Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Food authentication: truffle species classification by non-targeted lipidomics analyses using mass spectrometry assisted by ion mobility separation†
Marina
Creydt
 * and
Markus
Fischer
* and
Markus
Fischer

Hamburg School of Food Science - Institute of Food Chemistry, University of Hamburg, Grindelallee 117, 20146 Hamburg, Germany. E-mail: marina.creydt@uni-hamburg.de; markus.fischer@uni-hamburg.de; Tel: +49-40-42838-4357
First published on 26th May 2022
Abstract
Truffles are appreciated as food all over the world because of their extraordinary aroma. However, quantities are limited and successful cultivation in plantations is very labor-intensive and expensive, or even impossible for some species. These factors make truffles a very valuable food, which is why it is particularly rewarding and tempting to declare inferior species of truffles as more expensive species and thereby counterfeit them. The various species differ in their aroma and thus in their culinary value, but the adulterations cannot be detected on the basis of pure morphology. For this reason, the objective of the present study was to develop a non-targeted lipidomics approach using ion mobility spectrometry-mass spectrometry to distinguish between the white truffle species Tuber magnatum and T. borchii as well as the black truffle species T. melanosporum, T. aestivum and T. indicum. Several hundred features were detected, which were present in significantly different concentrations in the studied truffle species. The most important of them were identified using MS/MS spectra and collision cross section (CCS) values. Some compounds were detected whose CCS values have not yet been published and may facilitate identification by other researchers in the future. Just a few marker substances are sufficient to be able to distinguish both black and white truffle species with 100% accuracy. These results can be used for the development of rapid tests, which in the best case will allow truffle analysis directly on-site.
1. Introduction
Truffles are among the most valuable edible mushrooms. Specifically, the white Alba truffle from Italy (T. magnatum) is one of the most expensive foods in the world. On average, one kilogram of this species can cost up to 5000 USD. The high price, as well as numerous attempts to cultivate this species on plantations, make this food particularly attractive for adulteration with the comparatively inexpensive truffle species T. borchii (syn. T. albidum), whose price is only a tenth that of T. magnatum. Fluctuations in sales prices are often accompanied by climatic influences, which have a high impact on the truffle yield. Morphologically, the two species are difficult to distinguish from each other. Based on their aroma profile, both species can be distinguished, but this requires extensive experience.1–3Furthermore, there are also adulterations of black truffle species again and again. In this regard, falsifications mainly concern the species T. melanosporum, which grows in France, Spain and Italy. Depending on the yield of the truffle collection, T. melanosporum is traded with a market value of 1000–2000 USD kg−1. Among the most favorable black truffle species are T. indicum, T. himalayense and T. sinense, which are very often imported from China to Europe and are therefore also referred to as Chinese truffles.4,5 The Chinese species are much cheaper than T. melanosporum and are also difficult to distinguish from T. melanosporum with the naked eye. When the different species are stored together, the low-grade truffle species often absorb aroma components from T. melanosporum making sensory differentiation even more difficult. In this way, counterfeiting is also relatively easy and high profit margins can be achieved.6 The black truffle species T. aestivum, which is also traded in the variant T. uncinatum depending on the time of harvest, belongs to the medium-priced truffle species. This species is used for falsifications of T. melanosporum, too. Partly, however, it is adulterated even with the Chinese species itself.
Due to the high relevance of truffle falsifications as well as the high profit margins that can be achieved and the difficulty of distinguishing between the different species, there is a great need for objective, analytical methods to detect possible fakes. For this reason, research efforts at various cellular levels have been massively advanced in recent years. For example, there are approaches to detect truffle species using genomics-,7–11 proteomics-12–14 or isotopolomics-based15–18 methods. However, all of these approaches are quite time-consuming or require special laboratory infrastructure. The only exception is the recording of near infrared (NIR) spectra,19,20 which, though, cannot unambiguously identify potential marker compounds that can be used to develop a rapid test.
Therefore, the aim of the present study was to be able to distinguish the white truffle species T. magnatum and T. borchii as well as the black truffle species T. melanosporum, T. aestivum and T. indicum using a non-targeted lipidomics-based approach. In addition, this procedure allows the identification of some marker compounds. Thus, following this non-targeted study, rapid, targeted methods can be developed specifically for these selected key metabolites to ensure easy transfer to industry (e.g. incoming inspection) and governmental inspection agencies.
In this study, a non-targeted lipidomics approach using mass spectrometry was chosen because we have had very good experiences with this strategy in previous studies, e.g. in determining the geographical origin of asparagus (Asparagus officinalis) and maize (Zea mays).21,22 Furthermore, as far as we know, there is still no knowledge about the lipidome composition of truffles. A liquid chromatography-electrospray ionization-ion mobility-quadrupole-time of flight mass spectrometer instrument (LC-ESI-IM-QTOF-MS) was selected for the presented approach. LC-ESI-QTOF-MS devices have already been used many times for comparable aims and objectives. The additional implementation of an ion mobility cell provides a further separation level, especially for isobaric compounds, as well as an additional identification parameter, so that the assignment of the marker compound is correspondingly facilitated and can be better validated.23
2. Materials and methods
2.1 Chemicals
Acetonitrile, isopropanol, methanol (all LC-MS grade) as well as chloroform (HPLC grade), and ammonium formate (≥95% puriss.) were obtained from Carl Roth GmbH (Karlsruhe, Germany). Water was purified by using a Merck Millipore water purification system (Direct-Q 3 UV-R system) with a resistance of 18 MΩ x cm (Darmstadt, Germany). Hexakis(1H,1H,3H-perfluoropropoxy)phosphazene, purine and LC/MS calibration standard for ESI-TOF were bought from Agilent Technologies (Santa Clara, CA, USA).2.2 Truffle samples
In total, 78 truffle samples from the years 2017–2020 were measured. These included five white and black species: T. magnatum (n = 18), T. borchii (n = 7), T. melanosporum (n = 10), T. indicum (n = 11) and T. aestivum/uncinatum (n = 32). For sample acquisition, care was taken to procure samples from the most relevant economic growing regions. Accordingly, the samples came from: Italy (n = 26), Romania (n= 14), China (n = 11), Spain (n = 5), Bulgaria (n = 5), France (n = 3), Croatia (n = 2), Hungary (n = 2), Australia (n = 2), Iran (n = 1), Slovenia (n = 1), Moldova (n = 1) and of five samples the geographical origin was not known. The majority of the samples were purchased directly from the cultivation sites by an experienced truffle trader (La Bilancia, Trüffelhandels GmbH (Munich, Germany)) in order to ensure the highest possible level of authenticity and the specification of the species. In addition, the species information was checked by means of DNA analyses.7The truffles were sent to the research institute at +5 °C or −20 °C, where they were cleaned of adhering soil, brushed, and washed with ultrapure water. Subsequently, the samples were frozen in liquid nitrogen and stored at −80 °C. At least 75 g of each sample was ground in a knife mill (Grindomix GM 300, Retsch, Haan, Germany) with the addition of dry ice at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The powder obtained was freeze-dried for 72 h with regular mixing and stored at −80 °C until further processing.
