Open Access Article
Open Access ArticleRiboflavin–citrate conjugate multicore SPIONs with enhanced magnetic responses and cellular uptake in breast cancer cells†
Wid
Mekseriwattana
 ab,
Pablo
Guardia
b,
Beatriz Torres
Herrero
ab,
Pablo
Guardia
b,
Beatriz Torres
Herrero
 c,
Jesus M.
de la Fuente
c,
Chutima
Kuhakarn
c,
Jesus M.
de la Fuente
c,
Chutima
Kuhakarn
 d,
Anna
Roig
d,
Anna
Roig
 b and
Kanlaya Prapainop
Katewongsa
b and
Kanlaya Prapainop
Katewongsa
 *ae
*ae
aSchool of Materials Science and Innovation, Faculty of Science, Mahidol University, Bangkok 10400, Thailand
bInstitut de Ciència de Materials de Barcelona (ICMAB-CSIC), Campus UAB, Bellaterra 08193, Spain
cInstituto de Nanociencia y Materiales de Aragón (INMA), CSIC-Universidad de Zaragoza, Centro de Investigación Biomédica en Red de Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN), 50018 Zaragoza, Spain
dDepartment of Chemistry, Center of Exellence for Innovation in Chemistry (PERCH-CIC), Faculty of Science, Mahidol University, Bangkok 10400, Thailand
eDepartment of Biochemistry, Faculty of Science, Mahidol University, Bangkok 10400, Thailand. E-mail: kanlaya.pra@mahidol.edu
First published on 2nd March 2022
Abstract
Breast cancer accounts for up to 10% of the newly diagnosed cancer cases worldwide, making it the most common cancer found in women. The use of superparamagnetic iron oxide nanoparticles (SPIONs) has been beneficial in the advancement of contrast agents and magnetic hyperthermia (MH) for the diagnosis and treatment of cancers. To achieve delivery of SPIONs to cancer cells, surface functionalization with specific ligands are required. Riboflavin carrier protein (RCP) has been identified as an alternative target for breast cancer cells. Here, we report a novel riboflavin (Rf)-based ligand that provides SPIONs with enhanced colloidal stability and high uptake potential in breast cancer cells. This is achieved by synthesizing an Rf-citrate ligand. The ligand was tested in a multicore SPION system, and affinity to RCP was assessed by isothermal titration calorimetry which showed a specific, entropy-driven binding. MRI and MH responses of the coated Rf-SPIONs were tested to evaluate the suitability of this system as a theranostic platform. Finally, interaction of the Rf-SPIONs with breast cancer cells was evaluated by in vitro cellular uptake in MCF-7 breast cancer cells. The overall characterization of the Rf-SPIONs highlighted the excellent performance of this platform for theranostic applications in breast cancer.
Introduction
Breast cancer is the most common cancer in women, accounting for up to 10% of the newly diagnosed cancer cases worldwide. According to the World Health Organization (WHO), 2.3 million women were diagnosed with breast cancer, and 0.7 million died globally in 2020.1 To increase the survival rate, early diagnosis of the cancer is crucial. The WHO recommends mammography screening once every 2 years; however, its low accuracy and sensitivity strongly compromise the early detection of small tumours, especially in young women.2 In this regard, magnetic resonance imaging (MRI) emerges as an alternative diagnosis technique with higher resolution.3In the last decade, nanotechnology has been instrumental in the advancement of contrast agents used in diagnosis,4 specifically for MRI where superparamagnetic iron oxide nanoparticles (SPIONs) have been extensively studied and exploited as contrast agents.5 SPIONs generate hypointense contrast in MRI imaging at the areas where they accumulate, and thus provide higher sensitivity in locating the cancerous regions, which helps in the detection of cancer at early stages.
Following diagnosis, surgery is the most effective treatment for breast cancer. Nonetheless, recurrence is prone to occur depending on each cancer subtype.6 With the high recurrence rate, adjuvant and neo-adjuvant treatments are required for effective therapy. These include, but are not limited to chemotherapy, hormonal therapy and radiation therapy. However, most of the said treatments are disruptive and can cause severe side effects.7 Recently, magnetic hyperthermia (MH) therapy has become an interesting alternative adjuvant treatment for cancers. By utilizing SPIONs, local heating can be induced by an external oscillating magnetic field, thus, promoting cell apoptosis with minimal, if any, side effects.8
Accordingly, use of SPIONs makes an excellent platform for both diagnosis and therapy. To be efficiently utilized, the amount of SPIONs used must be minimal to comply with the safety limit. Therefore, SPIONs must possess a high magnetic response and be able to efficiently accumulate in the cancer regions. To achieve the first requirement, SPIONs must have appropriate sizes and high crystallinity, those characteristics being strongly dependent on the synthesis method. Advanced approaches involving the decomposition of iron complexes at high temperatures provide a higher quality of SPIONs, but in a rather small amount. In contrast, conventional co-precipitation methods provide a large amount of nanoparticles (NPs) but with an average quality in terms of crystallinity or the magnetic response.9,10 The use of SPIONs in nanomedicine requires large-scale production with high quality of SPIONs. Our efforts aim to improve the quality of SPIONs produced by this approach to fulfill the requirement for biomedical applications and demand of future use in vivo and pre-clinical tests.
Regardless of the synthetic approach, saturation magnetization (Ms) values close to the bulk counterpart are highly desired. To improve the magnetic response of SPIONs, structural factors such as the tuning composition or shape have been explored.11 Recently, interparticle interaction has been pointed out as another strategy to improve the magnetic response of SPIONs. This can be enhanced by particle assembly or by the formation of multicore structures.12
To achieve effective delivery of SPIONs to cancer cells, surface functionalization with specific ligands is required. Ligands designed to target particular cancer cells involve biomolecules, human antibodies or peptides which are stable only in aqueous solutions.13 As such, the suitable synthetic routes are limited to those in which SPIONs are produced in water or by organic phase synthesis following a ligand exchange.
For the particular case of breast cancer, HER2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 14,15 and folate receptors16 have been often targeted due to their over-expression, by exploiting folic acid, cell-targeting peptides,17 or anti-EGFR monoclonal antibodies.18,19 However, targeting breast cancer cells is challenging due to the difference in receptor expression among diverging cellular subtypes.6 For this reason, discovering new strategies to broaden the selection for a specific treatment, in response to the cancer characteristics of individual patients, is in high demand for breast cancer types with a high risk of relapse.
14,15 and folate receptors16 have been often targeted due to their over-expression, by exploiting folic acid, cell-targeting peptides,17 or anti-EGFR monoclonal antibodies.18,19 However, targeting breast cancer cells is challenging due to the difference in receptor expression among diverging cellular subtypes.6 For this reason, discovering new strategies to broaden the selection for a specific treatment, in response to the cancer characteristics of individual patients, is in high demand for breast cancer types with a high risk of relapse.
Riboflavin carrier protein (RCP) has been identified as an alternative target for breast cancer cells.20,21 The serum RCP level is elevated in prostate and breast cancer patients with localized overexpression in cancer tissues.22–24 To make use of the overexpression, riboflavin (Rf) derivatives have been studied as drug delivery ligands by exploiting the binding function of RCP toward the vitamin. For instance, Bareford et al. modified polymers with Rf and achieved enhanced cellular uptake in MCF-7 breast cancer cells.25 Similarly, Beztsinna and co-workers reported Rf-functionalized liposomes with specific delivery to PC3 prostate cancer cells.26 Also recently, our group demonstrated the enhanced delivery of poly(lactic-co-glycolic) acid NPs coated with a Rf-chitosan layer in MDA-MB-231 breast cancer cells.27
Rf has been also successfully implemented on SPIONs, using its derivatives (flavin mononucleotide and flavin adenine dinucleotide).28,29 Both systems showed enhanced cellular uptake in prostate cancer cells, and thus emerged as a definitive ligand towards breast cancer.30 However, both systems showed stability issues and required additional stabilizing agents.31 Also, the exploitation of Rf derivative ligands for SPIONs is challenging due to the lack of reactive functional groups and high polarity.
