Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Proteomic and metabolomic profiling of acute and chronic stress events associated with military exercises†
Justin
McKetney‡
ab,
Conor C.
Jenkins‡
c,
Catie
Minogue
ab,
Phillip M.
Mach
c,
Erika K.
Hussey
de,
Trevor G.
Glaros
 *cf,
Joshua
Coon
*abgh and
Elizabeth S.
Dhummakupt
*cf,
Joshua
Coon
*abgh and
Elizabeth S.
Dhummakupt
 *c
*c
aDepartment of Biomolecular Chemistry, University of Wisconsin, Madison, WI 53706, USA. E-mail: jcoon@chem.wisc.edu
bNational Center for Quantitative Biology of Complex Systems, Madison, WI 53706, USA
cDEVCOM Chemical Biological Center, Aberdeen Proving Grounds, MD 21010, USA. E-mail: tglaros@lanl.gov; elizabeth.s.dhummakupt.civ@army.mil
dDEVCOM Soldier Center, Natick, MA 01760, USA
eDefense Innovation Unit, Mountain View, CA 94043, USA
fLos Alamos National Laboratory, Los Alamos, NM 87545, USA
gMorgridge Institute for Research, Madison, WI 53515, USA
hDepartment of Chemistry, University of Wisconsin, Madison, WI 53706, USA
First published on 3rd December 2021
Abstract
By characterizing physiological changes that occur in warfighters during simulated combat, we can start to unravel the key biomolecular components that are linked to physical and cognitive performance. Viable field-based sensors for the warfighter must be rapid and noninvasive. In an effort to facilitate this, we applied a multiomics pipeline to characterize the stress response in the saliva of warfighters to correlate biomolecular changes with overall performance and health. In this study, two different stress models were observed – one of chronic stress and one of acute stress. In both models, significant perturbations in the immune, metabolic, and protein manufacturing/processing systems were observed. However, when differentiating between stress models, specific metabolites associated with the “fight or flight” response and protein folding were seen to be discriminate of the acute stress model.
Introduction
The human ‘fight or flight’ stress response serves an important evolutionary purpose in preparing and mobilizing physiological systems to react to altered homeostasis or a perceived threat.1,2 When stressor events occur, sympathetic stimulation of the autonomic nervous system releases adrenaline and noradrenaline.1,3 Such signaling molecules induce many physiological changes associated with acute stress, like vasoconstriction, rapid glucose breakdown and elevated heart rate. A slower, secondary response also occurs from the stimulation of the hypothalamic–pituitary–adrenal (HPA) axis. This releases glucocorticoids, such as cortisol, which regulate metabolism, energy availability, inflammation, and cognitive function.4Humans experience a variety of stress-inducing events daily. These stressors, which fluctuate in type, timing, and severity, have consequences ranging from mild alterations in homeostasis to death.5 Individuals in particular occupations, such as warfighters, medics, airline pilots, and athletes encounter ‘high-stress’ events on a regular basis.6–11 Identification of when and how acute and chronic stress impact these individuals, especially in comparison to a ‘normal’ person, warrants investigation. Stress can inhibit optimal performance and negatively impact physiology and health; especially since chronic stress can cause the development of long-term issues such as post-traumatic stress disorder,12–17 metabolic syndromes, cardiovascular disease, and immune dysfunction.18–20
Warfighters encounter multiple types of stress (acute and chronic) depending on particular situations (i.e. skirmishes, infantry tactics, and training).9 Understanding how these different situations affect warfighter performance and their long-term health are key to sustainment, readiness, and ensuring wellbeing post-service. Detangling this complicated system requires a deeper knowledge of how cellular systems are altered when experiencing warfighter-specific stress. A starting point to achieving such a mechanistic understanding is to characterize the molecules and biological signatures of these conditions. Previous work has identified peptides,21 proteins,22 and metabolites23 associated with physical and mental fatigue in a variety of contexts including both physicians23 and athletes.24 Beyond mechanistic understanding, the ability to monitor the chemical signatures of stress and fatigue could play a vital role in both strategic decisions regarding warfighters in the field, as well as interventions that could reduce or mitigate the negative health consequences associated with stress events.
