Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanoconfined anti-oxidizing RAFT nitroxide radical polymer for reduction of low-density lipoprotein oxidation and foam cell formation†
Suman
Basak
 ae,
Harshvardhan Ajay
Khare
ae,
Harshvardhan Ajay
Khare
 ab,
Paul J.
Kempen
ab,
Paul J.
Kempen
 ac,
Nazila
Kamaly
ac,
Nazila
Kamaly
 d and
Kristoffer
Almdal
d and
Kristoffer
Almdal
 *e
*e
aDepartment of Health Technology, DTU Health Tech, Technical University of Denmark, Kgs. Lyngby, 2800, Denmark
bDepartment of Clinical Physiology, Nuclear Medicine & PET and Cluster for Molecular Imaging, Rigshospitalet and University of Copenhagen, Copenhagen, 2200, Denmark
cNational Centre for Nano Fabrication and Characterization, DTU Nanolab, Technical University of Denmark, Kgs. Lyngby, 2800, Denmark
dDepartment of Chemistry, Molecular Sciences Research Hub, Imperial College London, London, W12 0BZ, UK
eDepartment of Chemistry, Technical University of Denmark, Kgs. Lyngby, 2800, Denmark. E-mail: kral@dtu.dk
First published on 4th January 2022
Abstract
Atherosclerosis is a leading cause of death worldwide. Antioxidant therapy has been considered a promising treatment modality for atherosclerosis, since reactive oxygen species (ROS) play a major role in the pathogenesis of atherosclerosis. We developed ROS-scavenging antioxidant nanoparticles (NPs) that can serve as an effective therapy for atherosclerosis. The newly developed novel antioxidant ROS-eliminating NPs were synthesized via reversible addition–fragmentation chain-transfer (RAFT) polymerization and act as a superoxide dismutase (SOD) mimetic agent. SOD is an anti-ROS enzyme which is difficult to use for passive delivery due to its low half-life and stability. Copolymers were synthesized using different feed ratios of 2,2,6,6-tetramethyl-4-piperidyl methacrylate (PMA) and glycidyl methacrylate (GMA) monomers and an anti-ROS nitroxyl radical polymer was prepared via oxidation. The copolymer was further conjugated with a 6-aminofluorescein via a oxirane ring opening reaction for intracellular delivery in RAW 264.7 cells. The synthesized copolymers were blended to create NPs (∼150 nm size) in aqueous medium and highly stable up to three weeks. The NPs were shown to be taken up by macrophages and to be cytocompatible even at high dose levels (500 μg mL−1). Finally, the nitroxide NPs has been shown to inhibit foam cell formation in macrophages by decreasing internalization of oxidized low-density lipoproteins.
Introduction
Atherosclerosis, a chronic inflammatory disease, is a major cause of coronary heart disease and stroke in humans.1 The disease is characterized by intimal plaques and cholesterol accumulation in the arterial walls.2 During atherogenesis, the primary process of inflammation is accompanied by oxidative stress, which is exemplified by the overproduction of reactive oxygen species (ROS) and oxidized low-density lipoprotein (ox-LDL). Hence an imbalance between radical production (reactive oxygen) and radical scavenging systems (the antioxidant defense system) is observed.3 High levels of ROS can induce oxidative stress that is closely associated with the pathogenesis of atherosclerosis. ROS can lead to NFκB activation, cell apoptosis, protein modification, and oxidative damage to other biomolecules.4,5 ROS also disrupts redox dependent signaling in the vessel wall to promote progress of atherosclerosis,6,7 involving signal transduction pathways,8 regulatory genes associated with vascular function,7 inflammatory components of atherosclerosis,9 and clearance of apoptotic cells by macrophages.10Therefore, suppressing systemic oxidative stress, preventing vascular oxidative stress, and reducing ROS generation in plaques represent promising strategies for the treatment of atherosclerosis. In this respect, antioxidants have a crucial role in the prevention and treatment of atherosclerosis through different mechanisms. These include: the inhibition of low density lipoprotein (LDL) oxidation,11 the reduction of reactive oxygen species (ROS) generation,12 the inhibition of cytokine secretion, the prevention of atherosclerotic plaque formation and platelet aggregation, the preclusion of mononuclear cell infiltration, the prevention of endothelial dysfunction and vasodilation, the augmentation of nitric oxide (NO) bioavailability, the modulation of the expression of adhesion molecules such as vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) on endothelial cells, and the suppression of foam cell formation.13 In this aspect, various types of antioxidants have been investigated,14 such as vitamins E and C,15 coenzyme Q,16 proanthocyanidin,17 4-hydroxy-TEMPO,18 and NADPH oxidase.19 Although these preclinical studies have substantiated protective effects of antioxidants on atherosclerosis, clinical trials did not afford positive effects.20 Lewis et al. nicely demonstrated sugar based amphiphilic NPs to prevent oxidized lipid uptake and suppress scavenger receptor expression in macrophages.21 Annexin A1 actions were mimicked by Ac2-26 fragment containing NPs in reducing oxidative stress, promoting collagen buildup, and increasing the anti-inflammatory cytokine interleukin-10 and stabilize the plaque.22 In addition, recently developed 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO) containing NPs significantly inhibited the development of atherosclerosis in ApoE−/− mice after i.v. delivery and the therapy afforded stabilized plaques with less cholesterol crystals and a smaller necrotic core.23 Thus, TEMPO and its derivatives are potentially powerful candidates for ROS scavenging.24 TEMPO decorated redox micelles have been used to suppress inflammation and treat drug-resistant in carcinoma cells.25,26
