 Open Access Article
Open Access ArticleSite-specific recognition of SARS-CoV-2 nsp1 protein with a tailored titanium dioxide nanoparticle – elucidation of the complex structure using NMR data and theoretical calculation†
Peter
Agback
*a,
Tatiana
Agback
 a,
Francisco
Dominguez
b,
Elena I.
Frolova
b,
Gulaim A.
Seisenbaeva
a,
Francisco
Dominguez
b,
Elena I.
Frolova
b,
Gulaim A.
Seisenbaeva
 a and
Vadim G.
Kessler
a and
Vadim G.
Kessler
 *a
*a
aDepartment of Molecular Sciences, Swedish University of Agricultural Sciences, Box 7015, SE-75007, Uppsala, Sweden. E-mail: peter.agback@slu.se; vadim.kessler@slu.se
bDepartment of Microbiology, University of Alabama at Birmingham, 1720 2nd Ave South Birmingham, AL 35294, USA
First published on 17th February 2022
Abstract
The ongoing world-wide Severe Acute Respiratory Syndrome coronavirus 2 (SARS-CoV-2) pandemic shows the need for new potential sensing and therapeutic means against the CoV viruses. The SARS-CoV-2 nsp1 protein is important, both for replication and pathogenesis, making it an attractive target for intervention. In this study we investigated the interaction of this protein with two types of titania nanoparticles by NMR and discovered that while lactate capped particles essentially did not interact with the protein chain, the aminoalcohol-capped ones showed strong complexation with a distinct part of an ordered α-helix fragment. The structure of the forming complex was elucidated based on NMR data and theoretical calculation. To the best of our knowledge, this is the first time that a tailored titanium oxide nanoparticle was shown to interact specifically with a unique site of the full-length SARS-CoV-2 nsp1 protein, possibly interfering with its functionality.
SARS-CoV-2 is a member of the Betacoronavirus (β-CoV) genus. Its genome is represented by a single-stranded RNA of ∼30 kb in length.1–3 The genomic RNA is directly translated into two very long polyproteins, which are further processed into the individual non-structural proteins nsp1 to nsp16 by the encoded protease activities.4 These proteins are viral components of the replication complexes, and also induce modification in the intracellular environment required for efficient viral replication. Nsp1 protein of coronaviruses is the major determinant of viral pathogenesis.5–7 It is known to efficiently inhibit cellular translation, to induce degradation of cellular mRNAs and, thus, to prevent the development of antiviral response.8–20 Nsp1 is also required for viral replication, buts its exact role remains elusive. These different roles make it an attractive target for development of therapeutic means against SARS-CoV-2 infection. Nsp1 is a relatively small protein (20 kDa) and contains a folded domain (aa 10 to 124) and two intrinsically disordered domains. The last 26-aa-long fragment of nsp1 is responsible for binding with the 40S ribosome subunit. Function and interactive partner of the folded domain remains unknown. Recently, structural analysis of SARS-CoV-2 nsp1 has been performed by our and other groups. Particularly, two X-ray structures of the folded domain (7K7P and 7K3N), and secondary structure determination of the full-length nsp1 based on NMR assignment of the backbone resonances have been published.21–24 In our previous study, we have also made partial side-chain assignment.24 Availability of the structural information allows the analysis of nsp1 interaction with cellular and viral proteins and small molecules, making it a useful model for revealing potential interaction mechanisms.