1. The powder obtained was freeze-dried for 72 h with regular mixing and stored at −80 °C until further processing.
2.3 Metabolite extraction
Extraction of the lipid fraction was performed according to the method of Bligh&Dyer,24 which has shown promising results in previous studies.25,26 Briefly, 50 mg of each lyophilizate was mixed with 750 μL of ice-cold chloroform/methanol mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) in a 2.0 mL reaction tube (Eppendorf, Hamburg, Germany). Cell disruption was performed using a ball mill (Omni International IM, GA, USA) with two steel balls (3 mm in diameter) for 1 minute at 3 m s−1. Then 250 μL chloroform and 500 μL water were added, and the samples were homogenized again with the ball mill for a further 2 minutes. The extracts obtained were centrifuged for 20 minutes at 16
2, v/v) in a 2.0 mL reaction tube (Eppendorf, Hamburg, Germany). Cell disruption was performed using a ball mill (Omni International IM, GA, USA) with two steel balls (3 mm in diameter) for 1 minute at 3 m s−1. Then 250 μL chloroform and 500 μL water were added, and the samples were homogenized again with the ball mill for a further 2 minutes. The extracts obtained were centrifuged for 20 minutes at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g and 4 °C (Sigma, Osterode, Germany). After this step, 100 μL of the lower phase was taken and diluted with 900 μL of eluent B of the chromatographic method. The solutions were then centrifuged again at 16
000 g and 4 °C (Sigma, Osterode, Germany). After this step, 100 μL of the lower phase was taken and diluted with 900 μL of eluent B of the chromatographic method. The solutions were then centrifuged again at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g and 4 °C for 10 minutes. From the liquid phase, 500 μL was taken and transferred into a glass vial (Macherey-Nagel GmbH & Co. KG, Düren, Germany). For each measurement, 4 μL were injected. All steps were performed under ice cooling and with ice-cold solvent to avoid changes of the metabolites as far as possible as well as to ensure a reproducible analysis.
000 g and 4 °C for 10 minutes. From the liquid phase, 500 μL was taken and transferred into a glass vial (Macherey-Nagel GmbH & Co. KG, Düren, Germany). For each measurement, 4 μL were injected. All steps were performed under ice cooling and with ice-cold solvent to avoid changes of the metabolites as far as possible as well as to ensure a reproducible analysis.
2.4 Liquid chromatography and mass spectrometry conditions
Measurements were performed using an UHPLC system (1290 Infinity II, Agilent Technologies) coupled to an Agilent 6560 IM-QTOF-MS instrument (Agilent Technologies), equipped with an ESI source (Dual JetStream, Agilent Technologies) and a gas kit (Alternate Gas Kit, Agilent Technologies). Chromatographic separation of the extracts was performed with a reversed phase C18 column (1.7 μm, 150 × 2.1 mm, Phenomenex, Aschaffenburg, Germany). The column was maintained at 50 °C at a flow rate of 0.3 mL min−1. Prior to the measurements, the mass spectrometer was calibrated in the low mass range mode (m/z 50–1700) and extended dynamic range mode using the Agilent Technologies ESI tune mix. During the measurements, the two lock masses Hexakis(1H,1H,3H-perfluoropropoxy)phosphazene ([M + H]+, m/z 922.0098) as well as purine ([M + H]+, m/z 121.0509) were infused via a second sprayer in order to be able to calibrate the data sets.The mobile phases consisted of water (A) and isopropanol:acetonitrile (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) (B), both containing 0.1 mmol L−1 ammonium formate. The following gradient program was used: 0–2 min, 55% (B), 2–4 min, 55–80% (B); 4–22 min, 80–100% (B); 22–23 min, 100% (B); 23–24 min, 100–55% (B); 24–30 min, 55% (B). The mass spectrometer was operated in positive ionization mode and the mass range recorded was in the range of m/z 50–1700. The sample groups were measured randomly to avoid possible bias. Furthermore, the stability of the analytical system was checked using quality control (QC) samples, which were regularly injected every 13 measurements and consisted of aliquots of all sample extracts. The ESI parameters were chosen based on the work of Reisdorph et al. and were as follows: gas temperature 300 °C; drying gas flow rate 12 L min−1; nebulizer 35 psi; sheath gas temperature 275 °C; sheath gas flow rate 12 L min−1; capillary voltage 3500 V, nozzle voltage 250 V.27
1, v/v) (B), both containing 0.1 mmol L−1 ammonium formate. The following gradient program was used: 0–2 min, 55% (B), 2–4 min, 55–80% (B); 4–22 min, 80–100% (B); 22–23 min, 100% (B); 23–24 min, 100–55% (B); 24–30 min, 55% (B). The mass spectrometer was operated in positive ionization mode and the mass range recorded was in the range of m/z 50–1700. The sample groups were measured randomly to avoid possible bias. Furthermore, the stability of the analytical system was checked using quality control (QC) samples, which were regularly injected every 13 measurements and consisted of aliquots of all sample extracts. The ESI parameters were chosen based on the work of Reisdorph et al. and were as follows: gas temperature 300 °C; drying gas flow rate 12 L min−1; nebulizer 35 psi; sheath gas temperature 275 °C; sheath gas flow rate 12 L min−1; capillary voltage 3500 V, nozzle voltage 250 V.27
In addition, the corresponding drift times were measured in IM-TOF mode in order to be able to calculate the CCS values later. The parameters for measuring the drift times were also set following the publication by Reisdorph et al.27 Nitrogen served as drift gas and was adjusted to a pressure of approx. 3.95 Torr. The other settings were as follows: frame rate 1 frame per s; IM transient rate 19 IM transients/frame; max drift time 50 ms; trap fill time 3200 μs; trap release time 250 μs; multiplexing pulse sequence length 4 bit. Deviating from the tune file values, the drift tube entrance voltage was changed to 1574 V, the drift tube exit voltage to 224 V, the rear funnel entrance voltage to 217.5 V and the rear funnel exit voltage to 45 V. For calibration of drift times, the Agilent Technologies ESI tune mix was also infused into the mass spectrometer with the same parameters for 1 minute. In addition, several truffle samples were injected with 8 μL, 1 μL and also 1 μL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution. In this way, saturation effects can be avoided, and it can be ensured that the most accurate CCS values possible are obtained, which have not been influenced by space charge effects. MS/MS fragment spectra were recorded at 10, 20, 40 and 60 eV in QTOF mode to identify the most relevant marker compounds.
10 dilution. In this way, saturation effects can be avoided, and it can be ensured that the most accurate CCS values possible are obtained, which have not been influenced by space charge effects. MS/MS fragment spectra were recorded at 10, 20, 40 and 60 eV in QTOF mode to identify the most relevant marker compounds.
