Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Biotechnological synthesis of Pd-based nanoparticle catalysts
Christopher
Egan-Morriss
 a,
Richard L.
Kimber
a,
Richard L.
Kimber
 b,
Nigel A.
Powell
b,
Nigel A.
Powell
 c and
Jonathan R.
Lloyd
c and
Jonathan R.
Lloyd
 *a
*a
aDepartment of Earth and Environmental Sciences, Williamson Research Centre for Molecular Environmental Science, University of Manchester, UK. E-mail: jon.lloyd@manchester.ac.uk
bDepartment of Environmental Geosciences, Centre for Microbiology and Environmental Systems Science, University of Vienna, 1090, Vienna, Austria
cJohnson Matthey Technology Centre, Reading, RG4 9NH, UK
First published on 21st December 2021
Abstract
Palladium metal nanoparticles are excellent catalysts used industrially for reactions such as hydrogenation and Heck and Suzuki C–C coupling reactions. However, the global demand for Pd far exceeds global supply, therefore the sustainable use and recycling of Pd is vital. Conventional chemical synthesis routes of Pd metal nanoparticles do not meet sustainability targets due to the use of toxic chemicals, such as organic solvents and capping agents. Microbes are capable of bioreducing soluble high oxidation state metal ions to form metal nanoparticles at ambient temperature and pressure, without the need for toxic chemicals. Microbes can also reduce metal from waste solutions, revalorising these waste streams and allowing the reuse of precious metals. Pd nanoparticles supported on microbial cells (bio-Pd) can catalyse a wide array of reactions, even outperforming commercial heterogeneous Pd catalysts in several studies. However, to be considered a viable commercial option, the intrinsic activity and selectivity of bio-Pd must be enhanced. Many types of microorganisms can produce bio-Pd, although most studies so far have been performed using bacteria, with metal reduction mediated by hydrogenase or formate dehydrogenase enzymes. Dissimilatory metal-reducing bacteria (DMRB) possess additional enzymes adapted for extracellular electron transport that potentially offer greater control over the properties of the nanoparticles produced. A recent and important addition to the field are bio-bimetallic nanoparticles, which significantly enhance the catalytic properties of bio-Pd. In addition, systems biology can integrate bio-Pd into biocatalytic processes, and processing techniques may enhance the catalytic properties further, such as incorporating additional functional nanomaterials. This review aims to highlight aspects of enzymatic metal reduction processes that can be bioengineered to control the size, shape, and cellular location of bio-Pd in order to optimise its catalytic properties.
1. Introduction
Catalysts are vital in modern industry; they are responsible for 30% of Europe's gross domestic product, and are required in the processing of 80% of all manufactured products.1 Supported palladium nanoparticles are important catalysts that can catalyse a wide array of industrially important reactions, such as hydrogenation, hydrogenolysis, dehydrogenation, and C–C couplings such as Heck, Suzuki, and Sonogashira reactions.2–4 These reactions are used to synthesise fine chemicals, which are essential feed stocks for the pharmaceutical, agrochemical and lifestyle product industries. Some reactions can only be performed with a catalyst, and in many cases catalysts offer shortcuts in chemical synthesis routes that replace a significant number of stoichiometric steps. A vast amount of waste is produced from stoichiometric reactions, for example, a kilogram of product can also produce up to 100 kg of by products such as inorganic salts, metal hydrides and mineral acids.5 Palladium catalysis also has drawbacks however, such as the high cost of the metal and the need to avoid catalyst poisoning by sulfur, which can inhibit reactant surface adsorption at low degrees of surface coverage.6 Furthermore, the demand for palladium far outstrips the global supply, leading to high prices. Pd has applications as a catalyst in autocatalytic converters, and in the chemicals industry, as well as having uses in electronics, dental implants, and jewellery.7 Therefore technologies that enable the sustainable reuse of Pd are of utmost importance. In order to reduce waste and increase the sustainability of fine chemical production, there is an ongoing need for new catalysts with higher catalytic activity, better selectivity, longer life time, and minimal loss in activity with ageing.8,9Catalysts are broadly separated into two classes, homogeneous catalysts that exist in the same phase as the reactants they are catalysing and heterogeneous catalysts that exist in a separate phase. Homogeneous Pd catalysts typically consist of soluble Pd complexes stabilised in solution by a variety of ligands.10 Heterogeneous Pd catalysts are typically nanoparticles stabilised on a support material such as activated carbon, zeolites, metal oxides, earth metal salts, or polymers.11,12 Homogeneous Pd catalysts are often preferred for industrial applications, in part due to the stereoselectivity they provide, which cannot be obtained with heterogeneous catalysts. However, the ligands required whilst popular and efficient, are also toxic and/or expensive.13 Homogeneous catalysts can be difficult to separate from a reaction mixture, making their reuse or recycling challenging. The metal or ligand may end up in the final product, such as in pharmaceuticals, and must be removed.14 Heterogeneous catalysts are more sustainable; they can be separated more easily from a reaction system by simple filtration, enabling them to be reused several times, and in the absence of leaching, the product is not contaminated with transition metals or ligands.15 Supported Pd catalysts make up around 30% of the fine chemicals sector, and are most commonly used for hydrogenation reactions.16 However, supported Pd nanoparticles have limitations including lower (or absent) stereoselectivity and lower activity. This may be compensated for with higher reaction temperatures, but thermal degradation of the substrates or products may occur.17
Palladium metal nanoparticles produced by microorganisms (bio-Pd), under environmentally benign conditions, offer a potentially sustainable solution to the issues highlighted above. Bio-Pd is synthesised via enzymatic reduction at ambient temperature and pressure, simply requiring inexpensive buffer solutions and an electron donor, such as organic acids or hydrogen.18,19 The formation of metallic nanoparticles via enzymatic reduction from industrial waste solutions has been demonstrated, potentially revalorising these waste streams and allowing the recovery and reuse of precious metals.20,21 Microbial cells contain proteins and other biomolecules with high affinities for metals that help to prevent agglomeration, and control the size of nanoparticles without the need for toxic and expensive chemicals, such as chemical capping agents.22,23 Bio-Pd can also be reused several times whilst retaining catalytic activity and selectivity.24–26
Bacteria were first shown to bind cationic metal species via electrostatic interaction to their anionic cell surface in the common Gram-positive soil bacterium Bacillus subtilis.27 After binding metals to their surface, many different species of microbe are capable of reducing the metal ions to produce metal nanoparticles, for example from radionuclides, and transition metals.28 A two-step mechanism was proposed for the bioreduction of metals via bacterial cells; first the metal ion adsorbs to an active site on the cell surface, which then acts as a nucleation site for the reduction of metal ions from solution and their subsequent growth.29 Microbes generate the electrons for bioreduction from the enzymatic oxidation of an electron donor, and these electrons are transferred through cellular enzymatic pathways to a terminal reductase, where the metal species act as an electron acceptor.
Lloyd et al. first reported the bioreduction of soluble Pd(II) to form Pd(0) nanoparticles in the Gram-negative sulfate-reducing bacterium Desulfovibrio desulfuricans.18 Bio-Pd has since been produced by many different microbes, and its catalytic properties widely explored. Bio-Pd has performed comparably to commercial supported Pd catalysts, such as Pd/C, for a range of reactions, including hydrogenation reactions, C–C coupling reactions, and remediative reactions such as the reduction of Cr(VI) to Cr(III), or the dehalogenation of environmental pollutants.26,30–32 Recently, with the addition of precious and noble metals, microbially produced bimetallic nanoparticles have also been produced that possess improved catalytic properties over bio-Pd.33–36
Microbially supported metal nanoparticles are promising green catalysts, however, in order to be commercially viable they must be bioengineered to have higher catalytic activity and/or selectivity, be more durable, less expensive at scale, or catalyse a wider range of reactions than current commercially available catalysts.37 This review will aim to highlight the research into bio-Pd synthesis required to achieve these goals, and assess the catalytic properties achieved so far from microbial bionanocatalyst systems.
2. Design of supported palladium catalysts
2.1 Size and shape of nanoparticles
The electron configuration of Pd makes it a very stable catalyst, as reactants are able to bind to the metal surface and form intermediate states that can react to form the final product. Pd nanoparticles are excellent catalysts as they have a high proportion of highly reactive surface atoms. The particle size determines the ratio of surface-to-bulk metal atoms, with most industrial catalysts requiring metal nanoparticles in the 1–20 nm range to provide high enough specific surface areas.38 On the other hand, the particle shape is related to the types of facets on the particle surface, which affects their reactivity.39 Atoms with lower coordination numbers, i.e. fewer bonded atoms, are generally more reactive, and surface atoms tend to have a decreasing coordination number in the order of face > edge > vertex. For example, in fcc metals such as Pd, {111} facets have a coordination number of 9, {110} and {100} facets have coordination numbers of 8 and 7, and defects such as steps or kinks can have a coordination number of 6.40 Nanoparticles can catalyse reactions that generate multiple potential products. The selectivity towards a certain product is determined by reactants binding to the metal surface in a specific configuration that allows a certain atom in a molecule to react.Pd catalysed selective hydrogenations are important industrial reactions, for example converting alkynes to alkenes for the synthesis of polymers. Rosenmund, and later Lindlar, developed Pd catalysts poisoned with barium, lead or sulfur to lower their catalytic activity and increase their selectivity.41,42 Modern catalysts avoid using these toxic metals, instead focusing on the effects of particle size, the role of facets and atomic surface sites, and using modifiers to specify stereochemistry and enhance selectivity.43–45 For example, the hydrogenation of benzene on a Pt{111} single-crystal surface gave a mix of cyclohexane and cyclohexene, while only cyclohexene was generated on Pt{100}.46 Larger Pd nanoparticles (18 nm) and plane surface atoms dominated by {100} and {111} facets were shown to favour semi-hydrogenation; whereas smaller nanoparticles (6 nm) containing edge and corner sites favoured full hydrogenation.47 The hydrogenation of pentene was shown to experience a strong particle size effect, whereas ethene hydrogenation was structure insensitive.48 Therefore, although decreasing the particle size increases activity, selectivity may not be favourable, and controlling the size and monodispersity of bio-Pd, along with the shape and surface facets available, would be highly desirable to tailor its catalytic properties. Although some control over size has been established, fairly little characterisation of the shape of bio-Pd nanoparticles has yet been reported.26,49
The abiotic formation of nanoparticles from solution is generally divided into two steps; nucleation of seed particles, and their subsequent growth. The crystal structure of seed particles often controls the shape of nanoparticles, along with the capping agents used and the rate of atomic deposition.39 The size of nanoparticles depends on their growth and is often controlled by the type and concentration of capping agent and solvent, and the reduction rate.50 For example, shape and size control of Ag nanoparticles was achieved by altering the ratio between the growth rates of {100} and {111} planes using different seeds and types of capping agents.22 In terms of bio-Pd, the rate of metal bioreduction will depend on the type of organism and electron donor used. The Pd(II) complexes that adsorb to the cell in the initial biosorption step may act as seeds that contribute to the final size and shape of the nanoparticles. Cell biomolecules contain functional groups that bind metals,51 and peptides have been shown to act as capping agents that may direct or inhibit growth of nanoparticles, controlling their size and function.52,53 In addition, these biomolecules will be responsible for stabilising bio-Pd nanoparticles during multiple reaction cycles, as the adsorption of reaction species during catalytic operation can alter nanoparticle size. For example the average Pd crystallite size was shown to increase from 2.4 nm to 23 nm in 5% Pd/C, after use in the Heck reaction.54
2.2 Support material
The catalytic effect of supported metal nanoparticles is mainly governed by the chemical properties of the active metal phase. The support distributes the active phase over a large surface area, however, further interactions involving the support material must not be overlooked.55 Studies of chemically synthesised metal nanoparticles show that the support material can be critical to the efficiency and stability of the catalytic system, affecting the appearance of specific active sites at the metal–support boundary, variations in metal nanoparticle shape and crystal structure, and changes in the metal particle charging effects.56 Cooperative catalysis can occur between functional groups present on the support material that operate alongside the catalytic metal surface during a chemical reaction, assisting in the adsorption and desorption of reactants.57 For example, a hydrophilic carbon support enabled high selectivity for the hydrogenation of phenol to cyclohexanone by Pd nanoparticles.58,59 In contrast removing oxygen surface groups from various carbon materials assisted the selective hydrogenation of cinnamaldehyde towards cinnamyl alcohol for various platinum group metal (PGM) catalysts.60,61 The biomolecules within the cellular envelope that bind and stabilise bio-Pd nanoparticles contain a high density of functional groups, which may play a role in a given reaction. For example, in an early study by De Windt et al. the dechlorination of the hydrophobic polychlorinated biphenyl (PCB) 2,3,4-trichlorobiphenyl was most effectively catalysed by bio-Pd when large Pd(0) nanoparticles covered most of the hydrophilic cell surface.30 However, when nanoparticles up to 100-fold smaller were present, embedded within the hydrophilic cell surface, the PCB was not significantly degraded, possibly due to the lack of binding sites at the cell surface. Conversely, the opposite relationship was observed for the dechlorination of the anionic contaminant perchlorate, which presumably would find favourable binding sites on protonated cell surface functional groups.Other important aspects of heterogeneous catalysts are their recovery and reusability. The support material must allow separation from the reaction mixture; for carbon supports this means high attrition resistance in order to avoid generating extremely fine particles that make filtration.16 Bio-Pd can be simply filtered from reaction mixtures and reused, and may be more resistant to Pd nanoparticle aggregation on reuse than Pd/C.26 The cell support material is amenable to various processing techniques that could alter the cell surface chemistry, enhancing the support's role in certain reactions, and making the bionanoparticle more reusable (see Section 8).
3. Pd biosorption by microbes
Microbes are excellent biosorbents of metal species due to their high surface-to-volume ratios and high contents of potentially active chemisorption sites.62 The biosorption of precious metals by microbes, or biomaterials derived from them, has been of great interest to recover metals from waste solutions.63–65 Biosorption is the first step in the bioreduction process, and likely contributes to the size, shape and localisation of bio-Pd nanoparticles. Biosorption is the passive sequestration by biomass of metal species from solution; this is an abiotic process that occurs on cells whether live or dead.63 Bioaccumulation can follow in some examples, and is the metabolically active transport of metal species into the cell. Bioreduction of metal species to a potentially less mobile state may occur in the cytoplasm, at the cell surface, or extracellularly via secretion of metabolites.66 Pd(II) bioreduction also occurs abiotically such as in the presence of H2 or formate. Microbial cell surfaces contain a wide array of biomolecules such as chitin (in fungi), and peptidoglycan and lipopolysaccharide (in bacteria), which in turn contain many different functional groups that can bind metals.67,68 In addition, PGMs in solution can form an array of anionic, neutral, and cationic complexes depending on the pH, ligands, and ions in solution, all of which in turn affects the cell surface chemistry. Biosorption can therefore occur through different mechanisms, including ionic interactions such as ion exchange, electrostatic forces, hydrogen bonding or van der Waals forces, or through covalent interactions such as coordination.66Although the mechanism of Pd(II) biosorption can vary across microbial species, Pd(II) biosorption has been reported to occur at amine, carboxyl, hydroxyl and phosphoryl groups.69–75 The N and O atoms of these groups can act as ligands that coordinate to Pd(II). These functional groups can also become protonated, or deprotonated, with changing pH, which changes their ionic interactions with Pd(II) complexes. Many microbes display strongly pH-dependent Pd(II) biosorption due to ionic interactions. In the yeast Saccharomyces sp. optimal Pd(II) biosorption occurred at pH = 2–3, due to electrostatic interactions between anionic [PdCl4]2− and [PdCl3(H2O)]− complexes and protonated functional groups.76 However, the biosorption capacity was much lower outside this narrow pH range. At strongly acidic pH (pH < 2) biosorption decreased due to Cl− ions competing with [PdCl4]2− for protonated cell surface sites. At pH >3.5, hydrolysis of Pd(II) complexes leads to different aquachloro- (e.g. [PdCl3(H2O)]−) and chlorohydroxo- (e.g. [PdCl3(OH)]2−) complexes forming, which may have low affinity for the metal binding functional groups available. Similar biosorption behaviour was also observed for the alga Chlorella vulgaris, with optimal biosorption at pH = 1–2.77 On the other hand, in the fungus Aspergillus sp., optimal Pd(II) biosorption was recorded at pH = 4–11.76 Given that Pd(II) biosorption was mostly pH-independent, Pd(II) complexes likely formed covalent interactions, such as complexation, with the various functional groups in the fungal cell wall. The amine groups of chitosan, chitin and glucan, and amine groups more generally, have been shown to effectively complex Pd(II).68,78–81
Yong et al. showed that the choice of Pd(II) precursor impacted biosorption by D. desulfuricans. Biosorption of [PdCl4]2− was pH-dependent from pH = 1–7, suggesting an ionic biosorption mechanism, with optimal biosorption occurring at pH = 4. On the other hand, the biosorption of [Pd(NH3)4]2+ was pH-independent, maintaining around 20% biosorption across the measured pH = 1–7, suggesting a covalent mechanism.82 The pH would affect the cell surface charge in both cases. However, whereas pH would have a significant effect on the speciation of the aquachloropalladate complex formed, the tetraamine complex would be largely unaffected by pH. Bioreduction of [Pd(NH3)4]2+ was optimal at pH = 7, and the rate of Pd(II) reduction fell as the pH decreased.82 The reduction rate decreased faster at pH < 7 with formate as electron donor compared to H2, because H2 is a stronger abiotic reducing agent and formate driven reduction relies more on the cell's enzymatic bioreduction.