Addressing the above-mentioned challenges, this work reports a novel Rf-based ligand that provides SPIONs with enhanced colloidal stability and enhanced efficiency toward interaction with breast cancer cells. This is achieved by synthesizing a Rf-citrate conjugate. The ligand was tested in a multicore SPION system, synthesized through a co-precipitation method. Affinity to RCP was assessed by isothermal titration calorimetry which showed a specific, entropy-driven binding. MRI and MH responses of the coated SPIONs were assessed to evaluate the suitability of this system as a theranostic platform. Finally, interaction with breast cancer was evaluated by in vitro cellular uptake assays with MCF-7 cells. The overall characterization of the Rf-SPIONs highlighted the excellent performance of this platform for theranostic applications in breast cancer.
Experimental
Materials
Ferrous chloride tetrahydrate (FeCl2·4H2O, ≥99%), ferric chloride hexahydrate (FeCl3·6H2O, 97%), neutral red (≥90%), potassium hexacyanoferrate, riboflavin and human serum albumin (HSA, ≥96%) were purchased from Sigma-Aldrich. Ammonia solution (NH4OH, 25%) and tri-sodium citrate dihydrate (99%) were obtained from Merck. Hydrochloric acid was obtained from Riedel-de Haen. Citric acid was purchased from Vidhyasom Co. Ltd. 3-(4,5-Dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, 98%) was obtained from PanReac AppliChem. Dimethyl sulfoxide (DMSO) was obtained from Fisher Scientific. Sodium chloride (99.9%), potassium chloride (99.8%), di-sodium hydrogen phosphate dihydrate (99.5%), and potassium dihydrogen phosphate (99.8%) were obtained from VWR Chemicals. All chemicals were used as received without further purification. All aqueous solutions were prepared using Milli-Q water (MQ-water) obtained from a Plus water purification system (Millipore, Milford, MA, USA).MCF-7 cells (HTB22) were purchased from ATCC. All cell culture supplies, including: Dulbecco's Modified Eagle's Medium (DMEM), fetal bovine serum (FBS), penicillin/streptomycin solution and trypsin–EDTA solution were obtained from Gibco, UK.
Purification of RCP
To isolate RCP, homogenized chicken egg white was saturated with Rf (final Rf concentration of circa 8 μM). NaCl was then added to the solution to get the final concentration of 20% w/v, and the solution was stirred for 30 min. An equal volume of 5% phenol solution in water was added to induce protein precipitation. The precipitated proteins were removed by vacuum filtration, and the final solution was dialyzed twice against MQ-water (24 h each) followed by 24 h against 50 mM Tris–HCl pH 7.5 buffer. The solution was loaded to a DEAE-sepharose column and eluted with 200 mM NaCl solution in 50 mM Tris–HCl pH 7.5 buffer. RCP-Rf complexes were collected and dialyzed twice against 6 mM HCl solution (24 h each) to remove the bounded Rf and next against 50 mM Tris–HCl pH 7.5 buffer (12 h). Finally, the obtained apoprotein was concentrated by using a stirred cell concentrator and stored at −80 °C. Purity of the protein was determined by performing SDS-PAGE and its concentration was measured by a Micro BCA protein assay (ThermoFisher Scientific, USA).Synthesis and characterization of Rf-citrate
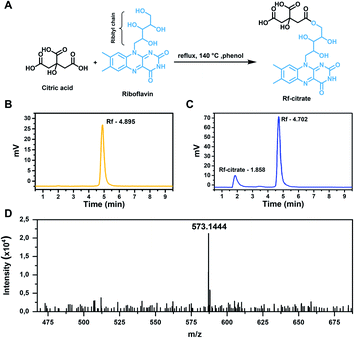
For the synthesis of Rf-citrate, 3.0 g (8.0 mmol) of Rf and 1.7 g (8.8 mmol) of citric acid were dissolved in 30 mL of liquified phenol. The solution was then heated up to the reflux temperature (140 °C) and kept at that temperature for 4 h before cooling down to room temperature. Products were precipitated by adding the reaction mixture to 200 mL of diethyl ether and collected by vacuum filtration; the obtained orange precipitates were dried under reduced pressure.The formation of Rf-citrate was analyzed by high-resolution mass spectrometry (Bruker Micro TOF spectrometer, US) and high-pressure liquid chromatography (HPLC, Waters 2695, Waters Corporation, US) on a reverse phase C18 column (150 mm × 4.6 mm, Acclaim™ 120, ThermoFisher Scientific, US) using a methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (40
water (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60) mobile phase and a fluorescence detector (excitation wavelength, 450 nm; emission wavelength, 510 nm).32
60) mobile phase and a fluorescence detector (excitation wavelength, 450 nm; emission wavelength, 510 nm).32
Synthesis of SPIONs
SPIONs were synthesized by following a reported co-precipitation method with minor modifications.10 Briefly, 0.75 g of FeCl2·4H2O (3.8 mmol) and 2.5 g of FeCl3·6H2O (9.2 mmol) were dissolved in 100 mL of MQ-water. The mixture was heated up to 80 °C with vigorous magnetic stirring under a N2 atmosphere. Next, 5 mL of 25% ammonia solution (67 mmol) were added dropwise to the solution which rapidly turned black in color. After 30 min at 80 °C, the solution was cooled down to room temperature, and SPIONs were collected by magnetic separation. Then, 15 mL of fresh MQ-water was added to the precipitate and vortexed until the SPIONs resuspend back in the solution. The NPs were then collected by magnetic separation. This procedure was repeated at least twice. The final precipitate was dispersed in 100 mL MQ-water (circa 10 mg mL−1).Synthesis of citrate-coated SPIONs (C-SPIONs)
The synthesis of C-SPIONs was done following a method reported by Nigam et al.9 Briefly, 3.7 g of FeCl2·4H2O (18.7 mmol) and 0.87 g of FeCl3·6H2O (4.4 mmol) were dissolved in 40 mL of MQ-water under vigorous stirring. The solution was then heated up to 70 °C under a nitrogen gas flow and kept at this temperature. After 30 min, 10 mL of 25% NH4OH solution (134 mmol) were added, and the reaction was maintained at this temperature for another 30 minutes. Finally, 2 mL of a tri-sodium citrate dihydrate solution (0.75 g mL−1) were added, and the temperature was increased up to 90 °C. After 1 h at this temperature, the solution was cooled down to room temperature, and NPs were washed and collected using the same procedure as described above. The resulting NPs were dispersed in 100 mL of fresh MQ-water (10 mg mL−1) and stored for further use.Synthesis of Rf-functionalized SPIONs (Rf-SPIONs)
To obtain Rf-SPIONs, 50 mg each of Rf-citrate and SPIONs were mixed in 40 mL of MQ-water and stirred under a N2 atmosphere for 90 min at 90 °C. The reaction was then cooled down, and NPs were collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The NPs were dispersed in 15 mL of fresh MQ-water, and the separation process was carried out at least three times to remove excess ligand molecules. The final NPs were dispersed in 10 mL of MQ-water (circa 5 mg mL−1) and stored for further characterization.