Metal oxide nanoparticles (NP), i.e., nanoparticles of common sand minerals, are abundant in ground water and watercourses, due to both physical and chemical erosion via dissolution and re-crystallization of ground minerals.25 Their surface can be rather chemically reactive, known to be able to produce or destroy reactive oxygen species by catalyzing redox reactions, especially in daylight via photochemical mechanisms.26 On the other hand, they are also known to interact strongly and specifically with biomolecules, especially proteins. Pristine NP are practically unknown in biological systems, because of the “protein corona” phenomenon, aiming to describe the assembly of protein molecules on the surface of larger NP. Smaller NP are suspected to interact specifically with selected fragments in the structure of e.g., blood proteins.27 The nature of proteins, specifically adsorbed by NP is related not only to their size, but, most importantly, to the chemical composition of the NP. The effects of interactions may be drastically different. Some NP, like those of alumina, Al2O3, Boehmite, AlO(OH), or zirconia, ZrO2, have been reported to catalyze hydrolysis of proteins, while other, like TiO2, were proven to cause protein coagulation.28–30 Molecular interactions between NP and proteins have until recently been very scarcely characterized. It has been noticed that proteins and NP can co-assemble into micro-sized colloid crystals.31 An attractive possibility to investigate such interactions would be the studies of complexes derived from individual poly oxo-metalate species (POMs). Importance of possible specific POM–protein interactions for potential biological applications has been outlined in a recent review.32 POMs can be considered as individual molecular species but are at the same time nanoparticles by definition as the size of, for example, Keggin POMs [PM12O40]3− (M = Mo, W and partially substituted for Ti, Zr, Nb) varies in the range 1.02–1.04 nm.32 Until recently, only very few structures of proteins with POMs were reported, suffering from rather poor resolution of the area surrounding often strongly disordered POM species.33
In this study we have applied nuclear magnetic resonance (NMR) spectroscopy in combination with tuning of the chemical composition and surface structure of titanium oxide nanoparticles (NP) to probe their affinity to the full-length of SARS-CoV-2 nsp1 protein.
For investigation of interaction with SARS-CoV-2 nsp1 protein we selected two kinds of well-characterized small titanium dioxide nanoparticles, possessing anatase structure.‡ One of them NP1 with hydrodynamic diameter 3.8 nm and zeta-potential −8.2 mV was obtained from industrially produced TiBALDH precursor and was proved to be terminated by lactate ligands,34i.e. inner-sphere surface complexation with chelate bonding on the surface as proved by NMR spectroscopy. Surface-capping with a chelating carboxylate ligand such as, for example, citrate or lactate, is a common approach to stabilization of colloid solutions of inorganic nanoparticles via enhanced surface charging (zeta-potential).35 The other material NP2 was produced from triethanolamine modified precursor by acidic hydrolysis and proved to contain particles with hydrodynamic diameter 3.2 nm and zeta-potential −11.4 mV.36 The measured negative potential was indicating that the ligand was in aqueous medium essentially desorbed from the particles, leaving them with surface protected by outer sphere ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) TiO− (HOC2H4)3NH+ complexes in statistics with electrically neutral
TiO− (HOC2H4)3NH+ complexes in statistics with electrically neutral ![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) TiOH sites. The zeta-potential of this second type of titania, −11.4 mV, was quite close to that of sol–gel TiO2 obtained by just hydrolysis with water of pure alkoxide precursor with subsequent peptization and equilibration with water, −11.7 mV,37 where the counter-ions usually are ammonium-cations. Both types of particles are forming clear colloid solutions stabilized by their double electric layers manifested by the measured zeta-potentials. The behavior of the particles upon titration with the protein turned rather different: while addition of NP1 in 1
TiOH sites. The zeta-potential of this second type of titania, −11.4 mV, was quite close to that of sol–gel TiO2 obtained by just hydrolysis with water of pure alkoxide precursor with subsequent peptization and equilibration with water, −11.7 mV,37 where the counter-ions usually are ammonium-cations. Both types of particles are forming clear colloid solutions stabilized by their double electric layers manifested by the measured zeta-potentials. The behavior of the particles upon titration with the protein turned rather different: while addition of NP1 in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio did not produce any measurable effect on the 1H-15N TROSY NMR spectrum of nsp1, that of NP2 resulted in very distinct signal perturbation for several amide resonances of the second half of the rigid α-helix fragment of nsp1 (L39, E41 – G49) as well as that of H110 (Fig. 1).