In an impressive study by Deplanche et al., seven different species of bacteria were assessed for their Pd(II) biosorption capacity, bioreduction, and the subsequent activity of the resulting bio-Pd in catalytic reactions.83 The biosorption capacity was strongly correlated to the bioreduction rate, and Gram-negative species possessed far superior Pd(II) biosorption capacity over the two Gram-positive species at the acidic pH (pH = 2.3) used in this study. Anionic Pd complexes would have been the dominant Pd(II) species under these conditions, which would be electrostatically attracted to protonated functional groups. Pd(II) reportedly coordinates to the carboxyl groups of the S-layer of Gram-positive bacteria, which may have been deteriorated significantly by the acidic pH,70 whereas coordination to the carboxyl and amine groups of peptidoglycan in the Gram-negative bacteria was less affected.83 Several studies have demonstrated that competing ions in solution can significantly decrease Pd(II) biosorption, and consequently bioreduction. [PdCl4]2− biosorption by D. desulfuricans was significantly lower in the presence of Cl− ions than NO3− or SO42− ions, from their respective acids at pH = 1–4.84 Increasing the Cl− concentration at pH = 2 roughly halved the biosorption capacity and significantly reduced the bioreduction rate; whereas increasing NO3− concentration had no effect on biosorption capacity or bioreduction rate.84,85 The effect of competing ions on biosorption of [Pd(NH3)4]2+ was not reported. However, the bioreduction rate of the ammonia complex was significantly reduced by increasing NO3− concentration, and was completely inhibited at 100 mM of Cl−.85 The solution chemistry therefore has a significant impact on Pd(II) biosorption and bioreduction; as has been shown to be the case for the bioreduction of other metals. For example, using different buffers drastically affected the products of Tc(VII) bioreduction by S. oneidensis.86
4. Bio-Pd from microbial bioreduction
Metal bioreduction is performed by many different microorganisms on a broad range of metal species, and contributes significantly to the biogeochemical cycling of trace elements. Metal-reducing bacteria couple the oxidation of an electron donor to the reduction of oxidized metal species, transferring electrons from an organic or inorganic substrate to the metal. In many examples, these processes support anaerobic metabolism and growth. Hydrogen is a ubiquitous electron donor in these bacteria, which use hydrogenase enzymes to catalyse the oxidation of H2 to form protons and electrons. Different groups of hydrogenases are classified by the metal atoms associated with their active centres, for example [NiFe]-hydrogenases have been identified to perform metal reduction.87 Formate is also a common electron donor, with formate dehydrogenase (FDH) enzymes used to catalyse the oxidation of formate to CO2 and H+. These enzymes can reduce metals directly, or in tandem, for example forming a formate hydrogenlyase (FHL) complex, as found in E. coli.88 An important development in the field has been the discovery and application of specialist dissimilatory metal-reducing bacteria (DMRB). In addition to possessing hydrogenase and FDH enzymes, DMRB are adapted to utilise a wider range of organic substrates as electron donors for metal reduction, and possess complex protein pathways to transport electrons from reactions in the cytoplasm to the outer cell surface.894.1 Bio-Pd from H2/formate oxidation
The bioreduction of Pd(II) to Pd(0) was first studied in the Gram-negative sulfate-reducing bacterium (SRB) Desulfovibrio desulfuricans, using hydrogen as the electron.18 Pd(0) particles were localised to the periplasm, the location of the [NiFe]-hydrogenase in this bacterium. Many other species of bacteria have since been shown to reduce Pd(II) using either formate or H2 as the electron donor (Table 1).| Bacteria | Electron donor | Proposed reduction pathway | Catalytic application | Reference |
|---|---|---|---|---|
| Arthrobacter oxidans | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Bacillus sphaericus | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 126 |
| Cupriavidus metallidurans | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Cupriavidus necator | Formate | FDH + hydrogenase + abiotic | H2 from hypophosphite | 97 |
| Desulfovibrio desulfuricans | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83, 94, 126 and 127 |
| Desulfovibrio vulgaris | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 127 |
| Enterococcus faecalis | Formate | FDH + hydrogenase + abiotic | Cr(VI) reduction | 128 |
| Escherichia coli (hydrogenase mutants) | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 91 |
| Escherichia coli | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Geobacter sulfurreducens | Formate | FDH + hydrogenase + abiotic | Cr(VI) reduction | 129 |
| Klebsiella oxytoca | Glucose | N/A | Azo dye reduction | 130 |
| Micrococcus luteus | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Paracoccus denitrificans | Formate | FDH + hydrogenase + abiotic | H2 from hypophosphite | 97 |
| Pseudomonas putida | Formate | FDH + hydrogenase + abiotic | H2 from hypophosphite | 97 |
| Serratia sp. | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Shewanella loihica PV-4 | Lactate | Omc (MtrCAB) | Azo dye reduction | 131 |
| Shewanella loihica PV-4 | Formate | FDH + hydrogenase + abiotic | Cr(VI) reduction | 120 |
| Shewanella oneidensis | Formate | FDH + hydrogenase + abiotic | 4-Nitrophenol reduction | 81 |
| Shewanella oneidensis | Formate | FDH + hydrogenase + abiotic | Perchlorate reduction | 30 |
| Shewanella oneidensis | H2 | Hydrogenase + abiotic | Cr(VI) reduction | 83 |
| Shewanella oneidensis | Formate | FDH + hydrogenase + abiotic | Reduction of nitrobenzene & 4-chlorophenol | 96 |
| Shewanella oneidensis | Lactate | Omc (MtrCAB) | 4-Nitrophenol reduction | 119 |
| Shewanella sp. CNZ-1 | Lactate | N/A | 4-Nitrophenol reduction | 119 |
Studies on gene knockout mutants have established the role of hydrogenases as a terminal reductase of Pd(II) in bacterial model systems. In the Gram-negative SRB D. fructosivorans, a wild-type strain was compared to a periplasmic [NiFe]-hydrogenase deletion mutant, a periplasmic [Fe]-hydrogenase deletion mutant, and a double deletion mutant missing both hydrogenases.90 All of the mutants reduced Pd(II) at a comparable rate to the wild type and deposited Pd(0) in the periplasm, with the exception of the [NiFe]-mutant, which deposited Pd(0) on the cytoplasmic side of the inner membrane, the location of the remaining hydrogenases. Pd was therefore shown to be deposited at the periplasmic site of the [NiFe]-hydrogenase enzyme, implicating it as the terminal reductase in Pd(II) bioreduction.90 Similarly, Pd(0) deposition was found to occur at the site of hydrogenases in knockout studies of E. coli, when using formate as the electron donor.91E. coli contains at least three [NiFe]-hydrogenases; Hyd-1 and Hyd-2 are periplasmic inner membrane-bound enzymes, whereas Hyd-3 is associated with a cytoplasmic FHL complex on the inner membrane.88 When only Hyd-3 was present bio-Pd was mostly located on the cytoplasmic side of the inner membrane. When double deletion mutants left only periplasmic Hyd-1 active in the reduction of Pd(II), bio-Pd was localised to, and spanned the width of, the periplasm. When only Hyd-2 was present bio-Pd was associated with the inner membrane. The mutant lacking all three hydrogenases did not appear to enzymatically reduce Pd(II), producing large extracellular Pd deposits that were likely due to abiotic reduction by formate.91 The parent strain and the mutant containing Hyd-1 both produced a 5% Pd/E. coli (wt/dry wt) catalyst with comparable activity to a 5% Pd/C commercial catalyst for the reduction of Cr(VI). In comparison the mutants without Hyd-1 produced bio-Pd with lower catalytic activity, potentially due to the bio-Pd being less accessible at the inner membrane.91
The choice of electron donor in bioreduction leads to bio-Pd with different nanostructures and catalytic properties. Three different SRBs produced bio-Pd with superior activity for the reductive dehalogenation of chlorinated aromatic compounds, when supplied with formate as electron donor over hydrogen.92 Intracellular bio-Pd nanoparticles were mainly crystalline icosahedrons enclosed by {111} facets when formate was supplied as electron donor. When hydrogen was supplied the intracellular particles were amorphous.49 Formate driven bio-Pd in E. coli was reported to be smaller and more dispersed than hydrogen-driven bio-Pd.93 A key reason for these differences, as discussed above, are that hydrogen and formate are metabolised differently by cells, and result in different Pd(II) bioreduction rates. The ability to control the size of bio-Pd nanoparticles is a necessary component in tailoring their catalytic properties.
One method for controlling the particle size of bio-Pd is to vary the concentrations of cells relative to the initial concentration of Pd(II); typically expressed as the cell dry weight (CDW) to Pd(II) ratio, or Pd loading of the cell as a weight percentage (wt/wt). Yong et al. demonstrated that increasing the concentration of biomass, or decreasing the initial Pd(II) concentration, increased the relative amount of Pd(II) biosorption to the cell.82 Mabbett and Mackaskie demonstrated that this metal loading affected the catalytic properties of bio-Pd. Pd/D. desulfuricans prepared with H2 as the electron donor at a 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CDW
1 CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio outperformed the same catalyst prepared at a 1
Pd ratio outperformed the same catalyst prepared at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CDW
1 CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio for the reduction of Cr(VI), reducing 90% and 70%, respectively, after 2 hours.94 The superior catalytic activity was attributed to smaller particle size, however Pd/D. desulfuricans prepared at a 10
Pd ratio for the reduction of Cr(VI), reducing 90% and 70%, respectively, after 2 hours.94 The superior catalytic activity was attributed to smaller particle size, however Pd/D. desulfuricans prepared at a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio displayed negligible Cr(VI) reduction. The total metal loading impacts on the nanoparticle dimensions, and cellular location. For example, Attard et al. noted that at a 20 wt% loading Pt/D. desulfuricans nanoparticles were mostly observed at the cell surface with a mean size of 4.5 nm. In contrast, at a 1 wt% loading Pt/D. desulfuricans nanoparticles were embedded further within the cell envelope, with a smaller mean size of 2.3 nm.95 Hou et al. also showed that the CDW
1 ratio displayed negligible Cr(VI) reduction. The total metal loading impacts on the nanoparticle dimensions, and cellular location. For example, Attard et al. noted that at a 20 wt% loading Pt/D. desulfuricans nanoparticles were mostly observed at the cell surface with a mean size of 4.5 nm. In contrast, at a 1 wt% loading Pt/D. desulfuricans nanoparticles were embedded further within the cell envelope, with a smaller mean size of 2.3 nm.95 Hou et al. also showed that the CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio affected the extracellular distribution of bio-Pd, which in turn impacted the catalytic activity of bio-Pd nanoparticles.96 Pd/S. oneidensis nanoparticles were synthesised using formate as the electron donor at five different CDW
Pd ratio affected the extracellular distribution of bio-Pd, which in turn impacted the catalytic activity of bio-Pd nanoparticles.96 Pd/S. oneidensis nanoparticles were synthesised using formate as the electron donor at five different CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratios: 6
Pd ratios: 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 3
1, 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 1
1, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 and 1
3 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6, which increased the average particle size from <10 nm to >50 nm. When tested as a suspension catalyst for the reduction of nitrobenzene and 4-chlorophenol, the smallest nanoparticles gave some of the lowest rates of reaction. The FDH and hydrogenase enzymes that reduce Pd(II) are periplasmic in S. oneidensis, therefore the smallest nanoparticles (<10 nm) synthesised at 6
6, which increased the average particle size from <10 nm to >50 nm. When tested as a suspension catalyst for the reduction of nitrobenzene and 4-chlorophenol, the smallest nanoparticles gave some of the lowest rates of reaction. The FDH and hydrogenase enzymes that reduce Pd(II) are periplasmic in S. oneidensis, therefore the smallest nanoparticles (<10 nm) synthesised at 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CDW
1 CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio were embedded in the periplasm and had the lowest cell surface coverage, exposed surface area, and extracellular distribution. The Pd nanoparticles synthesised at the 1
Pd ratio were embedded in the periplasm and had the lowest cell surface coverage, exposed surface area, and extracellular distribution. The Pd nanoparticles synthesised at the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 CDW
3 CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio gave the highest rates of reaction. These particles possessed a larger average size of 25.8 nm, but with high exposed surface area, extracellular distribution, and cell surface coverage, indicating that these properties were more favourable than size.96 At lower CDW
Pd ratio gave the highest rates of reaction. These particles possessed a larger average size of 25.8 nm, but with high exposed surface area, extracellular distribution, and cell surface coverage, indicating that these properties were more favourable than size.96 At lower CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratios there were fewer cell nucleation sites and the cells were exposed to more metal toxicity. The particles were larger and more often extracellular, as they nucleated in the periplasm then grew beyond the cell surface; in addition damaged cells would release enzymes providing additional extracellular nucleation sites, and abiotic reduction would occur. Similarly, Bunge et al. observed that increasing the biomass:Pd(II) ratio in Pd/Cupriavidus necator resulted in a higher fraction of periplasmic Pd(0) compared to extracellular Pd(0). This corresponded to a loss of catalytic activity in the production of hydrogen from hypophosphite.97 De Windt et al. observed that the highest CDW
Pd ratios there were fewer cell nucleation sites and the cells were exposed to more metal toxicity. The particles were larger and more often extracellular, as they nucleated in the periplasm then grew beyond the cell surface; in addition damaged cells would release enzymes providing additional extracellular nucleation sites, and abiotic reduction would occur. Similarly, Bunge et al. observed that increasing the biomass:Pd(II) ratio in Pd/Cupriavidus necator resulted in a higher fraction of periplasmic Pd(0) compared to extracellular Pd(0). This corresponded to a loss of catalytic activity in the production of hydrogen from hypophosphite.97 De Windt et al. observed that the highest CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio (10
Pd ratio (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) tested produced the smallest nanoparticles, mean size <20 nm, with the highest catalytic activity for perchlorate reduction.30 The perchlorate reduction rate fell sharply when catalysed by larger nanoparticles synthesised at lower CDW
1) tested produced the smallest nanoparticles, mean size <20 nm, with the highest catalytic activity for perchlorate reduction.30 The perchlorate reduction rate fell sharply when catalysed by larger nanoparticles synthesised at lower CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratios. Some authors have suggested that bio-Pd nanoparticles are susceptible to catalyst poisoning by sulfur containing amino acids, particularly in smaller particles prepared at higher CDW
Pd ratios. Some authors have suggested that bio-Pd nanoparticles are susceptible to catalyst poisoning by sulfur containing amino acids, particularly in smaller particles prepared at higher CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd.81,97 On the other hand, due to the relatively low proportion of sulfhydryl binding sites in live cells, Pd–S formation in bio-Pd may actually be relatively low and this could even contribute to enhanced catalytic activity or selectivity.98–101
Pd.81,97 On the other hand, due to the relatively low proportion of sulfhydryl binding sites in live cells, Pd–S formation in bio-Pd may actually be relatively low and this could even contribute to enhanced catalytic activity or selectivity.98–101
4.2 Bio-Pd in dissimilatory metal-reducing bacteria (DMRB)
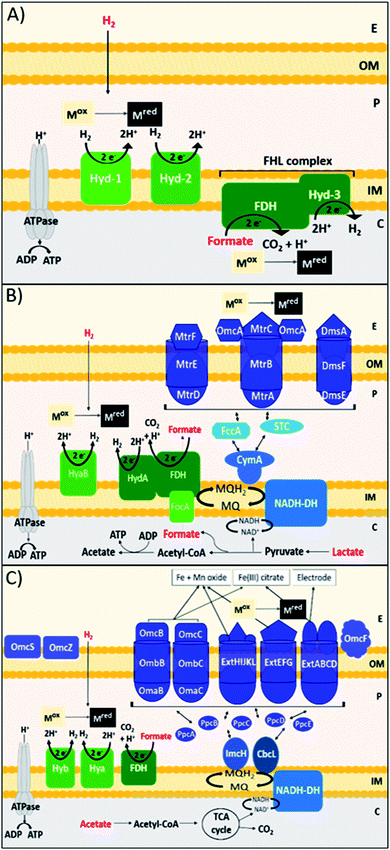
DMRB obtain energy for growth in anaerobic sedimentary environments, by coupling the oxidation of organic electron donors to the reduction of metal species.102 DMRB naturally utilise minerals, such as insoluble Fe(III) and Mn(IV) oxides, as terminal electron acceptors, and are directly involved in the biogeochemical cycling of carbon and other elements, including many metals.103 DMRB have been used to remediate metal and radionuclide contaminants, such as Cr(VI), U(VI), Tc(VII), from soil and aqueous environments, and can even be used in electricity production via the reduction of an anode surface in a microbial fuel cell.104–107 DMRB have been shown to reduce precious metal ions, such as Au(III), Ag(I), Pt(IV) and Pd(II), forming zerovalent metal nanoparticles.19,108–111 DMRB synthesise electron conductive pathways that run from the inner membrane to the surface of the outer membrane and beyond; these pathways are comprised primarily of c-type cytochromes.112 In terms of Pd(II) bioreduction, DMRB can oxidise a wide diversity of electron donors, and this can affect the reduction pathway, nanoparticle localisation, and the rate of reduction for a particular metal species, potentially affecting the properties of nanoparticles produced. Shewanella species are marine facultative anaerobic bacteria that thrive at redox interfaces.113Shewanella species perform the recovery and formation of nanoparticles of precious metals such as Au, Ag, Pt, Rh and In,108,114–117 and have produced catalytically active bio-Pd nanoparticles.30,96,118–120 A key model Shewanella species for study is Shewanella oneidensis MR-1, which utilises lactate, formate and hydrogen most effectively as electron donors for metal reduction, via reasonably well-understood metabolic pathways (Fig. 1B).121–123 H2 and formate consistently produce faster rates of metal reduction in S. oneidensis than lactate, through the periplasmic [NiFe]-hydrogenase hyaB, and FDH enzymes.106,124,125 When lactate is utilised as the electron donor, electrons are passed to the outer membrane through the Mtr pathway, in particular the MtrCAB conduit that spans the outer membrane.132,133S. oneidensis utilises these outer membrane cell surface c-type cytochromes, MtrC and OmcA, as well as extracellular nanowires, to transfer electrons to oxidised metal species.107,134,135 Another model genus of DMRB is Geobacter, exemplified by G. sulfurreducens, an obligatory anaerobic Gram-negative bacterium found in the subsurface.136G. sulfurreducens transfers electrons to extracellular metal species via a diverse array of c-type cytochrome protein pathways, as well as through conductive extracellular pili nanowires (Fig. 1C).137,138G. sulfurreducens commonly utilises H2, formate and acetate as electron donors for bioreduction.139De Windt et al. found that S. oneidensis is capable of reducing Pd(II) using H2, formate, lactate, pyruvate and ethanol as electron donors. H2 was the most effective electron donor resulting in complete removal of Pd(II) from solution with no significant decrease in the viability of cells. Formate and lactate were also effective, removing over 90% of Pd(II).19G. sulfurreducens has been shown to reduce Pd(II) using H2, formate, and acetate.111,129,140 Whereas H2 and formate driven reduction likely occurs due to the periplasmic [NiFe]-hydrogenase HyB, and periplasmic FDH enzymes,129,139,140 the outer membrane terminal reductase of acetate-driven Pd(II) bioreduction in G. sulfurreducens has not yet been identified. A transcriptional analysis found that many of the outer membrane cytochromes in G. sulfurreducens that are important in Fe(III) reduction, such as OmcB, OmcC, OmcS and OmcZ were downregulated during Pd(II) reduction.141–143
Studies have shown that metal nanoparticles can be reduced at both the inner and outer membranes of these bacteria. For example, in G. sulfurreducens the acetate driven bioreduction of Ag(I) produced Ag(0) nanoparticles at the outer membrane, whereas the H2 driven bioreduction of Au(III) produced Au(0) nanoparticles at the inner membrane.144 Dundas et al. used a S. oneidensis ΔHydA/ΔHyaB double hydrogenase mutant, with lactate as the electron donor, to confine bio-Pd to the outer membrane, whereas a ΔMtrC/ΔOmcA double deletion mutant confined bio-Pd to the periplasm.145 Yang et al. found that exposing S. oneidensis to the NADH-dehydrogenase inhibitor rotenone during formate driven Pd(II) bioreduction also confined bio-Pd to the periplasm.146 In general, periplasmic Pd(0) nanoparticles are smaller than outer membrane Pd(0) NPs, for example Dundas et al. measured periplasmic Pd(0) to be <25 nm, compared to extracellular particles that were mostly 30–60 nm.145 Therefore the apparent trade-off between nanoparticle localisation, size, and exposed surface area due to choice of electron donor requires further study.