It appears that the NP based targeting strategies are effective and promising for molecular imaging and therapy of atherosclerosis.27,28 In atherosclerosis nano-delivery could be achieved in different ways such as: (i) nanoparticles that can target plaques by direct infiltration via the injured endothelium or the dysfunctional neovessels.29,30 (ii) Intravenous or intraperitoneal injection that can be endocytosed by circulating phagocytes, followed by translocation to atherosclerotic lesions.31,32 Therefore, nanoparticles serve as vehicles for targeted delivery of different therapeutics to atherosclerotic plaques and are a most promising way for next generation atherosclerotic drug delivery.22,33,34
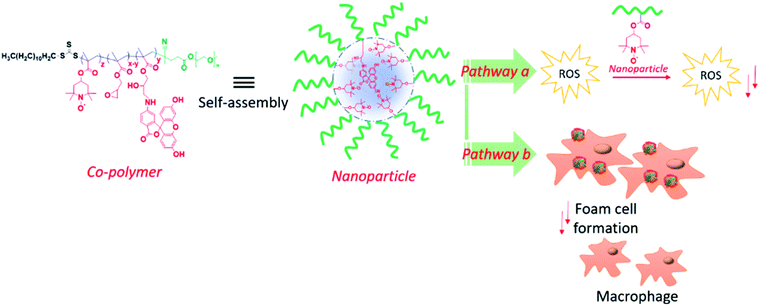
The nanogel NP based redox protein delivery plays an important role for the reduction of foam cell formation, as reported elsewhere.35 In redox protein therapy, it is challenging and difficult to protect the protein active site under physiological conditions. However, SOD enzyme delivery is well established and has several limitations, which include high production costs, short catalytic half-lives, limited active sites, and environmental sensitivity. We introduce a new type of RAFT mediated antioxidant NP for atherosclerosis therapy, which can mimic the SOD enzyme. The NPs are composed of diblock copolymers, consisting of polyethylene glycol together with PMA and GMA (Fig. 1). We hypothesize that the NPs will perform via two different mechanistic pathways: (a) NPs quench free radicals and reduce the oxidation of LDL by transition metal (e.g. Cu2+); (b) NPs induces the cellular uptake of ox-LDL and reduce the foam cell formation. Moreover, the RAFT nitroxide NPs have superior properties as they offer: (1) biocompatibility even at high dose level (500 μg mL−1), (2) efficiently mimicking SOD enzyme and very sensitive at low concentration limit, (3) NP stability in aqueous solution up to 3 weeks, (4) facile RAFT synthesis providing molecular weight control and monodispersity and easy NPs creation in aqueous medium, and (5) finally, softness, ∼100 nm PEGylated NPs potentially are useful as a passive delivery platform. Hydrophilic polyethylene glycol and hydrophobic GMA or 6-aminofluorescein (6-AF) renders the polymer amphiphilic and aids the self-assembly in aqueous solution. Moreover, polyethylene glycol improves the cytocompatibility of the NPs. The nitroxyl radical containing NPs facilitate the antioxidant ability and reduce the LDL oxidation. This NP could be easily up taken by the macrophages and inhibit ox-LDL induced foam cell formation that is closely related to the pathogenesis of atherosclerosis. Thus, these antioxidant NPs could be a promising therapy for atherosclerosis.
Materials and methods
Materials
All the materials were used without any further purification. 2,2,6,6-Tetramethyl-4-piperidyl methacrylate (>98%; PMA) was purchased from TCI Chemicals Pvt. Ltd. Glycidyl methacrylate (≥98%; GMA) and poly(ethyleneglycol)methylether 4-cyano 4[(dodecylsulfanylthiocarbonyl)sulfanyl] pentanoate (PEGmacro-CTA; Mn = 10 kDa) were purchased from Sigma. We measured Mp = 8300 g mol−1 by SEC see Fig. 2 and Mn = 9.8 kg mol−1 by NMR. 6-Aminofluorescein (≥95%; 6-AF), 3-chloroperbenzoic acid (≥78%; mCPBA) and 2,2′-azobis(2-methylpropionitrile) (≥98%; AIBN) were purchased from Sigma. Human LDL (Sigma-Aldrich) and Oil Red O solution (0.5% in isopropanol, Sigma-Aldrich) were purchased from Sigma. Hoechst 33342 Solution (20 mM) (Thermo Scientific™), ProLong® Gold mount media (Thermo Fisher Scientific), Nunc™ MicroWell™ 96-Well Microplates – Thermo Fisher Scientific, Total Antioxidant Capacity Assay Kit (Sigma-Aldrich), Cell Counting Kit – 8 (Sigma-Aldrich), Dulbecco's Phosphate Buffered Saline (PBS) (Cell culture grade, Sigma-Aldrich), DMEM (Dulbecco's Modified Eagle's medium) (Sigma-Aldrich), chloroform-d 99.8 atom % D (≥99.8%, Sigma-Aldrich), dichloromethane ≥99.8% stabilized. | ||
| Fig. 2 (a) Outline of RAFT nitroxide polymer synthesis. (b) FTIR spectra of copolymer before (black) and after oxidation (red). (c) SEC of macro-CTA, before and after fluorophore grafting polymer. For calibration curve – see Fig. SI2.† (d) UV-vis spectra of polymer before and after oxidation and after fluorophore (6-AF) grafting. | ||
Instruments and characterizations
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PEG macro-CTA
PEG macro-CTA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AIBN = 50/100/200/
AIBN = 50/100/200/![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (reported in ESI†).
0.5 (reported in ESI†).| [M1 = PMA; M2 = glycidyl methacrylate] |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (equiv.) ratio in anhydrous dichloromethane (DCM). The mCPBA solution was added drop-wise to the polymer solution and then allowed to stir at room temperature under nitrogen for 12 hours. The oxidized polymer was precipitated in hexane, filtered, and dried under reduced pressure overnight to get the final polymer (SBTG1). Note that the polymer changed from a white powder to an orange powder after oxidation. The successful oxidation of the nitroxide polymer was analyzed using UV-vis spectroscopy. The UV-vis spectroscopy was used as a complementary technique to measure the absorbance of nitroxyl group at 460 nm wavelength and to characterize the conversion.
1 (equiv.) ratio in anhydrous dichloromethane (DCM). The mCPBA solution was added drop-wise to the polymer solution and then allowed to stir at room temperature under nitrogen for 12 hours. The oxidized polymer was precipitated in hexane, filtered, and dried under reduced pressure overnight to get the final polymer (SBTG1). Note that the polymer changed from a white powder to an orange powder after oxidation. The successful oxidation of the nitroxide polymer was analyzed using UV-vis spectroscopy. The UV-vis spectroscopy was used as a complementary technique to measure the absorbance of nitroxyl group at 460 nm wavelength and to characterize the conversion.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DMSO (3
DMSO (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v)) at room temperature overnight to remove unreacted 6-aminofluorescein (6-AF). The pure 6-AF conjugated copolymer (SBTGF1) was freeze dried and obtained as an orange powder and used for further characterization.