1 molar ratio did not produce any measurable effect on the 1H-15N TROSY NMR spectrum of nsp1, that of NP2 resulted in very distinct signal perturbation for several amide resonances of the second half of the rigid α-helix fragment of nsp1 (L39, E41 – G49) as well as that of H110 (Fig. 1).
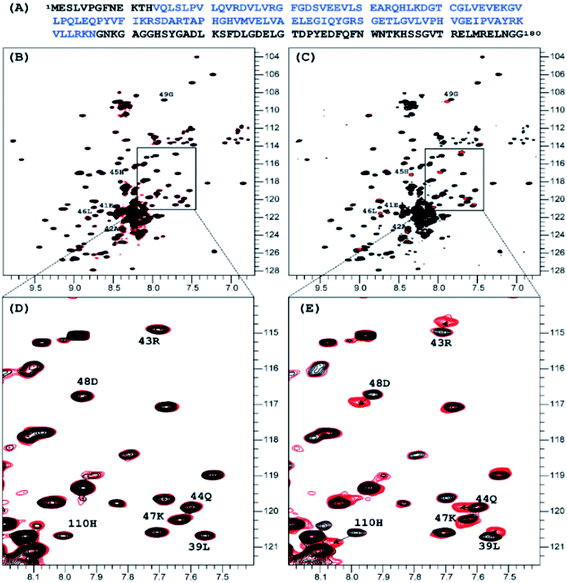 | ||
| Fig. 1 Sequence of the full length of SARS-CoV-2 nsp1 protein is presented on top in panel, in blue color is the sequence used in crystallization 7K7P (A). Superposition of the 1H-15N TROSY spectra of SARS-CoV-2 nsp1 protein apo form (black color) and apo with added nano particles (red color) are shown in panels (B)–(E). For the added NP1 particle, ratio nsp1 to NP1 is 1 to 10, spectra presented in (B) and its expanded part in (D) panels are identical to the apo form of nsp1 indicating that there is no interaction between NP1 and nsp1. For the added NP2 particle, ratio nsp1 to NP2 is 1 to 2, spectra presented in (C) and its expanded part in (E) panels are different. The chemical shift assignment of NH backbone resonances with induced chemical shift perturbation by binding of NP2 to nsp1 are shown by the number and symbols corresponding to the sequence (A). | ||
A further addition of NP1 to the sample to increase the ratio to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 did not result in any changes as well. Addition of NP2, to increase the ratio to 2
1 did not result in any changes as well. Addition of NP2, to increase the ratio to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 resulted in only minor changes for the amides mentioned earlier. The results clearly show site specific binding of NP2 to the α-helix of nsp1. This difference between the NPs originated most probably from the fact that the surface of NP1 was blocked by strongly bound chelate-forming lactate ligands, while that of NP2 remained highly reactive. The major driving force in the interaction of NP2 with the nsp1 protein was most likely the formation of inner sphere carboxylate complexes on the surface achieved by binding of the side chain carboxylate groups of aspartic and glutamic acids. The structure of such complexes always involves binding of the two oxygen atoms of the carboxylate groups to two adjacent Ti atoms connected via a double oxygen bridge Ti(μ-O)2Ti (see Fig. 2a).
1 resulted in only minor changes for the amides mentioned earlier. The results clearly show site specific binding of NP2 to the α-helix of nsp1. This difference between the NPs originated most probably from the fact that the surface of NP1 was blocked by strongly bound chelate-forming lactate ligands, while that of NP2 remained highly reactive. The major driving force in the interaction of NP2 with the nsp1 protein was most likely the formation of inner sphere carboxylate complexes on the surface achieved by binding of the side chain carboxylate groups of aspartic and glutamic acids. The structure of such complexes always involves binding of the two oxygen atoms of the carboxylate groups to two adjacent Ti atoms connected via a double oxygen bridge Ti(μ-O)2Ti (see Fig. 2a).