Another factor to consider in DMRB is the use of electron shuttles. S. oneidensis can perform indirect electron transfer via the secretion of flavins, such as riboflavin (RF) and flavin mononucleotide (FMN), which enhance extracellular electron transfer.147,148 Similarly, Okamoto et al. have shown that G. sulfurreducens cells secrete and utilise flavins as a bound cofactor in outer membrane c-type cytochromes.149 Dundas et al. found that either RF or FMN caused the number of Pd nanoparticles at the outer membrane to increase and the Pd nanoparticle size to decrease, although the bioreduction rate was slightly slower than the parent strain.145 The electron shuttle anthraquinone 2,6-disulfonate (AQDS) has been utilised to enhance the rate of Pd(II) bioreduction in both S. oneidensis and G. sulfurreducens, producing smaller nanoparticles with enhanced catalytic activity.35,72,129
5. Industrial applications of bio-Pd
Pd/C is the most commonly used heterogeneous catalyst in industry, and recently the applications of Pd/C catalysts have been expanded.4 Bio-Pd shares some common features with abiotically produced Pd/C; both can be synthesised via the reduction of a Pd(II) salt in the presence of the support, both contain crystallites roughly in the size range 2 to >20 nm, and Pd may vary in oxidation state in both depending on the degree of reduction.15 With respect to catalyst testing, so far bio-Pd studies have focused on hydrogenation and C–C coupling reactions.5.1 Hydrogenation reactions
Hydrogenation reactions typically involve the addition of a pair of hydrogen atoms to a molecule via the reduction of double or triple bonds, which requires the presence of a catalyst. The chemistry is longstanding; Sabatier won the Nobel Prize in Chemistry in 1912 for using “finely disintegrated metals for the hydrogenation of organic compounds”,150 and currently, catalytic hydrogenations are responsible for the production of around 10–20% of industrial chemicals.151 Palladium nanoparticles are effective catalysts used for hydrogenation, selective hydrogenation, and hydrogenolysis reactions (Table 2) due to their high activity and hydrogen adsorption capacity.11 Industrially, catalytic hydrogenations are used to produce margarine,152 vitamins,153–155 and pharmaceuticals.156 Amongst many other examples, Pd/C catalysts are used in the N-debenzylation step of the synthesis of the anti-depressant nebivolol, and in the carbenzyloxy removal step of the antiviral drug valaciclovir.157| Bacteria | Electron donor | Proposed reduction pathway | Catalytic application | Reaction time (hours) | Conversion (%) | Yield (%) | Reference |
|---|---|---|---|---|---|---|---|
| Arthrobacter oxidans | H2 | Hydrogenase + abiotic | Selective hydrogenation of 2-butyne-1,4-diol | 4 | ∼75 | ∼99 | 160 |
| Bacillus sphaericus | H2 | Hydrogenase + abiotic | Hydrogenation of itaconic acid | 1 | 92.3 | n/a | 158 |
| Desulfovibrio desulfuricans | H2 | FDH + hydrogenase + abiotic | Dechlorination of 2-chlorophenol | 2 | ∼16 | n/a | 92 |
| 2 | ∼30 | ||||||
| Formate | Dechlorination of 2,3,4,5-tetrachlorobiphenyl | 2 | 10.4 | ||||
| Desulfovibrio desulfuricans | H2 | Hydrogenase + abiotic | Hydrogenation of itaconic acid | 1 | 93.3 | n/a | 158 |
| Hydrogenation of 4-azidoaniline | 3 | 84 | n/a | 126 | |||
| Selective hydrogenation of 3-nitrostyrene | 2 | 81 | 74 | 126 | |||
| Hydrogenolysis of 1-bromo-2-nitrobenzene | 2.25 | 10 | 10 | 126 | |||
| Selective hydrogenation of 2-butyne-1,4-diol | 4 | ∼90 | ∼80 | 159 | |||
| Selective hydrogenation of 2-pentyne | 5 | 100 | ∼55 | 25 | |||
| Upgrading crude bio-oil | 4 | 77 | n/a | 162 | |||
| Desulfovibrio vulgaris | H2 | FDH + hydrogenase + abiotic | Dechlorination of 2-chlorophenol | 2 | ∼9 | n/a | 92 |
| Formate | Dechlorination of 2,3,4,5-tetrachlorobiphenyl | 2 | ∼18 | ||||
| 2 | 3 | ||||||
| Desulfovibrio sp. ‘O7’ | H2 | FDH + hydrogenase + abiotic | Dechlorination of 2-chlorophenol | 2 | ∼16 | n/a | 92 |
| Formate | 2 | ∼15 | |||||
| Escherichia coli (bio-Pt) | H2 | Hydrogenase + abiotic | Selective hydrogenation of 2-butyne-1,4-diol | 5 | 100 | ∼10 | 163 |
| Selective hydrogenation of 2-methyl-3-butyn-2-ol | 3 | ∼55 | ∼38 | 161 | |||
| Selective hydrogenation of 4-octyne | 4.15 | ∼90 | ∼63 | ||||
| Selective hydrogenation of 2-pentyne | 100 | ∼71 | ∼60 | ||||
| Escherichia coli | H2 | Hydrogenase + abiotic | Selective hydrogenation of 2-pentyne | 0.83 | 100 | 55 | 164 |
| Selective hydrogenation of soybean oil | 5 | 45.5 | ∼41 | ||||
| 78.4 | |||||||
| Rhodobacter capsulatus | H2 | Hydrogenase + abiotic | Selective hydrogenation of 2-butyne-1,4-diol | 5 | 100 | ∼65 | 160 |
| Shewanella oneidensis | Formate | FDH + hydrogenase + abiotic | Dechlorination of 2,3,4-trichloro biphenyl | 5 | 100 | 32 | 30 |
| Dechlorination of lindane (γ-hexachlorocyclohexane) | 24 | 100 | 100 | 165 |
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond, whereas Pd/C reduced the –NO2 group. In a follow-up study, Pd/D. desulfuricans gave superior selectivity over a 5% Pd/Al2O3 (wt/wt) commercial catalyst for the selective hydrogenation of 2-butyne-1,4-diol, performed in 2-propanol.159 A 20% Pd/D. desulfuricans (wt/dry wt) catalyst converted over 80% of the alkyne to the desired alkene product, 2-butene-1,4-diol. In comparison, the commercial catalyst produced over 80% of the undesired alkane product, 2-butane-1,4-diol. Interestingly, 5% Pd/D. desulfuricans converted less of the starting material and produced less alkene (∼60%) than 20% Pd/D. desulfuricans, both at a catalyst loading of 0.205 mol% Pd.159 The same reaction was catalysed by bio-Pd from the Gram-positive soil bacterium Arthrobacter oxidans and the Gram-negative photosynthetic bacterium Rhodobacter capsulatus.160 The bio-Pd samples achieved maximum selectivities of 0.98 and 1.0 for 5% Pd/A. oxidans and 5% Pd/R. capsulatus, respectively. A 5% Pd/Al2O3 commercial catalyst gave a fast rate of reaction but was not selective, achieving 0.67 selectivity at 73% conversion, compared to ∼0.85 for 5% Pd/A. oxidans at the same conversion.160 The hydrogenation rate in bio-Pd may have been slower due to the mass transfer limitations of reactants diffusing through the outer membrane, which may have helped to prevent overhydrogenation. In addition, 25% Pd/A. oxidans gave a slower reaction rate but higher selectivity at 100% conversion than 5% Pd/A. oxidans, which was likely due to size effects. Larger bio-Pd particles at the higher metal loading would have lower activity, possessing fewer low coordination number sites such as terraces and edges, preventing overhydrogenation. Bio-Pt nanoparticles on E. coli were shown to possess comparable selectivity to a reduced Pt/C catalyst for the selective hydrogenation of various alkynes.161 In the selective hydrogenation of 2-butyne-1,4-diol using a 20% Pt/E. coli (wt/dry wt) catalyst, the butenediol selectivity was increased significantly when the bio-Pt was chemically processed to remove most of the biomass, although the reaction rate fell considerably.163 These results suggest that removing biomass from the nanoparticles may liberate defect sites on the metal surface that favour formation of the alkene product. Bio-Pd was also investigated for the selective hydrogenation of 2-pentyne, where it displayed superior selectivity over a commercial Pd/Al2O3 catalyst. Pd/D. desulfuricans retained a higher selectivity for 2-pentene than Pd/Al2O3 at alkyne conversions above 70%, although 5% Pd/D. desulfuricans (wt/dry wt) only produced 30% of the reaction rate of 5% Pd/Al2O3 (wt/wt).25 At 92.5% alkyne conversion, 5% Pd/D. desulfuricans gave a pentene/pentane ratio of 3.3 and a cis/trans ratio of 2.5. In comparison, Pd/Al2O3 achieved lower ratios of 2.0 and 2.0, respectively. At a higher Pd(II) loading, 25% Pd/D. desulfuricans achieved a faster rate of reaction and higher selectivity than 5% Pd/D. desulfuricans. Alumina may adsorb excess hydrogen that acts as a local hydrogen reservoir, causing the overhydrogenation of 2-pentene to pentane.166 Cell biomass, on the other hand, may be more inert to hydrogen adsorption and would not experience this affect.
C bond, whereas Pd/C reduced the –NO2 group. In a follow-up study, Pd/D. desulfuricans gave superior selectivity over a 5% Pd/Al2O3 (wt/wt) commercial catalyst for the selective hydrogenation of 2-butyne-1,4-diol, performed in 2-propanol.159 A 20% Pd/D. desulfuricans (wt/dry wt) catalyst converted over 80% of the alkyne to the desired alkene product, 2-butene-1,4-diol. In comparison, the commercial catalyst produced over 80% of the undesired alkane product, 2-butane-1,4-diol. Interestingly, 5% Pd/D. desulfuricans converted less of the starting material and produced less alkene (∼60%) than 20% Pd/D. desulfuricans, both at a catalyst loading of 0.205 mol% Pd.159 The same reaction was catalysed by bio-Pd from the Gram-positive soil bacterium Arthrobacter oxidans and the Gram-negative photosynthetic bacterium Rhodobacter capsulatus.160 The bio-Pd samples achieved maximum selectivities of 0.98 and 1.0 for 5% Pd/A. oxidans and 5% Pd/R. capsulatus, respectively. A 5% Pd/Al2O3 commercial catalyst gave a fast rate of reaction but was not selective, achieving 0.67 selectivity at 73% conversion, compared to ∼0.85 for 5% Pd/A. oxidans at the same conversion.160 The hydrogenation rate in bio-Pd may have been slower due to the mass transfer limitations of reactants diffusing through the outer membrane, which may have helped to prevent overhydrogenation. In addition, 25% Pd/A. oxidans gave a slower reaction rate but higher selectivity at 100% conversion than 5% Pd/A. oxidans, which was likely due to size effects. Larger bio-Pd particles at the higher metal loading would have lower activity, possessing fewer low coordination number sites such as terraces and edges, preventing overhydrogenation. Bio-Pt nanoparticles on E. coli were shown to possess comparable selectivity to a reduced Pt/C catalyst for the selective hydrogenation of various alkynes.161 In the selective hydrogenation of 2-butyne-1,4-diol using a 20% Pt/E. coli (wt/dry wt) catalyst, the butenediol selectivity was increased significantly when the bio-Pt was chemically processed to remove most of the biomass, although the reaction rate fell considerably.163 These results suggest that removing biomass from the nanoparticles may liberate defect sites on the metal surface that favour formation of the alkene product. Bio-Pd was also investigated for the selective hydrogenation of 2-pentyne, where it displayed superior selectivity over a commercial Pd/Al2O3 catalyst. Pd/D. desulfuricans retained a higher selectivity for 2-pentene than Pd/Al2O3 at alkyne conversions above 70%, although 5% Pd/D. desulfuricans (wt/dry wt) only produced 30% of the reaction rate of 5% Pd/Al2O3 (wt/wt).25 At 92.5% alkyne conversion, 5% Pd/D. desulfuricans gave a pentene/pentane ratio of 3.3 and a cis/trans ratio of 2.5. In comparison, Pd/Al2O3 achieved lower ratios of 2.0 and 2.0, respectively. At a higher Pd(II) loading, 25% Pd/D. desulfuricans achieved a faster rate of reaction and higher selectivity than 5% Pd/D. desulfuricans. Alumina may adsorb excess hydrogen that acts as a local hydrogen reservoir, causing the overhydrogenation of 2-pentene to pentane.166 Cell biomass, on the other hand, may be more inert to hydrogen adsorption and would not experience this affect.
In a following study, 2% Pd/E. coli (wt/dry wt) achieved a cis/trans 2-pentene ratio of 2.8 at 100% 2-pentyne conversion, compared to 0.7 for a 2% Pd/Al2O3 commercial catalyst.164 A 5% Pd/E. coli catalyst gave a threefold higher rate of reaction than 2% Pd/E. coli, but with a lower cis/trans ratio of 1.6. The hydrogenation rate constant of the C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C was calculated to be 7.6 times higher than of the C
C was calculated to be 7.6 times higher than of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond, and 2-pentyne adsorption was measured to be much stronger than 2-pentene adsorption to the Pd surface.
C bond, and 2-pentyne adsorption was measured to be much stronger than 2-pentene adsorption to the Pd surface.
Zhu et al. also investigated the solvent-free selective hydrogenation of soybean oil by bio-Pd.164 A 5 wt% Pd/E. coli catalyst required over 5 hours to convert around 50% of the reactant oil substrate, whereas 5 wt% Pd/Al2O3 converted over 60% in 1 hour. The Pd/Al2O3 catalyst reached a peak concentration of 1.07 mol L−1 of the desired alkene product, cis-C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, but this fell to less than half by the end of the reaction, with the major product being the fully hydrogenated alkane. On the other hand, 5% Pd/E. coli recorded a peak concentration of 1.03 mol L−1 of cis-C18
1, but this fell to less than half by the end of the reaction, with the major product being the fully hydrogenated alkane. On the other hand, 5% Pd/E. coli recorded a peak concentration of 1.03 mol L−1 of cis-C18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which did not fall throughout the reaction, and only low concentrations of undesired products were produced. When 5% Pd/D. desulfuricans was tested for the same reaction it produced very similar hydrogenation products to Pd/E. coli.
1, which did not fall throughout the reaction, and only low concentrations of undesired products were produced. When 5% Pd/D. desulfuricans was tested for the same reaction it produced very similar hydrogenation products to Pd/E. coli.
5.2 C–C coupling reactions
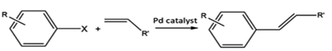
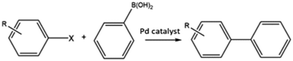
C–C bond forming coupling reactions have become an indispensable tool in organic synthesis and have led to a step change in the synthesis of complex drug molecules, enabling the parallel synthesis of key precursor components that can be coupled together at a later stage.171 Heck and Mizoroki discovered the coupling of aryl or vinyl halides with activated alkenes in the presence of a base, forming a C–C bond to produce a substituted alkene (Scheme 1).172,173 The Heck reaction has been used in industrial applications to synthesise products such as the anti-inflammatory drug naproxen,174 the asthma drug Singulair175 and the herbicide prosulfuron.176 The Suzuki–Miyaura C–C coupling reaction was later developed in 1979, which couples a boronic acid or ester with organic halides to produce a biaryl derivative (Scheme 2).177 The Suzuki reaction has also been used extensively on an industrial scale, such as to manufacture the fungicide Boscalid,178 and for a key step in the natural product (+)-discodermolide, a potent cancer cell growth inhibitor.179Although Heck used homogeneous Pd(II) acetate as his original catalyst, it was noted that heterogeneous Pd/C catalysts were active for the reaction, albeit with lower rate and yields; later studies confirmed this and showed some activity for aryl chlorides.180 Pd/C catalysts with high activity for Heck reactions of nonactivated bromobenzene were reported, with optimized conditions allowing Pd concentrations as low as 0.005 mol%.54 The authors found that high Pd dispersion, low degree of Pd reduction, high water content, and uniform Pd impregnation combined to give the most active Pd/C catalyst. Pd/C was first shown to catalyse Suzuki coupling reactions by Marck et al.181 and have been reported for the construction of biaryl derivatives in good to excellent yields, and with good reusability.182,183 The C–C coupling of aryl bromides was demonstrated in Heck, Suzuki, and Sonogashira couplings.184
| Microbe | Electron donor | Proposed reduction pathway | Aryl halide | Coupling partner | Reaction time (hours) | Conversion (%) | Reference |
|---|---|---|---|---|---|---|---|
| Arthrobacter oxidans | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | 51 | 83 |
| Styrene | 4 | ∼65 | |||||
| Cupriavidus metallidurans | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | ∼70 | 83 |
| Styrene | 4 | ∼70 | |||||
| Cupriavidus necator | H2 or formate | FDH + hydrogenase + abiotic | Iodobenzene | n-Butyl acrylate | 12 | 97 | 24 |
| 4-Iodobenzonitrile | 24 | 100 | |||||
| 4-Chloro-iodobenzene | 24 | 100 | |||||
| 4-Iodobenzaldehyde | 24 | 88 | |||||
| 4-Iodotoluene | 24 | 98 | |||||
| Methyl 4-iodobenzoate | 24 | 86 | |||||
| 2-Iodobenzaldehyde | 24 | 86 | |||||
| 5-Iodo-2-methoxybenzoate | 24 | 81 | |||||
| 1-Iodo-4-methoxybenzene | 24 | 96 | |||||
| 4-Bromobenzonitrile | 24 | 97 | |||||
| 1-Bromo-4-nitrobenzene | 24 | 88 | |||||
| 1-Bromo-4-(trifluoromethyl)benzene | 24 | 53 | |||||
| Desulfovibrio desulfuricans | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | 98 | 26, 83 and 159 |
| 5 | 96.4 | ||||||
| Styrene | 2 | ∼80 | |||||
| 4 | 80 | ||||||
| Escherichia coli | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | 78 | 83 |
| Styrene | 4 | 75 | |||||
| 4-Bromoacetophenone | Ethyl acrylate | 5 | 54 | ||||
| 3-Chlorotoluene | Ethyl acrylate | 5 | 0 | ||||
| Micrococcus luteus | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | ∼65 | 83 |
| Styrene | 4 | ∼65 | |||||
| Pseudomonas putida | Formate | FDH + hydrogenase + abiotic | Iodobenzene | n-Butyl acrylate | 24 | 99 | 24 |
| 1-Iodo-4-methoxybenzene | 24 | 96 | |||||
| 4-Bromobenzonitrile | 24 | 88 | |||||
| Phanerochaete chrysosporium | None | Putative fungal enzymes | Iodobenzene | Styrene | 3 | 95 | 68 |
| Serratia sp. | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | ∼85 | 83 |
| Styrene | 4 | ∼65 | |||||
| Shewanella oneidensis | H2 | Hydrogenase + abiotic | Iodobenzene | Ethyl acrylate | 2 | >90 | 83 |
| Styrene | 4 | 60 |
Søbjerg et al. tested bio-Pd as a catalyst for the Heck couplings of various aryl halides with n-butylacrylate (Table 3) in the presence of DMF and Na2CO3, at a catalyst loading of <1 mol%, at 80 °C for 12–24 hours.24 Bio-Pd was synthesised on cells of two Gram-negative Proteobacteria, Cupriavidus necator, which formed mostly small, well dispersed particles <10 nm, and Pseudomonas putida, which formed large Pd aggregates >100 nm. Both bio-Pd catalysts successfully converted activated and nonactivated aryl iodides in high yields, performing comparably to a commercial Pd nanopowder (<25 nm). Neither bio-Pd catalyst catalysed the coupling of non-activated aryl bromides or activated aryl chlorides. The significant size differences between Pd/C. necator and Pd/P. putida affected their activities for the Heck reaction, achieving conversions of 97% and 88%, respectively, for the coupling of the activated aryl bromide 4-bromobenzonitrile with n-butylacrylate. It is worth noting the reaction was only catalysed with the addition of tetrabutylammonium bromide (TBAB), which can stabilise Pd nanoparticles in solution and acts as a phase transfer catalyst.24 The requirement of TBAB indicates active Pd species may leach into solution from bio-Pd, a common mechanism for supported Pd catalysts in Heck and Suzuki reactions.12
Deplanche et al. showed that the species of bacteria had a significant effect on the ability of bio-Pd to catalyse the Heck coupling of phenyl iodide with styrene, and with ethylacrylate.83 Gram-negative species generally outperformed Gram-positive species, with Pd/E. coli, Pd/D. desulfuricans, and Pd/C. metallidurans achieving the highest conversions.