000 rpm for 15 min. The NPs were dispersed in 15 mL of fresh MQ-water, and the separation process was carried out at least three times to remove excess ligand molecules. The final NPs were dispersed in 10 mL of MQ-water (circa 5 mg mL−1) and stored for further characterization.
Characterization of SPIONs
SPIONs were first characterized by X-ray diffraction; 2θ scans were collected from 10° to 80° using a Cu-Kα radiation source operating at 40 kV/15 mA and equipped with a Ni-Kβ filter (Aeris benchtop X-ray diffractometer, Malvern Panalytical, UK). The sample was prepared by placing powdered SPIONs in a zero-diffraction sample holder.Hydrodynamic diameter (DH) and zeta potential values of the SPIONs were measured using a Zetasizer Nano ZS90 (Malvern Panalytical, UK). For both measurements, samples were diluted in MQ-water to a concentration of approximately 0.05 mg SPION per mL. Measurements were performed in individual triplicates, and the results are reported as mean values.
Thermogravimetric analysis was carried out by recording mass changes of the samples loaded into a platinum pan upon heating at a rate of 10 °C min−1, under a N2 atmosphere to avoid oxidation.
Magnetic properties of the SPIONs were studied using a superconducting quantum interference device (SQUID, MPMS5XL magnetometer, Quantum Design, CA, USA). The samples were prepared by dropping a dispersion of particles into a SQUID capsule, pre-filled with cotton wool. The amount of the samples added was between 0.2 and 0.5 mg SPION. Hysteresis loops at 10 and 300 K under a maximum field of ±40 kOe were measured. Zero-field and field cooled magnetization curves were recorded from 5–275 K under a constant magnetic field of 50 Oe. Magnetization values were normalized by SPION mass, determined by a previously reported UV-vis quantification method.33
Morphology of the SPIONs was studied using transmission electron microscopy (TEM, JEOL JEM-1210) and cryogenic TEM (cryo-TEM, JEOL JEM-2011). Crystal orientation of the SPION clusters was analysed by high-resolution TEM (HRTEM, FEI Tecnai G2 F20 S-TWIN). The samples were prepared by dropping and drying a diluted dispersion of the SPION dispersion on copper grids. For cryo-TEM, the samples were diluted to a concentration of 0.05 mg SPION per mL and blotted for 1 s after dropping, and then rapidly quenched in liquid ethane and transferred to a TEM microscope while temperature was kept at −175 °C.
Isothermal titration calorimetry (ITC)
The binding affinity of RCP with the SPIONs was measured using the ITC technique (MicroCal PEAQ-ITC, Malvern Panalytical, UK). Experiments were performed in distilled water using the following parameters: 25 °C, 264 rpm, 500 s between injections, and 30 injections (10 μL each). Briefly, 2 mL of a solution containing the proteins (12.5 μM) was added to the sample cell. After equilibration, 1 mL of an aqueous dispersion of SPIONs (30 mM) was injected into the cell, using the described settings.Magnetic resonance imaging (MRI)
Longitudinal (T1) and transversal (T2) relaxation times were measured by using a Bruker Biospec 7T MRI scanner. SPIONs were diluted in MQ-water to different concentrations ranging from 0.08–0.45 mM of Fe. Relaxivity values were calculated from the slope of linear fitting of either T1−1 or T2−1 plotted against the corresponded Fe concentration.Calorimetric measurements of the specific absorption rate
The heating performances of the samples were characterized by using a commercially available magnetic nano-heating device (D5 Series, CoilSet CAL2, nanoscale Biomagnetics). The specific absorption rate (SAR) values were measured on 1 ± 0.1 mg SPION per mL−1 solutions (100 μL) exposed to an alternating magnetic field of different amplitudes (8–24 kA m−1) and frequencies (163, 491 and 763 kHz). All measurements were performed under quasi-adiabatic conditions, and the SAR values were calculated according to the following equation: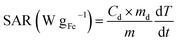 | (1) |
In vitro cytotoxicity test
MCF-7 cells were cultured in DMEM and supplemented with 10% FBS and 1% penicillin/streptomycin. Cells were maintained at 37 °C with 5% CO2 in a humidified incubator. Subculture was done at 80% confluency using 0.25% trypsin–EDTA, and the media were changed every four days.Cells were seeded onto a 96-well plate with a cell density of 8 × 103 cells per well and incubated for 24 h. Three concentrations of Rf-SPIONs (50, 100 and 200 μg mL−1) dispersed in DMEM were added to the cells (200 μL per well). After incubation (4, 12 and 24 h), the media were removed and cells were rinsed with PBS and then 100 μL of MTT solution (0.5 mg mL−1 in DMEM) was added and incubated for 3 h at 37 °C under a 5% CO2 atmosphere in a humidified incubator. After incubation, the media were removed, and 100 μL of DMSO was added to each well. Cell viability was determined by measuring the absorbance of the dissolved formazan at 540 nm using a microplate reader (Tecan Spark 10 M).
Prussian blue staining
Cellular uptake of the SPIONs by MCF-7 cells was visualized by Prussian blue staining. The cells were seeded and incubated for 24 h in a 24-well plate containing circular coverslips (0.5 mL, 2 × 105 cells per mL). The media were discarded and replaced with fresh DMEM containing 200 μg mL−1 of either C-SPIONs or Rf-SPIONs. After incubation for 24 h, the cells were rinsed once with DMEM followed by thrice with PBS and fixed with 3.6% formaldehyde. Finally, cells were immersed in an aqueous solution containing 2% ferrocyanide and 2% HCl for staining of iron, followed by counterstaining in 0.5% neutral red solution. The coverslips were mounted on a glass slide and allowed to dry overnight. The stained cells were observed under an optical microscope (Nikon Eclipse T E2000-U).Flow cytometry analysis
As described above, MCF-7 cells were first seeded onto a 6-well plate (2.5 mL, 2 × 105 cells per mL). After incubation for 24 h, the media were discarded and replaced with fresh DMEM containing 200 μg mL−1 of either C-SPIONs or Rf-SPIONs. After 24 h incubation, the cells were washed once with DMEM followed by twice with PBS and trypsinized before fresh DMEM was added. Then the cells were transferred to a microtube and collected by centrifugation (5000 rpm, 5 min). The collected cells were rinsed with ice-cold PBS followed by fixation with 3.6% formaldehyde on ice. After fixation, the cells were separated by centrifugation and redispersed in PBS. Flow cytometry was performed using an Attune NxT flow cytometer (ThermoFisher Scientific, MA, USA) where a fixed number of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 gated events were collected and analysed.
000 gated events were collected and analysed.
Results and discussion
Synthesis of Rf-citrate
The synthesis was done by a one-step esterification reaction under refluxing conditions (Fig. 1A). The presence of the conjugate in the crude product was determined by HPLC where a minor peak was detected at a retention time of 1.8 min, along with a peak at 5 min ascribed to Rf (Fig. 1B and C). The faster elution time of Rf-citrate (1.8 against 5 min) indicates a higher polarity of the conjugate compared to free Rf. This is expected, considering the non-symmetrical structure of the conjugate. The structure of Rf-citrate was confirmed by mass spectrometry where a peak with an m/z value of 573.1444 was found, matching the calculated value (calculated [M]+: 550.1547; calculated [M + Na]+: 573.1445; Fig. 1D). It is worth mentioning that as observed in Fig. 1C, the final product contains both the conjugated and free Rf. Note that purification processes are challenging as both molecules show high polarity. Therefore, the ligand was kept as a crude product and used for the functionalization of SPIONs.Here, Rf was designed to conjugate with citric acid to obtain the ability to bind to SPIONs, as Rf alone does not contain suitable functional groups to facilitate the interaction. On the other hand, citric acid has been widely reported as an excellent stabilizing agent for metal oxide NPs.34,35 The citric molecules bind tightly to the SPION via the formation of hydrogen bonds with the amphoteric –OH groups on the surfaces,36 which were formed when put in an aqueous medium due to the Lewis acid nature of the Fe atoms.37 As such, the formulated Rf-citrate is expected to bind to SPIONs through a similar interaction (this point is further discussed in the sections below). Hence, while the citric acid moiety provides colloidal stability, Rf ensures enhanced interaction towards breast cancer cells.