Saliva presents an accessible biofluid that can be obtained in a non-invasive manner25,26 without specialized training or a clinical setting. In addition to ease of collection, saliva provides a safer alternative due to lower risks of disease transmission when compared to blood or other biofluids.27 The molecular composition of saliva is impacted by both physical and psychological factors including age, circadian rhythm, pain level, and stress.27,28 The dynamic composition of saliva along with its ease of collection make it an ideal diagnostic and investigatory tool. Proteomic and metabolomic analysis of saliva has identified biomarkers for cancer, multiple sclerosis, diabetes, and heart disease.29–31 Additionally, salivary compounds associated with fetal development,32 hypoxic conditions,33 and stress6 have also been identified. In an effort to more holistically understand the molecular changes occurring on multiple molecule types (i.e. metabolites and proteins), multiomic analysis has been employed in this study. This type of analysis integrates multiple data streams to allow for more comprehensive understanding of biological processes under different conditions.34,35
For this study, we collected saliva from members of the 82nd Airborne's 2nd Battalion, 505th Parachute Infantry Regiment of the U.S. Army at discrete time points (Fig. 1) before, during, and after a 72 hour combat simulation event. The three-day “Battle Ready” period was a series of baselining activities for various metrics (e.g. fitness testing, strength assessments, cognitive/social-emotional/physical surveys) that were collected during the “Mission” period. The 72 hour “Mission” period began with a weight-loaded six mile infiltration march, receiving operational orders, moving on foot to the operational objective and performing the objective, moving to a secondary objective, receiving a change of orders, then moving to a third objective, performing the objective, and then performing a weight-loaded six mile exfiltration march. In addition to the general chronic physical and cognitive stress associated with the “Mission” period, soldiers also participated in two instances of acute stress involving short live combat firefights. The “Recovery” period was similar to the “Battle Ready” period in that metrics (e.g. fitness testing, strength assessments, and cognitive/social-emotional/physical surveys) were collected as final data points. Salivary samples obtained throughout the mission were subjected to proteomic and metabolomic characterization to elucidate biomarkers and biochemical pathways that are most affected from these stressors. Repeated measurements of the same subject allowed for individualized baselines to be established, an essential feature of the study design. While a number of well-known bio-indicators such as dopamine, cortisol, and serotonin are perturbed during stress,36 herein we unveil a cadre of proteins and metabolites dynamically changing in conjunction with each other and stress level.
Experimental
Saliva collection – chronic stress & acute stress studies
Salivary samples were collected from 30 participants at discrete times before, during and after a training event. Up to 5 mL of saliva was collected by passive drool and frozen in the field on dry ice, then stored at −80 °C until analysis. Participants refrained from eating, drinking, and nicotine consumption thirty minutes prior to each collection. For the chronic stress analysis, only 20 participant's samples were selected for analysis due to absent or inadequate sample volumes. Secondary sampling occurred during acute stress events (ASE) occurring directly before and after the mission. Sampling procedure followed the scheme detailed above. All collection protocols were approved for human subjects’ research by the local designated Combat Capabilities Development Command Review Board (IRB). In accordance with the Declaration of Helsinki, all participants provided written informed consent before the completion of any study procedures. Demographic information for participants is available in ESI,† Supplemental File 1.Metabolomics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RCF for 20 minutes at 5 °C. A 100 μL aliquot of saliva was added to a centrifuge tube, along with 20 μL of internal standard and 800 μL of 8
000 RCF for 20 minutes at 5 °C. A 100 μL aliquot of saliva was added to a centrifuge tube, along with 20 μL of internal standard and 800 μL of 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v/v) acetonitrile:methanol:acetone. The internal standard is prepared by making working stocks of each solution at 2 mg mL−1 by dissolving 10 mg of each standard in 5 mL of 90
1 (v/v/v) acetonitrile:methanol:acetone. The internal standard is prepared by making working stocks of each solution at 2 mg mL−1 by dissolving 10 mg of each standard in 5 mL of 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 water