All three catalysts gave strongly biphasic reaction profiles, for example Pd/E. coli catalysed 75% of the coupling of phenyl iodide and ethylacrylate in the first 15 minutes, followed by a plateau in conversion. Bio-Pd could also show significant disparities between reactions. Pd/S. oneidensis displayed a 30 minute lag phase before converting 90% of the coupling of phenyl iodide and ethylacrylate, but gave one of the lowest conversions of 60% for the coupling of phenyl iodide and styrene. 2.5% Pd/E. coli (wt/dry wt) gave a 78% conversion for the coupling of phenyl iodide and ethyl acrylate, achieved a 54% conversion of the aryl bromide, 4-bromoacetophenone, but was not able to catalyse the coupling of the aryl chloride, 3-chlorotoluene.83 Recently, the fungus Phanerochaete chrysosporium synthesised bio-Pd of 10–14 nm, which was assessed for the Heck coupling of iodobenzene and styrene.68 Pd/P. chrysosporium produced 95% conversion to stilbene after 120 minutes, which was comparable to a 5% Pd/C commercial catalyst.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratios, and at the lowest ratio almost all of the bio-Pd(0) nanoparticles were measured to be >100 nm. These larger agglomerated particles had the highest catalytic activity, achieving complete substrate conversions. Conversely, at the highest CDW
Pd ratios, and at the lowest ratio almost all of the bio-Pd(0) nanoparticles were measured to be >100 nm. These larger agglomerated particles had the highest catalytic activity, achieving complete substrate conversions. Conversely, at the highest CDW![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Pd ratio all the particles were measured to be <25 nm, and no catalytic activity was observed.
Pd ratio all the particles were measured to be <25 nm, and no catalytic activity was observed.
| Bacteria | Electron donor | Proposed reduction pathway | Aryl halide | Reaction time (hours) | Conversion (%) | Reference |
|---|---|---|---|---|---|---|
| Cupriavidus necator | Formate | FDH + hydrogenase + abiotic | Iodobenzene | 6 | 86 | 24 |
| 4-Iodoanisole | 6 | 100 | ||||
| 4-Iodotoluene | 6 | 96 | ||||
| 4-Chloro-iodobenzene | 6 | 60 | ||||
| 4-Iodobenzonitrile | 6 | 84 | ||||
| 4-Iodoacetophenone | 6 | 97 | ||||
| 4-Iodobenzaldehyde | 6 | 79 | ||||
| 2-Iodobenzaldehyde | 6 | 75 | ||||
| 3-Iodo-N-methylbenzamide | 16 | 97 | ||||
| 4-Methoxy-iodobenzene | 6 | 100 | ||||
| Methyl 4′-iodo-4-methoxybiphenyl-3-carboxylate | 6 | 89 | ||||
| 4-Bromobenzonitrile | 16 | 0 | ||||
| Cupriavidus necator | H2 | Hydrogenase + abiotic | 4-Iodoanisole | 20 | 100 | 31 |
| Escherichia coli | H2 | Hydrogenase + abiotic | 4-Bromoanisole | 18 | 62 | 83 |
| 1-Bromo-4-(trifluoromethyl)benzene | 18 | 90 | ||||
| 4-Bromoacetophenone | 18 | 23 | ||||
| 2-Bromopyridine | 18 | 0 | ||||
| 4-Chloroanisole | 24 | 17 | ||||
| 3-Chlorotoluene | 18 | 0 | ||||
| Pseudomonas putida | Formate | FDH + hydrogenase + abiotic | 4-Iodoanisole | 6 | 100 | 24 |
| 4-Bromobenzonitrile | 6 | 0 | ||||
| Staphylococcus sciuri | H2 | Hydrogenase + abiotic | 4-Iodoanisole | 20 | 100 | 31 |
Deplanche et al. assessed 2.5% Pd/E. coli (wt/dry wt) for the Suzuki coupling of different aryl halides with phenylboronic acid in the presence of Na2CO3, water, ethanol, and John-Phos ligand, at a catalyst loading of 0.1 mol%, for 18 hours at 80 °C.83 The Suzuki couplings were unreactive without a monophosphine (John-Phos) ligand present, which can stabilise Pd species leached into solution, which may suggest a homogeneous reaction mechanism. Pd/E. coli displayed good activity for the activated substrate 1-bromo-4-(trifluoromethyl)benzene, and for 4-bromoanisole, whereas the activated substrate 4-bromoacetophenone produced only a moderate conversion. No reaction was observed for 2-bromopyridine or for the aryl chloride 3-chlorotoluene, and a modest 17% yield was achieved for 4-chloroanisole.
6. Microbial bimetallic nanoparticles
Bimetallic nanoparticles have been shown to possess superior catalytic activity and selectivity over monometallic nanoparticles in industrially relevant reactions such as C–C coupling reactions and hydrogenations.185–187 Introducing a second metal phase to the nanoparticle structure can change its properties, such as the surface composition, the geometry of surface adsorption sites, and the electronic properties of the catalyst.188 Furthermore, altering the ratio of the two metals enables fine-tuning of these properties.Microbial bimetallic nanoparticles are a relatively new addition to the field with significant potential, and have demonstrated far superior catalytic properties to monometallic bio-Pd (Table 5). Fine tuning of the nanostructure of these particles, such as developing well-defined core–shell structures, along with better control of particle size, and identification of the catalytically active species provides the possibility of achieving even better catalytic performance.
| Bacteria | Metals | Electron donor | Proposed reduction pathway | Catalytic application | Reference |
|---|---|---|---|---|---|
| Bacillus benzeovorans | Pd + Pt | H2 | Hydrogenase + abiotic | Upgrading heavy oil | 195 |
| Pd + Ru | Transfer hydrogenation of 5-HMF | 196 | |||
| Cupriavidus necator | Pd + Au | Formate | FDH + hydrogenase + abiotic | 4-Nitrophenol reduction | 197 |
| Desulfovibrio desulfuricans | Pd + Au | H2 | Hydrogenase + abiotic | Benzyl alcohol oxidation | 34 and 191 |
| Pd + Pt | Upgrading heavy oil | 195 | |||
| Escherichia coli | Pd + Au | H2 | Hydrogenase + abiotic | Benzyl alcohol oxidation | 34 and 192 |
| Pd + Pt | Cr(VI) reduction | 20 | |||
| Selective hydrogenation of 2-pentyne | 20 | ||||
| Selective hydrogenation of soybean oil | 21 | ||||
| Pd + Ru | Transfer hydrogenation of 5-HMF | 198 | |||
| Shewanella oneidensis | Pd + Au | H2 | Hydrogenase + abiotic | Dechlorination of trichloroethene and diclofenac | 33 and 199 |
| Formate | FDH + hydrogenase + abiotic | Suzuki coupling | 32 | ||
| Lactate | OMC (MtrCAB) | Suzuki coupling | 36 | ||
| OMC (MtrCAB) (biomagnetite) | Reduction of nitroaromatics | 200 | |||
| Pd + Ag | Lactate | OMC (MtrCAB) (reduced GO) | 4-Nitrophenol reduction | 201 | |
| OMC (MtrCAB) | Suzuki coupling | 36 | |||
| Pd + Pt | Formate | FDH + hydrogenase + abiotic | 4-Nitrophenol reduction | 35 and 202 |
6.1 Bio-PdAu
Bio-PdAu was the first microbial bimetallic system reported. De Corte et al. found that bio-PdAu possessed different catalytic properties when synthesised via simultaneous or sequential reduction by S. oneidensis.33 Bio-PdAu produced from the simultaneous reduction of Pd(II) and Au(III) (PdAu/S. oneidensis) degraded 77.8% of the environmental contaminant diclofenac after 24 hours. Bio-PdAu produced from the sequential reduction of Au(III) followed by Pd(II) (AuPd/S. oneidensis), only degraded 36.5% of diclofenac. However, bio-PdAu produced from the sequential reduction of Pd(II) followed by Au(III) (PdAu/S. oneidensis) produced little catalytic activity. Simultaneously reduced PdAu/S. oneidensis also dechlorinated trichloroethylene (TCE) in 40 minutes, vastly outperforming sequentially reduced Au–Pd/S. oneidensis. Thin section TEM showed that simultaneously reduced PdAu/S. oneidensis possessed on average larger nanoparticles than sequentially reduced AuPd/S. oneidensis, with a mean particle size of 11.0 nm and 6.9 nm, respectively. μXRD showed a significant contraction of the simultaneously reduced PdAu/S. oneidensis lattice relative to Pd/S. oneidensis, suggesting the formation of unique crystalline domains of alloyed PdAu. This alloyed phase was only weakly present in sequentially reduced AuPd/S. oneidensis, which predominantly contained a mixture of monometallic Pd and Au. Therefore it appears the enhanced catalytic performance of PdAu/S. oneidensis resulted from structural rather than size differences. Chemically synthesised core–shell Au–Pd nanoparticles were shown to catalyse TCE degradation with rate constants up to 1000 times higher than for the alloyed structure of PdAu/S. oneidensis.189 The core–shell structure may allow Au atoms to withdraw electron density from Pd more effectively than an alloyed structure, thereby increasing the interaction potential of Pd with reactants.190Deplanche et al. synthesised bio-PdAu using cells of D. desulfuricans and E. coli via the sequential reduction of Pd(II) followed by Au(III).34 When tested for the oxidation of benzyl alcohol, PdAu/E. coli gave a 35% conversion and 96% selectivity to the desired product benzaldehyde, and PdAu/D. desulfuricans gave 38% conversion and 95% selectivity. In comparison, a 5% Pd/C commercial catalyst only gave 25% conversion and 70% selectivity. In following studies characterising bio-PdAu,191,192 the authors found evidence of core–shell structure within bio-PdAu (Fig. 2d). They suggested the structure was formed from a sacrificial hydrogen mechanism, whereby Pd(0) surface atoms are oxidized to Pd(II) ions by undergoing galvanic exchange with Au(III) ions, which are then reduced to Au(0). The Pd(II) ions would then be re-reduced to form an outer Pd(0) shell. However, this mechanism has been shown to form a ‘cluster-in-cluster’ structure (Fig. 2c) as opposed to core–shell.193 PdAu/G. sulfurreducens was synthesised by simultaneous reduction of Pd(II) and Au(III), which contained some larger particles with core–shell Au–Pd structure.194 The core–shell structure was attributed to the higher reduction potential of Au(III), as slow reduction kinetics may have resulted in Au seeds initially forming onto which Pd shells can subsequently grow.
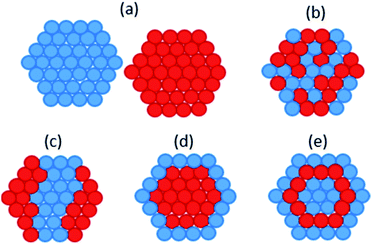 | ||
| Fig. 2 Different possible atomic structures of bimetallic nanoparticles (a) separate monometallic nanoparticles; (b) mixed alloys; (c) cluster-in-cluster; (d) core–shell; (e) multishell. | ||
Kimber et al. synthesised PdAu/S. oneidensis via simultaneous reduction of Pd(II) and Au(III) and tested the resulting nanoparticles for Suzuki coupling reactions.36 EXAFS revealed that the bio-PdAu nanoparticles displayed increased bond lengths suggesting limited alloying of the metals, and there was also some indication of non-uniform core–shell structure in a minority of particles. PdAu/S. oneidensis outperformed monometallic Pd/S. oneidensis for the Suzuki coupling of phenylboronic acid and 4-bromoacetophenone, converting >99% after 24 hours. These results are in agreement with previous results, where PdAu/S. oneidensis was shown to be more active for the Suzuki coupling of various aryl iodides, and an aryl bromide, with arylboronic acids than Pd/S. oneidensis.32 The effect of different electron donors in S. oneidensis was apparent as lactate driven particles displayed a narrower size distribution and smaller average particle size than H2 driven PdAu/S. oneidensis, likely due to abiotic reduction by H2.33,36 In addition to structural effects the composition of bio-PdAu nanoparticles has been shown to affect its catalytic properties. De Corte et al. synthesised PdAu/S. oneidensis with varying Pd(II) and Au(III) concentrations, where a low Pd![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Au ratio (50
Au ratio (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) produced the fastest removal rate of diclofenac.33 Deplanche et al. reported that a 2.5/2.5 wt% Au/Pd metal loading on cells of E. coli and D. desulfuricans gave the best conversion and selectivity for the oxidation of benzyl alcohol, over a 1/4 wt% Au/Pd loading.34
1) produced the fastest removal rate of diclofenac.33 Deplanche et al. reported that a 2.5/2.5 wt% Au/Pd metal loading on cells of E. coli and D. desulfuricans gave the best conversion and selectivity for the oxidation of benzyl alcohol, over a 1/4 wt% Au/Pd loading.34
6.2 Bio-PdAg
Han et al. used S. oneidensis to simultaneously reduce graphene oxide (GO), Pd(II), and Ag(I) from solution to form PdAg/rGO/S. oneidensis, which significantly outperformed Pd/S. oneidensis for the reduction of 4-nitrophenol.201 The optimal catalytic rate constant for 4-nitrophenol reduction of 0.241 min−1 was obtained at a Pd/Ag ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and a 2 mg catalyst loading, which produced a 99% yield of 4-aminophenol. The authors postulated that the transfer of electrons from Ag to Pd occurred through the conductive rGO sheets, to form electron-rich regions in the rGO sheets between the two metals, which offered additional active sites for adsorption and reaction. Kimber et al. produced PdAg/S. oneidensis via simultaneous reduction of Pd(II) and Ag(I), which significantly outperformed bio-PdAu and bio-Pd for the Suzuki coupling of phenylboronic acid and 4-bromoacetophenone.36 Bio-PdAg converted >99% after 5 hours, compared to 70% and 11% for bio-PdAu and Pd, respectively. Monometallic bio-Ag and bio-Au nanoparticles were not active for the reaction. PdAg/S. oneidensis coupled a broad range of arylboronic acids and aryl bromides in good to excellent yields, even achieving a double coupling of 3,5-dibromobenzaldehyde with phenylboronic acid. PdAg/S. oneidensis nanoparticles had a larger average particle size (11.2 nm) and size distribution than PdAu/S. oneidensis. Less bimetallic alloying was observed in the bio-PdAg than bio-PdAu, and EXAFS suggested that multiple Pd species were present in PdAg/S. oneidensis, including Pd–O and Pd–S shells.
1 and a 2 mg catalyst loading, which produced a 99% yield of 4-aminophenol. The authors postulated that the transfer of electrons from Ag to Pd occurred through the conductive rGO sheets, to form electron-rich regions in the rGO sheets between the two metals, which offered additional active sites for adsorption and reaction. Kimber et al. produced PdAg/S. oneidensis via simultaneous reduction of Pd(II) and Ag(I), which significantly outperformed bio-PdAu and bio-Pd for the Suzuki coupling of phenylboronic acid and 4-bromoacetophenone.36 Bio-PdAg converted >99% after 5 hours, compared to 70% and 11% for bio-PdAu and Pd, respectively. Monometallic bio-Ag and bio-Au nanoparticles were not active for the reaction. PdAg/S. oneidensis coupled a broad range of arylboronic acids and aryl bromides in good to excellent yields, even achieving a double coupling of 3,5-dibromobenzaldehyde with phenylboronic acid. PdAg/S. oneidensis nanoparticles had a larger average particle size (11.2 nm) and size distribution than PdAu/S. oneidensis. Less bimetallic alloying was observed in the bio-PdAg than bio-PdAu, and EXAFS suggested that multiple Pd species were present in PdAg/S. oneidensis, including Pd–O and Pd–S shells.
6.3 Bio-PdPt
Tuo et al. synthesised PdPt/S. oneidensis via the simultaneous reduction of Pd(II) and Pt(IV), at pH = 7 in mineral salts medium.35 Two populations of PdPt/S. oneidensis nanoparticles were produced; small spheres with an average size of 4.41 nm, or larger flower-shaped 60 nm particles. PdPt/S. oneidensis catalysed the reduction of 4-nitrophenol with a rate constant of 0.0316 min−1, and after 6 cycles retained over 92% conversion of 4-nitrophenol, despite the rate falling to 0.0078 min−1. Xu et al. produced PdPt/S. oneidensis using a similar protocol, however the bioreduction was performed in solutions of the metals dissolved in HCl at pH = 2.202 These PdPt/S. oneidensis nanoparticles consisted of predominantly small spheres with an average size of 13.2 nm, and no flower-shaped particles were produced. A much improved rate constant of 0.743 min−1 was observed for 4-nitrophenol reduction. XPS detected Pt(0) along with mainly Pd(0) and some Pd(II). The enhanced catalytic properties observed were likely due to changes in the lattice structure, and enhanced electron transport between the two metals. Omajali et al. demonstrated that bio-Pd and bio-PdPt nanoparticles gave comparable results to a commercial NiMo/Al2O3 catalyst for the catalytic upgrading of heavy oil.195 Gram-negative D. desulfuricans and the Gram-positive bacterium Bacillus benzeovorans first reduced Pd(II) to form bio-Pd, followed by the reduction of Pt(IV). The commercial catalyst produced a higher American Petroleum Institute (API) gravity than the bio-PdPt catalysts but was also hindered by more coking. In addition, bio-PdPt produced higher volumes of liquid fractions on average.6.4 Bio-PdRu
Macaskie et al. also produced bio-PdRu bimetallic nanoparticles for the catalytic transfer hydrogenation of 5-hydroxymethyl furfural (HMF) to produce the “drop in” fuel 2,5-dimethylfuran (DMF).196,203 PdRu/B. benzeovorans and PdRu/E. coli were both synthesised by sequential reduction, first pre-seeding cells with bio-Pd, followed by reduction of Ru(III). Multiple species of both metals were identified including: Pd(0), Pd(IV), Ru(III), Ru(IV), Ru(VI), but not Ru(0). 2.5/2.5 wt% PdRu/B. benzeovorans (wt/dry wt) gave a 94.7% conversion and 55.5% yield of HMF and DMF, respectively.196 In comparison, 5/5 wt% PdRu/E. coli (wt/dry wt) gave a 100% conversion and 54.4% yield of HMF and DMF, respectively, which was comparable to a commercial 5% Ru/C (wt/wt) catalyst.203 Similarly to bio-PdAu nanoparticles, core–shell structures of bio-PdRu were identified, but were a minority population.7. Recovery and recyclability of bio-Pd
7.1 Bio-Pd from acidic waste
A promising feature of microbial bioreduction is the ability to synthesise bio-Pd catalysts from waste materials. High concentrations of precious metals such as Pd, Pt and Au, are recoverable from secondary sources. The Mackaskie group have made considerable progress in recovering precious metals from waste electrical and electronic equipment, such as printed circuit boards, and from spent industrial and automotive waste leachates.204–206 When first assessed for recovering Pd(II) from acid leachate of spent automotive catalysts, cells of D. desulfuricans only recovered around 15% of Pd(II) due to high chloride concentration.82 However, an effective way around this was to pre-seed the cells with a low Pd loading (1 wt%) before exposing them to the waste solution. The Pd(0) pre-seeded cells provided additional nucleation sites for the autocatalytic reduction of dissolved Pd species.20,21,207–209 Mabbett et al. used pre-seeded 1% Pd/E. coli to recover Pd from industrial waste leachates (pH = 1.5); which contained a mixture of metals including Pd, Pt, Rh, as well as very high Cl− concentration from dissolving in aqua regia.207 The recovered Pd/E. coli catalysts were used for the reduction of 500 μM Cr(VI), and the reduction rate was dependent on the Cl− concentration of the initial metal waste solution. When the Cl− concentration was doubled the Cr(VI) reduction rate fell from of 0.030 to 0.005 mmol h−1 g−1, respectively. In comparison, 5% Pd/E. coli (wt/dry wt) synthesised from a non-waste Pd(II) salt solution reduced Cr(VI) at a much faster rate of 0.158 mmol h−1 g−1. Murray et al. found that pre-seeding cells of E. coli with either Pd or Pt resulted in 65% recovery of PGMs from diluted acid leachate (pH = 1.6).208 On the other hand, blank cells gave negligible recovery, suggesting the metal was recovered mainly through abiotic reduction onto the cell-supported Pd(0) seeds. Bimetallic PdPt/E. coli nanoparticles were successfully recovered from model acidic solutions via the pre-seeding method. Pre-seeded 2% Pd/E. coli (wt/dry wt) resulted in the recovery of a 16 wt% total PdPt/E. coli from a model solution, with higher activity for Cr(VI) reduction than 2% Pd/E. coli.20 Likewise, a recovered 7.6/8.4 wt% PdPt/E. coli (wt/dry wt) bimetallic catalyst fully reduced 0.5 mM Cr(VI) in 3 hours.21 The recovered PdPt/E. coli catalyst also catalysed the hydrogenation of soybean oil, converting 91% of linoleic acid after 2 hours, compared to 100% conversion by a commercial 2% Pd/Al2O3 catalyst.Søbjerg et al. produced Pd/C. necator from a spent Pd/C catalyst used for a hydrogenation reaction, which was dissolved in aqua regia and then neutralised.24C. necator recovered half of the waste Pd and Pd/C. necator catalysed the Heck coupling of p-iodoanisole with n-butylacrylate, furnishing a 90% isolated yield. C. metallidurans and C. necator also recovered precious metals from a mixed metal acidic leachate (pH = 1.4) containing Ag, Au, Pt, Pd, Rh, Ir, Ru, Pb, and Cu.209 Both pre-palladized and fresh cells were incubated with the leachate for 24 hours, and Pd was recovered from the solution at the same rate. The bio-Pd produced catalysed the Heck coupling of 4-iodoanisole and n-butylacrylate, in the presence of TBAB. For both species of bacteria the pre-palladized cells gave 100% conversions, whereas blank cells only achieved 50% conversion.