2.5 Data processing and identification
IM-TOF data files were demultiplexed by the PNNL PreProcessor software (version 2020.03.23).28,29 The settings were: demultiplexing checked; moving average smoothing checked; m/z not used; drift 3; chromatography/infusion 3, signal intensity lower threshold 20 counts, remove spikes checked, saturation repair not checked. Subsequently, CCS calibration was performed by IM-MS Browser software (version 10.0, Agilent Technologies). Four-dimensional feature finding was carried out using Mass Profiler software (version 10.0, Agilent Technologies) with these parameters: restrict RT to 0.0–23.0 min; ion intensity > 150.0 counts; isotope model common organic (no halogens); limit charge states to a range of 1–2; report single-ion features with charge state z = 1; RT tolerance = ±10.0% + 0.50 min; DT tolerance = ± 1.5%; tolerance = ± 20.0 ppm + 2.0 mDa; Q-Score > 70.0. A feature had to be detectable in at least 50% of all samples. The obtained bucket table was exported as.xls files and transferred to MetaboAnalyst 5.0 software.30 The percentage of missing values was about 25%. The missing values were replaced by the smallest value with which a feature could still be detected. Furthermore, a sum normalization and an autoscaling were performed. Plots of principal component analysis (PCA) or partial least squares discriminant analysis (PLS-DA) were calculated to estimate variances and homologies between sample groups. The most relevant marker substances were extracted with a t-test (for two groups) or analysis of variance (ANOVA, for more than two groups) using false discovery rates (FDRs) according to Benjamini–Hochberg.31 In addition, receiver operating characteristic (ROC) curves and area under curve (AUC) values were calculated for the evaluation of potential suitable marker compounds. MS/MS fragment spectra were recorded for each of the 60 most relevant marker compounds. The identification was partially supported by the Lipid Annotator software (Agilent Technologies) and LipidBlast (FiehnLab-Metabolomics UC Davis Genome Center, Davis, CA, USA) as well as by the databases LipidMaps32 and FooDB.33 In addition, tentative identification suggestions were checked using CCS values with the LipidCCS database or LipidCCS Predictor.34,353. Results and discussion
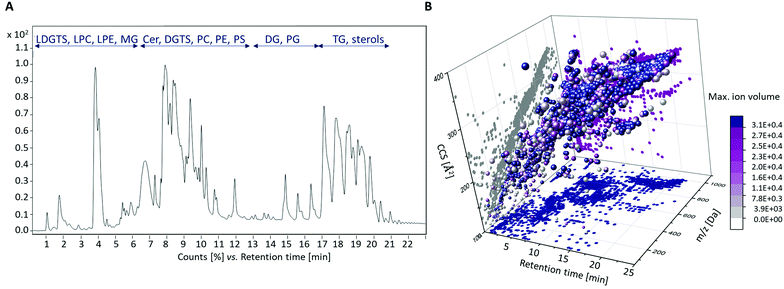
All data sets of the black and white truffle samples, including the QC samples, were first analyzed together to assess the quality of the measurements and to identify possible outliers. The bucket table obtained contained 1191 features. The two PCA scores plots (Fig. S1 in the ESI†) show that the measurements were reproducible, and no interference occurred, as the QC samples showed almost no variances. In addition, first differences and homologies according to the species affiliations of the samples become clear.Fig. 1A shows an example of the total ion chromatogram (TIC) of a sample of the species T. magnatum and the different chemical substance classes that could be detected. These mainly include: Lyso-diacylglyceryl-O-4′-(N,N,N-trimethyl) homoserines (LDGTSs), lyso-glycerophosphocholines (LPCs), lyso-glycerophosphoethanolamines (LPEs), monoacylglycerols (MGs), ceramides (Cers), diacylglyceryl-O-4′-(N,N,N-trimethyl) homoserines (DGTSs), glycerophosphocholines (PCs), glycerophosphoethanolamines (PEs), glycerophosphoserines (PSs), diacylglycerols (DGs), glycerophosphoglycerols (PGs), triacylglycerols (TGs) and sterols. Fig. 1B illustrates the features detected after four-dimensional separation in terms of retention time (RT), m/z ratio, calculated CCS values and max ion volume.
The identified analyte classes were mostly in line with our expectations from previous studies performed on foods using comparable methods.21,22,36,37 However, LDGTS and DGTS derivatives could be detected comparatively rarely, since their occurrence is mainly restricted to fungi, bacteria, amoebae, algae and nonvascular plants.38 To our best knowledge, this class of compounds was detected in truffles for the first time in this study.
Since clear and also many differences within the various sample groups became apparent early on during the data evaluation, the analysis was focused on the measurements in positive ionization mode. If it had turned out that these differences did not allow sufficient distinction, the investigation of the samples in negative mode or of polar analytes would have been an alternative, since other analyte windows are detected in this way.
3.1 Non-targeted lipidomics analysis of white truffle samples
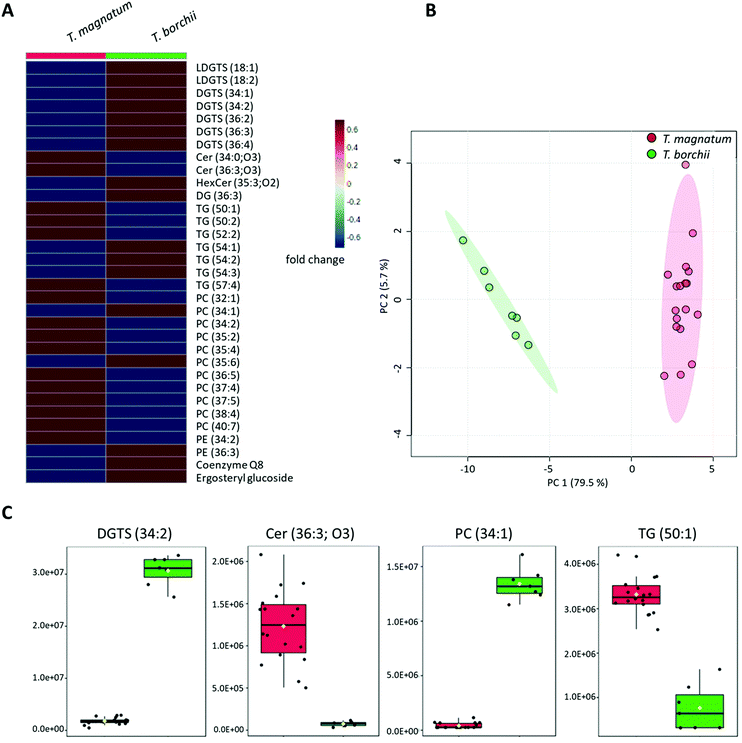
The bucket table obtained for the white truffle species T. magnatum and T. borchii contained 1521 features, of which 459 were found to be significantly different in the sample groups using a t-test calculation, as the FDR values were <0.05. This large number of potential marker compounds in the two truffle species is also reflected in the PCA scores plot (Fig. S2A in the ESI†), which shows a clear separation of the two sample groups. The result of a PLS-DA calculation and the associated leave-one-out cross validation (LOOCV) is correspondingly good. The Q2-value for two components is 0.95, indicating excellent separation of the two sample groups.MS/MS fragment spectra were recorded from the 60 most significant features and CCS values were compared with the LipidCCS database when possible (Table S1 and Fig. S4–S11 in the ESI†). In general, the deviation of the measured CCS values from the calculated CCS values of the LipidCCS database was ±1–2%, indicating a good result. In the case of phospholipids, several isobaric compounds can often be considered. The resolving power of the ion mobility is not yet sufficient for a clearer distinction, which is why a range was given here. In these cases, the metabolites must be confirmed, if necessary, either by measurements in the negative ion mode or by means of standards. In total, a preliminary identification could be carried out for 38 features. Due to different adduct-ions, individual substances were assigned multiple times. This particularly affected some TGs, which mainly appeared as [M + NH4]+-adducts, but also partly formed [M + Na]+-adducts, so that the number was reduced to a total of 33 metabolites. The relative concentration ratios of these identified marker compounds in the two sample groups are shown in Fig. 2A. All identified compounds have an FDR < 0.001 and therefore are highly significant. The resulting PCA plot based on these identified marker compounds is shown in Fig. 2B. However, for a good separation of the two sample groups, only a few marker compounds are sufficient to be able to achieve a reliable distinction. This aspect is illustrated in Fig. 2C using a few selected marker compounds. Since the results shown in Fig. 2 are based on a greatly reduced data set, no sum normalization was performed. In addition, the substitution of missing values was omitted because the most relevant features could be detected with comparatively high signal intensities, which is why this step was no longer necessary.