1 (v/v)) at room temperature overnight to remove unreacted 6-aminofluorescein (6-AF). The pure 6-AF conjugated copolymer (SBTGF1) was freeze dried and obtained as an orange powder and used for further characterization.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000, 1 mL per well), incubated for 15 minutes, 37 °C, 5% CO2 and washed with PBS buffer. Finally, cover slip was fixed on the glass slides using mounting medium (that can add favorable properties such as optimizing the refractive index to match that of glass, preventing photo-bleaching) and microscope Zeiss LSM 710 was used with 63× magnification to investigated the cellular uptake at the fluorescence intensity; λex: 490 nm/λem: 520 nm.
5000, 1 mL per well), incubated for 15 minutes, 37 °C, 5% CO2 and washed with PBS buffer. Finally, cover slip was fixed on the glass slides using mounting medium (that can add favorable properties such as optimizing the refractive index to match that of glass, preventing photo-bleaching) and microscope Zeiss LSM 710 was used with 63× magnification to investigated the cellular uptake at the fluorescence intensity; λex: 490 nm/λem: 520 nm.
| Sa/Sv = concentration of antioxidant in each sample |
Furthermore, the anti-ROS efficacy of the RAFT nitroxide NPs was examined by malondialdehyde (MDA) lipid peroxidation assay (reported in ESI†).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 minutes. LDL was oxidized with 5 μM Cu2+ for 4 h at 37 °C and Cu2+ was removed by dialysis (with and without treated SBTG1 nitroxide NPs). The top layer was collected and mixed with 2 μL of BHT in 100 μL of deionized water and the final volume was adjusted to 200 μL. Finally, 600 μL of TBA solution was mixed together with the 200 μL analyte solution and incubated at 95 °C for 60 minutes and the UV absorbance of the solution measured at 532 nm.
000g for 5 minutes. LDL was oxidized with 5 μM Cu2+ for 4 h at 37 °C and Cu2+ was removed by dialysis (with and without treated SBTG1 nitroxide NPs). The top layer was collected and mixed with 2 μL of BHT in 100 μL of deionized water and the final volume was adjusted to 200 μL. Finally, 600 μL of TBA solution was mixed together with the 200 μL analyte solution and incubated at 95 °C for 60 minutes and the UV absorbance of the solution measured at 532 nm.
Results and discussion
Design and synthesis of reversible addition fragmentation chain transfer (RAFT) copolymer
An amphiphilic copolymer was synthesized by the copolymerization of PMA (M1) and glycidyl methacrylate (M2) using poly(ethylene glycol) methyl ether 4-cyano-4-[(dodecylsulfanylthiocarbonyl)sulfanyl]pentanoate (PEG macro-CTA)as chain transfer agent as shown in Scheme 1. The linear diblock copolymers are designed to have a PEG hydrophilic block that displays biocompatibility, followed by a random sequence of free radical scavenging 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO) and glycidyl methacrylate block that, at appropriate condition, will direct the self-assembly towards particle in aqueous medium (Fig. 1). Free radical scavenger 2,2,6,6-tetramethylpiperidine-N-oxyl (TEMPO) was chosen due to the known SOD mimicking activity and reduction of LDL oxidation.23 Copolymerization reactions of M1 and M2 were carried out using azobisisobutyronitrile (AIBN) as an initiator and PEG macro-CTA as chain transfer agent (CTA) in dry tetrahydrofuran (THF) at 70 °C via RAFT polymerization (Scheme 1). | ||
| Scheme 1 Synthesis of copolymer via RAFT polymerization, and grafting of 6-aminofluorescein via oxirane ring-opening reaction. | ||
For the synthesis of the RAFT nitroxide copolymer, PEGylated macro-CTA was used in order to produce a hydrophilic nanostructure, and ensure biocompatibility.38 After purification by precipitation in n-hexane, the copolymers were characterized by 1H NMR spectroscopy. The RAFT polymerization offer the synthesis of water soluble amphiphilic architectures with precise molecular weight and relatively narrow PDI.39 The copolymers were named as SBPGx where x represents the series of different feed ratios of M1 and M2 to the macro-CTA (SBPG1–SBPG3, Table 1). The macro-CTA to initiator ratio [macro-CTA]/[AIBN] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 was kept constant during all the polymerization reactions. After purification by precipitation in n-hexane, the copolymers were characterized by 1H NMR spectroscopy. The 1H NMR spectrum of the macro-CTA and SBPG1 are shown and discussed in the ESI.† The number-average molecular weight (Mn,SEC) and molecular weight distribution (dispersity, Đ) values of the copolymers were evaluated by size-exclusion chromatography (SEC) in N,N-dimethylformamide (DMF) and results are summarized (Table 1).
0.5 was kept constant during all the polymerization reactions. After purification by precipitation in n-hexane, the copolymers were characterized by 1H NMR spectroscopy. The 1H NMR spectrum of the macro-CTA and SBPG1 are shown and discussed in the ESI.† The number-average molecular weight (Mn,SEC) and molecular weight distribution (dispersity, Đ) values of the copolymers were evaluated by size-exclusion chromatography (SEC) in N,N-dimethylformamide (DMF) and results are summarized (Table 1).
| Polymer | [M1]/[M2]/[CTA]a | Conv.b (%) | M n,SECc (g mol−1) | M p,SEC | Đ | M n,theo (g mol−1) |
|---|---|---|---|---|---|---|
a [macro-CTA]/[AIBN] = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 for all polymerization reactions. Polymerization time = 24 h.
b Determined gravimetrically.
c Measured using SEC analysis in DMF. Mp is the molecular weight based on PEG calibration at the peak of the elution curve.
d
M
n,theo = ([M1 + M2]/[macro-CTA] × (Mw of M1 + Mw of M2) × conversion% + Mw of macro-CTA), where Mw = molecular weight of the components. 0.5 for all polymerization reactions. Polymerization time = 24 h.
b Determined gravimetrically.
c Measured using SEC analysis in DMF. Mp is the molecular weight based on PEG calibration at the peak of the elution curve.
d
M
n,theo = ([M1 + M2]/[macro-CTA] × (Mw of M1 + Mw of M2) × conversion% + Mw of macro-CTA), where Mw = molecular weight of the components.