The harshly acidic leachates used to recover precious metals typically range from pH = 0–2.5. Neutrophilic bacteria can successfully recover the metals, however the enzymes used for bioreduction, such as hydrogenases, become denatured at low pH after only a few hours.207,208 Metal-reducing acidophilic extremophile microbes are an interesting alternative for the recovery of Pd and the formation of catalytic bio-Pd in one step. Okibe et al. showed that the Gram-negative aerobic acidophilic Fe(III)-reducing bacteria, Acidocella aromatica and Acidiphilium cryptum, reduce Pd(II) using formate, H2 or monosaccharides (glucose and fructose) as electron donors.210 Bio-Pd was first produced from synthetic solutions, and then from an acidic leachate of spent commercial Pd catalysts (pH = 2.5) containing 20 mg L−1 Pd(II), 2240 mg L−1 Al3+ and 187 mM Cl−. Pd/Ac. aromatica from the acid leachate performed comparably to Pd/Ac. aromatica from the synthetic Pd(II) solution for the catalytic reduction of Cr(VI). In addition, the two catalysts displayed similar median nanoparticle sizes of 18.1 and 19.6 nm, respectively. In a following study, Ac. aromatica was shown to reduce Au(III) to Au(0) nanoparticles at pH = 2.5.211 The thermo-acidophilic archaeon Sulfolobus tokodaii was shown to reduce Pd(II) from acidic solutions (pH = 2).212 The archaeon showed increased resistance to chloride concentration, successfully reducing Pd(II) in the presence of up to 500 mM Cl−. The mean size of bio-Pd nanoparticles decreased from 25 nm to 8.7 nm when the Cl− concentration increased from 0 to 50 mM; but above 50 mM Cl− the size increased again. The smallest Pd/S. tokodaii nanoparticles were the most catalytically active, producing the highest specific Cr(VI) reduction rate of 2.0 mg Cr(VI)/L h−1 mg−1 Pd(0). Pd/S. tokodaii outperformed a Pd/C commercial catalyst, which recorded a rate of 0.5 mg Cr(VI)/L h−1 mg−1 Pd(0).
7.2 Recyclability of bio-Pd
Bio-Pd catalysts have been shown to possess superior recyclability in some studies compared to conventional Pd catalysts. The recyclability of Pd/D. desulfuricans was superior to a commercial 5% Pd/C catalyst for the Heck reaction.26 After 6 cycles the final conversion of 5% Pd/D. desulfuricans fell from 99% to 93%, its rate decreasing from 0.31 to 0.22 mmol min−1, whereas the final conversion of 5% Pd/C dropped from 98% to 77% and its rate fell from 0.28 to 0.0090 mmol min−1. Pd leaching from bio-Pd was found to be negligible over the 6 cycles. Bennett et al. also found that 25% Pd/D. desulfuricans maintained comparable selectivity for 2-pentene on its second cycle, albeit with a lower rate of reaction.25 Pd/C. necator achieved yields of 91%, 97%, 97% and 95% for successive cycles of the Suzuki coupling of p-iodotoluene and phenylboronic acid.24G. sulfurreducens supported biomagnetite abiotically reduced Pd(II) to form a 5% Pd/Fe3O4/G. sulfurreducens (wt/dry wt) catalyst, which gave a 100% yield for the Heck coupling of iodobenzene with both ethyl acrylate and styrene, and was easily recoverable with a magnet. After four cycles the catalyst maintained >90% conversion, whereas a colloidal Pd catalyst could only be used twice.213 Tuo et al. produced a bimetallic catalyst on a biomagnetite support using S. oneidensis.200 The PdAu/Fe3O4/S. oneidensis nanoparticles were mostly 4–9.4 nm, and gave higher conversions and catalytic rate constants than Pd alone for the reduction of 4-nitrophenol, and seven other nitroaromatic compounds. The biomagnetite supported bimetallic particles were highly reusable, and after 8 cycles retained 87% of their initial activity.8. Further processing and applications of bio-Pd
8.1 Processing
Microbial cells are robust and amenable to processing techniques such as immobilisation, pyrolysis, and the incorporation of nanomaterials, which may significantly alter the catalytic properties of bio-Pd and improve its ease of use. Bio-Pd can simply be added to liquid phase reactions as a suspension catalyst, for example to catalyse the reduction of various nitroaromatic compounds.35,129,200,214 However, long term storage of bio-Pd containing cell suspensions has been shown to result in Pd leaching from the cell.215 The preparation method of Mackaskie et al. involves washing bio-Pd with acetone before air drying, and grinding into a fine powder.25,26,83,164,192 Wood et al. observed that no Pd leaching occurred when washing in acetone, and that the dried palladized cells were more resistant to attrition under shear than suspended cell.160 This process has drastically improved the catalytic properties of H2/formate driven bio-Pd, as the acetone disrupts the outer cell membrane so the periplasmic Pd(0) particles are more accessible.94,216 Ultrasonication of the cells may aid in this, including with the addition of NaOH to further release nanoparticles from the cell.118,128,163,192 In addition, palladized cells can be freeze-dried for later use in reactions.202,217 Bio-Pd can be encapsulated in polymeric beads, which are advantageous over cell suspensions in terms of separating for reuse from a reaction mixture, preventing Pd leaching into solution, and immobilising in continuous flow reactors. However, encapsulated bio-Pd suffers mass transfer limitations, resulting in slower reaction rates.127,218,219The pyrolysis (or carbonisation) of bio-Pd produces a carbon support material from the biomass whilst retaining dispersed Pd(0) nanoparticles with strong catalytic properties.220 Ng et al. pyrolysed a Pd/S. oneidensis biofilm resulting in a high surface area Pd/C nanocomposite, containing residual phosphorous and nitrogen dopants from the biofilm.221 The nanocomposite outperformed a commercial Pd nanopowder for the reduction of Cr(VI), and retained >90% of its Cr(VI) reduction capacity after five cycles. Carbon nanomaterials have been integrated into bio-Pd. Microbes can simultaneously reduce Pd(II) and graphene oxide (GO) to form nanocomposites, in order to increase electrochemical conductivity and catalytic activity.201,222–224 A Pd/rGO/S. oneidensis catalyst produced a five-fold higher Cr(VI) reduction rate than Pd/S. oneidensis.225 The enhanced Cr(VI) reduction was attributed to the co-reduction of GO resulting in enhanced Pd immobilisation and cell viability.
Electrochemical catalysis is beyond the scope of this review, however effective fuel cell catalysts have been synthesised from bio-Pd, for example, to enhance the rate limiting oxygen reduction reaction in microbial fuel cells.226,227 Bio-Pd commonly undergoes modification via pyrolysis and/or the incorporation of rGO for use in fuel cells.228–231
Recent studies have incorporated various nanomaterials with bacteria, such as metal–organic frameworks (MOFs) and TiO2 nanotubes, which enhanced the adsorption and reduction of various environmental contaminants.232–234 Another study integrated the synthesis of a photoactive polymer onto microbial surfaces through bio-Pd catalysed Sonogashira polymerisation.235
8.2 Biocatalysis
The studies discussed so far have focused on the abiotic catalytic properties of bio-Pd nanoparticles. However, the catalytic properties of bio-Pd have been effectively combined with the enzymatic activity of microbial cells (from the field of “biocatalysis”), creating powerful whole cell catalysts capable of cascade reactions. In their seminal paper, Foulkes et al. produced bio-Pd in E. coli containing a genetically engineered monoamine oxidase to create a whole cell biocatalyst for the enantioselective deracemization of a cyclic.217 After five catalytic cycles, comprising of a monoamine oxidase catalysed resolution of a racemic mix of (R) and (S) 1-methyltetrahydroisoquinoline (MTQ), followed by bio-Pd conversion of the amine, resulting in an enantiomeric excess of 96%. Pd was recovered by a microbial community in a H2 driven membrane biofilm reactor to form bio-Pd nanoparticles.236,237 Aerobic denitrification by the biofilm reactor was significantly enhanced by the presence of bio-Pd, with nitrate reduction accomplished via bacterial respiration, and nitrite reduction via bacterial respiration and Pd(0) catalysis.238 In a following study the same system was effectively applied to the degradation of the toxic contaminant 4-chlorophenol (4-CP).239 Pd(0) catalysed the reductive dechlorination of 4-CP, with over 90% selectivity to cyclohexanone. Bacteria within the biofilm then utilised cyclohexanone as an electron donor to accelerate nitrate reduction. Similarly, superior aerobic denitrification was reported by Bacillus megatarium containing bio-Pd, which enhanced the activity of the denitrifying enzyme, amplified the electron transfer flux to nitrate, and abiotically catalysed the reduction of nitrite.240 Palladized cells of P. putida displayed enhanced aerobic degradation of phenol. Bio-Pd enhanced electron transfer along the enzymatic pathway that allows P. putida to use phenol as a growth substrate, resulting in enhanced proton motive force driving ATP production.2419. Conclusions and future directions
Microbially supported Pd nanoparticle catalysts have been widely studied, yet contrasting results are often produced depending on the process conditions. This review gives a synopsis of the diffuse literature, and highlights the key criteria that must be considered in order to control nanoparticle formation and catalytic activity. Optimising the following parameters in lab systems may allow synthesis of bio-Pd that catalyses industrially relevant reactions with comparable activity and selectivity to commercial heterogeneous Pd catalysts.Firstly, the species of microorganism used to produce bio-Pd is vital, as the Pd(II) bioreduction capacity varies depending on the cell structure and enzymatic reduction systems available. In addition, the role of the cell as support material is an important factor that warrants further study. The choice of electron donor can dictate the enzymatic reduction system used, which affects size, crystallinity, monodispersity, and cellular location of bio-Pd. The degree of metal loading relative to the cell density gives rough control over the size of bio-Pd nanoparticles, and affects the cellular localisation of particles, in particular the extracellular distribution of Pd. The solution chemistry determines the Pd(II) species that can interact with the cell, which undoubtedly plays a role in the structure and properties of the nanoparticles formed from Pd(II) biosorption and bioreduction. Close attention needs to be paid to the: choice of buffer, pH, complexing ligands, and ions in solution. The impact of all of these parameters must be considered when comparing data from various studies of bio-Pd.
An important development is the biosynthesis of bimetallic nanoparticles, which offer drastically improved catalytic performance over monometallic bio-Pd. For example, bio-PdAg and bio-PdAu were proficient at catalysing the Suzuki coupling of aryl bromides, whereas bio-Pd gave little reactivity and often required the assistance of phase transfer catalysts. The changes in nanostructure of these bimetallic nanoparticles appear to be responsible for their enhanced catalytic properties, although more in-depth characterisation is required.
Microbial processes are of particular interest for the recovery of precious metals from industrial waste solutions, and the recovery of bio-Pd-based catalysts has been demonstrated. This early research has focused on acidic leachates, which are a challenging feedstock due to their high chloride concentrations, and wide array of metal contents, which often results in bio-Pd with inferior catalytic properties compared to bio-Pd from metal salt solutions. Optimising the bioreduction of acidic leachates may overcome this, creating highly active catalysts from waste solutions in a single step process. Here the presence of other metals in waste streams needs careful consideration, as they may have deleterious or positive impacts on catalytic capabilities. Bio-Pd catalysts are highly reusable, albeit generally with reduced reaction rates. The microbial cell makes a robust support material, as such cells are amenable to many processing treatments such as pyrolysis, freeze drying or encapsulation. Other nanomaterials may also be incorporated into the bioreduction process to create new nanocomposite materials with enhanced properties. In addition, bio-Pd can be integrated into more complex hybrid “biometallic” systems using the tools of systems biology to create powerful multifunctional biocatalysts. In summation, microbially supported Pd nanoparticles are still a relatively new sustainable biotechnology with promising applications in catalysis.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank EPSRC and Johnson Matthey for funding of the PhD iCASE studentship award for CEM, and BBSRC for funding from the grants BB/L013711/1 and BB/R010412/1.Notes and references
- C. Richard Catlow, M. Davidson, C. Hardacre and G. J. Hutchings, Catalysis making the world a better place, Philos. Trans. R. Soc., A, 2016, 374, 20150089 CrossRef PubMed.
- J.-L. Malleron, J.-C. Fiaud and J.-Y. Legros, WACKER Process, in Handbook of Palladium-Catalyzed Organic Reactions, 1997 Search PubMed.
- D. C. Harrowven, Handbook of Organopalladium Chemistry for Organic Synthesis, Stuttg, 2003 Search PubMed.
- X. Liu and D. Astruc, Development of the Applications of Palladium on Charcoal in Organic Synthesis, Adv. Synth. Catal., 2018, 360, 3426–3459 CrossRef CAS.
- C. Torborg and M. Beller, Recent applications of palladium-catalyzed coupling reactions in the pharmaceutical, agrochemical, and fine chemical industries, Adv. Synth. Catal., 2009, 351(18), 3022–3043 CrossRef.
- J. Oudar, Sulfur Adsorption and Poisoning of Metallic Catalysts, Catal. Rev., 1980, 22, 171–195 CrossRef CAS.
- J. Matthey, PGM Market Report February 2021, 2021 Search PubMed.
- D. Astruc, Nanoparticles and Catalysis, 2008 Search PubMed.
- J. G. De Vries, Palladium-catalysed coupling reactions, Top. Organomet. Chem., 2012, 42, 1–34 CrossRef CAS.
- G. C. Fortman and S. P. Nolan, N-Heterocyclic carbene (NHC) ligands and palladium in homogeneous cross-coupling catalysis: A perfect union, Chem. Soc. Rev., 2011, 40, 5151–5169 RSC.
- H. U. Blaser, A. Indolese, A. Schnyder, H. Steiner and M. Studer, Supported palladium catalysts for fine chemicals synthesis, J. Mol. Catal. A: Chem., 2001, 173, 3–18 CrossRef CAS.
- N. T. S. Phan, M. van der Sluys and C. W. Jones, On the nature of the active species in palladium catalyzed Mizoroki–Heck and Suzuki–Miyaura couplings - Homogeneous or heterogeneous catalysis, a critical review, Adv. Synth. Catal., 2006, 348(6), 609–679 CrossRef CAS.
- C. Deraedt and D. Astruc, “Homeopathic” palladium nanoparticle catalysis of cross carbon-carbon coupling reactions, Acc. Chem. Res., 2014, 47, 494–503 CrossRef CAS PubMed.
- J. P. Corbet and G. Mignani, Selected patented cross-coupling reaction technologies, Chem. Rev., 2006, 106, 2651–2710 CrossRef CAS PubMed.
- F. X. Felpin, T. Ayad and S. Mitra, Pd/C: An old catalyst for new applications - Its use for the Suzuki–Miyaura reaction, Eur. J. Org. Chem., 2006, 2679–2690 CrossRef CAS.
- E. Auer, A. Freund, J. Pietsch and T. Tacke, Carbons as supports for industrial precious metal catalysts, Appl. Catal., A, 1998, 173, 259–271 CrossRef CAS.
- L. Yin and J. Liebscher, Carbon-carbon coupling reactions catalyzed by heterogeneous palladium catalysts, Chem. Rev., 2007, 107, 133–173 CrossRef CAS PubMed.
- J. R. Lloyd, P. Yong and L. E. Macaskie, Enzymatic recovery of elemental palladium by using sulfate-reducing bacteria, Appl. Environ. Microbiol., 1998, 64(11), 4607–4609 CrossRef CAS PubMed.
- W. De Windt, P. Aelterman and W. Verstraete, Bioreductive deposition of palladium (0) nanoparticles on Shewanella oneidensis with catalytic activity towards reductive dechlorination of polychlorinated biphenyls, Environ. Microbiol., 2005, 7(3), 314–325 CrossRef CAS PubMed.
- A. J. Murray, J. Zhu, J. Wood and L. E. Macaskie, A novel biorefinery: Biorecovery of precious metals from spent automotive catalyst leachates into new catalysts effective in metal reduction and in the hydrogenation of 2-pentyne, Miner. Eng., 2017, 113, 102–108 CrossRef CAS.
- A. J. Murray, J. Zhu, J. Wood and L. E. Macaskie, Biorefining of platinum group metals from model waste solutions into catalytically active bimetallic nanoparticles, Microbiol. Biotechnol., 2018, 11, 359–368 CrossRef CAS PubMed.
- X. Xia, J. Zeng, Q. Zhang, C. H. Moran and Y. Xia, Recent developments in shape-controlled synthesis of silver nanocrystals, J. Phys. Chem. C, 2012, 116(41), 21647–21656 CrossRef CAS PubMed.
- N. I. Hulkoti and T. C. Taranath, Biosynthesis of nanoparticles using microbes-A review, Colloids Surf., B, 2014, 121, 474–483 CrossRef CAS PubMed.
- L. S. Søbjerg, D. Gauthier, A. T. Lindhardt, M. Bunge, K. Finster and R. L. Meyer, et al., Bio-supported palladium nanoparticles as a catalyst for Suzuki–Miyaura and Mizoroki–Heck reactions, Green Chem., 2009, 11(12), 2041–2046 RSC.
- J. A. Bennett, N. J. Creamer, K. Deplanche, L. E. Macaskie, I. J. Shannon and J. Wood, Palladium supported on bacterial biomass as a novel heterogeneous catalyst: A comparison of Pd/Al2O3 and bio-Pd in the hydrogenation of 2-pentyne, Chem. Eng. Sci., 2010, 65(1), 282–290 CrossRef CAS.
- J. A. Bennett, I. P. Mikheenko, K. Deplanche, I. J. Shannon, J. Wood and L. E. Macaskie, Nanoparticles of palladium supported on bacterial biomass: New re-usable heterogeneous catalyst with comparable activity to homogeneous colloidal Pd in the Heck reaction, Appl. Catal., B, 2013, 140–141, 700–707 CrossRef CAS.
- T. J. Beveridge and R. G. E. Murray, Uptake and retention of metals by cell walls of Bacillus subtilis, J. Bacteriol., 1976, 127(3), 1502–1518 CrossRef CAS PubMed.