Among the identified marker compounds, a whole series of DGTS and LDGTS derivatives were conspicuous. A comparison of the measured CCS values with the lipids CCS database could not be carried out for this substance class, as there are currently no entries. Furthermore, according to our research, no CCS values for DGTS derivatives have been published in the literature so far. Nonetheless, the CCS values measured in the present study for the identified LDGTS and DGTS derivatives can be found in Tables S1 and S2 in the ESI.† In general, the identification of LDGTS and DGTS from MS/MS fragment spectra as [M + H]+-adducts was relatively straightforward as these compounds show two characteristic fragments at m/z 144.10 (C7H14NO2+) and m/z 236.15 (C10H22NO5+) in positive ionization mode (Fig. S4 and S5 in the ESI†).39
We exclude a potential contamination of the T. borchii samples with DGTS or LDGTS, because we have not noticed these compounds in comparable measurements so far and these compound classes are also not very frequently represented (see above), so that possible cross-contaminations are unlikely. In addition, all samples were extracted and measured in a randomized order to exclude systematic errors as far as possible.
Compounds from the group of phospholipids also proved suitable for species differentiation. The detected PCs as [M + H]+-adducts could be assigned mainly on the basis of the fragment m/z 184.07 (C5H15NO4P+) in the MS/MS spectra, which originates from the head group of this class of molecules (Fig. S9 in the ESI†).39 In addition, two PEs could also be identified as [M + H]+-adducts, notably by a neutral loss of the head group of m/z 141.02 (C2H8NO4P).40 The assignment of both the PCs and the PEs could be confirmed by comparison with the calculated CCS values of the LipidCCS database.
As already mentioned above, DGTS derivatives can be detected mainly in lower organisms. Although DGTS do not contain a phosphorus atom, they take over similar functions as PCs and PEs in living organisms due to their amiphilic properties and are therefore primarily components of cell membranes. They also frequently occur as direct substituents of phospholipids.41 In the present study, the identified DGTSs and LDGTSs could only be detected in higher concentrations in the T. borchii samples, while the identified phospholipids were present in the samples of T. magnatum in most cases at higher signal intensities. Therefore, it can be assumed that these differences in the two white truffle species are also due to the fact that PCs and PEs can be replaced by DGTS and LDGTS, respectively. However, the extent to which these differences are due to genetic variability or the geographical locations, such as variable phosphorus content in the soil, is difficult to assess. The cultivation of truffles is demanding and T. magnatum cannot be grown at all, so no specific experiments can be carried out, e.g. with different fertilizers. The majority of the white truffle samples analyzed in this study came from Italy, and the collection areas of the two species overlap, so that no conclusions can be drawn here either.
In addition, some Cers proved to be conspicuous as marker substances that could also be detected as [M + H]+-adducts. The identification on the basis of MS/MS spectra was mainly based on the neutral loss of two water molecules, which is typical for this class of compounds (Fig. S6 in the ESI†).39 While the identification of Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0;O3) and Cer (36
0;O3) and Cer (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3;O3) could be confirmed on CCS values and comparison with the LipidCCS database, it was not possible to check the CCS value of the detected glucosyl-Cer, as no entry was available. Cers have numerous functions. They serve as membrane components, but also influence the growth of organisms and take part in numerous cellular processes as intermediates. Their occurrence has been extensively documented by NMR in black truffles42 and in various mushrooms.43 Due to their diverse functions and widespread occurrence, it is plausible that Cers could also be detected in this study as marker substances in the differentiation of white truffle species.
3;O3) could be confirmed on CCS values and comparison with the LipidCCS database, it was not possible to check the CCS value of the detected glucosyl-Cer, as no entry was available. Cers have numerous functions. They serve as membrane components, but also influence the growth of organisms and take part in numerous cellular processes as intermediates. Their occurrence has been extensively documented by NMR in black truffles42 and in various mushrooms.43 Due to their diverse functions and widespread occurrence, it is plausible that Cers could also be detected in this study as marker substances in the differentiation of white truffle species.
Furthermore, DG (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) was detected as a relevant marker substance in the form of various adducts ([M + H]+, [M + NH4]+, and [M + Na]+) as well as numerous TGs as [M + NH4]+- and [M + Na]+-adducts. DGs and TGs can usually be assigned based on the neutral loss of the different acyl side-chains and a water molecule (Fig. S7 and S8 in the ESI†).39,44 In addition, corresponding entries regarding the CCS values were available for confirmation in the LipidCCS database. Both substance groups are very widespread in eukaryotic organisms. As intermediates and second messengers, DGs are part of numerous biochemical processes, while TGs mainly serve as storage for fatty acyl chains.45–47
3) was detected as a relevant marker substance in the form of various adducts ([M + H]+, [M + NH4]+, and [M + Na]+) as well as numerous TGs as [M + NH4]+- and [M + Na]+-adducts. DGs and TGs can usually be assigned based on the neutral loss of the different acyl side-chains and a water molecule (Fig. S7 and S8 in the ESI†).39,44 In addition, corresponding entries regarding the CCS values were available for confirmation in the LipidCCS database. Both substance groups are very widespread in eukaryotic organisms. As intermediates and second messengers, DGs are part of numerous biochemical processes, while TGs mainly serve as storage for fatty acyl chains.45–47
In addition, differences between the two white truffle species result from variances in the concentration of ergosteryl glucoside and coenzyme Q8. Sommer et al. have recently published on almost the same set of samples that different truffle species have different sterol fingerprints suitable for authentication.48,49 In yeast and fungi, ergosterol is one of the most abundant sterols.50 It is therefore obvious that a marker substance from this substance class could be identified in this study, too.