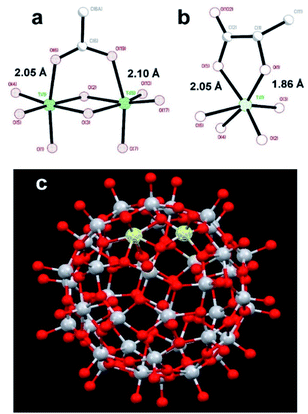 | ||
| Fig. 2 Molecular structures of (a) fragment, demonstrating the binding of a carboxylate ligand to TiO2 (anatase type) surface;38 (b) fragment showing commonly observed chelated bonding of a lactate ligand to a Ti atom;34 (c) the reported titanium-oxygen core in the structure of H6[Ti42(μ3-O)60(OiPr)42(OH)12] structure.39 A Ti(μ-O)2Ti fragment on the surface capable to binding a carboxylate group is highlighted. | ||
The attachment of the carboxylic group appears quite strong and is surprisingly uniform with T–O distances ca. 2.05 and ca. 2.10 Å respectively.38
This binding appears, however, to be substantially weaker than that of a lactate ligand. Lactate units, as revealed by the literature data, and the study of the new compound reported in this work, are always bound in a chelating mode to a single titanium atom.34 It features a Ti–O distance to a single-bound oxygen atom in the carboxylate typical for such fragments, ca. 2.05 Å (see Fig. 2a), and with a much shorter (and thus stronger) Ti–O bond to the alkoxide oxygen in the lactate structure of ca. 1.86 Å (see Fig. 2b). Inner sphere bonding corresponding to the mode typical for carboxylate binding is thus possible to a pair of adjacent Ti atoms connected via a pair of O-bridges as indicated in Fig. 2c. This may provide explanation for the absence of interaction between lactate terminated TiO2 and the protein. Still the binding of NP2 is complete, indicated by the fact that further addition did not induce any further chemical shift perturbations. Most likely other interactions than Ti–O are involved.
In order to provide closer molecular insight into the interaction of the NP2 material with SARS-CoV-2 nsp1 protein, we decided to investigate possibility of geometrical matching between a model TiO2 particle and the structure of the SARS-CoV-2 nsp1 protein reported from the X-ray single crystal study (7K7P).22 To represent the curved anatase structure we used the model of spherically shaped anatase monolayer in the recently reported H6[Ti42(μ3-O)60(OiPr)42(OH)12] structure.39 The cif-file was obtained from the ESI† to the publication and was lacking both carbon and hydrogen atom positions making it ideal for geometrical matching by using the program Chimera.40 It was converted into PDB-format and used for docking. What we observe in the 1H-15N TROSY spectra is of course chemical shift changes of the backbone amide groups. It is very unlikely that of the amides would directly interact with the nanoparticle without major structural changes of nsp1. Most likely these are the sidechains that interact with the titanium-oxide nanoparticle. One of possible docking configurations consistent with the NMR spectrum in Fig. 1 is displayed in Fig. 3. The conclusion that could be obtained from matching the surface geometries was that the TiO2 particle was experiencing a specific attraction resulting in inner sphere complex formation with one carboxylate fragment, originating from D48 and additional possible hydrogen bonding to the sidechains of E41 (carboxylate), Q44 (amide carbonyl) and H45 (Nδ1). The role of H110 is unclear, further studies are necessary to understand its role. The results of geometric matching are in good agreement with the observed changes in the NMR spectrum. This indicates that the structure of H6[Ti42(μ3-O)60(OiPr)42(OH)12] has a good resemblance with the structure of small anatase NP. One should note that the NP2 used in this study is larger than the nanoparticle used in the modelling with a presumably flatter surface making more interactions with the α-helix possible.