- J. R. Lloyd, Microbial reduction of metals and radionuclides, FEMS Microbiol. Rev., 2003, 27, 411–425 CrossRef CAS PubMed.
- T. J. Beveridge and W. S. Fyfe, Metal fixation by bacterial cell walls, Can. J. Earth Sci., 1985, 22(12), 1893–1898 CrossRef CAS.
- W. De Windt, N. Boon, J. van den Bulcke, L. Rubberecht, F. Prata and J. Mast, et al., Biological control of the size and reactivity of catalytic Pd(0) produced by Shewanella oneidensis, Antonie van Leeuwenhoek, Int. J. Gen. Mol. Microbiol., 2006, 90(4), 377–389 CrossRef PubMed.
- L. S. Søbjerg, A. T. Lindhardt, T. Skrydstrup, K. Finster and R. L. Meyer, Size control and catalytic activity of bio-supported palladium nanoparticles, Colloids Surf., B, 2011, 85(2), 373–378 CrossRef PubMed.
- T. S. A. Heugebaert, S. De Corte, T. Sabbe, T. Hennebel, W. Verstraete and N. Boon, et al., Biodeposited Pd/Au bimetallic nanoparticles as novel Suzuki catalysts, Tetrahedron Lett., 2012, 53(11), 1410–1412 CrossRef CAS.
- S. De Corte, T. Hennebel, J. P. Fitts, T. Sabbe, V. Bliznuk and S. Verschuere, et al., Biosupported bimetallic Pd-Au nanocatalysts for dechlorination of environmental contaminants, Environ. Sci. Technol., 2011, 45(19), 8506–8513 CrossRef CAS PubMed.
- K. Deplanche, I. P. Mikheenko, J. A. Bennett, M. Merroun, H. Mounzer and J. Wood, et al., Selective oxidation of benzyl-alcohol over biomass-supported Au/Pd bioinorganic catalysts, Top. Catal., 2011, 54, 1110–1114 CrossRef CAS.
- Y. Tuo, G. Liu, B. Dong, H. Yu, J. Zhou and J. Wang, et al., Microbial synthesis of bimetallic PdPt nanoparticles for catalytic reduction of 4-nitrophenol, Environ. Sci. Pollut. Res., 2017, 24, 5249–5258 CrossRef CAS PubMed.
- R. L. Kimber, F. Parmeggiani, T. S. Neill, M. L. Merroun, G. Goodlet and N. A. Powell, et al., Biotechnological synthesis of Pd/Ag and Pd/Au nanoparticles for enhanced Suzuki–Miyaura cross-coupling activity, Microbiol. Biotechnol., 2021, 14(6), 2435–2447 CrossRef PubMed.
- J. R. Lloyd, V. S. Coker, R. L. Kimber, C. I. Pearce, M. P. Watts and J. B. Omajali, et al., New Frontiers in Metallic Bio-nanoparticle Catalysis and Green Products from Remediation Processes, RSC Green Chem. Ser., 2020, 244–265 CAS.
- G. A. Somorjai and R. M. Rioux, High technology catalysts towards 100% selectivity: Fabrication, characterization and reaction studies, Catal. Today, 2005, 100, 201–215 CrossRef CAS.
- S. Xie, S. I. Choi, X. Xia and Y. Xia, Catalysis on faceted noble-metal nanocrystals: Both shape and size matter, Curr. Opin. Chem. Eng., 2013, 2(2), 142–150 CrossRef.
- T. Bligaard and J. K. Nørskov, Ligand effects in heterogeneous catalysis and electrochemistry, Electrochim. Acta, 2007, 52(18), 5512–5516 CrossRef CAS.
- K. W. Rosenmund, Über eine neue Methode zur Darstellung von Aldehyden. 1. Mitteilung, Ber. Dtsch. Chem. Ges., 1918, 51, 585–593 CrossRef CAS.
- H. Lindlar, Hydrogenation of acetylenic bond utilizing a palladium-lead catalyst, US Pat., 2681938, 1954 Search PubMed.
- Á. Molnár, A. Sárkány and M. Varga, Hydrogenation of carbon-carbon multiple bonds: Chemo-, regio- and stereo-selectivity, J. Mol. Catal. A: Chem., 2001, 173, 185–221 CrossRef.
- A. Binder, M. Seipenbusch, M. Muhler and G. Kasper, Kinetics and particle size effects in ethene hydrogenation over supported palladium catalysts at atmospheric pressure, J. Catal., 2009, 268, 150–155 CrossRef CAS.
- P. E. Garcia, A. S. Lynch, A. Monaghan and S. D. Jackson, Using modifiers to specify stereochemistry and enhance selectivity and activity in palladium-catalysed, liquid phase hydrogenation of alkynes, Catal. Today, 2011, 164, 548–551 CrossRef CAS.
- K. M. Bratlie, C. J. Kliewer and G. A. Somorjai, Structure effects of benzene hydrogenation studied with sum frequency generation vibrational spectroscopy and kinetics on Pt(111) and Pt(100) single-crystal surfaces, J. Phys. Chem. B, 2006, 110(36), 17925–17930 CrossRef CAS PubMed.
- M. Crespo-Quesada, A. Yarulin, M. Jin, Y. Xia and L. Kiwi-Minsker, Structure sensitivity of alkynol hydrogenation on shape- and size-controlled palladium nanocrystals: Which sites are most active and selective?, J. Am. Chem. Soc., 2011, 133, 12787–12794 CrossRef CAS PubMed.
- A. M. Doyle, S. K. Shaikhutdinov and H. J. Freund, Surface-bonded precursor determines particle size effects for alkene hydrogenation on palladium, Angew. Chem., Int. Ed., 2005, 44, 629–631 CrossRef CAS PubMed.
- J. B. Omajali, I. P. Mikheenko, M. L. Merroun, J. Wood and L. E. Macaskie, Characterization of intracellular palladium nanoparticles synthesized by Desulfovibrio desulfuricans and Bacillus benzeovorans, J. Nanopart. Res., 2015, 17(6), 1–17 CrossRef CAS PubMed.
- T. Teranishi and M. Miyake, Size Control of Palladium Nanoparticles and Their Crystal Structures, Chem. Mater., 1998, 10(2), 594–600 CrossRef CAS.
- V. Chabert, L. Babel, M. P. Füeg, M. Karamash, E. S. Madivoli and N. Herault, et al., Kinetics and Mechanism of Mineral Respiration: How Iron Hemes Synchronize Electron Transfer Rates, Angew. Chem., Int. Ed., 2020, 59(30), 12331–12336 CrossRef CAS PubMed.
- R. Coppage, J. M. Slocik, B. D. Briggs, A. I. Frenkel, R. R. Naik and M. R. Knecht, Determining peptide sequence effects that control the size, structure, and function of nanoparticles, ACS Nano, 2012, 6(2), 1625–1636 CrossRef CAS PubMed.
- R. Coppage, J. M. Slocik, M. Sethi, D. B. Pacardo, R. R. Naik and M. R. Knecht, Elucidation of peptide effects that control the activity of nanoparticles, Angew. Chem., Int. Ed., 2010, 49(22), 3767–3770 CrossRef CAS PubMed.
- K. Köhler, R. G. Heidenreich, J. G. E. Krauter and J. Pietsch, Highly active palladium/activated carbon catalysts for heck reactions: Correlation of activity, catalyst properties, and Pd leaching, Chem.–Eur. J., 2002, 8(3), 622–631 CrossRef.
- H. Jüntgen, Activated carbon as catalyst support. A review of new research results, Fuel, 1986, 65(10), 1436–1446 CrossRef.
- A. Y. Stakheev and L. M. Kustov, Effects of the support on the morphology and electronic properties of supported metal clusters: Modern concepts and progress in 1990s, Appl. Catal., A, 1999, 188, 3–35 CrossRef CAS.
- I. C. Gerber and P. Serp, A Theory/Experience Description of Support Effects in Carbon-Supported Catalysts, Chem. Rev., 2020, 120, 1250–1349 CrossRef CAS PubMed.
- P. Makowski, R. Demir Cakan, M. Antonietti, F. Goettmann and M. M. Titirici, Selective partial hydrogenation of hydroxy aromatic derivatives with palladium nanoparticles supported on hydrophilic carbon, Chem. Commun., 2008, 999–1001 RSC.
- Y. Wang, J. Yao, H. Li, D. Su and M. Antonietti, Highly Selective Hydrogenation of Phenol and Derivatives over a Pd carbon nitride catalyst in aqueous media, J. Am. Chem. Soc., 2011, 133(8), 2362–2365 CrossRef CAS PubMed.
- M. L. Toebes, F. F. Prinsloo, J. H. Bitter, A. J. Van Dillen and K. P. De Jong, Influence of oxygen-containing surface groups on the activity and selectivity of carbon nanofiber-supported ruthenium catalysts in the hydrogenation of cinnamaldehyde, J. Catal., 2003, 214(1), 78–87 CrossRef CAS.
- J. Zhu, M. Dou, M. Lu, X. Xiang, X. Ding and W. Liu, et al., Thermo-responsive polymer grafted carbon nanotubes as the catalyst support for selective hydrogenation of cinnamaldehyde: Effects of surface chemistry on catalytic performance, Appl. Catal., A, 2019, 575, 11–19 CrossRef CAS.
- T. Beveridge, Role of Cellular Design In Bacterial Metal Accumulation And Mineralization, Annu. Rev. Microbiol., 1989, 43(1), 147–171 CrossRef CAS PubMed.
- C. Mack, B. Wilhelmi, J. R. Duncan and J. E. Burgess, Biosorption of precious metals, Biotechnol. Adv., 2007, 25(3), 264–271 CrossRef CAS PubMed.
- S. W. Won, J. Mao, I. S. Kwak, M. Sathishkumar and Y. S. Yun, Platinum recovery from ICP wastewater by a combined method of biosorption and incineration, Bioresour. Technol., 2010, 101(4), 1135–1140 CrossRef CAS PubMed.
- X. Ju, K. Igarashi, S.-i Miyashita, H. Mitsuhashi, K. Inagaki and S.-i Fujii, et al., Effective and selective recovery of gold and palladium ions from metal wastewater using a sulfothermophilic red alga, Galdieria sulphuraria, Bioresour. Technol., 2016, 211, 759–764 CrossRef CAS PubMed.
- P. Kotrba, M. Mackova and T. Macek, Microbial Biosorption of Metals, 2011 Search PubMed.
- S. Schultze-Lam, D. Fortin, B. S. Davis and T. J. Beveridge, Mineralization of bacterial surfaces, Chem. Geol., 1996, 132, 171–181 CrossRef CAS.
- S. Tarver, D. Gray, K. Loponov, D. B. Das, T. Sun and M. Sotenko, Biomineralization of Pd nanoparticles using Phanerochaete chrysosporium as a sustainable approach to turn platinum group metals (PGMs) wastes into catalysts, Int. Biodeterior. Biodegrad., 2019, 143, 104724 CrossRef CAS.
- Z. Lin, C. Zhou, J. Wu, H. Cheng, B. Liu and Z. Ni, et al., Adsorption and reduction of palladium (Pd2+) by Bacillus licheniformis R08, Chin. Sci. Bull., 2002, 47, 1262–1266 CrossRef CAS.
- K. Fahmy, M. Merroun, K. Pollmann, J. Raff, O. Savchuk and C. Hennig, et al., Secondary structure and Pd(II) coordination in S-layer proteins from Bacillus sphaericus studied by infrared and X-ray absorption spectroscopy, Biophys. J., 2006, 91(3), 996–1007 CrossRef CAS PubMed.
- A. Sari, D. Mendil, M. Tuzen and M. Soylak, Biosorption of palladium(II) from aqueous solution by moss (Racomitrium lanuginosum) biomass: Equilibrium, kinetic and thermodynamic studies, J. Hazard. Mater., 2009, 162, 874–879 CrossRef CAS PubMed.
- A. M. Pat-Espadas, E. Razo-Flores, J. R. Rangel-Mendez and F. J. Cervantes, Direct and quinone-mediated palladium reduction by Geobacter sulfurreducens: Mechanisms and modeling, Environ. Sci. Technol., 2014, 48(5), 2910–2919 CrossRef CAS PubMed.
- J. Cui, N. Zhu, N. Kang, C. Ha, C. Shi and P. Wu, Biorecovery mechanism of palladium as nanoparticles by Enterococcus faecalis: From biosorption to bioreduction, Chem. Eng. J., 2017, 328, 1051–1057 CrossRef CAS.
- L. Tan, H. Dong, X. Liu, J. He, H. Xu and J. Xie, Mechanism of palladium(II) biosorption by: Providencia vermicola, RSC Adv., 2017, 7(12), 7060–7072 RSC.
- H. Xu, L. Tan, H. Cui, M. Xu, Y. Xiao and H. Wu, et al., Characterization of Pd(II) biosorption in aqueous solution by Shewanella oneidensis MR-1, J. Mol. Liq., 2018, 255, 333–340 CrossRef CAS.
- B. Godlewska-Zyłkiewicz, S. Sawicka and J. Karpińska, Removal of platinum and palladium from wastewater by means of biosorption on fungi Aspergillus sp. and yeast Saccharomyces sp., Water, 2019, 11, 1522 CrossRef.
- B. Godlewska-Zyłkiewicz, Biosorption of platinum and palladium for their separation/preconcentration prior to graphite furnace atomic absorption spectrometric determination, Spectrochim. Acta, Part B, 2003, 58(8), 1531–1540 CrossRef.
- J. Park, S. W. Won, J. Mao, I. S. Kwak and Y. S. Yun, Recovery of Pd(II) from hydrochloric solution using polyallylamine hydrochloride-modified Escherichia coli biomass, J. Hazard. Mater., 2010, 181, 794–800 CrossRef CAS PubMed.
- S. W. Won, J. Park, J. Mao and Y. S. Yun, Utilization of PEI-modified Corynebacterium glutamicum biomass for the recovery of Pd(II) in hydrochloric solution, Bioresour. Technol., 2011, 102(4), 3888–3893 CrossRef CAS PubMed.
- A. E. Rotaru, W. Jiang, K. Finster, T. Skrydstrup and R. L. Meyer, Non-enzymatic palladium recovery on microbial and synthetic surfaces, Biotechnol. Bioeng., 2012, 109(8), 1889–1897 CrossRef CAS PubMed.
- S. De Corte, S. Bechstein, A. R. Lokanathan, J. Kjems, N. Boon and R. L. Meyer, Comparison of bacterial cells and amine-functionalized abiotic surfaces as support for Pd nanoparticle synthesis, Colloids Surf., B, 2013, 102, 898–904 CrossRef CAS PubMed.
- P. Yong, N. A. Rowson, J. P. G. Farr, I. R. Harris and L. E. Macaskie, Bioaccumulation of palladium by Desulfovibrio desulfuricans, J. Chem. Technol. Biotechnol., 2002, 7, 593–601 CrossRef.
- K. Deplanche, J. A. Bennett, I. P. Mikheenko, J. Omajali, A. S. Wells and R. E. Meadows, et al., Catalytic activity of biomass-supported Pd nanoparticles: Influence of the biological component in catalytic efficacy and potential application in “green” synthesis of fine chemicals and pharmaceuticals, Appl. Catal., B, 2014, 147, 651–665 CrossRef CAS.
- I. De Vargas, L. E. Macaskie and E. Guibal, Biosorption of palladium and platinum by sulfate-reducing bacteria, J. Chem. Technol. Biotechnol., 2004, 79(1), 49–56 CrossRef CAS.
- P. Yong, N. A. Rowson, J. P. G. Farr, I. R. Harris and L. E. Macaskie, Bioreduction and biocrystallization of palladium by Desulfovibrio desulfuricans NCIMB 8307, Biotechnol. Bioeng., 2002, 80(4), 369–379 CrossRef CAS PubMed.
- L. Shi, S. M. Belchik, A. E. Plymale, S. Heald, A. C. Dohnalkova and K. Sybirna, et al., Purification and characterization of the [NiFe]-Hydrogenase of Shewanella oneidensis MR-1, Appl. Environ. Microbiol., 2011, 77(16), 5584–5590 CrossRef CAS PubMed.
- N. S. Sickerman and Y. Hu, Hydrogenases, Methods Mol. Biol., 2019, 65–88 CrossRef CAS PubMed.
- J. R. Lloyd, J. A. Cole and L. E. Macaskie, Reduction and removal of heptavalent technetium from solution by Escherichia coli, J. Bacteriol., 1997, 179(6), 2014–2021 CrossRef CAS PubMed.
- J. E. Butler, N. D. Young and D. R. Lovley, Evolution of electron transfer out of the cell: Comparative genomics of six Geobacter genomes, BMC Genomics, 2010, 11, 40 CrossRef PubMed.
- I. P. Mikheenko, M. Rousset, S. Dementin and L. E. Macaskie, Bioaccumulation of palladium by Desulfovibrio fructosivorans wild-type and hydrogenase-deficient strains, Appl. Environ. Microbiol., 2008, 74(19), 6144–6146 CrossRef CAS PubMed.
- K. Deplanche, I. Caldelari, I. P. Mikheenko, F. Sargent and L. E. Macaskie, Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains, Microbiology, 2010, 156(9), 2630–2640 CrossRef CAS PubMed.
- V. S. Baxter-Plant, I. P. Mikheenko and L. E. Macaskie, Sulphate-reducing bacteria, palladium and the reductive dehalogenation of chlorinated aromatic compounds, Biodegradation, 2003, 14, 83–90 CrossRef CAS PubMed.
- J. Courtney, K. Deplanche, N. V. Rees and L. E. Macaskie, Biomanufacture of nano-Pd(0) by Escherichia coli and electrochemical activity of bio-Pd(0) made at the expense of H2 and formate as electron donors, Biotechnol. Lett., 2016, 38(11), 1903–1910 CrossRef CAS PubMed.
- A. N. Mabbett and L. E. Macaskie, A new bioinorganic process for the remediation of Cr(VI), J. Chem. Technol. Biotechnol., 2002, 77(10), 1169–1175 CrossRef CAS.
- G. Attard, M. Casadesús, L. E. MacAskie and K. Deplanche, Biosynthesis of platinum nanoparticles by Escherichia coli MC4100: Can such nanoparticles exhibit intrinsic surface enantioselectivity?, Langmuir, 2012, 28(11), 5267–5274 CrossRef CAS PubMed.
- Y. N. Hou, B. Zhang, H. Yun, Z. N. Yang, J. L. Han and J. Zhou, et al., Palladized cells as suspension catalyst and electrochemical catalyst for reductively degrading aromatics contaminants: Roles of Pd size and distribution, Water Res., 2017, 125, 288–297 CrossRef CAS PubMed.
- M. Bunge, L. S. Søbjerg, A. E. Rotaru, D. Gauthier, A. T. Lindhardt and G. Hause, et al., Formation of palladium(0) nanoparticles at microbial surfaces, Biotechnol. Bioeng., 2010, 107(2), 206–215 CrossRef CAS PubMed.
- J. K. Dunleavy, Sulfur as a catalyst poison, Platinum Met. Rev., 2006, 50(2), 110 CrossRef CAS.
- B. Mishra, M. Boyanov, B. A. Bunker, S. D. Kelly, K. M. Kemner and J. B. Fein, High- and low-affinity binding sites for Cd on the bacterial cell walls of Bacillus subtilis and Shewanella oneidensis, Geochim. Cosmochim. Acta, 2010, 74(15), 4219–4233 CrossRef CAS.
- A. J. McCue and J. A. Anderson, Sulfur as a catalyst promoter or selectivity modifier in heterogeneous catalysis, Catal.: Sci. Technol., 2014, 4, 272–294 RSC.