Synthesis of Rf-coated SPIONs
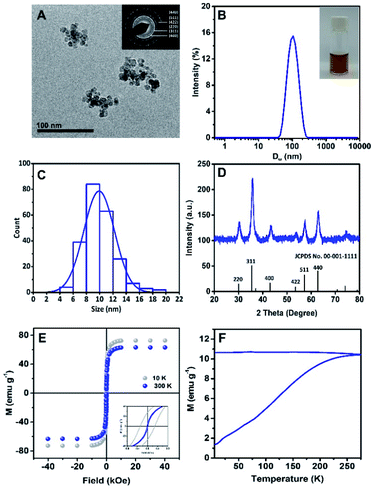
The morphology of Rf-SPIONs was investigated by cryo-TEM. The images revealed that Rf-SPIONs appeared as irregular-shaped clusters, formed by small cores with sizes around 10 nm (Fig. 2A and C). Dynamic light scattering (DLS) measurement showed a DH of 100 nm with narrow distribution (PdI < 0.2) which indicated that the Rf-SPIONs formed stabilized clusters when dispersed in aqueous media. The synthesized SPIONs also showed good crystallinity as shown by narrow rings and peaks in the SAED (inset of Fig. 2A) and XRD spectra (Fig. 2D). Diffraction patterns confirmed the presence of a spinel structure likely ascribed to maghemite/magnetite iron oxide. The crystalline size obtained by XRD was in good agreement with the core size measured by TEM (8 nm compared to 10 nm, Table 1).| Samples | D H (nm) | PdI | Zeta-potential (mV) | TEM core size (nm) | XRD crystallite core size (nm) | M s (emu g−1) | H c (Oe) | M r (emu g−1) | T B (K) |
|---|---|---|---|---|---|---|---|---|---|
| Rf-SPIONs | 100 | 0.1 | −28 ± 2 | 9.9 ± 2.3 | 8.1 ± 0.6 | 72 ± 5 | 280 | 27 ± 2 | < 300 |
| C-SPIONs | 100 | 0.2 | −27 ± 10 | 7.6 ± 2.5 | 7.1 ± 1.2 | 68 ± 5 | 260 | 25 ± 2 | 210 |
Clustering of SPIONs is usual for co-precipitation synthesis, due to the rapid, uncontrollable seeding and growing of the NPs. Such a size and degree of clustering can be partially tuned by the concentration and addition process of the stabilizer, which in this case can be compared to citric acid.34,38 Here, the Rf-SPIONs were synthesized through a two-step process, which allowed the SPIONs to form clusters prior to introduction of Rf-citrate, which then work as a stabilizer to separate the SPIONs into smaller multicore structures.
Magnetic properties of Rf-SPIONs were evaluated by measuring hysteresis loops at 10 and 300 K (Fig. 2E). At 10 K, Rf-SPIONs showed a ferromagnetic response with a Ms value of 72 emu g−1 and a coercive field (Hc) of 280 Oe, while at 300 K, the NPs showed a near superparamagnetic behaviour (Hc = 0) with a Ms value of 63 emu g−1 (Fig. 2E). Zero-field and field cooled magnetization measurement was performed to determine the SPION blocking temperature (TB) (Fig. 2F). Rf-SPIONs showed high TB with an estimated value of close to 300 K, despite the core size of 10 nm. This high TB is typically found in SPIONs with sizes closer to 20 nm.39,40 This deviation results from dipole interaction between magnetic cores inside the cluster.41 Overall, the Ms value at 10 K for Rf-SPIONs was close to those reported for the bulk maghemite or magnetite (83.5 emu g−1),42 indicating high quality SPIONs. The magnetic properties of C-SPIONs were similar as shown in Fig. S2.†
At 300 K, conventional co-precipitation synthesis yields SPIONs with Ms values below 60 emu g−1.9,10,43,44 Nonetheless, recent investigations reported Ms values well close to 70–75 emu g−1. The origin of the high magnetic response is ascribed to core-to-core interaction within the multicore structures.45 Indeed, multicore nanostructures showing high Ms values and superparamagnetic behavior at room temperature have been recently identified as outstanding candidates for biomedical applications,46,47 in particular, as theranostic platforms for MRI and MH.48 For SPIONs to be used in biomedicine, good magnetic properties are required (e.g. high Ms or superparamagnetic behavior at room temperature).49 Such properties do strongly depend on the size and crystallinity of the SPIONs, these also being strongly dependent on the synthetic procedure.50 For some particular applications such as MH, NPs' size has to lie between the superparamagnetic and the ferromagnetic regime at room temperature (e.g. 18 to 25 nm for maghemite NPs51). This results in the use of large particles, which show poor colloidal stability due to particle aggregation.52 Hence, multicore nanostructures represent an advanced solution to address this issue. Although the single multicore structure shows a large magnetic moment under an applied magnetic field due to the collective behavior, the small individual core still shows superparamagnetic behavior at room temperature (TB < 300 K, Fig. 2F). Thus, when the field is removed, the magnetization of each domain is reduced back to zero (or close to zero) and so the one of the multicore clusters. This results in reduction of the cluster-to-cluster dipolar magnetic interaction and reversing the multicore aggregation. All in all, the procedure reported here provides a feasible route to produce colloidal solutions of multicore SPIONs on a large scale (1 g of SPIONs per batch).
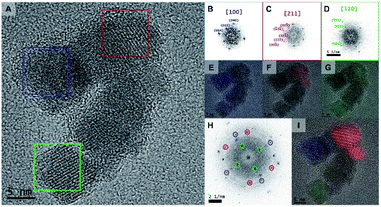
HRTEM was carried out to further investigate the structure of the Rf-SPIONs (Fig. 3). Focusing on a multicore structure (Fig. 3A), three different cores were analyzed, revealing distinct zone axes corresponding to the [100], [211], and [110] families (Fig. 3B–D). This suggests different orientations of the cores in a cluster, confirming that crystalline continuity does not spread across the entire structure. However, filtered Fourier transform analysis allowed the identification of preferential orientation between the adjacent cores as shown by the colored areas in Fig. 3E–G. Indeed, sharing of the same zone axis was observed between the adjoining cores, which could facilitate exchange interaction resulting in a collective magnetic response leading to a high TB.46,48
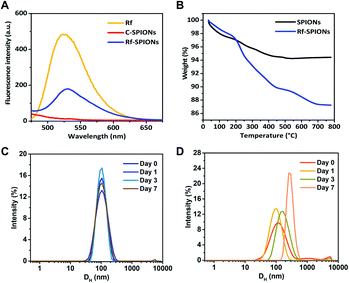
The presence of Rf ligands in Rf-SPIONs was confirmed by fluorescence spectrometry (Fig. 4A). A fluorescence emission peak at around 530 nm was observed for the Rf-SPIONs matching that of free Rf, while C-SPIONs do not present fluorescence. Thermogravimetric analysis was performed to evaluate the ligand amount on the SPIONs (Fig. 4B).53 Compared to non-coated SPIONs, a 13% weight loss was measured for Rf-SPIONs. Extrapolating the presence of moisture from the SPIONs (6% weight loss), the amount of the ligand attached to the SPIONs can be estimated to be circa 7% wt.