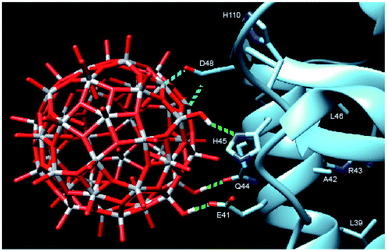 | ||
| Fig. 3 Model showing the docking of the metal–oxygen core of H6[Ti42(μ3-O)60(OiPr)42(OH)12] with the structure of nsp1 taken from 7K7P.22,39 The possible interactions are show in cyan: carboxylate of D48 with titanium, forming an inner sphere complex, and green: hydrogen bonding of OH-groups from the nanoparticle to E41, Q44 and H45. The amino acids of the other amides showing chemical shift perturbation are marked. | ||
The full-length nsp1 protein contains several structural elements: one α-helix, β-sheets, turns and one short and one long disordered domain. It is not fully clear as to why we only see site specific binding to the second half of the α-helix. There are 12 glutamic acids and 4 aspartic acids in the sequence, all presumably possible interaction points, if one assumes that the carboxylate binding is the main driving force. Most likely the distance between them and the possibility of other hydrogen bond partners are a good fit for the surface of the titanium oxide nanoparticle. It is not clear as to why H110 shows significant perturbation as well. The sidechain seems to be too far away for hydrogen bonding, but it might possibly be affected by changes in stacking interaction with H45 induced by the nanoparticle binding. Further investigations of the nature of this interaction are being planned. The studies will necessarily have to interweave the disciplines of inorganic chemistry and structural biology which has been considered to hold great interest in the future. As a starting point we are now looking to map the changes in the sidechains of the nsp1 protein upon nanoparticle binding by NMR spectroscopy. By this we hope to precisely define the nature of the interaction between the titanium oxide nanoparticle and the nsp1 protein. More detailed docking studies are also necessary to understand the interactions between the nanoparticle and the nsp1 protein. The calculation by either quantum or molecular mechanics of a protein structure together with a metal-oxide nanoparticle will be a challenging task.
It should be mentioned that antiviral activity of metal oxides has been traced recently in a number of cases and an effect of complex cerium oxide NP specifically against SARS-Cov-2 was observed.41,42 However, no mechanism of action for this have been suggested yet. Our reported finding in this study could provide a possible mechanism for this effect. The interest in titania nanoparticles studied in this work was motivated by their proved biocompatibility as they have earlier been reported in reversible bio-encapsulation of both bacterial and human single cells.36,43–45 Recent reports indicated that TiO2 particles with sizes of the range 3–5 nm, as those of NP1 and NP2 materials applied in this work, could improve penetration of the cell membrane for small molecule medicines.46 The observed effect implied thus a possibility for titania NP to enter the human cells, making nsp1 protein a potentially relevant model. The present study using the SARS-CoV-2 nsp1 as an example of a target, is, however, reporting only the chemical mechanism of interaction between oxide nanoparticles and proteins with a principal new insight that a nanoparticle can act as a specific ligand, binding to a protein. Nevertheless, in this stage of our research, it is too early to make any definite claims about actual possibility of their application as anti-viral medicines. The debate on potential adverse effects of nanoparticles is also continuing and has not reached a consensus yet. Our on-going study of the structure of protein and nanoparticle complexes will reveal the mechanism underlying their activity and can be developed into a tool for selective control of viral enzymes.
Conclusions
Possibility of site-specific binding of a metal oxide nanoparticle to a viral protein has been revealed in this work and possible chemical mechanism for such molecular recognition has been outlined using the data of NMR spectroscopy and theoretical calculation. This finding also shows that nanoparticles could be used as a tool for mapping of the structure-dynamic landscape of active proteins, especially if they could be tailor-made to find specific structural motifs.Possibility of specific geometrical matching with resulting strong binding between NP and the protein, opens prospects for the chemical tuning of NPs. This includes modifying the chemical composition and the surface structure of the NPs. The goal is then to make them a potential tool for blocking protein recognition sites with a broad area of possible applications ranging from new techniques of rapid detection of viral antigens to creating new anti-viral media, such as coatings, filters, and, in perspective, possibly even medicines.