- Y. Liu, Y. Li, J. A. Anderson, J. Feng, A. Guerrero-Ruiz and I. Rodríguez-Ramos, et al., Comparison of Pd and Pd4S based catalysts for partial hydrogenation of external and internal butynes, J. Catal., 2020, 383, 51–59 CrossRef CAS.
- D. Lovley, Dissimilatory Metal Reduction, Annu. Rev. Microbiol., 1993, 47(1), 263–290 CrossRef CAS PubMed.
- D. R. Lovley, D. E. Holmes and K. P. Nevin, Dissimilatory Fe(III) and Mn(IV) reduction, Adv. Microb. Physiol., 2004, 49, 219–286 CrossRef CAS PubMed.
- J. R. Lloyd, V. A. Sole, C. V. G. Van Praagh and D. R. Lovley, Direct and Fe(II)-mediated reduction of technetium by Fe(III)-reducing bacteria, Appl. Environ. Microbiol., 2000, 66(9), 3743–3749 CrossRef CAS PubMed.
- D. R. Lovley, Bug juice: Harvesting electricity with microorganisms, Nat. Rev. Microbiol., 2006, 4, 497–508 CrossRef CAS PubMed.
- M. J. Marshall, A. S. Beliaev, A. C. Dohnalkova, D. W. Kennedy, L. Shi and Z. Wang, et al., c-Type cytochrome-dependent formation of U(IV) nanoparticles by Shewanella oneidensis, PLoS Biol., 2006, 4(8), 1324–1333 CrossRef CAS PubMed.
- S. M. Belchik, D. W. Kennedy, A. C. Dohnalkova, Y. Wang, P. C. Sevinc and H. Wu, et al., Extracellular reduction of hexavalent chromium by cytochromes MtrC and OmcA of Shewanella oneidensis MR-1, Appl. Environ. Microbiol., 2011, 77(12), 4035–4041 CrossRef CAS PubMed.
- Y. Konishi, K. Ohno, N. Saitoh, T. Nomura, S. Nagamine and H. Hishida, et al., Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae, J. Biotechnol., 2007, 128(3), 648–653 CrossRef CAS PubMed.
- N. Law, S. Ansari, F. R. Livens, J. C. Renshaw and J. R. Lloyd, Formation of nanoscale elemental silver particles via enzymatic reduction by Geobacter sulfurreducens, Appl. Environ. Microbiol., 2008, 74(22), 7090–7093 CrossRef CAS PubMed.
- S. De Corte, T. Hennebel, S. Verschuere, C. Cuvelier, W. Verstraete and N. Boon, Gold nanoparticle formation using Shewanella oneidensis: A fast biosorption and slow reduction process, J. Chem. Technol. Biotechnol., 2011, 86(4), 547–553 CrossRef CAS.
- A. M. Pat-Espadas, E. Razo-Flores, J. R. Rangel-Mendez and F. J. Cervantes, Reduction of palladium and production of nano-catalyst by Geobacter sulfurreducens, Appl. Microbiol. Biotechnol., 2013, 97, 9553–9560 CrossRef CAS PubMed.
- M. J. Edwards, D. J. Richardson, C. M. Paquete and T. A. Clarke, Role of multiheme cytochromes involved in extracellular anaerobic respiration in bacteria, Protein Sci., 2020, 29, 830–842 CrossRef CAS PubMed.
- J. K. Fredrickson, M. F. Romine, A. S. Beliaev, J. M. Auchtung, M. E. Driscoll and T. S. Gardner, et al., Towards environmental systems biology of Shewanella, Nat. Rev. Microbiol., 2008, 6, 592–603 CrossRef CAS PubMed.
- T. Ogi, K. Tamaoki, N. Saitoh, A. Higashi and Y. Konishi, Recovery of indium from aqueous solutions by the Gram-negative bacterium Shewanella algae, Biochem. Eng. J., 2012, 63, 129–133 CrossRef CAS.
- C. K. Ng, K. Sivakumar, X. Liu, M. Madhaiyan, L. Ji and L. Yang, et al., Influence of outer membrane c-type cytochromes on particle size and activity of extracellular nanoparticles produced by Shewanella oneidensis, Biotechnol. Bioeng., 2013, 110(7), 1831–1837 CrossRef CAS PubMed.
- K. Tamaoki, N. Saito, T. Nomura and Y. Konishi, Microbial recovery of rhodium from dilute solutions by the metal ion-reducing bacterium Shewanella algae, Hydrometallurgy, 2013, 139, 26–29 CrossRef CAS.
- J. W. Wu and I. S. Ng, Biofabrication of gold nanoparticles by Shewanella species, Bioresour. Bioprocess., 2017, 4, 50 CrossRef.
- K. Tamaoki, N. Saito, T. Ogi, T. Nomura and Y. Konishi, Microbial reduction and recovery of palladium using metal ion-reducing bacterium Shewanella algae, Kagaku Kogaku Ronbunshu, 2010, 36(4), 288–292 CrossRef CAS.
- H. Zhang and X. Hu, Rapid production of Pd nanoparticle by a marine electrochemically active bacterium: Shewanella sp. CNZ-1 and its catalytic performance on 4-nitrophenol reduction, RSC Adv., 2017, 7(65), 41182–41189 RSC.
- W. Wang, B. Zhang, Q. Liu, P. Du, W. Liu and Z. He, Biosynthesis of palladium nanoparticles using: Shewanella loihica PV-4 for excellent catalytic reduction of chromium(VI), Environ. Sci.: Nano, 2018, 22(8), 919–929 Search PubMed.
- G. Meshulam-Simon, S. Behrens, A. D. Choo and A. M. Spormann, Hydrogen metabolism in Shewanella oneidensis MR-1, Appl. Environ. Microbiol., 2007, 73(4), 1153–1165 CrossRef CAS PubMed.
- G. E. Pinchuk, O. V. Geydebrekht, E. A. Hill, J. L. Reed, A. E. Konopka and A. S. Beliaev, et al., Pyruvate and lactate metabolism by Shewanella oneidensis MR-1 under fermentation, oxygen limitation, and fumarate respiration conditions, Appl. Environ. Microbiol., 2011, 77(23), 8234–8240 CrossRef CAS PubMed.
- A. L. Kane, E. D. Brutinel, H. Joo, R. Maysonet, C. M. VanDrisse and N. J. Kotloski, Formate metabolism in Shewanella oneidensis generates proton motive force and prevents growth without an electron acceptor, J. Bacteriol., 2016, 198(8), 1337–1346 CrossRef CAS PubMed.
- C. Liu, Y. A. Gorby, J. M. Zachara, J. K. Fredrickson and C. F. Brown, Reduction kinetics of Fe(III), Co(III), U(VI), Cr(VI), and Tc(VII) in cultures of dissimilatory metal-reducing bacteria, Biotechnol. Bioeng., 2002, 80(6), 637–649 CrossRef CAS PubMed.
- M. J. Marshall, A. E. Plymale, D. W. Kennedy, L. Shi, Z. Wang and S. B. Reed, Hydrogenase- and outer membrane c-type cytochrome-facilitated reduction of technetium(VII) by Shewanella oneidensis MR-1, Environ. Microbiol., 2008, 10(1), 125–136 CAS.
- N. J. Creamer, K. Deplanche, T. J. Snape, I. P. Mikheenko, P. Yong and D. Samyahumbi, A biogenic catalyst for hydrogenation, reduction and selective dehalogenation in non-aqueous solvents, Hydrometallurgy, 2008, 94, 138–143 CrossRef CAS.
- A. C. Humphries, I. P. Mikheenko and L. E. Macaskie, Chromate reduction by immobilized palladized sulfate-reducing bacteria, Biotechnol. Bioeng., 2006, 94(1), 81–90 CrossRef CAS PubMed.
- C. Ha, N. Zhu, R. Shang, C. Shi, J. Cui and I. Sohoo, et al., Biorecovery of palladium as nanoparticles by Enterococcus faecalis and its catalysis for chromate reduction, Chem. Eng. J., 2016, 288, 246–254 CrossRef CAS.
- Y. Tuo, G. Liu, J. Zhou, A. Wang, J. Wang and R. Jin, Microbial formation of palladium nanoparticles by Geobacter sulfurreducens for chromate reduction, Bioresour. Technol., 2013, 133, 606–611 CrossRef CAS PubMed.
- P. T. Wang, Y. H. Song, H. C. Fan and L. Yu, Bioreduction of azo dyes was enhanced by in-situ biogenic palladium nanoparticles, Bioresour. Technol., 2018, 266, 176–180 CrossRef CAS PubMed.
- E. Ahmed, S. Kalathil, L. Shi, O. Alharbi and P. Wang, Synthesis of ultra-small platinum, palladium and gold nanoparticles by Shewanella loihica PV-4 electrochemically active biofilms and their enhanced catalytic activities, J. Saudi Chem. Soc., 2018, 22(8), 919–929 CrossRef CAS.
- D. Coursolle and J. A. Gralnick, Reconstruction of extracellular respiratory pathways for iron(III) reduction in Shewanella oneidensis strain MR-1, Front. Microbiol., 2012, 3(56), 1–11 Search PubMed.
- L. Shi, K. M. Rosso, J. M. Zachara and J. K. Fredrickson, Mtr extracellular electron-transfer pathways in Fe(III)-reducing or Fe(II)-oxidizing bacteria: A genomic perspective, Biochem. Soc. Trans., 2012, 40(6), 1261–1267 CrossRef CAS PubMed.
- S. Pirbadian, S. E. Barchinger, K. M. Leung, H. S. Byun, Y. Jangir and R. A. Bouhenni, et al., Shewanella oneidensis MR-1 nanowires are outer membrane and periplasmic extensions of the extracellular electron transport components, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(35), 12883–12888 CrossRef CAS PubMed.
- C. L. Reardon, A. C. Dohnalkova, P. Nachimuthu, D. W. Kennedy, D. A. Saffarini and B. W. Arey, et al., Role of outer-membrane cytochromes MtrC and OmcA in the biomineralization of ferrihydrite by Shewanella oneidensis MR-1, Geobiology, 2010, 8(1), 56–68 CrossRef CAS PubMed.
- F. Caccavo, D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz and M. J. McInerney, Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism, Appl. Environ. Microbiol., 1994, 60(10), 3752–3759 CrossRef CAS PubMed.
- G. Reguera, Harnessing the power of microbial nanowires, Microb. Biotechnol., 2018, 11(6), 979–994 CrossRef CAS PubMed.
- G. Reguera, K. D. McCarthy, T. Mehta, J. S. Nicoll, M. T. Tuominen and D. R. Lovley, Extracellular electron transfer via microbial nanowires, Nature, 2005, 435, 1098–1101 CrossRef CAS PubMed.
- M. Mollaei, P. H. A. Timmers, M. Suarez-Diez, S. Boeren, A. H. van Gelder and A. J. M. Stams, et al., Comparative proteomics of Geobacter sulfurreducens PCAT in response to acetate, formate and/or hydrogen as electron donor, Environ. Microbiol., 2020, 23(1), 299–315 CrossRef PubMed.
- M. D. Yates, R. D. Cusick and B. E. Logan, Extracellular palladium nanoparticle production using Geobacter sulfurreducens, ACS Sustainable Chem. Eng., 2013, 1(9), 1165–1171 CrossRef CAS.
- T. Mehta, M. V. Coppi, S. E. Childers and D. R. Lovley, Outer membrane c-type cytochromes required for Fe(III) and Mn(IV) oxide reduction in Geobacter sulfurreducens, Appl. Environ. Microbiol., 2005, 71(12), 8634–8641 CrossRef CAS PubMed.
- Y. Liu, J. K. Fredrickson, J. M. Zachara and L. Shi, Direct involvement of ombB, omaB, and omcB genes in extracellular reduction of Fe(III) by Geobacter sulfurreducens PCA, Front. Microbiol., 2015, 6, 1075 Search PubMed.
- A. Hernández-Eligio, A. M. Pat-Espadas, L. Vega-Alvarado, M. Huerta-Amparán, F. J. Cervantes and K. Juárez, Global transcriptional analysis of Geobacter sulfurreducens under palladium reducing conditions reveals new key cytochromes involved, Appl. Microbiol. Biotechnol., 2020, 104(9), 4059–4069 CrossRef PubMed.
- R. M. Jarvis, N. Law, I. T. Shadi, P. O’Brien, J. R. Lloyd and R. Goodacre, Surface-enhanced Raman scattering from intracellular and extracellular bacterial locations, Anal. Chem., 2008, 80(17), 6741–6746 CrossRef CAS PubMed.
- C. M. Dundas, A. J. Graham, D. K. Romanovicz and B. K. Keitz, Extracellular Electron Transfer by Shewanella oneidensis Controls Palladium Nanoparticle Phenotype, ACS Synth. Biol., 2018, 7(12), 2726–2736 CrossRef CAS PubMed.
- Z. N. Yang, Y. N. Hou, B. Zhang, H. Y. Cheng, Y. C. Yong and W. Z. Liu, Insights into palladium nanoparticles produced by Shewanella oneidensis MR-1: Roles of NADH dehydrogenases and hydrogenases, Environ. Res., 2020, 191, 110196 CrossRef CAS PubMed.
- H. Von Canstein, J. Ogawa, S. Shimizu and J. R. Lloyd, Secretion of flavins by Shewanella species and their role in extracellular electron transfer, Appl. Environ. Microbiol., 2008, 74(3), 615–623 CrossRef CAS PubMed.
- E. D. Brutinel and J. A. Gralnick, Shuttling happens: Soluble flavin mediators of extracellular electron transfer in Shewanella, Appl. Microbiol. Biotechnol., 2012, 93, 41–48 CrossRef PubMed.
- A. Okamoto, K. Saito, K. Inoue, K. H. Nealson, K. Hashimoto and R. Nakamura, Uptake of self-secreted flavins as bound cofactors for extracellular electron transfer in Geobacter species, Energy Environ. Sci., 2014, 7(4), 1357–1361 RSC.
- P. Sabatier, Hydrogénations et déshydrogénations par catalyse, Ber. Dtsch. Chem. Ges., 1911, 44(3), 1984–2001 CrossRef CAS.
- F. Nerozzi, Heterogeneous catalytic hydrogenation, Platinum Met. Rev., 2012, 56(4), 236–241 CrossRef CAS.
- V. I. Savchenko and I. A. Makaryan, Palladium catalyst for the production of pure margarine: Catalyst and new non-filtration technology improve production and quality, Platinum Met. Rev., 1999, 43(2), 74–82 CAS.
- W. Bonrath, T. Netscher, J. Schutz and B. Wustenberg, Process for Manufacture of TMHQ, 2012, WO2012025587.
- O. Isler and K. Doebel, Synthesen in der Vitamin-K-Reihe I. Über totalsynthetisches Vitamin K1, Helv. Chim. Acta, 1954, 37(1), 225–233 CrossRef CAS.
- G. M. Wolf and L. H. Sternbach, Synthesis of biotin, US Pat., 2489235, 1949 Search PubMed.
- D. Schils, F. Stappers, G. Solberghe, R. Van Heck, M. Coppens and D. van den heuvel, et al., Ligandless Heck coupling between a halogenated aniline and acrylonitrile catalyzed by Pd/C: Development and optimization of an industrial-scale Heck process for the production of a pharmaceutical intermediate, Org. Process Res. Dev., 2008, 12(3), 530–536 CrossRef CAS.
- B. Chen, U. Dingerdissen, J. G. E. Krauter, H. G. J. Lansink Rotgerink, K. Möbus and D. J. Ostgard, New developments in hydrogenation catalysis particularly in synthesis of fine and intermediate chemicals, Appl. Catal., A, 2005, 280(1), 17–46 CrossRef CAS.
- N. J. Creamer, I. P. Mikheenko, K. Deplanche, P. Yong, J. Wood and K. Pollmann, et al., A Novel Hydrogenation and Hydrogenolysis Catalyst Using Palladized Biomass of Gram-negative and Gram-positive Bacteria, Adv. Mater. Res., 2007, 20–21, 603–606 CAS.
- I. P. Mikheenko, J. A. Bennett, I. J. Shannon, J. Wood and L. E. Macaskie, Biomineralised palladium is an effective hydrogenation catalyst, Adv. Mater. Res., 2009, 71–73, 725–728 CAS.
- J. Wood, L. Bodenes, J. Bennett, K. Deplanche and L. E. Macaskie, Hydrogenation of 2-butyne-1, 4-diol using novel bio-palladium catalysts, Ind. Eng. Chem. Res., 2010, 49, 980–988 CrossRef CAS.
- G. A. Attard, J. A. Bennett, I. Mikheenko, P. Jenkins, S. Guan and L. E. Macaskie, et al., Semi-hydrogenation of alkynes at single crystal, nanoparticle and biogenic nanoparticle surfaces: The role of defects in Lindlar-type catalysts and the origin of their selectivity, Faraday Discuss., 2013, 162, 57–75 RSC.
- B. Kunwar, S. D. Deilami, L. E. Macaskie, J. Wood, P. Biller and B. K. Sharma, Nanoparticles of Pd supported on bacterial biomass for hydroprocessing crude bio-oil, Fuel, 2017, 209, 449–456 CrossRef CAS.
- J. A. Bennett, G. A. Attard, K. Deplanche, M. Casadesus, S. E. Huxter and L. E. Macaskie, Improving selectivity in 2-butyne-1,4-diol hydrogenation using biogenic Pt catalysts, ACS Catal., 2012, 2, 504–511 CrossRef CAS.
- J. Zhu, J. Wood, K. Deplanche, I. Mikheenko and L. E. Macaskie, Selective hydrogenation using palladium bioinorganic catalyst, Appl. Catal., B, 2016, 199, 108–122 CrossRef CAS.
- B. Mertens, C. Blothe, K. Windey, W. De Windt and W. Verstraete, Biocatalytic dechlorination of lindane by nano-scale particles of Pd(0) deposited on Shewanella oneidensis, Chemosphere, 2007, 66(1), 99–105 CrossRef CAS PubMed.
- D. Bianchi, G. E. E. Gardes, G. M. Pajonk and S. J. Teichner, Hydrogenation of ethylene on alumina after hydrogen spillover, J. Catal., 1975, 38, 135–146 CrossRef CAS.
- F. J. Urbano and J. M. Marinas, Hydrogenolysis of organohalogen compounds over palladium supported catalysts, J. Mol. Catal. A: Chem., 2001, 173, 329–345 CrossRef CAS.
- S. Harrad, M. Robson, S. Hazrati, V. S. Baxter-Plant, K. Deplanche and M. D. Redwood, Dehalogenation of polychlorinated biphenyls and polybrominated diphenyl ethers using a hybrid bioinorganic catalyst, J. Environ. Monit., 2007, 9(4), 314–318 RSC.
- T. Hennebel, H. Simoen, W. De Windt, M. Verloo, N. Boon and W. Verstraete, Biocatalytic dechlorination of trichloroethylene with bio-palladium in a pilot-scale membrane reactor, Biotechnol. Bioeng., 2009, 102(4), 995–1002 CrossRef CAS PubMed.
- L. E. MacAskie, A. C. Humphries, I. P. Mikheenko, V. S. Baxter-Plant, K. Deplanche and M. D. Redwood, et al., Use of Desulfovibrio and Escherichia coli Pd-nanocatalysts in reduction of Cr(VI) and hydrogenolytic dehalogenation of polychlorinated biphenyls and used transformer oil, J. Chem. Technol. Biotechnol., 2012, 87(10), 1430–1435 CrossRef CAS.
- C. Barnard, Palladium-catalysed C–C coupling: Then and now, Platinum Met. Rev., 2008, 52(1), 38–45 CrossRef CAS.
- T. Mizoroki, K. Mori and A. Ozaki, Arylation of Olefin with Aryl Iodide Catalyzed by Palladium, Bull. Chem. Soc. Jpn., 1971, 44(2), 581 CrossRef CAS.