Coenzyme Q8 belongs to the substance class of ubiquinones, which play an important role in electron transport. This compound could be detected in the white truffle samples as [M + Na]+-adduct. Ubiquinones usually show a characteristic fragment at m/z 197.08 (C10H13O4+) in the MS/MS spectrum (Fig. S11 in the ESI†).51 This fragment could not initially be detected in the white truffle samples, but was present in the MS/MS measurements of the black truffle samples (Tables S1 and S2 in the ESI†). A confirmation based on the CCS values could not be made because, to the best of our knowledge, no reference data can currently be found in the literature in this case either.
A geographical differentiation of the samples was not possible. However, the sample data set was not designed for this either, since a higher number of samples must then be available per location in order to achieve reliable results (Fig. S3A in the ESI†).
3.2 Non-targeted lipidomics analysis of black truffle samples
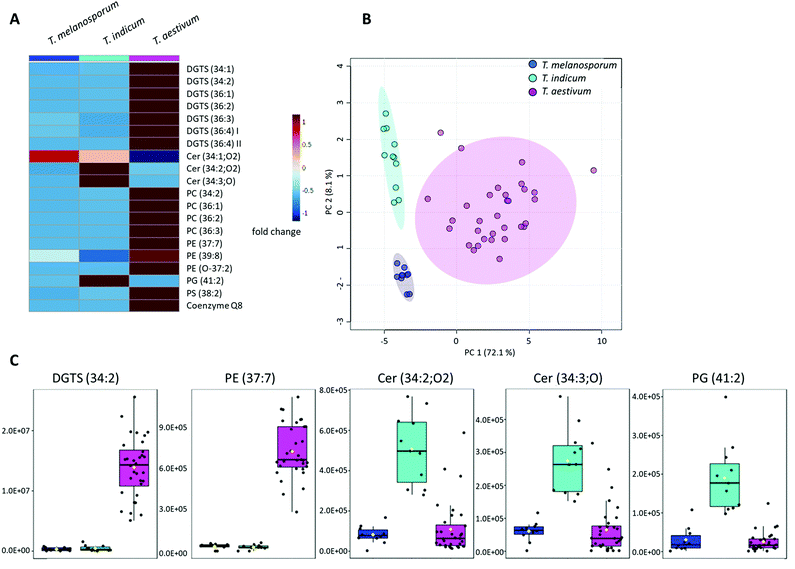
The bucket table of non-targeted measurements of the black truffle species T. melanosporum, T. indicum and T. aestivum contained 1211 features. Using ANOVA calculation, 465 features could be classified as significantly different in the three sample groups based on the FDRs. However, in contrast to the white truffle species, the clustering of the sample groups in the PCA scores plot was less clear (Fig. S2B in the ESI†). Although tendencies for a differentiation are evident, the 95% confidence regions overlap significantly. Analysis of other principal components (not shown in the figures) also failed to improve the results. Nevertheless, the Q2-value of the LOOCV, which was determined using PLS-DA, was 0.80 for two components. This value indicates that there were definitely differences within the three sample groups. Further assessment of the individual features revealed that the T. aestivum samples were easy to separate from the other two groups. However, distinguishing the T. melanosporum from the T. indicum samples was more challenging. For this reason, all T. aestivum samples were excluded from the data set and a t-test was performed with the remaining T. melanosporum and T. indicum samples. Only three features proved to be significantly different, but they were sufficient to allow a complete separation of these two groups of samples.Similar to the white truffle samples, MS/MS fragment spectra of the 60 features with the smallest FDRs were recorded (Table S2 and Fig. S4–S11 in the ESI†). The three features that made it possible to distinguish the T. melanosporum samples from the T. indicum samples were also treated in the same way. For a total of 23 features, a preliminary identification could be carried out. Due to multiple assignments of different adducts, the number was reduced to a total of 20 compounds (Fig. 3A). The compound DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) was identified twice, but at different RTs (8.0 min and 9.3 min) and with different CCS-values, so that these must be isomers. For this reason, these two isomers were labeled I and II in Fig. 3A.
4) was identified twice, but at different RTs (8.0 min and 9.3 min) and with different CCS-values, so that these must be isomers. For this reason, these two isomers were labeled I and II in Fig. 3A.
The PCA scores plot obtained, which could be calculated using the 20 identified compounds, is shown in Fig. 3B. The three groups form clearly defined clusters and thus illustrate the chemical differences in the sample groups. In addition, the marker compounds selected as examples in Fig. 3C show the relatively large differences that also exist within the black truffle species. The compounds Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2;O2), Cer (34
2;O2), Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3;O) and PG (41
3;O) and PG (41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) are the three compounds previously identified to distinguish the T. melanosporum from the T. indicum samples.
2) are the three compounds previously identified to distinguish the T. melanosporum from the T. indicum samples.
Compared to the white truffle samples, almost the same substance classes proved to be relevant, partly even the identical marker compounds. These included: DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DGTS (34
1), DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), DGTS (36
2), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), DGTS (36
2), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), DGTS (36
3), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PC (34
4), PC (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) as well as coenzyme Q8. Certainly, further overlaps would result on the basis of other significant features. Furthermore, it was noticeable that numerous TGs were significant in the differentiation of the white truffle species, whereas these played no role in the distinction between the black truffle species. In addition to the substance classes already described in Section 3.1, other phospholipids such as PG (41
2) as well as coenzyme Q8. Certainly, further overlaps would result on the basis of other significant features. Furthermore, it was noticeable that numerous TGs were significant in the differentiation of the white truffle species, whereas these played no role in the distinction between the black truffle species. In addition to the substance classes already described in Section 3.1, other phospholipids such as PG (41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and PS (38
2) and PS (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) were identified.
2) were identified.
For both substances, and for some PEs, no unique MS/MS fragments could be detected and assigned, neither manually nor software-supported. In these cases, in addition to the high-resolution mass, the CCS values proved to be particularly useful for identification, as an alternative parameter was available for assigning the structures.
Nevertheless, it was particularly noticeable, especially in the dataset of the black truffle species, that numerous features could not be assigned either with the help of the fragment spectra or with the CCS values. This is probably mainly due to the fact that numerous metabolites have not yet been published, which makes clear identification difficult. In addition, relatively few research groups have dealt with lipidomics analyses of fungi and truffles in particular, or the focus has been on the polar compounds. Consequently, there is currently still a lack of reference data overall, so that further research in this regard will certainly worthwhile in the future.
As with the white truffle species, it was not possible to distinguish the origin (Fig. S3B in the ESI†), which in turn could be due to the sample data set.
3.3 Possible strategies for transferring the results into economic application
Measurement times on high-resolution mass spectrometers are often very limited, but the implementation of IM cells can usually reduce analysis times. This is a first starting point for the transfer from academic research to routine application. However, such devices are unavailable in industry and governmental food control agencies in most cases. For this reason, non-targeted metabolomics methods are not currently widely used for routine applications. Consequently, in order to use the methods economically, alternative strategies need to be pursued, and the results and procedures presented in this study should be simplified. The example presented here for the differentiation of truffle species offers particularly high potential, as basically only very few marker compounds need to be analyzed in order to obtain reliable results.