|
||||||
| SBPG1 | 50/50/1 | 30 | 8510 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
1.13 | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 010 010 |
| SBPG2 | 100/100/1 | 35 | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 780 780 |
1.62 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 690 690 |
| SBPG3 | 200/200/1 | 33 | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 990 990 |
1.27 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 444 444 |
Differences between theoretical and experimental molecular weight can be attributed to the differences in hydrodynamic volume between SBPGx and the polyethylene glycol (PEG) calibration standard used (Fig. SI1†). Increase in the molecular weight with respect to the PEG macro-CTA confirmed the attachment of both M1 and M2 comonomers into the polymer chain (Fig. SI2†). SEC plots (see Fig. SI2†) of SBPGx polymers showed unimodal molecular weight distributions. The molecular weight of the copolymers increases with the increase of [monomer (M1) + (M2)]/[macro-CTA] ratio in the feed. Therefore, the number-average molecular weight (Mn,NMR) of the copolymer (SBPG1) was evaluated by proton NMR analysis (see Fig. SI4, SI5, Tables SI1 and SI2†), and the Mn,NMR = 19.8 kg mol−1 for SBPG1 is comparable to the Mn,theo. All copolymers solubility were tested in order to create NPs in aqueous medium. We concluded that the relatively hydrophobic nature of the side chain combined with the increased molecular weight lead to insolubility in aqueous medium for SBPG2 and SBPG3 (Fig. SI6†). Furthermore, the control of Đ is less satisfactory at high ratios. Thus, copolymer SBPG1 appear most attractive for the investigation of the potential to generate stable NPs useful in drug delivery.
Oxidation of the PMA block using mCPBA in dry DCM (Fig. 2a) was carried out at different time intervals (2 h, 4 h, 6 h, 12 h) and the reaction monitored using UV-spectrophotometry (Fig. SI7†). The conversion is linear in time and, thus the highest yield (%) was recovered after 12 h reaction. The oxidized polymer was precipitated in cold hexane and dried overnight to obtain an orange colour polymer named SBTG1 (copolymer containing nitroxide). The N–O˙ and +N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching after oxidation (SBTG1) was examined by FTIR spectroscopy with a characteristic peak at 1360 cm−1 and 1555 cm−1, respectively (Fig. 2b).40 The epoxy group is intact with the polymer backbone and the asymmetric vibrational modes associated to the epoxy ring at 926 and 843 cm−1. The trithiocarbonate (C
O stretching after oxidation (SBTG1) was examined by FTIR spectroscopy with a characteristic peak at 1360 cm−1 and 1555 cm−1, respectively (Fig. 2b).40 The epoxy group is intact with the polymer backbone and the asymmetric vibrational modes associated to the epoxy ring at 926 and 843 cm−1. The trithiocarbonate (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S) unit at 1060 cm−1 is observed after oxidation and it is noted that the RAFT end functionality shown in (Scheme 1) is present after oxidation. The fluorophore (6-aminofluorescein, 6-AF) was grafted to SBTG1 polymer using oxirane ring-opening reaction, and was named SBTGF1 (copolymer containing both nitroxyl radical and fluorophore) (Scheme 1). The fluorophore (6-aminofluorescein, 6-AF) grafting was confirmed by SEC and UV-vis spectroscopy (Fig. 2c and d). We note that spectrophotometric quantification of both oxidation and fluorophore grafting is hampered by turbidity of the aqueous samples (see Fig. SI8†). The SEC shows considerable increase in the hydrodynamic volume on oxidation and the following fluorophore grafting. The Mp-values of CTA, SBPG1, and SBTGF1 are 8300 g mol−1, 10
S) unit at 1060 cm−1 is observed after oxidation and it is noted that the RAFT end functionality shown in (Scheme 1) is present after oxidation. The fluorophore (6-aminofluorescein, 6-AF) was grafted to SBTG1 polymer using oxirane ring-opening reaction, and was named SBTGF1 (copolymer containing both nitroxyl radical and fluorophore) (Scheme 1). The fluorophore (6-aminofluorescein, 6-AF) grafting was confirmed by SEC and UV-vis spectroscopy (Fig. 2c and d). We note that spectrophotometric quantification of both oxidation and fluorophore grafting is hampered by turbidity of the aqueous samples (see Fig. SI8†). The SEC shows considerable increase in the hydrodynamic volume on oxidation and the following fluorophore grafting. The Mp-values of CTA, SBPG1, and SBTGF1 are 8300 g mol−1, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 g mol−1 and 19
300 g mol−1 and 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 180 g mol−1, respectively.
180 g mol−1, respectively.
Nanoparticles (NPs) preparation and characterization
Amphiphilic copolymers with densely grafted macromolecular architectures in selective solvents can possess a variety of morphologies such as spherical micelles,41 cylindrical nanostructures,42 multimolecular micelle43etc. However, these molecular assemblies are vastly dependent on the hydrophobicity/hydrophilicity (fb/fc) ratios of the copolymers.44 We have designed three different nanoparticles (NPs) named SBPG1, SBTG1 and SBTGF1 polymer based on the native, oxidized and fluorophore grafted structure. The nanoparticles were fabricated via the self-assembly of the copolymer by dialyzing their solutions in DMSO against deionized water (DI water). Because of the presence of long PEG side chains, the synthesized polymers form micelles in aqueous media with the M1/M2 random copolymer as core and the hydrophilic PEG as corona. First, the self-assembly behavior was investigated by dynamic light scattering (DLS) (Fig. 3a). The hydrodynamic diameter (Dh) values were found to be 85 ± 5.2 (PDI = 0.24 ± 0.02), 120 ± 7.3 (PDI = 0.15 ± 0.03), 157 ± 2.1 nm (PDI = 0.32 ± 0.04) for SBPG1, SBTG1, and SBTGF1, respectively, with a monomodal size distribution and low PDI (Table 2). Furthermore, SBPG2 and SPBG3 show tendencies to form multimodal size distribution of the formed nanoparticles. This fact supports the choice of copolymer SBPG1 for the remainder of the study (see Fig. SI8a–c†).| Nanoparticle | Size (nm) | PDI | Zeta potential (mV) | |
|---|---|---|---|---|
| SBPG1 | In DI water | 85 ± 5.2 | 0.24 ± 0.02 | −3.4 ± 0.12 |
| In PBS | 92 ± 1.2 | 0.37 ± 0.05 | ||
| SBTG1 | In DI water | 120 ± 7.3 | 0.15 ± 0.03 | +14.4 ± 1.43 |
| In PBS | 126 ± 6.7 | 0.34 ± 0.01 | ||
| SBTGF1 | In DI water | 157 ± 2.1 | 0.32 ± 0.04 | +27 ± 2.09 |
| In PBS | 159 ± 3.2 | 0.31 ± 0.03 | ||
The significant increase in hydrodynamic size (Dh), upon self-assembly in water of the three different SBPGx polymers ranging from 85–312 nm (Fig. SI8a–c†) as a function of [monomer (M1) + (M2)]/[macro-CTA] ratio in the feed is rationalized by the fact that the balance between hydrophilic (PEG) and hydrophobic (M1/M2 random copolymer) block is pushed towards a larger hydrophobic block. Further, the increasing size from SBPG1 to SBTGF1 micelles is due to stretching of the hydrophobic block in accordance with the size observed by SEC.