To evaluate colloidal stability, the evolution of the DH by the DLS technique was monitored for 7 days after dispersing the samples in water. This method is a robust tool to determine the colloidal stability of the NPs in the liquid phase.54,55 The results demonstrate that DH of Rf-SPIONs remained constant at 100 nm with no significant shifting or broadening of the distribution. Instead, for C-SPIONs, a size increase was observed starting from day 3, showing continuous aggregation with a final DH of 350 nm. These results supported our hypothesis that by incorporating Rf with citric acid, the conjugates could interact with the SPION surfaces and provide stability to the particle clusters. The superior stabilizing character of Rf-citrate as compared to citric acid could be due to the steric hindrance of the Rf moiety acting in synergy with electrostatic repulsion from the citrate part. Note that the same experiment was done for SPIONs incubated with Rf. In this case, the DLS spectra do clearly show higher aggregation of the NPs (Fig. S3†). This confirms that the coating of Rf on the SPIONs is achieved by incorporation of the Rf with citrate, and the latter promotes interaction with the SPION surfaces.
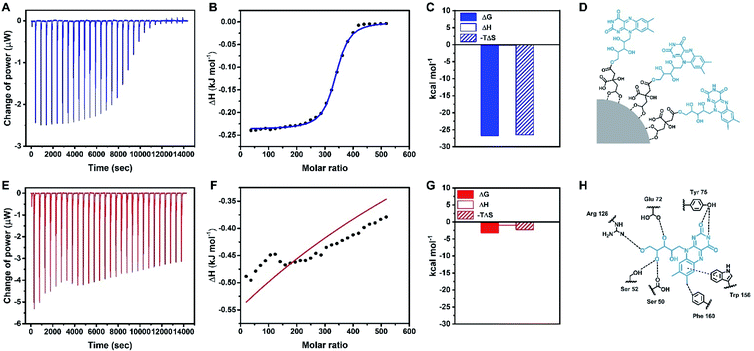
Thermodynamic measurements of the binding between Rf-SPIONs and RCP
ITC characterization was used to provide further information on the interaction of Rf-SPIONs with RCP. As a carrier of Rf,56 RCP binds strongly to free Rf molecules with a dissociation constant (Kd) of 127 nM (Fig. S4A and B†). Note that the affinity is calculated to be 1/Kd. In this regard, RCP did also bind to Rf-SPIONs but with 160 times greater Kd (Fig. 5A, B and Table 2). This represents a significant decrease of the binding affinity of RCP to Rf-SPIONs when compared to that of free Rf. Such a decrease has also been reported in Rf-conjugated dendrimers.57 In this particular case, the decrease in binding affinity is likely ascribed to steric hindrance when Rf moieties are anchored to the SPION surfaces. As Rf is attached to citric acid which is attached on to the SPION surface, the interaction of the ligand with the binding site of RCP is reduced (Fig. 5D and H). Thermodynamic parameters of the binding process revealed an energetically favourable binding driven by entropy changes (Fig. 5C), while the binding of free Rf is driven by enthalpy (Fig. S4C†). The difference in the binding mode is a consequence of the modification at the ribityl chain of the Rf molecule (see Fig. 1A) which reduces hydrophilicity of the structure, causing displacement of water molecules from the hydrophobic pocket of RCP upon binding and increasing the entropy of the system. Similar characterization was carried out for C-SPIONs. When compared with the Rf-SPIONs, the Kd of C-SPIONs with RCP was 4 orders of magnitude higher than that measured for Rf-SPIONs. This weak binding of RCP to C-SPIONs emphasizes the key role of Rf as a specific ligand toward RCP. Finally, control experiments were carried out with HSA to ensure specificity towards RCP. In this case, low binding affinity of HSA to Rf-SPIONs was observed (Table 2 and Fig. S4D–F†), while no binding was observed from C-SPIONs, which could be due to the fact that the experiment was performed at neutral pH since we previously demonstrated that HSA can form coronas with C-SPIONs under basic conditions.58 All data led us to conclude that the Rf-citrate conjugate promotes specific and strong binding between SPIONs and RCP. As such, the developed multicore Rf-SPIONs are excellent candidates as a platform for breast cancer cell drug delivery.| Protein | SPION | K d (μM) | ΔG (kJ mol−1) | ΔH (kJ mol−1) | −TΔS (kJ mol−1) |
|---|---|---|---|---|---|
| a NB: no binding. | |||||
| RCP | Rf-SPIONs | 21 | −26.7 | −0.237 | −26.5 |
| RCP | C-SPIONs | 270![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
−3.23 | −0.961 | −2.27 |
| HSA | Rf-SPIONs | 4800 | −13.2 | 205 | −218 |
| HSA | C-SPIONs | NB | NB | NB | NB |
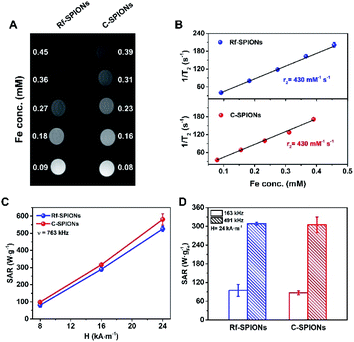
MRI and MH responses
Longitudinal and transversal relaxivities (r1 and r2 respectively) of the synthesized Rf- and C-SPIONs were measured to investigate the potential of the systems as MRI contrast agents. Both NPs showed very high r2 values of 430 mM−1 s−1 in water (Fig. 6A and B). For longitudinal relaxation, the r1 values for Rf- and C-SPIONs were found to be 0.8 and 1.3 mM−1 s−1 respectively (Fig. S5†). The high transversal relaxivity points towards Rf- and C-SPIONs as excellent T2 contrast agents, the measured values being among the highest reported thus far for iron oxide NPs.59,60 Such high performance is directly related to the clustered morphology of the samples. On the one hand, single 9 nm SPIONs exhibit r2 values not exceeding 100 mM−1 s−1.58,61 On the other hand, studies on a cluster made of 10 nm citrate-coated SPIONs synthesized by the co-precipitation method reported an r2 value close to 400 mM−1 s−1.5,62 From these results, we can conclude that the samples reported here show excellent performances in terms of MRI contrast properties.The samples were also tested for MH therapy by measuring the SAR values on the diluted samples (1 mg mL−1) under different magnetic field conditions (Fig. 6C and D). As expected, the SAR values increased as the frequency and magnetic field amplitude increased. The SAR values for C-SPIONS were slightly higher than those of Rf-SPIONs (e.g. 580 ± 30 against 530 ± 20 W gFe−1 at 24 kA m−1 and 763 kHz). Nonetheless, deviations are within the experimental error, and the heating performance for both systems can be considered comparable. Both systems show performances well above those reported for NPs produced by co-precipitation approaches.60 This improvement is likely to be associated with the formation of a clustered or multicore structure.46,63 Even though the crystallographic orientation is not well-preserved throughout the entire core (Fig. 3), the TB values close to room temperature suggest the presence of magnetic coherence (Fig. 2F). This results in an enhancement of the hysteresis losses and hence an increase over the SAR values.
The performances could be compared with those of other multicore structures produced from similar approaches. For instance, a SAR value of 12 W gFe−1 was recorded for 20 nm clusters made of 9 nm cores at 100 kHz and 10 kA m−1.48 In this case, Rf-SPIONS showed a SAR value of 95 ± 20 W gFe−1 under slightly higher conditions (163 kHz and 24 kA m−1) which underlines an improvement of our systems to the reported values. However, the performances are still far below the values for iron oxide multicore NPs of 20 nm reported by Wilhelm et al. (2000 W gFe−1 at 700 kHz and 25 kA m−1).46 This difference could be the result of several factors. On the one hand, the measurements were performed at a much higher frequency. On the other hand, these multicore NPs were produced by a high-temperature hydrolysis approach, requiring long annealing times, (220 °C, 12 h) and showed spherical arrangement. In contrast, the domains in our SPIONs were produced under milder conditions and shorter times (90 °C, 1 h), hence, they appeared in random shapes.
Finally, it is worth underlining that performances between 85 and 95 W gFe−1 were achieved by applying an oscillating magnetic field with an amplitude of 24 kA m−1 and a frequency of 163 kHz, which is well below the biological limit (hν < 5 × 109 A m−1 s−1).64 These performances allow for efficient MH therapies at reasonably low SPION doses. Combined with excellent MRI responses, high colloidal stability and specific binding toward RCP of the Rf-SPIONs underline its potential as a theranostic agent for breast cancer therapy.
Cellular uptake
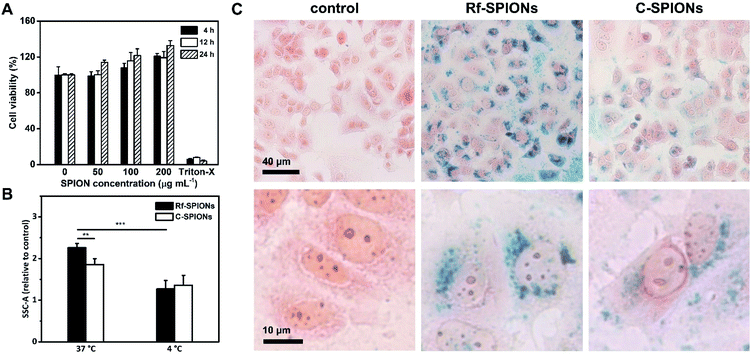
The MCF-7 cell line was chosen as a breast cancer cell model for evaluation of cellular uptake of Rf-SPIONs due to its retaining of estrogen response, making it a potent cell line for overexpression of RCP, which is an estrogen-regulated protein.65 Although the overexpression is not evidenced, MCF-7 cells have been used as a model in several studies regarding the uptake of Rf-functionalized NPs.25,31Biocompatibility of the Rf-SPIONs was ensured by MTT cell viability assay before the uptake experiments. Cell viability remained over 100% when treated at different concentrations, up to 200 μg mL−1 (Fig. 7A). As MTT assay measures cellular metabolism, the dose-dependent increase in cell viability could be due to the stimulating effect from the SPIONs on the cells. One possible mechanism is through reducing intracellular H2O2, which affects the cell cycle and promotes proliferation.66
To study the cellular uptake, cells were incubated with 200 μg mL−1 of Rf-SPIONs for 24 h. The same experiments were also carried out using C-SPIONs. Following incubation, cells were subjected to flow cytometry analysis where side scattering signals (SSC) were recorded. Plotting of SSC against forward scattering signals (FSC) is a reliable method to quantify the internalization level of the SPIONs in different cell lines.67,68 Rf-SPIONs showed significantly higher internalization into the cells at physiological temperature with an over 20% difference in cellular internalization (2.2 against 1.8 times the signal when compared to the control group, Fig. 7B).
The distribution of the internalized SPIONs was also observed by Prussian blue staining (Fig. 7C). After 24 h incubation, Rf-SPIONs showed notably higher internalization when compared to C-SPIONs as seen from the density of blue pigments. Furthermore, the images at higher magnification revealed that both Rf- and C-SPIONs tend to accumulate in perinuclear regions in vesicle-like distributions. It is worth mentioning that, with Rf-SPIONs, internalization was found in almost all the cells in the microscopic field. The same observation was observed from flow cytometry where the percentage of cells with higher SSC signals than the threshold value was 66% for cells treated with Rf-SPIONs, while for C-SPIONs, it was lower (Fig. S6†), which suggested that the Rf-SPIONs have a higher ability to internalize the cells.
Considering the results, the enhancement in cellular internalization is evidently due to the presence of the Rf moiety on the SPION surfaces. One possible mechanism is that the internalization is enhanced through binding interaction between Rf-citrate and RCP as demonstrated earlier. It is well established that cellular uptake of free Rf evolves from a saturable to a linear uptake kinetic profile when the temperature decreases from 37 to 4 °C.65 This is because the protein-mediated active transport mechanism is normally inhibited at low temperatures. As a result, both cellular uptake of Rf- and C-SPIONs decreased to a comparable level (Fig. 7B). The decrease percentage was calculated to be 40 and 26% for Rf- and C-SPIONs, respectively. This observation highlighted that the cellular uptake mechanism for Rf-SPIONs is mainly through a protein-assisted pathway which, considering their binding affinity, could be through binding with RCP. It is worth noting that at 4 °C, magnetic NP adhesion on the plasma membrane could be observed as demonstrated in HeLa cells.69 However, further investigations are needed to precisely clarify the internalization pathway.
Conclusions
We have successfully synthesized a novel Rf-based ligand by conjugation of Rf with citric acid which provides colloidal stabilization to the SPIONs, yielding aqueous-stabilized multicore SPION clusters. HRTEM analysis reveals some extent of preferred orientation between adjacent cores with good crystallinity, which results in high Ms and TB, showing both excellent MRI performance and MH response.We also demonstrated that the Rf-SPION system firmly binds to RCP, which is a carrier protein overexpressed in breast cancer tissues. Testing with MCF-7 breast cancer cells clearly showed the effect of Rf in enhancing cellular internalization, with high homogeneity. We hypothesize that the internalization was through the binding with RCP as the uptake level was decreased under reduced temperature conditions. However, extending systematic experiments is needed to clarify this mechanism of uptake. To this extent, the Rf-citrate ligand has been proven to provide both stabilization and breast cancer targeting effects to SPION clusters, which marks the potential of the system to be developed as a breast cancer theranostic agent.
Author contributions
W. M.: methodology, formal analysis, investigation, and writing the original draft. P. G.: formal analysis and writing the original draft. B. T. H.: investigation. J. M. F. methodology and resources. C. K.: methodology and supervision. A. R.: methodology, writing the original draft, supervision and review & editing. K. P. K.: conceptualization, methodology, supervision and review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This project was supported by Mahidol University under a New Discovery and Frontier Research Grant to K. P. K. This project is funded by National Research Council of Thailand (NRCT) and Mahidol University: N42A650357. Partial support was also provided by a Central Instrument Facility (CIF) grant and the Center for Nanoimaging, Center of Excellence for Innovation in Chemistry (PERCH-CIC), Ministry of Higher Education, Science, Research and Innovation, Faculty of Science, Mahidol University. W. M. was supported by the Science Achievement Scholarship of Thailand. The authors would like to thank Associate Professor Nuttawee Niamsiri, Department of Biotechnology, Faculty of Science, Mahidol University, for the use of the Zetasizer instrument. This research was also funded by the Spanish Government, through the “Severo Ochoa” Programme for Centers of Excellence in R&D (FUNFUTURE, CEX2019-000917 S) and DGA, through the “Fondo Social Europeo” (DGA E15_20R). B. T. H. is grateful for a predoctoral fellowship FPU (FPU19/01311). P. G. acknowledges financial support from the Spanish government (MICIU) through the Ramon y Cajal research program (RyC2019-028414-I).References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram, A. Jemal and F. Bray, Ca-Cancer J. Clin., 2021, 71, 209 CrossRef PubMed.
- M. Zeeshan, B. Salam, Q. S. B. Khalid, S. Alam and R. Sayani, Cureus, 2018, 10, e2448 Search PubMed.
- A. Bhushan, A. Gonsalves and J. U. Menon, Pharmaceutics, 2021, 13, 723 CrossRef CAS PubMed.
- A. Abbasi Kajani, S. Haghjooy Javanmard, M. Asadnia and A. Razmjou, ACS Appl. Bio Mater., 2021, 4, 5908 CrossRef CAS PubMed.
- E. Taboada, R. Solanas, E. Rodríguez, R. Weissleder and A. Roig, Adv. Funct. Mater., 2009, 19, 2319 CrossRef CAS.
- A. J. Lowery, M. R. Kell, R. W. Glynn, M. J. Kerin and K. J. Sweeney, Breast Cancer Res. Treat., 2012, 133, 831 CrossRef PubMed.
- C. L. Shapiro and A. Recht, N. Engl. J. Med., 2001, 344, 1997 CrossRef CAS PubMed.
- S. Kossatz, R. Ludwig, H. Dahring, V. Ettelt, G. Rimkus, M. Marciello, G. Salas, V. Patel, F. J. Teran and I. Hilger, Pharm. Res., 2014, 31, 3274 CrossRef CAS PubMed.
- S. Nigam, K. C. Barick and D. Bahadur, J. Magn. Magn. Mater., 2011, 323, 237 CrossRef CAS.
- K. Petcharoen and A. Sirivat, Mater. Sci. Eng., B, 2012, 177, 421 CrossRef CAS.
- E. C. Abenojar, S. Wickramasinghe, J. Bas-Concepcion and A. C. S. Samia, Prog. Nat. Sci.: Mater. Int., 2016, 26, 440 CrossRef CAS.
- D. Niculaes, A. Lak, G. C. Anyfantis, S. Marras, O. Laslett, S. K. Avugadda, M. Cassani, D. Serantes, O. Hovorka, R. Chantrell and T. Pellegrino, ACS Nano, 2017, 11, 12121 CrossRef CAS PubMed.
- M. Das, C. Mohanty and S. K. Sahoo, Expert Opin. Drug Delivery, 2009, 6, 285 CrossRef CAS PubMed.
- F. M. Kievit, Z. R. Stephen, O. Veiseh, H. Arami, T. Wang, V. P. Lai, J. O. Park, R. G. Ellenbogen, M. L. Disis and M. Zhang, ACS Nano, 2012, 6, 2591 CrossRef CAS PubMed.
- B. Smith, I. Lyakhov, K. Loomis, D. Needle, U. Baxa, A. Yavlovich, J. Capala, R. Blumenthal and A. Puri, J. Controlled Release, 2011, 153, 187 CrossRef CAS PubMed.
- K. Hayashi, K. Ono, H. Suzuki, M. Sawada, M. Moriya, W. Sakamoto and T. Yogo, ACS Appl. Mater. Interfaces, 2010, 2, 1903 CrossRef CAS PubMed.
- D. Smilowicz, J. C. Slootweg and N. Metzler-Nolte, Mol. Pharm., 2019, 16, 4572 CrossRef CAS PubMed.
- M. F. Attia, N. Anton, J. Wallyn, Z. Omran and T. F. Vandamme, J. Pharm. Pharmacol., 2019, 71, 1185 CrossRef CAS PubMed.
- M. Fernandez, F. Javaid and V. Chudasama, Chem. Sci., 2018, 9, 790 RSC.
- M. Darguzyte, N. Drude, T. Lammers and F. Kiessling, Cancers, 2020, 12, 295 CrossRef CAS PubMed.
- K. Thakur, S. K. Tomar, A. K. Singh, S. Mandal and S. Arora, Crit. Rev. Food Sci. Nutr., 2017, 57, 3650 CrossRef CAS PubMed.
- A. A. Karande, L. Sridhar, K. S. Gopinath and P. R. Adiga, Int. J. Cancer, 2001, 95, 277 CrossRef CAS PubMed.
- T. Johnson, A. Ouhtit, R. Gaur, A. Fernando, P. Schwarzenberger, J. Su, M. F. Ismail, H. I. El-Sayyad, A. Karande, Z. A. Elmageed, P. Rao and M. Raj, Front. Biosci., 2009, 14, 3634 CrossRef CAS PubMed.
- P. N. Rao, J. Crippin, E. Levine, J. Hunt, S. Baliga, L. Balart, L. Anthony, M. Mulekar and M. H. Raj, Hepatol. Res., 2006, 35, 83 CrossRef CAS PubMed.
- L. M. Bareford, B. R. Avaritt, H. Ghandehari, A. Nan and P. W. Swaan, Pharm. Res., 2013, 30, 1799 CrossRef CAS PubMed.
- N. Beztsinna, Y. Tsvetkova, M. Bartneck, T. Lammers, F. Kiessling and I. Bestel, Bioconjugate Chem., 2016, 27, 2048 CrossRef CAS PubMed.
- W. Mekseriwattana, A. Phungsom, K. Sawasdee, P. Wongwienkham, C. Kuhakarn, P. Chaiyen and K. P. Katewongsa, Photochem. Photobiol., 2021, 97, 1548 CrossRef CAS PubMed.
- J. Jayapaul, S. Arns, W. Lederle, T. Lammers, P. Comba, J. Gatjens and F. Kiessling, Biomaterials, 2012, 33, 8822 CrossRef CAS PubMed.
- J. Jayapaul, M. Hodenius, S. Arns, W. Lederle, T. Lammers, P. Comba, F. Kiessling and J. Gaetjens, Biomaterials, 2011, 32, 5863 CrossRef CAS PubMed.
- J. Jayapaul, S. Arns, M. Bunker, M. Weiler, S. Rutherford, P. Comba and F. Kiessling, Nano Res., 2016, 9, 1319 CrossRef CAS PubMed.
- Y. Tsvetkova, N. Beztsinna, J. Jayapaul, M. Weiler, S. Arns, Y. Shi, T. Lammers and F. Kiessling, Contrast Media Mol. Imaging, 2016, 11, 47 CrossRef CAS PubMed.
- X. Tang, D. A. Cronin and N. P. Brunton, J. Food Compos. Anal., 2006, 19, 831 CrossRef CAS.
- M. Torras, C. Moya, G. A. Pasquevich and A. Roig, Mikrochim. Acta, 2020, 187, 488 CrossRef CAS PubMed.
- L. Li, K. Y. Mak, C. W. Leung, K. Y. Chan, W. K. Chan, W. Zhong and P. W. T. Pong, Microelectron. Eng., 2013, 110, 329 CrossRef CAS.
- P. C. Hidber, T. J. Graule and L. J. Gauckler, J. Am. Ceram. Soc., 1996, 79, 1857 CrossRef CAS.
- M. E. de Sousa, M. B. Fernández van Raap, P. C. Rivas, P. Mendoza Zélis, P. Girardin, G. A. Pasquevich, J. L. Alessandrini, D. Muraca and F. H. Sánchez, J. Phys. Chem., 2013, 117, 5436 CAS.