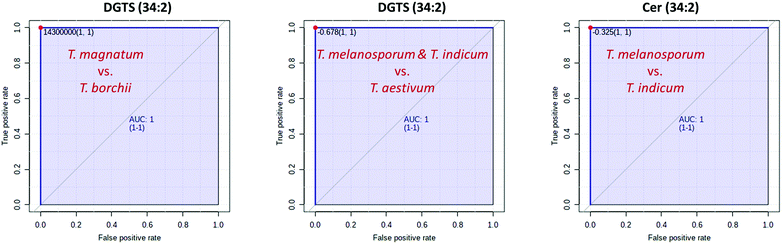
Fig. 2C and 3C show that just a few marker compounds are sufficient to distinguish the different truffle species from one another. This hypothesis was tested using univariate ROC analysis and the MetaboAnalyst 5.0 biomarker analysis tool. The AUC values obtained from the ROC analyses were used for assessment.52–54 Twenty-six of the 33 marker compounds that could be identified in this study as being suitable for distinguishing the white truffle species had an AUC value of 1, making them optimal classifiers for achieving 100% correct sample assignment. These substances included: LDGTS (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DGTS (34
1), DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DGTS (34
1), DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), DGTS (36
2), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), DGTS (36
2), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), DGTS (36
3), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), Cer (34
4), Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0;O3), Cer (36
0;O3), Cer (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3; O3), TG (50
3; O3), TG (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), TG (50
1), TG (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), TG 52
2), TG 52![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), TG (54
2), TG (54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), TG (57
3), TG (57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PC (32
4), PC (32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC (34
1), PC (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), PC (34
1), PC (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC (35
2), PC (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PC (35
2), PC (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PC (35
4), PC (35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6), PC (36
6), PC (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), PC (37
5), PC (37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PC (37
4), PC (37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), PC (38
5), PC (38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4), PC (40
4), PC (40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7), PE (34
7), PE (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and PE (36
2) and PE (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3).
3).
Since ROC analyses are only suitable for binary classification models, but three different black truffle species were present, the first step was to separate the T. indicum vs. the T. melanosporum and the T. aestivum samples. Five of the 20 identified marker compounds had AUC values of 1, illustrating the separation of the two sample groups, also with an accuracy of 100%. These marker compounds were: DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), DGTS (36
1), DGTS (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4) II, PC (36
4) II, PC (36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), PE (37
2), PE (37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) and PE (O-37
7) and PE (O-37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2). The procedure was then repeated to assess the possible separation of the T. melansporum from the T. aestivum samples. Cer (34
2). The procedure was then repeated to assess the possible separation of the T. melansporum from the T. aestivum samples. Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2;O2) and Cer (34
2;O2) and Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3;O) in particular proved to be suitable, both of which had an AUC value of 1.0 and can therefore also be used for a 100% separation of the two sample groups.
3;O) in particular proved to be suitable, both of which had an AUC value of 1.0 and can therefore also be used for a 100% separation of the two sample groups.
By combining several marker substances, for example DGTS (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) and Cer (34
2) and Cer (34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2;O2), both the white and the black truffle species could be reliably distinguished from one another (Fig. 4). Due to the small number of marker substances required, it would be obvious to develop a rapid test for the selected markers with which the truffle samples can be checked on-site. Possible tailor-made receptors could be aptamers, i.e. single-stranded DNA or RNA oligonucleotides that can also bind comparatively small molecules with high specificity due to their three-dimensional folding.55,56 Besides the rather expensive development of a rapid test, an alternative is to convert the non-targeted method into a targeted method. Recently, we were able to demonstrate this approach using a triple quadrupole mass spectrometer to prove the geographical origin of asparagus.57 Triple quadrupole mass spectrometers are now standard equipment in most laboratories, so the method can be implemented very easily in existing infrastructures. Since the commercially available triple quadrupole mass spectrometers are not equipped with an ion mobility cell, we also checked that there are no overlaps of the marker metabolites with potentially interfering analytes in the ion mobility dimension. However, this was not the case, so it should be possible to transfer the non-targeted approach to a targeted method.
2;O2), both the white and the black truffle species could be reliably distinguished from one another (Fig. 4). Due to the small number of marker substances required, it would be obvious to develop a rapid test for the selected markers with which the truffle samples can be checked on-site. Possible tailor-made receptors could be aptamers, i.e. single-stranded DNA or RNA oligonucleotides that can also bind comparatively small molecules with high specificity due to their three-dimensional folding.55,56 Besides the rather expensive development of a rapid test, an alternative is to convert the non-targeted method into a targeted method. Recently, we were able to demonstrate this approach using a triple quadrupole mass spectrometer to prove the geographical origin of asparagus.57 Triple quadrupole mass spectrometers are now standard equipment in most laboratories, so the method can be implemented very easily in existing infrastructures. Since the commercially available triple quadrupole mass spectrometers are not equipped with an ion mobility cell, we also checked that there are no overlaps of the marker metabolites with potentially interfering analytes in the ion mobility dimension. However, this was not the case, so it should be possible to transfer the non-targeted approach to a targeted method.
According to our experiments in this study, but also in comparable research questions, we could not prove any influence of the harvest year, so that the analytics should not be influenced in this respect.22 Nevertheless, especially when establishing such a method, we consider it useful to further investigate this parameter and to analyze authentic reference samples at regular intervals. In addition, it does not seem to be relevant whether frozen or non-frozen truffles are analyzed. In the present study, mainly freshly collected truffles were analyzed, which were initially frozen in the research center in order to be able to store them for a longer period of time. Nevertheless, eight samples came already frozen (−20 °C) directly from the retailer. However, these samples behaved identically to the other samples, so that no measures need to be taken in this respect.
4. Conclusions
In the present study, a successful differentiation of the white truffle species T. magnatum and T. borchii as well as the black species T. melanosporum, T. indicum and T. aestivum could be performed using a lipidomics-based approach. An LC-ESI-IM-QTOF-MS instrument was chosen as an analytical platform so that, in addition to the classical MS/MS fragment spectra, the CCS values of the compounds were also available as a further parameter for identification. Numerous compounds were identified whose CCS values have not yet been published and can be used as a reference by other researchers in the future. Nevertheless, it must be pointed out that these values can only serve as orientation, as confirmation with standard substances has not been carried out. In addition, the resolution of the currently available LC-ESI-IM-QTOF-MS instruments is not yet sufficient to reliably distinguish between isobaric substances based solely on the CCS values and the MS/MS spectra. However, there are currently strong efforts to further advance the developments and to improve the resolution of IM cells, for example by extending the drift distance, so that the performance in this regard will certainly increase in the coming years.58By means of t-test or ANOVA, numerous substances proved to be significant marker compounds and showed clear differences between the different truffle species. Just a few marker substances would be sufficient to distinguish the different truffle species with 100% accuracy. The most relevant key compounds included DGTS derivatives, Cers and numerous phospholipids as well as glycerides.