To gain more information on the morphology of the nanostructure of copolymers, cryo-TEM images were acquired. As shown in (Fig. 3c–e), SBPG1, SBTG1, and SBTGF1 copolymers self-assembled into spherical nanoparticles with an average size of 80 nm to 150 nm. The size increase from native nanoparticle (SBPG1) to fluorophore nanoparticle (SBTGF1) accommodates the chemical functionalization induced chain stretching of the micelle core forming block.
Stability study
Nanoparticle (NP) stability was investigated using DLS measurements. An ideal nanoparticle should be stable in an aqueous medium for a sufficiently long time for storage purposes without aggregation. Furthermore, a higher colloidal stability is also correlated with the long circulation time in vivo. Thus, PEGylated core–shell nanoparticles show higher colloidal stability in aqueous media and are potentially useful for drug delivery application.45 The particle size was evaluated in PBS and deionized water (DI water) at 25 °C (Table 2) and the stability of the particle was studied in deionized water (DI water). It was observed that storage up to three weeks did not lead to nanoparticles aggregation and the size remains the same (Fig. 4). As shown in the Fig. 4, the nanoparticles are completely stable in DI water at 25 °C.Cellular internalization
We investigated the cellular internalization of the 6-aminofluorescein (6-AF; λex: 490 nm/λem: 520 nm) containing nanoparticles (SBTGF1). RAW 264.7 cells were seeded in the 12-well plate on top of the cover slip with a cell density of 105 cells per mL for 24 h in an incubator at 37 °C, 5% CO2. The cells were treated with LPS (100 ng mL−1) and IFN-γ (100 IU mL−1) for 24 h. Next, RAW 264.7 cells were incubated with SBTGF1 NPs for 24 h at 37 °C. We observed respective signals at λem: 457 nm, 520 nm and 590 nm, which corresponds to DAPI, nanoparticles and cell membrane staining, respectively (Fig. 5a–d). Efficient internalization was observed without causing any pernicious effect to the RAW 264.7 cells. Thus, the confocal study suggests the efficient uptake of the SBTGF1 nanoparticles in the perinuclear region of the RAW 264.7 cells.Cytotoxicity analysis
The cytocompatibility of the polymeric nanoparticles is an important factor for a drug delivery system.46 In this regards, we employed CCK-8 assay to determine the cytotoxicity of the nitroxyl radical containing nanoparticles (SBTG1) after incubation with the RAW 264.7 cell line. RAW 264.7 cells were exposed to various RAFT nitroxide polymer (SBTG1) NP concentrations. The results are shown in Fig. 5e, where more than 80% cell viability is observed with SBTG1 NPs for concentration between 0 and 500 μg mL−1 for 24 h and 48 h at 37 °C indicating biocompatibility. Hence, the low toxicity of the SBTG1 NPs toward the RAW 264.7 cells provides a direct indication that the newly developed nitroxyl containing copolymer can be administered in sufficiently high dose to function as an antioxidant nanodrug.Inhibition of foam cell formation by Oil red O (ORO) assay
Macrophage foam cell formation in the atherosclerotic plaque is a major hallmark of this disease and the process is mediated by the uptake of ox-LDL.47 The excessive uptake of ox-LDL by macrophages leads to their conversion into foam cells.The LDL oxidation is mediated by superoxide (O2˙−) and hydroxyl radicals (˙OH) in the presence of transition metal ions (such as Cu2+).48 Furthermore, macrophages subsequently engulf oxidized low-density lipoprotein (ox-LDL) particles via scavenger receptors, mainly CD36 and scavenger receptor-A (SR-A), leading to the formation of foam cells. The hypothesis that treatment with nitroxide RAFT (SBTG1) nanoparticle is effective in inhibiting foam cell formation (see Fig. 6). Wang et al. reported that the reactive oxygen species scavenging nitroxide nanoparticles not only reduce the intracellular ROS production but also reduce the foam cell formation.23 It is reported that, nitro-oleic acid (NO2-OA) specifically interacts with CD36, thus limiting the binding and uptake of mLDL.49 Hence, antioxidant nano-therapy regulate the cellular ROS production and counteract the ox-LDL uptake.50 Therefore, we investigated the effect of SBTG1 NP treatment on the cellular uptake of ox-LDL in RAW264.7 cells. Anti-atherosclerosis effects of Mito-Tempol, an antioxidant, played a key role in autophagy restoration involving THP-1 macrophage-derived foam cells. Autophagy is restored by Mito-Tempol in ox-LDL loaded THP-1 macrophages via mTOR pathway and inhibits the ox-LDL induced foam cell formation.51 We examined whether treatment with SBTG1 NP effectively inhibit the ox-LDL uptake followed by foam cell formation. In this regards, we performed the oil red O (ORO) assay to identify the fat containing oil droplet by ORO staining. RAW 264.7 macrophages treated with 50 μg mL−1 ox-LDL for 24 h showed considerable intracellular lipid droplet formation (Fig. 6b, denoted by arrows on top right panel) and significant macrophage foam cell formation, as represented by staining with Oil Red O (ORO) (Fig. 6b). Pre-treatment with SBTG1 NP at either 10 or 25 μg mL−1 significantly reduced (at 10 μg mL−1) the formation of foam cells and notably suppressed (at 25 μg mL−1) foam cell formation. Control experiment was done without addition of ox-LDL. Quantification of intra-cellularly deposited ORO supported this microscopic observation (Fig. 6b). UV-vis spectra of the lysis solution provided the quantitative estimation of the oil droplet formation (Fig. 7a). Thus, these results demonstrated that SBTG1 NPs could attenuate the formation of foam cells from macrophages by reducing cellular internalization of ox-LDL almost to the level seen without ox-LDL exposure. A control experiment shows that nanoparticles without the nitroxy function does not induce the antioxidant function of the nanoparticles (see Fig. SI10†).