- P. I. Soares, F. Lochte, C. Echeverria, L. C. Pereira, J. T. Coutinho, I. M. Ferreira, C. M. Novo and J. P. Borges, Nanotechnology, 2015, 26, 425704 CrossRef PubMed.
- A. Omelyanchik, F. G. da Silva, G. Gomide, I. Kozenkov, J. Depeyrot, R. Aquino, A. F. C. Campos, D. Fiorani, D. Peddis, V. Rodionova and S. Jovanović, J. Alloys Compd., 2021, 883, 160779 CrossRef CAS.
- A. Lak, M. Kraken, F. Ludwig, A. Kornowski, D. Eberbeck, S. Sievers, F. J. Litterst, H. Weller and M. Schilling, Nanoscale, 2013, 5, 12286 RSC.
- J. Mohapatra, F. Zeng, K. Elkins, M. Xing, M. Ghimire, S. Yoon, S. R. Mishra and J. P. Liu, Phys. Chem. Chem. Phys., 2018, 20, 12879 RSC.
- L. Gutierrez, L. de la Cueva, M. Moros, E. Mazario, S. de Bernardo, J. M. de la Fuente, M. P. Morales and G. Salas, Nanotechnology, 2019, 30, 112001 CrossRef CAS PubMed.
- B. D. Cullity and C. D. Graham, in Introduction to Magnetic Materials, 2008, p. 175, DOI:10.1002/9780470386323.ch6.
- V. L. Kirillov, D. A. Balaev, S. V. Semenov, K. A. Shaikhutdinov and O. N. Martyanov, Mater. Chem. Phys., 2014, 145, 75 CrossRef CAS.
- S. Rani and G. D. Varma, Phys. B, 2015, 472, 66 CrossRef CAS.
- C. Blanco-Andujar, D. Ortega, P. Southern, Q. A. Pankhurst and N. T. Thanh, Nanoscale, 2015, 7, 1768 RSC.
- L. Lartigue, P. Hugounenq, D. Alloyeau, S. P. Clarke, M. Levy, J. C. Bacri, R. Bazzi, D. F. Brougham, C. Wilhelm and F. Gazeau, ACS Nano, 2012, 6, 10935 CrossRef CAS PubMed.
- H. Gavilan, S. K. Avugadda, T. Fernandez-Cabada, N. Soni, M. Cassani, B. T. Mai, R. Chantrell and T. Pellegrino, Chem. Soc. Rev., 2021, 50, 11614 RSC.
- D. F. Coral, P. A. Soto, V. Blank, A. Veiga, E. Spinelli, S. Gonzalez, G. P. Saracco, M. A. Bab, D. Muraca, P. C. Setton-Avruj, A. Roig, L. Roguin and M. B. Fernandez van Raap, Nanoscale, 2018, 10, 21262 RSC.
- M. Ramazanov, A. Karimova and H. Shirinova, Biointerface Res. Appl. Chem., 2020, 11, 8654 Search PubMed.
- Q. Li, C. W. Kartikowati, S. Horie, T. Ogi, T. Iwaki and K. Okuyama, Sci. Rep., 2017, 7, 9894 CrossRef PubMed.
- K. M. Krishnan, A. B. Pakhomov, Y. Bao, P. Blomqvist, Y. Chun, M. Gonzales, K. Griffin, X. Ji and B. K. Roberts, J. Mater. Sci., 2006, 41, 793 CrossRef CAS.
- S. P. Yeap, J. Lim, B. S. Ooi and A. L. Ahmad, J. Nanopart. Res., 2017, 19, 368 CrossRef.
- A. Nelson, Y. Zong, K. E. Fritz, J. Suntivich and R. D. Robinson, ACS Mater. Lett., 2019, 1, 177 CrossRef CAS.
- P. M. Carvalho, M. R. Felicio, N. C. Santos, S. Goncalves and M. M. Domingues, Front. Chem., 2018, 6, 237 CrossRef PubMed.
- J. Lim, S. P. Yeap, H. X. Che and S. C. Low, Nanoscale Res. Lett., 2013, 8, 381 CrossRef PubMed.
- P. R. Adiga, S. S. Visweswariah, A. Karande and N. Kuzhandhaivelu, J. Biosci., 1988, 13, 87 CrossRef CAS.
- A. B. Witte, C. M. Timmer, J. J. Gam, S. K. Choi, M. M. Banaszak Holl, B. G. Orr, J. R. Baker and K. Sinniah, Biomacromolecules, 2012, 13, 507 CrossRef CAS PubMed.
- C. Moya, R. Escudero, D. C. Malaspina, M. de la Mata, J. Hernandez-Saz, J. Faraudo and A. Roig, ACS Appl. Bio Mater., 2019, 2, 3084 CrossRef CAS PubMed.
- M. Salehipour, S. Rezaei, J. Mosafer, Z. Pakdin-Parizi, A. Motaharian and M. Mogharabi-Manzari, J. Nanopart. Res., 2021, 23, 48 CrossRef CAS.
- Q. L. Vuong, J. F. Berret, J. Fresnais, Y. Gossuin and O. Sandre, Adv. Healthcare Mater., 2012, 1, 502 CrossRef CAS PubMed.
- E. D. Smolensky, H. Y. Park, Y. Zhou, G. A. Rolla, M. Marjanska, M. Botta and V. C. Pierre, J. Mater. Chem. B, 2013, 1, 2818 RSC.
- S. M. Dadfar, D. Camozzi, M. Darguzyte, K. Roemhild, P. Varvara, J. Metselaar, S. Banala, M. Straub, N. Guvener, U. Engelmann, I. Slabu, M. Buhl, J. van Leusen, P. Kogerler, B. Hermanns-Sachweh, V. Schulz, F. Kiessling and T. Lammers, J. Nanobiotechnol., 2020, 18, 22 CrossRef CAS PubMed.
- P. Hugounenq, M. Levy, D. Alloyeau, L. Lartigue, E. Dubois, V. Cabuil, C. Ricolleau, S. Roux, C. Wilhelm, F. Gazeau and R. Bazzi, J. Phys. Chem. C, 2012, 116, 15702 CrossRef CAS.
- R. Hergt and S. Dutz, J. Magn. Magn. Mater., 2007, 311, 187 CrossRef CAS.
- A. B. Foraker, C. M. Khantwal and P. W. Swaan, Adv. Drug Delivery Rev., 2003, 55, 1467 CrossRef CAS PubMed.
- D. M. Huang, J. K. Hsiao, Y. C. Chen, L. Y. Chien, M. Yao, Y. K. Chen, B. S. Ko, S. C. Hsu, L. A. Tai, H. Y. Cheng, S. W. Wang, C. S. Yang and Y. C. Chen, Biomaterials, 2009, 30, 3645 CrossRef CAS PubMed.
- M. Youhannayee, S. Nakhaei-Rad, F. Haghighi, K. Klauke, C. Janiak, M. R. Ahmadian, R. Rabenalt, P. Albers and M. Getzlaff, J. Magn. Magn. Mater., 2019, 473, 205 CrossRef CAS.
- R. P. Friedrich, C. Janko, M. Poettler, P. Tripal, J. Zaloga, I. Cicha, S. Durr, J. Nowak, S. Odenbach, I. Slabu, M. Liebl, L. Trahms, M. Stapf, I. Hilger, S. Lyer and C. Alexiou, Int. J. Nanomed., 2015, 10, 4185 CrossRef CAS PubMed.
- C. Wilhelm and F. Gazeau, Biomaterials, 2008, 29, 3161 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2na00015f |
| This journal is © The Royal Society of Chemistry 2022 |