Abbreviations
| AUC | Area Under Curve |
| ANOVA | Analysis of Variance |
| CCS | Collision Cross Section |
| Cer | Ceramide |
| DG | Diacylglycerol |
| DGTS | Diacylglyceryl-O-4′-(N,N,N-trimethyl) homoserine |
| FDR | False Discovery Rate |
| LC-ESI-IM-QTOF-MS | Liquid Chromatography-Electrospray Ionization-Ion Mobility-Quadrupole-Time Of Flight Mass Spectrometer Instrument |
| LDGTS | Lyso-diacylglyceryl-O-4′-(N,N,N-trimethyl) homoserine |
| LPC | Lyso-glycerophosphocholine |
| LPE | Lyso-glycerophosphoethanolamine |
| LOOCV | Leave-One-Out Cross Validation |
| MG | Monoacylglycerol |
| NIR | Near Infrared |
| PC | Glycerophosphocholine |
| PCA | Principal Component Analysis |
| PE | Glycerophosphoethanolamine |
| PG | Glycerophosphoglycerol |
| PLS-DA | Partial Least Squares Discriminant Analysis |
| PS | Glycerophosphoserine |
| QC | Quality Control |
| ROC curve | Receiver Operating Characteristic curve |
| RT | Retention Time |
| TG | Triacylglycerols |
| TIC | Total Ion Chromatogram |
Data availability
The datasets supporting this article have been uploaded as part of the supplementary material.Author contributions
M. C., conceptualization, formal analysis, investigation, methodology, visualization, writing – original draft; M. F., funding acquisition, resources, supervision, writing – review & editing.Funding
This study was performed within the project “Food Profiling – Development of analytical tools for the experimental verification of the origin and identity of food”. This project (Funding reference number: 2816500914) is supported by means of the Federal Ministry of Food and Agriculture (BMEL) by a decision of the German Bundestag (parliament). Project support is provided by the Federal Institute for Agriculture and Food (BLE) within the scope of the program for promoting innovation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Stefanie Schelm, Nils Wax, Anne Haferkorn for carrying out the DNA analyses to verify the analysis material as well as Felix Burger, Stephan Burger, David Schütz, Malte Siegmund, Benjamin Wegner, and Alissa Dress for helpful discussions. We would also like to thank Alexander Möllers and Kerstin Blum for their support in processing the samples and Rudolf Pistorius for proofreading.References
- R. Splivallo and L. Culleré, in True Truffle (Tuber spp.) in the World: Soil Ecology, Systematics and Biochemistry, ed. A. Zambonelli, M. Lotti and C. Murat, Springer International Publishing, Cham, 2016, The Smell of Truffles: From Aroma Biosynthesis to Product Quality, pp. 393–407 Search PubMed.
- F. Le Tacon, in True Truffle (Tuber spp.) in the World: Soil Ecology, Systematics and Biochemistry, ed. A. Zambonelli, M. Lotti and C. Murat, Springer International Publishing, Cham, 2016, Influence of Climate on Natural Distribution of Tuber Species and Truffle Production, pp. 153–167 Search PubMed.
- A. Zambonelli, M. Iotti, F. Puliga and I. R. Hall, in Advances in Plant Breeding Strategies: Vegetable Crops: Volume 10: Leaves, Flowerheads, Green Pods, Mushrooms and Truffles, ed. J. M. Al-Khayri, S. M. Jain and D. V. Johnson, Springer International Publishing, Cham, 2021, Enhancing White Truffle (Tuber magnatum Picco and T. borchii Vittad.) Cultivation Through Biotechnology Innovation, 505–532 Search PubMed.
- German guidelines for mushrooms and mushrooms products, Deutsche- Lebensmittelbuch-Komission, Version from July 2nd 2020, BAnz AT 18.08.2020 B4, GMBl 2020 S. 547, https://www.bmel.de/SharedDocs/Downloads/DE/_Ernaehrung/Lebensmittel-Kennzeichnung/LeitsaetzePilze.pdf?_blob=publicationFile&v=2, (accessed February 2022).
- E. V. Trüffelverband, Trüffelverband stoppt Verbraucherverwirrung!, https://www.trüffelverband.de/index.php/wir-ueber-uns/lobbyarbeit, (accessed February 2022) Search PubMed.
- L. Culleré, V. Ferreira, M. E. Venturini, P. Marco and D. Blanco, Food Chem., 2013, 141, 105–110 CrossRef PubMed.
- S. Schelm, M. Siemt, J. Pfeiffer, C. Lang, H.-V. Tichy and M. Fischer, Foods, 2020, 9, 501 CrossRef CAS PubMed.
- A. Amicucci, C. Guidi, A. Zambonelli, L. Potenza and V. Stocchi, FEMS Microbiol. Lett., 2000, 189, 265–269 CrossRef CAS PubMed.
- N. Séjalon-Delmas, C. Roux, M. Martins, M. Kulifaj, G. Bécard and R. Dargent, J. Agric. Food Chem., 2000, 48, 2608–2613 CrossRef PubMed.
- E. Zampieri, A. Mello, P. Bonfante and C. Murat, FEMS Microbiol. Lett., 2009, 297, 67–72 CrossRef CAS PubMed.
- M. Leonardi, M. Iotti, A. Mello, A. Vizzini, A. Paz-Conde, J. Trappe and G. Pacioni, Cryptogam.: Mycol., 2021, 42, 149–170 Search PubMed.
- M. Klein, R. Kielhauser and T. Dilger, J. Mass Spectrom., 2020, 55, e4655 CAS.
- D. Krösser, B. Dreyer, B. Siebels, H. Voß, C. Krisp and H. Schlüter, Int. J. Mol. Sci., 2021, 22, 12999 CrossRef PubMed.
- K. El Karkouri, C. Couderc, P. Decloquement, A. Abeille and D. Raoult, Sci. Rep., 2019, 9, 17686 CrossRef PubMed.
- T. Segelke, K. von Wuthenau, G. Neitzke, M.-S. Müller and M. Fischer, J. Agric. Food Chem., 2020, 68, 14374–14385 CrossRef CAS PubMed.
- S. Krauß and W. Vetter, J. Agric. Food Chem., 2020, 68, 14386–14392 CrossRef PubMed.
- S. Hamzić Gregorčič, L. Strojnik, D. Potočnik, K. Vogel-Mikuš, M. Jagodic, F. Camin, T. Zuliani and N. Ogrinc, Molecules, 2020, 25, 2217 CrossRef PubMed.
- D. Sciarrone, A. Schepis, M. Zoccali, P. Donato, F. Vita, D. Creti, A. Alpi and L. Mondello, Anal. Chem., 2018, 90, 6610–6617 CrossRef CAS PubMed.
- T. Segelke, S. Schelm, C. Ahlers and M. Fischer, Foods, 2020, 9, 922 CrossRef CAS.
- C. Kappacher, B. Trübenbacher, K. Losso, M. Rainer, G. K. Bonn and C. W. Huck, Molecules, 2022, 27, 589 CrossRef CAS PubMed.