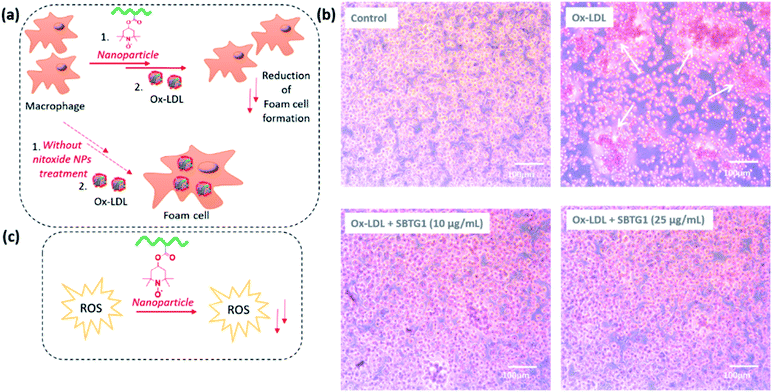 | ||
| Fig. 6 (a) Schematic illustration of anti-ROS, and reduction of foam cell formation effects of SBTG1 NPs. (b) Oil red assay in RAW 264.7 cell lines. Upper left panel: no ox-LDL; upper right panel: 50 μg mL−1 ox-LDL (24 h) – the white arrows point to lipid droplets – for an experiment that includes SBPG particles as well – see ESI Fig. S9†; lower left panel: 10 μg mL−1 SBTG1 followed by 50 μg mL−1 ox-LDL (24 h); lower right panel: 25 μg mL−1 SBTG1 followed by 50 μg mL−1 ox-LDL (24 h) (scale bar 100 μm). (c) Schematic illustration of anti ROS and free radical scavenging effects of SBTG1 NPs. | ||
Antioxidant effect of RAFT nitroxide nanoparticles
Oxidation of low-density lipoprotein (ox-LDL) is a key step in atherosclerosis, and transition metal ions (i.e. Cu2+) are implicated in this process. Interestingly, copper (Cu2+) levels are significantly increased in atherosclerotic lesions, compared with healthy, arterial walls, and promotes lipid peroxidation. A catalytic quantity of copper (Cu2+) together with superoxide (O2˙−) can stimulate LDL oxidation.52 (Cu2+) catalyzed breakdown of LOOH to reactive peroxyl (LOO˙) and alkoxyl (LO˙) radicals according to the reaction (1) & (2). Nitroxide radicals are effective to inhibit lipid peroxidation and scavenge free radicals. Nitroxide radicals have been found to protect cells against a variety of agents which cause oxidative stress,53 including superoxide,54 hydrogen peroxide,54 and oxidation of reduced metal ions, such as Cu(I) and Fe(II) a reaction that promote oxidation.55Therefore, we hypothesized that our RAFT nitroxide NPs could efficiently scavenge these free radicals and thereby reduce the LDL oxidation.
| LOOH + Cu(2+) = LOO˙ + Cu(1+) | (1) |
| Cu(1+) + LOOH = LO˙ + OH(−) + Cu(2+) | (2) |
The measurement of the total oxidant status (TOS)56 accurately reflects the oxidative stress markers of atherosclerotic environment. Nitric oxide (NO) acts as a free radical scavenger and contributes to host defence against oxidation. MDA is a reliable and commonly used marker of overall lipid peroxidation levels and the presence of oxidative stress.57 However, nitric oxide has been shown to reduce the lipid peroxidation by quenching the ROS.58 Therefore, this study aimed to determine oxidative stress levels that is associated with the overproduced ROS. The anti-ROS effectiveness of the RAFT nitroxide NPs was shown by the reduction of MDA formation (Fig SI10†) the antioxidant activity of SBTG1 NPs was evaluated using TAC (total antioxidant capacity assay),59 where the Cu2+/Cu+ system is used for antioxidant capacity quantification determined by UV spectrophotometry using Trolox standard. SBPG1 was considered as a control. SBPG1 and or, SBTG1 NPs were treated with different concentrations, and at higher concentration, the total antioxidant capacity (nM μL−1) increased (Fig. 7b). At 50 μg mL−1 the SBTG1 NPs showed almost double activity to the SBPG1 NPs. Thus, these results demonstrated that SBTG1 NPs at lower dose (50 μg mL−1) can serve as a potential antioxidant to reduce over expressed ROS and protect from oxidative damage.
Conclusion
In summary, we report the development of stable antioxidant and anti-ROS SOD mimicking polymeric nanoparticle. The nitroxyl radical containing NPs effectively show antioxidant property. The NPs could also significantly decrease cellular uptake of ox-LDL and thereby reduce foam cell formation in macrophages. The nanoparticles are efficiently internalized into the cells which remain viable up to high NP concentration. Thus, these properties of the synthesized NPs were realized by reducing oxidative stress and inflammation and thereby reducing the atherogenicity.Author contributions
Conceptualization: SB, KA, NK; formal analysis: SB, HK, PK, NK, KA; funding acquisition; investigation: SB, HK, PK; project administration: NK, KA; supervision: NK, KA; validation: NK, KA; writing – original draft: SB; writing – review & editing: NK, KA.Conflicts of interest
There are no conflicts to declare.Acknowledgements
S. B. acknowledges support from the Technical University of Denmark, Department of Health Technology. N. K. acknowledges funding from The Lundbeck Foundation (grant R215-2015-4190).References
- S. Daniel, Nat. Med., 2002, 8, 1211–1217 CrossRef PubMed.
- C. H. Hennekens and J. M. Gaziano, Clin. Cardiol., 1993, 16, 10–15 CrossRef PubMed.
- P. P. Singh, F. Mahadi, A. Roy and P. Sharma, Indian J. Clin. Biochem., 2009, 24, 324–342 CrossRef CAS PubMed.
- M. Mittal, M. R. Siddiqui, K. Tran, S. P. Reddy and A. B. Malik, Antioxid. Redox Signaling, 2014, 20, 1126–1167 CrossRef CAS PubMed.