- K. F. Heck and J. P. Nolley, Palladium-Catalyzed Vinylic Hydrogen Substitution Reactions with Aryl, Benzyl, and Styryl Halides, J. Org. Chem., 1972, 37(14), 2320–2322 CrossRef.
- W. Tse-Chong, Process for preparing aryl-substituted aliphatic carboxylic acids and their esters using cyclic phosphine catalysts, US Pat., 1994, 5,315,026 Search PubMed.
- I. Shinkai, A. O. King and R. D. Larsen, A practical asymmetric synthesis of LTD4 antagonist, Pure Appl. Chem., 1994, 66(7), 1551–1556 CAS.
- P. Baumeister and W. Scherrer.Process for the Preparation of Halogenated Aromatic Primary Amines, European Patent, EP0473552, 1991.
- N. A. S. Miyaura and S. Chem, Stereoselective Synthesis of Arylated (E)-Alkenes, J. Chem. Soc., Chem. Commun., 1979, 866–867 RSC.
- K. Eichen, M. Rack, F. Wetterich, E. Ammermann, G. Lorenz and S. Strathmann, New Bi:phenyl-Amide Derivatives Are Active against Wide Range of Phytopathogenic Fungi, German Patent, DE 19735224, 1999.
- S. J. Mickel, G. H. Sedelmeier, D. Niederer, F. Schuerch, M. Seger and K. Schreiner, et al., Large-Scale Synthesis of the Anti-Cancer Marine Natural Product (+)-Discodermolide. Part 4: Preparation of Fragment C7-24, Org. Process Res. Dev., 2004, 8(1), 113–121 CrossRef CAS.
- M. Julia and M. Duteil, Bull. Soc. Chim. Fr., 1973, 2790, 9–10 Search PubMed.
- G. Marck, A. Villiger and R. Buchecker, Aryl couplings with heterogeneous palladium catalysts, Tetrahedron Lett., 1994, 35(20), 3277–3280 CrossRef CAS.
- Y. Kitamura, S. Sako, A. Tsutsui, Y. Monguchi, T. Maegawa and Y. Kitade, Ligand-free and heterogeneous palladium on carbon-catalyzed hetero-Suzuki–Miyaura cross-coupling, Adv. Synth. Catal., 2010, 352(4), 718–730 CrossRef CAS.
- C. Liu, C. Liu, X. M. Li, Z. M. Gao and Z. L. Jin, Oxygen-promoted Pd/C-catalyzed Suzuki–Miyaura reaction of potassium aryltrifluoroborates, Chin. Chem. Lett., 2016, 27(5), 631–634 CrossRef CAS.
- R. G. Heidenreich, K. Köhler, J. G. E. Krauter and J. Pietsch, Pd/C as a highly active catalyst for Heck, Suzuki and Sonogashira reactions, Synlett, 2002, 1118–1122 CrossRef CAS.
- G. Schmid, H. West, J. O. Malm, J. O. Bovin and C. Grenthe, Catalytic properties of layered gold-palladium colloids, Chem.–Eur. J., 1996, 2(9), 1099–1103 CrossRef CAS.
- P. Dash, N. A. Dehm and R. W. J. Scott, Bimetallic PdAu nanoparticles as hydrogenation catalysts in imidazolium ionic liquids, J. Mol. Catal. A: Chem., 2008, 286, 114–119 CrossRef CAS.
- D. R. Pye and N. P. Mankad, Bimetallic catalysis for C–C and C-X coupling reactions, Chem. Sci., 2017, 8(3), 1705–1718 RSC.
- T. Chen and V. O. Rodionov, Controllable Catalysis with Nanoparticles: Bimetallic Alloy Systems and Surface Adsorbates, ACS Catal., 2016, 6, 4025–4033 CrossRef CAS.
- M. O. Nutt, K. N. Heck, P. Alvarez and M. S. Wong, Improved Pd-on-Au bimetallic nanoparticle catalysts for aqueous-phase trichloroethene hydrodechlorination, Appl. Catal., B, 2006, 69, 115–125 CrossRef CAS.
- M. R. Knecht, M. G. Weir, A. I. Frenkel and R. M. Crooks, Structural rearrangement of bimetallic alloy PdAu nanoparticles within dendrimer templates to yield core/shell configuration, Chem. Mater., 2008, 20(3), 1019–1028 CrossRef CAS.
- D. T. Tran, I. P. Jones, J. A. Preece, R. L. Johnston, K. Deplanche and L. E. MacAskie, Configuration of microbially synthesized Pd–Au nanoparticles studied by STEM-based techniques, Nanotechnology, 2012, 23(5), 55701 CrossRef CAS PubMed.
- K. Deplanche, M. L. Merroun, M. Casadesus, D. T. Tran, I. P. Mikheenko and J. A. Bennett, et al., Microbial synthesis of core/shell gold/palladium nanoparticles for applications in green chemistry, J. R. Soc., Interface, 2012, 9(72), 1705–1712 CrossRef CAS PubMed.
- N. Toshima and T. Yonezawa, Bimetallic nanoparticles – Novel materials for chemical and physical applications, New J. Chem., 1998, 22(11), 1179–1201 RSC.
- E. A. Lewis, H. Downie, R. F. Collins, E. Prestat, J. R. Lloyd and S. J. Haigh, Imaging the Hydrated Microbe-Metal Interface Using Nanoscale Spectrum Imaging, Part. Part. Syst. Charact., 2016, 33(11), 833–841 CrossRef.
- J. B. Omajali, A. Hart, M. Walker, J. Wood and L. E. Macaskie, In-situ catalytic upgrading of heavy oil using dispersed bionanoparticles supported on Gram-positive and Gram-negative bacteria, Appl. Catal., B, 2017, 203, 807–819 CrossRef CAS.
- J. B. Omajali, J. Gomez-Bolivar, I. P. Mikheenko, S. Sharma, B. Kayode and B. Al-Duri, et al., Novel catalytically active Pd/Ru bimetallic nanoparticles synthesized by Bacillus benzeovorans, Sci. Rep., 2019, 9(1), 1–12 CAS.
- B. Hosseinkhani, L. S. Søbjerg, A. E. Rotaru, G. Emtiazi, T. Skrydstrup and R. L. Meyer, Microbially supported synthesis of catalytically active bimetallic Pd-Au nanoparticles, Biotechnol. Bioeng., 2012, 109(1), 45–52 CrossRef CAS PubMed.
- J. Gomez-Bolivar, I. P. Mikheenko, L. E. Macaskie and M. L. Merroun, Characterization of palladium nanoparticles produced by healthy and microwave-injured cells of Desulfovibrio desulfuricans and Escherichia coli, Nanomaterials, 2019, 9, 857 CrossRef CAS PubMed.
- S. De Corte, T. Sabbe, T. Hennebel, L. Vanhaecke, B. De Gusseme and W. Verstraete, et al., Doping of biogenic Pd catalysts with Au enables dechlorination of diclofenac at environmental conditions, Water Res., 2012, 46(8), 2718–2726 CrossRef CAS PubMed.
- Y. Tuo, G. Liu, B. Dong, J. Zhou, A. Wang and J. Wang, et al., Microbial synthesis of Pd/Fe3O4, Au/Fe3O4 and PdAu/Fe3O4 nanocomposites for catalytic reduction of nitroaromatic compounds, Sci. Rep., 2015, 5, 13515 CrossRef CAS PubMed.
- R. Han, X. Song, Q. Wang, Y. Qi, G. Deng and A. Zhang, et al., Microbial synthesis of graphene-supported highly-dispersed Pd-Ag bimetallic nanoparticles and its catalytic activity, J. Chem. Technol. Biotechnol., 2019, 94(10), 3375–3383 CrossRef CAS.
- H. Xu, Y. Xiao, M. Xu, H. Cui, L. Tan and N. Feng, et al., Microbial synthesis of Pd-Pt alloy nanoparticles using Shewanella oneidensis MR-1 with enhanced catalytic activity for nitrophenol and azo dyes reduction, Nanotechnology, 2019, 30(6), 65607 CrossRef CAS PubMed.
- J. Gomez-Bolivar, I. P. Mikheenko, R. L. Orozco, S. Sharma, D. Banerjee and M. Walker, et al., Synthesis of Pd/Ru bimetallic nanoparticles by Escherichia coli and potential as a catalyst for upgrading 5-hydroxymethyl furfural into liquid fuel precursors, Front. Microbiol., 2019, 10, 1–17 CrossRef PubMed.
- P. Yong, L. E. Macaskie, N. A. Rowson, J. P. G. Farr and L. R. Harris, A novel electrobiotechnology for the recovery of precious metals from spent automotive catalysts, Environ. Technol., 2003, 24(3), 289–297 CrossRef CAS PubMed.
- P. Yong, N. A. Rowson, J. P. G. Farr, L. R. Harris, L. E. Macaskie and R. Harris, A novel electrobiotechnology for the recovery of precious metals from spent automotive catalysts a novel electrobiotechnology for the recovery of precious metals from spent automotive catalysts, Environ. Technol., 2017, 24(3), 289–297 CrossRef PubMed.
- N. J. Creamer, V. S. Baxter-Plant, J. Henderson, M. Potter and L. E. Macaskie, Palladium and gold removal and recovery from precious metal solutions and electronic scrap leachates by Desulfovibrio desulfuricans, Biotechnol. Lett., 2006, 28(18), 1475–1484 CrossRef CAS PubMed.
- A. N. Mabbett, D. Sanyahumbi, P. Yong and L. E. Macaskie, Biorecovered precious metals from industrial wastes: Single-step conversion of a mixed metal liquid waste to a bioinorganic catalyst with environmental application, Environ. Sci. Technol., 2006, 40(3), 1015–1021 CrossRef CAS PubMed.
- A. J. Murray, I. P. Mikheenko, E. Goralska, N. A. Rowson and L. E. Macaskie, Biorecovery of Platinum Group Metals from Secondary Sources, Adv. Mater. Res., 2007, 651–654 CAS.
- D. Gauthier, L. S. Sãbjerg, K. M. Jensen, A. T. Lindhardt, M. Bunge and K. Finster, Environmentally benign recovery and reactivation of palladium from industrial waste by using Gram-negative bacteria, ChemSusChem, 2010, 3(9), 1036–1039 CrossRef CAS PubMed.
- N. Okibe, D. Nakayama and T. Matsumoto, Palladium bionanoparticles production from acidic Pd(II) solutions and spent catalyst leachate using acidophilic Fe(III)-reducing bacteria, Extremophiles, 2017, 21(6), 1091–1100 CrossRef CAS PubMed.
- I. N. Rizki and N. Okibe, Size-controlled production of gold bionanoparticles using the extremely acidophilic fe(III)-reducing bacterium, Acidocella aromatica, Minerals, 2018, 8, 81 CrossRef.
- S. Kitjanukit, K. Sasaki and N. Okibe, Production of highly catalytic, archaeal Pd(0) bionanoparticles using Sulfolobus tokodaii, Extremophiles, 2019, 23(5), 549–556 CrossRef CAS PubMed.
- V. S. Coker, J. A. Bennett, N. D. Telling, T. Henkel, J. M. Charnock and G. van der Laan, et al., Microbial engineering of nanoheterostructures: Biological synthesis of a magnetically recoverable palladium nanocatalyst, ACS Nano, 2010, 4(5), 2577–2584 CrossRef CAS PubMed.
- W. Q. Kong, J. Y. Lin, X. He, Y. Y. Cheng, X. S. Zhang and G. Z. Deng, Reduction pathway and mechanism of chloronitrobenzenes synergistically catalyzed by bioPd and Shewanella oneidensis MR-1 assisted by calculation, Chemosphere, 2017, 187, 62–69 CrossRef CAS PubMed.
- S. De Corte, T. Hennebel, J. Segers, S. Van Nevel, S. Verschuere and W. Verstraete, et al., Influence of physicochemical parameters on stability and performance of biosupported Pd nanocatalysts, Nanomater. Environ., 2013, 1, 31–39 CAS.
- Y. O. Posokhov and A. Kyrychenko, Effect of acetone accumulation on structure and dynamics of lipid membranes studied by molecular dynamics simulations, Comput. Biol. Chem., 2013, 46, 23–31 CrossRef CAS PubMed.
- J. M. Foulkes, K. J. Malone, V. S. Coker, N. J. Turner and J. R. Lloyd, Engineering a biometallic whole cell catalyst for enantioselective deracemization reactions, ACS Catal., 2011, 1(11), 1589–1594 CrossRef CAS.
- T. Hennebel, P. Verhagen, H. Simoen, B. D. Gusseme, S. E. Vlaeminck and N. Boon, et al., Remediation of trichloroethylene by bio-precipitated and encapsulated palladium nanoparticles in a fixed bed reactor, Chemosphere, 2009, 76(9), 1221–1225 CrossRef CAS PubMed.
- A. Nordmeier, J. Woolford, L. Celeste and D. Chidambaram, Sustainable batch production of biosynthesized nanoparticles, Mater. Lett., 2017, 191, 53–56 CrossRef CAS.
- H. L. Parker, E. L. Rylott, A. J. Hunt, J. R. Dodson, A. F. Taylor and N. C. Bruce, Supported palladium nanoparticles synthesized by living plants as a catalyst for Suzuki–Miyaura reactions, PLoS One, 2014, 9(1), e87192 CrossRef PubMed.
- C. K. Ng, H. E. Karahan, S. C. J. Loo, Y. Chen and B. Cao, Biofilm-Templated Heteroatom-Doped Carbon-Palladium Nanocomposite Catalyst for Hexavalent Chromium Reduction, ACS Appl. Mater. Interfaces, 2019, 11(27), 24018–24026 CrossRef CAS PubMed.
- R. E. Priestley, A. Mansfield, J. Bye, K. Deplanche, A. B. Jorge and D. Brett, et al., Pd nanoparticles supported on reduced graphene-E. coli hybrid with enhanced crystallinity in bacterial biomass, RSC Adv., 2015, 5(102), 84093–84103 RSC.
- J. Liu, Y. Zheng, Z. Hong, K. Cai, F. Zhao and H. Han, Microbial synthesis of highly dispersed PdAu alloy for enhanced electrocatalysis, Sci. Adv., 2016, 2, e1600858 CrossRef PubMed.
- Y. N. Hou, S. Y. Sun, Z. N. Yang, H. Yun, T. t. Zhu and J. F. Ma, et al., Shewanella oneidensis MR-1 self-assembled Pd-cells-rGO conductive composite for enhancing electrocatalysis, Environ. Res., 2020, 184, 109317 CrossRef CAS PubMed.
- Y. X. Chen, X. Liu, Z. Fang, C. L. Zhang, S. Z. Abbas and Y. Y. Yu, Self-assembling of Shewanella@rGO@Pd bionanohybrid for synergistic bio-abiotic removal of Cr(VI), J. Chem. Technol. Biotechnol., 2020, 95(8), 2222–2228 CrossRef CAS.
- L. E. Macaskie, I. P. Mikheenko, J. B. Omajai, A. J. Stephen and J. Wood, Metallic bionanocatalysts: potential applications as green catalysts and energy materials, Microbiol. Biotechnol., 2017, 10(5), 1171–1180 CrossRef CAS PubMed.
- A. J. Stephen, N. V. Rees, I. Mikheenko and L. E. Macaskie, Platinum and palladium bio-synthesized nanoparticles as sustainable fuel cell catalysts, Front. Energy Res., 2019, 7, 1–13 CrossRef.
- W. Wang, J. L. Mi, Q. C. Shen and Y. C. Yong, Shewanella oneidensis Assisted Biosynthesis of Pd/Reductive-Graphene-Oxide Nanocomposites for Oxygen Reduction Reaction, ChemistrySelect, 2020, 5(11), 3179–3186 CrossRef CAS.
- S. Zhang, X. Deng, A. Chen, H. Zhou, Z. Xie and Y. Liang, Facile synthesis of Pd supported on Shewanella as an efficient catalyst for oxygen reduction reaction, Int. J. Hydrogen Energy, 2019, 44(39), 21759–21768 CrossRef CAS.
- L. Xiong, J. J. Chen, Y. X. Huang, W. W. Li, J. F. Xie and H. Q. Yu, An oxygen reduction catalyst derived from a robust Pd-reducing bacterium, Nano Energy, 2015, 12(1), 33–42 CrossRef CAS.
- B. Cao, Z. Zhao, L. Peng, H. Y. Shiu, M. Ding and F. Song, et al., Silver nanoparticles boost charge-extraction efficiency in Shewanella microbial fuel cells, Science, 2021, 373(6561), 1336–1340 CrossRef CAS PubMed.
- X. Song, X. Shi and M. Yang, Dual application of Shewanella oneidensis MR-1 in green biosynthesis of Pd nanoparticles supported on TiO2 nanotubes and assisted photocatalytic degradation of methylene blue, IET Nanobiotechnol., 2018, 12(4), 441–445 CrossRef PubMed.
- S. K. Springthorpe, C. M. Dundas and B. K. Keitz, Microbial reduction of metal-organic frameworks enables synergistic chromium removal, Nat. Commun., 2019, 10, 5212 CrossRef PubMed.
- M. Yang and X. Shi, Biosynthesis of Ag2S/TiO2 nanotubes nanocomposites by Shewanella oneidensis MR-1 for the catalytic degradation of 4-nitrophenol, Environ. Sci. Pollut. Res., 2019, 26(12), 12237–12246 CrossRef CAS PubMed.
- R. Qi, H. Zhao, X. Zhou, J. Liu, N. Dai and Y. Zeng, In situ Synthesis of Photoactive Polymers on a Living Cell Surface via Bio-Palladium Catalysis for Modulating Biological Functions, Angew. Chem., Int. Ed., 2021, 60(11), 5759–5765 CrossRef CAS PubMed.
- C. Zhou, A. Ontiveros-Valencia, Z. Wang, J. Maldonado, H. P. Zhao and R. Krajmalnik-Brown, Palladium Recovery in a H2-Based Membrane Biofilm Reactor: Formation of Pd(0) Nanoparticles through Enzymatic and Autocatalytic Reductions, Environ. Sci. Technol., 2016, 50(5), 2546–2555 CrossRef CAS PubMed.
- C. Zhou, Z. Wang, A. K. Marcus and B. E. Rittmann, Biofilm-enhanced continuous synthesis and stabilization of palladium nanoparticles (PdNPs), Environ. Sci.: Nano, 2016, 3(6), 1396–1404 RSC.
- C. Zhou, Z. Wang, A. Ontiveros-Valencia, M. Long, L. C. yu and H. P. Zhao, Coupling of Pd nanoparticles and denitrifying biofilm promotes H2-based nitrate removal with greater selectivity towards N2, Appl. Catal., B, 2017, 206, 461–470 CrossRef CAS.
- M. Long, X. Long, C. W. Zheng, Y. H. Luo, C. Zhou and B. E. Rittmann, Para-Chlorophenol (4-CP) Removal by a Palladium-Coated Biofilm: Coupling Catalytic Dechlorination and Microbial Mineralization via Denitrification, Environ. Sci. Technol., 2021, 55(9), 6309–6319 CrossRef CAS PubMed.
- Y. Jia, D. Qian, Y. Chen and Y. Hu, Intra/extracellular electron transfer for aerobic denitrification mediated by in situ biosynthesis palladium nanoparticles, Water Res., 2021, 189, 116612 CrossRef CAS PubMed.
- Z. Niu, Y. Jia, Y. Chen, Y. Hu, J. Chen and Y. Lv, Positive effects of bio-nano Pd (0) toward direct electron transfer in Pseudomona putida and phenol biodegradation, Ecotoxicol. Environ. Saf., 2018, 161, 356–363 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |