 2000266 CrossRef CAS PubMed.
- W. L. Hsu and H. Daiguji, Anal. Chem., 2016, 88, 9251–9258 CrossRef CAS PubMed.
- L. Mereuta, M. Roy, A. Asandei, J. K. Lee, Y. Park, I. Andricioaei and T. Luchian, Sci. Rep., 2014, 4, 3885 CrossRef PubMed.
- H. Wang, W. Huang, Y. Wang, W. Li, Q. Liu, B. Yin, L. Liang, D. Wang, X. Guan and L. Wang, ACS Sens., 2021, 6, 3781–3788 CrossRef CAS PubMed.
- M. Li, W. Li, Y. Xiao, Q. Liu, L. Liang, D. Wang, W. Huang and L. Wang, Biosens. Bioelectron., 2021, 194, 113602 CrossRef CAS PubMed.
- H. Chae, D. K. Kwak, M. K. Lee, S. W. Chi and K. B. Kim, Nanoscale, 2018, 10, 18423 RSC.
- W. Qiu and E. Skafidas, ACS Appl. Mater. Interfaces, 2014, 6, 16777–16781 CrossRef CAS PubMed.
- D. Rodriguez-Larrea and H. Bayley, Nat. Nanotechnol., 2013, 8, 288–295 CrossRef CAS PubMed.
- B. Cressiot, A. Oukhaled, L. Bacri and J. Pelta, Bionanoscience, 2014, 4, 111–118 CrossRef.
- G. Oukhaled, J. Mathe, A. L. Biance, L. Bacri, J. M. Betton, D. Lairez, J. Pelta and L. Auvray, Phys. Rev. Lett., 2007, 98, 158101 CrossRef CAS PubMed.
- E. L. Bonome, F. Cecconi and M. Chinappi, Nanoscale, 2019, 11, 9920–9930 RSC.
- W. Si and A. Aksimentiev, ACS Nano, 2017, 11, 7091–7100 CrossRef CAS PubMed.
- H. Y. Wang, Y. L. Ying, Y. Li, H. B. Kraatz and Y. T. Long, Anal. Chem., 2011, 83, 1746–1752 CrossRef CAS PubMed.
- Y. X. Hu, Y. L. Ying, Z. Gu, C. Cao, B. Y. Yan, H. F. Wang and Y. T. Long, Chem. Commun., 2016, 52, 5542–5545 RSC.
- A. B. Farimani, M. Heiranian and N. R. Aluru, npj 2D Mater. Appl., 2018, 2, 1–9 CrossRef.
- G. Di Muccio, A. E. Rossini, D. Di Marino, G. Zollo and M. Chinappi, Sci. Rep., 2019, 9, 6440 CrossRef PubMed.
- G. Zollo and A. E. Rossini, Nanoscale Adv., 2019, 1, 3547–3554 RSC.
- Z. Chen, Z. Wang, Y. Xu, X. Zhang, B. Tian and J. Bai, Chem. Sci., 2021, 12, 15750–15756 RSC.
- F. Piguet, H. Ouldali, M. Pastoriza-Gallego, P. Manivet, J. Pelta and A. Oukhaled, Nat. Commun., 2018, 9, 966 CrossRef PubMed.
- H. Ouldali, K. Sarthak, T. Ensslen, F. Piguet, P. Manivet, J. Pelta, J. C. Behrends, A. Aksimentiev and A. Oukhaled, Nat. Biotechnol., 2020, 38, 176–181 CrossRef CAS PubMed.
- B. Yuan, S. Li, Y. L. Ying and Y. T. Long, Analyst, 2020, 145, 1179–1183 RSC.
- S. Li, X. Y. Wu, M. Y. Li, S. C. Liu, Y. L. Ying and Y. T. J. S. M. Long, Small Methods, 2020, 4, 2000014 CrossRef CAS.
- A. Asandei, M. Chinappi, J. K. Lee, C. Ho Seo, L. Mereuta, Y. Park and T. Luchian, Sci. Rep., 2015, 5, 10419 CrossRef CAS PubMed.
- A. Asandei, M. Chinappi, H. K. Kang, C. H. Seo, L. Mereuta, Y. Park and T. Luchian, ACS Appl. Mater. Interfaces, 2015, 7, 16706–16714 CrossRef CAS PubMed.
- M. Chinappi, T. Luchian and F. Cecconi, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2015, 92, 032714 CrossRef PubMed.
- Z. Gu, Y. L. Ying, C. Cao, P. He and Y. T. Long, Anal. Chem., 2015, 87, 907–913 CrossRef CAS PubMed.
- N. Zhang, Y.-X. Hu, Z. Gu, Y.-L. Ying, P.-G. He and Y.-T. Long, Chin. Sci. Bull., 2014, 59, 4942–4945 CrossRef CAS.
- A. Asandei, A. E. Rossini, M. Chinappi, Y. Park and T. Luchian, Langmuir, 2017, 33, 14451–14459 CrossRef CAS PubMed.
- A. Asandei, G. Di Muccio, I. Schiopu, L. Mereuta, I. S. Dragomir, M. Chinappi and T. J. S. M. Luchian, Small Methods, 2020, 4, 1900595 CrossRef CAS.
- W. Li, Y. Wang, Y. Xiao, M. Li, Q. Liu, L. Liang, W. Xie, D. Wang, X. Guan and L. Wang, ACS Appl. Mater. Interfaces, 2022, 14, 32948–32959 CrossRef PubMed.
- Y. Xiao, J. Ren, Y. Wang, X. Chen, S. Zhou, M. Li, F. Gao, L. Liang, D. Wang, G. Ren and L. Wang, Biosens. Bioelectron., 2022, 212, 114415 CrossRef CAS PubMed.
- J. Li and D. S. Talaga, J. Phys.: Condens. Matter, 2010, 22, 454129 CrossRef PubMed.
- S. Redner, A Guide to First-Passage Processes, Cambridge University Press, 2001, vol. 8, p. 252 Search PubMed.
- S. Li, C. Cao, J. Yang and Y. T. Long, ChemElectroChem, 2019, 6, 126–129 CrossRef CAS.
- F. Hu, B. Angelov, S. Li, N. Li, X. Lin and A. Zou, Chembiochem, 2020, 21, 2467–2473 CrossRef CAS PubMed.
- C. Cao, Y. L. Ying, Z. L. Hu, D. F. Liao, H. Tian and Y. T. Long, Nat. Nanotechnol., 2016, 11, 713–718 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2na00190j |
| This journal is © The Royal Society of Chemistry 2022 |