- P. A. Grimsrud, H. Xie, T. J. Griffin and D. A. Bernlohr, J. Biol. Chem., 2008, 283, 21837–21841 CrossRef CAS PubMed.
- U. Förstermann, N. Xia and H. Li, Circ. Res., 2017, 120, 713–735 CrossRef PubMed.
- T. Münzel, G. G. Camici, C. Maack, N. R. Bonetti, V. Fuster and J. C. Kovacic, J. Am. Coll. Cardiol., 2017, 70, 212–229 CrossRef PubMed.
- K. K. Griendling, D. Sorescu, B. Lassègue and M. Ushio-Fukai, Arterioscler., Thromb., Vasc. Biol., 2000, 20, 2175–2183 CrossRef CAS PubMed.
- H. Li, S. Horke and U. Förstermann, Atherosclerosis, 2014, 237, 208–219 CrossRef CAS PubMed.
- Y. Wang, M. Subramanian, A. Yurdagul, V. C. Barbosa-Lorenzi, B. Cai, J. de Juan-Sanz, T. A. Ryan, M. Nomura, F. R. Maxfield and I. Tabas, Cell, 2017, 171, 331–345.e22 CrossRef CAS PubMed.
- S. Z. A. S. Shariat, S. A. Mostafavi and F. Khakpour, Iran. Biomed. J., 2013, 17, 22–38 CAS.
- L. He, T. He, S. Farrar, L. Ji, T. Liu and X. Ma, Cell. Physiol. Biochem., 2017, 44, 532–553 CrossRef PubMed.
- R. Asmis and J. Jelk, Arterioscler., Thromb., Vasc. Biol., 2000, 20, 2078–2086 CrossRef CAS PubMed.
- A. M. Gotto, J. Am. Coll. Cardiol., 2003, 41, 1205–1210 CrossRef CAS PubMed.
- J. T. Salonen, K. Nyyssönen, R. Salonen, H. M. Lakka, J. Kaikkonen, E. Porkkala-Sarataho, S. Voutilainen, T. A. Lakka, T. Rissanen, L. Leskinen, T. P. Tuomainen, V. P. Valkonen, U. Ristonmaa and H. E. Poulsen, J. Intern. Med., 2000, 248, 377–386 CrossRef CAS PubMed.
- S. R. Thomas, P. K. Witting and R. Stocker, BioFactors, 1999, 9, 207–224 CrossRef CAS PubMed.
- Y. Jun, K. Shigehiro, K. Takuro and A. Toshiaki, Atheroscler, 1999, 142, 139–149 CrossRef.
- C. H. J. Kim, J. B. Mitchell, C. A. Bursill, A. L. Sowers, A. Thetford, J. A. Cook, D. M. van Reyk and M. J. Davies, Atherosclerosis, 2015, 240, 234–241 CrossRef CAS PubMed.
- H. Langbein, C. Brunssen, A. Hofmann, P. Cimalla, M. Brux, S. R. Bornstein, A. Deussen, E. Koch and H. Morawietz, Eur. Heart J., 2016, 37, 1753–1761 CrossRef CAS PubMed.
- K. Sugamura and J. F. Keaney, Free Radicals Biol. Med., 2011, 51, 978–992 CrossRef CAS PubMed.
- D. R. Lewis, L. K. Petersen, A. W. York, K. R. Zablocki, L. B. Joseph, V. Kholodovych, R. K. Prud'Homme, K. E. Uhrich and P. V. Moghe, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 2693–2698 CrossRef CAS PubMed.
- G. Fredman, N. Kamaly, S. Spolitu, J. Milton, D. Ghorpade, R. Chiasson, G. Kuriakose, M. Perretti, O. Farokhzad, O. Farokzhad and I. Tabas, Sci. Transl. Med., 2015, 7, 275ra20 CAS.
- Y. Wang, L. Li, W. Zhao, Y. Dou, H. An, H. Tao, X. Xu, Y. Jia, S. Lu, J. Zhang and H. Hu, ACS Nano, 2018, 12, 8943–8960 CrossRef CAS PubMed.
- Z. Gao, Y. Horiguchi, K. Nakai, A. Matsumura, M. Suzuki, K. Ono and Y. Nagasaki, Biomaterials, 2016, 104, 201–212 CrossRef CAS PubMed.
- T. Yoshitomi, Y. Ozaki, S. Thangavel and Y. Nagasaki, J. Controlled Release, 2013, 172, 137–143 CrossRef CAS PubMed.
- S. Sha, L. B. Vong, P. Chonpathompikunlert, T. Yoshitomi, H. Matsui and Y. Nagasaki, Biomaterials, 2013, 34, 8393–8400 CrossRef CAS PubMed.
- A. Alaarg, C. Pérez-Medina, J. M. Metselaar, M. Nahrendorf, Z. A. Fayad, G. Storm and W. J. M. Mulder, Adv. Drug Delivery Rev., 2017, 119, 143–158 CrossRef CAS PubMed.
- W. J. M. Mulder, F. A. Jaffer, Z. A. Fayad and M. Nahrendorf, Sci. Transl. Med., 2014, 6, 1–12 Search PubMed.
- M. E. Lobatto, C. Calcagno, A. Millon, M. L. Senders, F. Fay, P. M. Robson, S. Ramachandran, T. Binderup, M. P. M. Paridaans, S. Sensarn, S. Rogalla, R. E. Gordon, L. Cardoso, G. Storm, J. M. Metselaar, C. H. Contag, E. S. G. Stroes, Z. A. Fayad and W. J. M. Mulder, ACS Nano, 2015, 9, 1837–1847 CrossRef CAS PubMed.
- X. Wei, M. Ying, D. Dehaini, Y. Su, A. V. Kroll, J. Zhou, W. Gao, R. H. Fang, S. Chien and L. Zhang, ACS Nano, 2018, 12, 109–116 CrossRef CAS PubMed.
- Y. Dou, Y. Chen, X. Zhang, X. Xu, Y. Chen, J. Guo, D. Zhang, R. Wang, X. Li and J. Zhang, Biomaterials, 2017, 143, 93–108 CrossRef CAS PubMed.
- S. Katsuki, T. Matoba, S. Nakashiro, K. Sato, J. I. Koga, K. Nakano, Y. Nakano, S. Egusa, K. Sunagawa and K. Egashira, Circulation, 2014, 129, 896–906 CrossRef CAS PubMed.