Author contributions
All authors contributed actively to discussion and writing of the manuscript. GAS has performed synthesis of applied nanomaterials, FD and EIF contributed with production and purification of the nsp1 protein, PA and TA contributed with NMR studies and PA also with theoretical matching, applying Chimera program. VGK contributed with X-ray single crystal study of the model compound and final editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to express their gratitude to Swedish Research Council (Vetenskapsrådet) for support of the Grant No. 2018-03811, Molecular mechanisms in interaction of mineral nanoparticles and proteins. This work was also supported by the Swedish Foundation for Strategic Research, Grant ITM17-0218, Innovative Experimental Modeling of Dynamic Protein States. EIF acknowledges the support of NIH grant R21AI146969 and UAB Research Acceleration Funds.Notes and references
- P. V’Kovski, A. Kratzel, S. Steiner, H. Stalder and V. Thiel, Nat. Rev. Microbiol., 2021, 19, 155–170 CrossRef.
- I. Sola, F. Almazan, S. Zuniga and L. Enjuanes, Annu. Rev. Virol., 2015, 2, 265 CrossRef CAS.
- L. Enjuanes, F. Almazan, I. Sola and S. Zuniga, Annu. Rev. Microbiol., 2006, 60, 211 CrossRef CAS PubMed.
- B. Malone, N. Urakova, E. J. Snijder and E. A. Campbell, Nat. Rev. Mol. Cell Biol., 2022, 23, 21–39 CrossRef CAS PubMed.
- K. Nakagawa and S. Makino, Cells, 2021, 10, 300 CrossRef CAS PubMed.
- M. Thoms, R. Buschauer, M. Ameismeier, L. Koepke, T. Denk, M. Hirschenberger, H. Kratzat, M. Hayn, T. Mackens-Kiani, J. D. Cheng, J. H. Straub, C. M. Stürzel, T. Fröhlich, O. Berninghausen, T. Becker, F. Kirchhoff, K. M. J. Sparrer and R. Beckmann, Science, 2020, 369, 1249 CrossRef CAS PubMed.
- K. Schubert, E. D. Karousis, A. Jomaa, A. Scaiola, B. Echeverria, L. Gurzeler, M. Leibundgut, V. Thiel, O. Mühlemann and N. Ban, Nat. Struct. Mol. Biol., 2020, 27, 959 CrossRef CAS PubMed.
- M. G. Wathelet, M. Orr, M. B. Frieman and R. S. Baric, J. Virol., 2007, 81, 11620 CrossRef CAS PubMed.
- R. Zust, L. Cervantes-Barragán, T. Kuri, G. Blakqori, F. Weber, B. Ludewig and V. Thiel, PLoS Pathog., 2007, 3, e109 CrossRef PubMed.
- C. P. Lapointe, R. Grosely, A. G. Johnson, J. Wang, I. S. Fernandez and J. D. Puglisi, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2017715118 CrossRef CAS PubMed.
- A. Tidu, A. Janvier, L. Schaffer, P. Sosnowski, L. Kuhn, P. Hammann, E. Westhof, G. Eriani and F. Martin, RNA, 2021, 27, 253–264 CrossRef PubMed.
- T. Tanaka, W. Kamitani, M. L. DeDiego, L. Enjuanes and Y. Matsuura, J. Virol., 2012, 86, 11128 CrossRef CAS PubMed.
- W. Kamitani, C. Huang, K. Narayanan, K. G. Lokugamage and S. Makino, Nat. Struct. Mol. Biol., 2009, 16, 1134 CrossRef CAS.
- K. G. Lokugamage, K. Narayanan, C. Huang and S. Makino, J. Virol., 2012, 86, 13598 CrossRef CAS PubMed.
- C. Huang, K. G. Lokugamage, J. M. Rozovics, K. Narayanan, B. L. Semler and S. Makino, PLoS Pathog., 2011, 7, e1002433 CrossRef CAS PubMed.