- D. Schütz, E. Achten, M. Creydt, J. Riedl and M. Fischer, Foods, 2021, 10, 2160 CrossRef PubMed.
- M. Creydt and M. Fischer, J. Agric. Food Chem., 2020, 68, 14343–14352 CrossRef CAS PubMed.
- A. Delvaux, E. Rathahao-Paris and S. Alves, Mass Spectrom. Rev., 2021 DOI:10.1002/mas.21685.
- E. Bligh and W. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- M. Creydt, M. Arndt, D. Hudzik and M. Fischer, J. Agric. Food Chem., 2018, 66, 12876–12887 CrossRef CAS PubMed.
- M. Creydt, L. Ludwig, M. Köhl, J. Fromm and M. Fischer, J. Chromatogr. A, 2021, 1641, 461993 CrossRef CAS PubMed.
- R. Reisdorph, C. Michel, K. Quinn, K. Doenges and N. Reisdorph, in Ion Mobility-Mass Spectrometry: Methods and Protocols, ed. G. Paglia and G. Astarita, Springer US, New York, NY, 2020, Untargeted Differential Metabolomics Analysis Using Drift Tube Ion Mobility-Mass Spectrometry, pp. 55–78 Search PubMed.
- A. Bilbao, B. C. Gibbons, S. M. Stow, J. E. Kyle, K. J. Bloodsworth, S. H. Payne, R. D. Smith, Y. M. Ibrahim, E. S. Baker and J. C. Fjeldsted, J. Proteome Res., 2022, 21, 798–807 CrossRef CAS PubMed.
- Pacific Northwest National Laboratory, PNNL Preprocessor software, https://pnnl-comp-mass-spec.github.io/PNNL-PreProcessor, (accessed February 2022).
- Z. Pang, J. Chong, G. Zhou, D. A. de Lima Morais, L. Chang, M. Barrette, C. Gauthier, P.-É. Jacques, S. Li and J. Xia, Nucleic Acids Res., 2021, 49, W388–W396 CrossRef CAS PubMed.
- Y. Benjamini and Y. Hochberg, J. R. Stat. Soc., 1995, 57, 289–300 Search PubMed.
- M. Sud, E. Fahy, D. Cotter, A. Brown, E. A. Dennis, C. K. Glass, A. H. Merrill Jr, R. C. Murphy, C. R.-H. Raetz, D. W. Russell and S. Subramaniam, Nucleic Acids Res., 2007, 35, D527–D532 CrossRef CAS PubMed.
- The Metabolomics Innovation Centre (TMIC), FooDB, https://foodb.ca, (accessed February 2022).
- Zhu Lab, University of Hong Kong, https://www.metabolomics-shanghai.org/LipidCCS/, (accessed February 2022).
- Z. Zhou, J. Tu, X. Xiong, X. Shen and Z.-J. Zhu, Anal. Chem., 2017, 89, 9559–9566 CrossRef CAS PubMed.
- S. Klockmann, E. Reiner, R. Bachmann, T. Hackl and M. Fischer, J. Agric. Food Chem., 2016, 64, 9253–9262 CrossRef CAS PubMed.
- N. Cain, O. Alka, T. Segelke, K. von Wuthenau, O. Kohlbacher and M. Fischer, Food Chem., 2019, 298, 125013 CrossRef CAS PubMed.
- W. R. Riekhof, S. Naik, H. Bertrand, C. Benning and D. R. Voelker, Eukaryotic Cell, 2014, 13, 749–757 CrossRef PubMed.
- PRIMe RIKEN Center for Sustainable Resource Science, https://prime.psc.riken.jp/compms/msdial/lipidnomenclature.html, (accessed February 2022).
- K. Vekey, A. Telekes and A. Vertes, Medical Applications of Mass Spectrometry, Elsevier Science, Amsterdam, 2008 Search PubMed.
- Y. Oishi, R. Otaki, Y. Iijima, E. Kumagai, M. Aoki, M. Tsuzuki, S. Fujiwara and N. Sato, Commun. Biol., 2022, 5, 19 CrossRef CAS PubMed.
- J.-M. Gao, A.-L. Zhang, H. Chen and J.-K. Liu, Chem. Phys. Lipids, 2004, 131, 205–213 CrossRef CAS PubMed.
- D. Warnecke and E. Heinz, Cell. Mol. Life Sci., 2003, 60, 919–941 CrossRef CAS PubMed.
- A. M. McAnoy, C. C. Wu and R. C. Murphy, J. Am. Soc. Mass Spectrom., 2005, 16, 1498–1509 CrossRef CAS PubMed.
- T. O. Eichmann and A. Lass, Cell. Mol. Life Sci., 2015, 72, 3931–3952 CrossRef CAS PubMed.
- Y. Yang and C. Benning, Curr. Opin. Biotechnol, 2018, 49, 191–198 CrossRef CAS PubMed.
- I. Hapala, P. Griac and R. Holic, Lipids, 2020, 55, 513–535 CrossRef CAS PubMed.
- K. Sommer, S. Krauß and W. Vetter, J. Agric. Food Chem., 2020, 68, 14393–14401 CrossRef CAS PubMed.
- A. Villares, A. García-Lafuente, E. Guillamón and Á. Ramos, J. Food Compos. Anal., 2012, 26, 177–182 CrossRef CAS.
- M. L. Rodrigues, mBio, 2018, 9, e01755 CrossRef PubMed.
- L. Fernández-Del-Río and C. F. Clarke, Metabolites, 2021, 11, 385 CrossRef PubMed.
- T. Fawcett, Pattern Recognit. Lett., 2006, 27, 861–874 CrossRef.
- V. Bewick, L. Cheek and J. Ball, Crit. Care, 2004, 8, 508–512 CrossRef PubMed.
- J. A. Hanley and B. J. McNeil, Radiology, 1982, 143, 29–36 CrossRef CAS PubMed.
- E. Frohnmeyer, N. Tuschel, T. Sitz, C. Hermann, G. T. Dahl, F. Schulz, A. J. Baeumner and M. Fischer, Analyst, 2019, 144, 1840–1849 RSC.
- C. Fischer, S. Klockmann, H. Wessels, T. Hünniger, J. Schrader, A. Paschke-Kratzin and M. Fischer, J. Biotechnol., 2016, 238, 30–34 CrossRef CAS PubMed.
- M. Creydt, B. Wegner, A. Gnauck, R. Hörner, C. Hummert and M. Fischer, Food Control, 2022, 135, 108690 CrossRef CAS.
- L. Deng, Y. M. Ibrahim, A. M. Hamid, S. V.-B. Garimella, I. K. Webb, X. Zheng, S. A. Prost, J. A. Sandoval, R. V. Norheim, G. A. Anderson, A. V. Tolmachev, E. S. Baker and R. D. Smith, Anal. Chem., 2016, 88, 8957–8964 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Further PCA scores plots, MS/MS fragments and CCS values of the identified key metabolites. See DOI: https://doi.org/10.1039/d2mo00088a |
| This journal is © The Royal Society of Chemistry 2022 |