- A. Kheirolomoom, C. W. Kim, J. W. Seo, S. Kumar, D. J. Son, M. K. J. Gagnon, E. S. Ingham, K. W. Ferrara and H. Jo, ACS Nano, 2015, 9, 8885–8897 CrossRef CAS PubMed.
- R. Duivenvoorden, J. Tang, D. P. Cormode, A. J. Mieszawska, D. Izquierdo-Garcia, C. Ozcan, M. J. Otten, N. Zaidi, M. E. Lobatto, S. M. Van Rijs, B. Priem, E. L. Kuan, C. Martel, B. Hewing, H. Sager, M. Nahrendorf, G. J. Randolph, E. S. G. Stroes, V. Fuster, E. A. Fisher, Z. A. Fayad and W. J. M. Mulder, Nat. Commun., 2014, 5, 1–12 Search PubMed.
- S. Basak, H. A. Khare, M. Roursgaard, P. J. Kempen, J. H. Lee, S. Bazban-Shotorbani, M. Kræmer, S. Chernyy, T. L. Andresen, K. Almdal and N. Kamaly, Biomacromolecules, 2021, 22, 386–398, DOI:10.1021/acs.biomac.0c01238.
- J. Li, J. Zhang, Y. Chen, N. Kawazoe and G. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 35683–35692 CrossRef CAS PubMed.
- Y. J. Garcia, A. J. Rodríguez-Malaver and N. Peñaloza, J. Neurosci. Methods, 2005, 144, 127–135 CrossRef CAS PubMed.
- R. Palao-Suay, M. R. Aguilar, F. J. Parra-Ruiz, S. Maji, R. Hoogenboom, N. A. Rohner, S. N. Thomas and J. San Román, Polym. Chem., 2016, 7, 838–850 RSC.
- N. J. Warren, O. O. Mykhaylyk, D. Mahmood, A. J. Ryan and S. P. Armes, J. Am. Chem. Soc., 2014, 136, 1023–1033 CrossRef CAS PubMed.
- Y. Zhang, A. Park, A. Cintora, S. R. McMillan, N. J. Harmon, A. Moehle, M. E. Flatté, G. D. Fuchs and C. K. Ober, J. Mater. Chem. C, 2017, 6, 111–118 RSC.
- J. Z. Du, L. Y. Tang, W. J. Song, Y. Shi and J. Wang, Biomacromolecules, 2009, 10, 2169–2174 CrossRef CAS PubMed.
- Z. Li, J. Ma, N. S. Lee and K. L. Wooley, J. Am. Chem. Soc., 2011, 133, 1228–1231 CrossRef CAS PubMed.
- J. Zou, Y. Yu, Y. Li, W. Ji, C. K. Chen, W. C. Law, P. N. Prasad and C. Cheng, Biomater. Sci., 2015, 3, 1078–1084 RSC.
- Z. H. Liu, W. J. Lv, S. L. Zhao, Y. Z. Shang, C. J. Peng, H. L. Wang and H. L. Liu, Comput. Condens. Matter, 2015, 2, 16–24 CrossRef.
- T. Gillich, C. Acikgöz, L. Isa, A. D. Schlüter, N. D. Spencer and M. Textor, ACS Nano, 2013, 7, 316–329 CrossRef CAS PubMed.
- S. Łukasiewicz, K. Szczepanowicz, E. Błasiak and M. Dziedzicka-Wasylewska, Langmuir, 2015, 31, 6415–6425 CrossRef PubMed.
- J. W. Gofman, F. Lindgren, H. Elliott, W. Mantz, B. Strisower, V. Herring, T. P. Lyon, J. W. Gofman, F. Lindgren, H. Elliott, W. Mantz, J. Hewitt, B. Strisower and V. Herring, Science, 1950, 111, 166–171 CrossRef CAS PubMed.
- S. M. Lynch and B. Frei, J. Lipid Res., 1993, 34, 1745–1753 CrossRef CAS.
- M. M. Vazquez, M. V. Gutierrez, S. R. Salvatore, M. Puiatti, V. A. Dato, G. A. Chiabrando, B. A. Freeman, F. J. Schopfer and G. Bonacci, Redox Biol., 2020, 36, 101591 CrossRef CAS PubMed.
- R. A. Chmielowski, D. S. Abdelhamid, J. J. Faig, L. K. Petersen, C. R. Gardner, K. E. Uhrich, L. B. Joseph and P. V. Moghe, Acta Biomater., 2017, 57, 85–94 CrossRef CAS PubMed.
- Y. Ma, Z. Huang, Z. Zhou, X. He, Y. Wang, C. Meng, G. Huang and N. Fang, Free Radicals Biol. Med., 2018, 129, 463–472 CrossRef CAS PubMed.
- J. K. Lodge, M. G. Traber and S. J. Sadler, Lipids, 2000, 35, 1087–1092 CrossRef CAS PubMed.
- Y. Miura, H. Utsumi and A. Hamada, Arch. Biochem. Biophys., 1993, 300, 148–156 CrossRef CAS PubMed.
- E. Lubos, D. E. Handy and J. Loscalzo, Role of Oxidative Stress and Nitric Oxide in Atherothrombosis, 2008, vol. 13 Search PubMed.
- H. He, T. L. Oo, W. Huang, L. F. He and M. Gu, Sci. Rep., 2019, 9, 1–12 Search PubMed.
- U. Singh and I. Jialal, Pathophysiology, 2006, 13, 129–142 CrossRef CAS PubMed.
- R. P. Maurya, M. K. Prajapat, V. P. Singh, M. Roy, R. Todi, S. Bosak, S. K. Singh, S. Chaudhary, A. Kumar and S. R. Morekar, Clin. Ophthalmol., 2021, 15, 871–879 CrossRef PubMed.
- M. D'Ischia, A. Palumbo and F. Buzzo, Nitric Oxide - Biol. Chem., 2000, 4, 4–14 CrossRef PubMed.
- R. Apak, K. Güçlü, B. Demirata, M. Özyürek, S. E. Çelik, B. Bektaşoǧlu, K. I. Berker and D. Özyurt, Molecules, 2007, 12, 1496–1547 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00631b |
| This journal is © The Royal Society of Chemistry 2022 |