- S. Yuan, L. Peng, J. J. Park, Y. X. Hu, S. C. Devarkar, M. B. Dong, Q. Shen, S. P. Wu, S. Chen, I. B. Lomakin and Y. Xiong, Mol. Cell, 2020, 80, 1055–1066 CrossRef CAS PubMed.
- K. Nakagawa, K. Narayanan, M. Wada, V. L. Popov, M. Cajimat, R. S. Baric and S. Makino, J. Virol., 2018, 92, e01157 CAS.
- W. Kamitani, K. Narayanan, C. Huang, K. Lokugamage, T. Ikegami, N. Ito, H. Kubo and S. Makino, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12885 CrossRef CAS PubMed.
- G. N. Gomez, F. Abrar, M. P. Dodhia, F. G. Gonzalez and A. Nag, Biochem. Cell Biol., 2019, 97, 758 CrossRef CAS PubMed.
- K. Zhang, L. Miorin, T. Makio, I. Dehghan, S. G. Gao, Y. H. Xie, H. L. Zhong, M. Esparza, T. Kehrer, A. Kumar, T. C. Hobman, C. Ptak, B. N. Gao, J. D. Minna, Z. J. Chen, A. García-Sastre, Y. Ren, R. W. Wozniak and B. M. A. Fontoura, Sci. Adv., 2021, 7, eabe7386 CrossRef CAS PubMed.
- C. Semper, N. Watanabe and A. Savchenko, iScience, 2021, 24, 101903 CrossRef CAS PubMed.
- L. K. Clark, T. J. Green and C. M. Petit, J. Virol., 2021, 95, e02019 CAS.
- Y. Wang, J. Kirkpatrick, S. zur Lage, S. M. Korn, K. Neißner, H. Schwalbe, A. Schlundt and T. Carlomagno, Biomol. NMR Assignments, 2021, 15(2), 287–295 CrossRef CAS PubMed.
- T. Agback, F. Dominguez, I. Frolov, E. I. Frolova and P. Agback, PLoS One, 2021, 16, e0251834 CrossRef CAS PubMed.
- H. Zhang and J. F. Banfield, Rev. Mineral. Geochem., 2001, 44, 1 CrossRef.
- Z. J. Yu, Q. Li, J. Wang, Q. H. Zhou and P. F. Li, Nanoscale Res. Lett., 2020, 15, 115 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050 CrossRef CAS PubMed.
- V. V. Vinogradov, A. V. Vinogradov, V. E. Sobolev, I. P. Dudanov and V. V. Vinogradov, J. Sol-Gel Sci. Technol., 2015, 73, 510 CrossRef.
- G. Absillis and T. N. Parac-Vogt, Inorg. Chem., 2012, 51, 9902 CrossRef CAS PubMed.
- G. A. Seisenbaeva, K. Fromell, V. V. Vinogradov, A. N. Terekhov, A. V. Pakhomov, B. Nilsson, K. N. Ekdahl, V. V. Vinogradov and V. G. Kessler, Sci. Rep., 2017, 7, 15448 CrossRef PubMed.
- M. A. Kostiainen, P. Hiekkataipale, A. Laiho, V. Lemieux, J. Seitsonen, J. Ruokolainen and P. Ceci, Nat. Nanotechnol., 2013, 8, 52 CrossRef CAS PubMed.
- N. I. Gumerova and A. Rompel, Interweaving Disciplines to Advance Chemistry: Applying Polyoxometalates in Biology, Inorg. Chem., 2021, 60, 6109 CrossRef CAS.
- A. Bijelic and A. Rompel, Coord. Chem. Rev., 2015, 22 CrossRef CAS PubMed.
- G. A. Seisenbaeva, G. Daniel, J. M. Nedelec and V. G. Kessler, Nanoscale, 2013, 5, 3330 RSC.
- H. Al-Johani, E. Abou-Hamad, A. Jedidi, C. M. Widdifield, J. Viger-Gravel, S. S. Sangaru, D. Gajan, D. H. Anjum, S. Ould-Chikh, M. N. Hedhili, A. Gurinov, M. J. Kelly, M. El Eter, L. Cavallo, L. Emsley and J. M. Basset, Nat. Chem., 2017, 9, 890–895 CrossRef CAS PubMed.
- V. G. Kessler, G. A. Seisenbaeva, S. Håkansson and M. Unell, Angew. Chem., 2008, 47, 8506 CrossRef CAS.
- A. V. Agafonov, D. A. Afanasyev, T. V. Gerasimova, A. S. Krayev, M. A. Kashirin, V. V. Vinogradov, A. V. Vinogradov and V. G. Kessler, ACS Sustainable Chem. Eng., 2016, 4, 2814 CrossRef CAS.
- G. A. Seisenbaeva, E. Ilina, S. Håkansson and V. G. Kessler, J. Sol-Gel Sci. Technol., 2010, 55, 1 CrossRef CAS.
- M. Y. Gao, F. Wang, Z. G. Gu, D. X. Zhang, L. Zhang and J. Zhang, J. Am. Chem. Soc., 2016, 138, 2556 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 13, 1605 CrossRef PubMed.
- T. Ito, K. Sunada, T. Nagai, H. Ishiguro, R. Nakano, Y. Suzuki, A. Nakano, H. Yano, T. Isobe, S. Matsushita and A. Nakajima, Mater. Lett., 2021, 290, 129510 CrossRef CAS PubMed.
- T. Yadavalli and D. Shukla, Nanomedicine, 2017, 13, 219 CrossRef CAS PubMed.
- S. H. Yang, E. H. Ko and I. S. Choi, Langmuir, 2012, 28(4), 2151–2155 CrossRef CAS PubMed.
- W. Youn, E. H. Ko, M. H. Kim, M. Park, D. Hong, G. A. Seisenbaeva, V. G. Kessler and I. S. Choi, Angew. Chem., Int. Ed., 2017, 56, 10702–10706 CrossRef CAS PubMed.
- H. Lee, N. Kim, H. B. Rheem, B. J. Kim, J. H. Park and I. S. Choi, Adv. Healthcare Mater., 2021, 10, 2100347 CrossRef CAS PubMed.
- N. V. Shatskaya, A. S. Levina, M. N. Repkova, S. I. Baiborodin, N. V. Shikinac, Z. R. Ismagilov and V. F. Zarytova, Nanotechnol. Russ., 2013, 8, 277–282 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Details on synthesis of applied nanoparticles, protein expression and purification, NMR experiments, theoretical calculations and results of X-ray single crystal studies of the model compound. See DOI: 10.1039/d1na00855b |
| ‡ Titania nanoparticles. Concentrated solution of TiBALDH, 50 wt% with respect to (NH4)8Ti4O4(OCOCHOCH3)8·4H2O was obtained from Sigma Aldrich and used for producing lactate-capped TiO2 NP by dilution with MilliQ water. The starting concentration of TiO2 was estimated according to established solution equilibrium as 45 mg mL.34 The starting solution of triethanolammonium capped TiO2 NP was obtained following the procedure described in ref. 36 by dissolving Ti(OEt)4 (5 mL) in anhydrous EtOH (5 mL), adding first 1.5 mL of triethanolamine and then 1 mL of hydrolyzing solution, prepared by mixing 0.5 M HNO3 (0.5 mL) with EtOH (2.0 mL). This procedure was resulting in a starting solution with concentration of TiO2 NP equal to 120 mg g−1 as confirmed earlier by TGA. Dilution was carried out with MilliQ water. Formal molar concentrations of TiO2 NP were calculated by dividing the TiO2 mass content in solution by estimated mass of a single particle, assuming the hydrodynamic sizes of lactate-capped TiO2 NP as spheres 3.8 nm and triethanolammonium capped TiO2 NP as 3.2 nm in diameter respectively.34,36 Both kinds of NP have earlier been proved to possess anatase structures. |
| This journal is © The Royal Society of Chemistry 2022 |