10 water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile. A working stock solution was prepared by combining the following volumes of each internal standard stock into a single vial containing 4715 μL of Fisher Optima gold label water with 0.1% formic acid (final volume 5000 μL): d3-creatine (10 μL), d10-leucine (10 μL), d3-L-tryptophan (10 μL), 13C6-citric acid (20 μL), 13C11-tryptophan (100 μL), 13C6-leucine (10 μL), 13C6-L-phenylalanine (10 μL), T-BOC-L-tert-leucine (10 μL), and T-BOC-L-aspartic acid (5 μL upon addition of the extraction solution, each sample was vortexed and refrigerated for at least 60 minutes at 5 °C to precipitate proteins. This solution was centrifuged at 20
acetonitrile. A working stock solution was prepared by combining the following volumes of each internal standard stock into a single vial containing 4715 μL of Fisher Optima gold label water with 0.1% formic acid (final volume 5000 μL): d3-creatine (10 μL), d10-leucine (10 μL), d3-L-tryptophan (10 μL), 13C6-citric acid (20 μL), 13C11-tryptophan (100 μL), 13C6-leucine (10 μL), 13C6-L-phenylalanine (10 μL), T-BOC-L-tert-leucine (10 μL), and T-BOC-L-aspartic acid (5 μL upon addition of the extraction solution, each sample was vortexed and refrigerated for at least 60 minutes at 5 °C to precipitate proteins. This solution was centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RCF for ten minutes at 5 °C to pellet the proteins. A 750 μL aliquot of supernatant was transferred to a new, labeled centrifuge tube and evaporated via Speedvac. Immediately prior to analysis by liquid chromatography mass spectrometry (LC-MS), sample was reconstituted in 100 μL of Optima grade water with 0.1% formic acid (Fischer Scientific, LS118) and vortexed briefly. Samples were placed at 4 °C for 10 minutes to allow for resuspension. Finally, each sample is centrifuged at 20
000 RCF for ten minutes at 5 °C to pellet the proteins. A 750 μL aliquot of supernatant was transferred to a new, labeled centrifuge tube and evaporated via Speedvac. Immediately prior to analysis by liquid chromatography mass spectrometry (LC-MS), sample was reconstituted in 100 μL of Optima grade water with 0.1% formic acid (Fischer Scientific, LS118) and vortexed briefly. Samples were placed at 4 °C for 10 minutes to allow for resuspension. Finally, each sample is centrifuged at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to removed insoluble particulates. The supernatant was transferred to glass autosampler vials (Agilent Technologies, Santa Clara, CA) for analysis.
000g for 10 minutes to removed insoluble particulates. The supernatant was transferred to glass autosampler vials (Agilent Technologies, Santa Clara, CA) for analysis.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 with a scan range of m/z 70–1000. AGC target was set to 3E6 with a maximum injection time of 100 ms. All metabolomics data were acquired in positive and negative ionization modes using the heated electrospray source ionization (HESI). The source settings were as follows: spray voltage: +4.0/−3.7 kV, capillary temperature: 325 °C, sheath gas (N2): 30 arbitrary units (AU), auxiliary gas (N2): 10 AU, S-Lens RF: 50 AU and probe heater: 350 °C. In addition to the samples, blanks of Fisher Optima gold label water with 0.1% formic acid were collected (three blanks at the beginning of the sequence and a blank after every ten sample injections), as well as quality control standards using the previously described internal standard mixture (collected once at the beginning of the sequence and after every ten sample injections and a blank).
000 with a scan range of m/z 70–1000. AGC target was set to 3E6 with a maximum injection time of 100 ms. All metabolomics data were acquired in positive and negative ionization modes using the heated electrospray source ionization (HESI). The source settings were as follows: spray voltage: +4.0/−3.7 kV, capillary temperature: 325 °C, sheath gas (N2): 30 arbitrary units (AU), auxiliary gas (N2): 10 AU, S-Lens RF: 50 AU and probe heater: 350 °C. In addition to the samples, blanks of Fisher Optima gold label water with 0.1% formic acid were collected (three blanks at the beginning of the sequence and a blank after every ten sample injections), as well as quality control standards using the previously described internal standard mixture (collected once at the beginning of the sequence and after every ten sample injections and a blank).
Proteomics
Protein precipitate was resuspended in reducing and alkylating buffer (10 mM TCEP, 40 mM chloroacetamide, 8 M urea, 100 mM Tris, pH 8) to a total protein concentration between 2–4 mg ml−1. Endoprotease Lys-C was added to each well at an approximate ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w protease/total protein. Plates were incubated at room temperature for 4 hours with slow rocking. Reducing and alkylating buffer was diluted to 25% concentration with 100 mM Tris, pH 8. Trypsin was added to sample wells at an approximate ratio of 50
1 w/w protease/total protein. Plates were incubated at room temperature for 4 hours with slow rocking. Reducing and alkylating buffer was diluted to 25% concentration with 100 mM Tris, pH 8. Trypsin was added to sample wells at an approximate ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 w/w protease/total protein. Samples were incubated overnight at room temperature.
1 w/w protease/total protein. Samples were incubated overnight at room temperature.
Digestion reactions were quenched when samples were brought to 0.5% TFA. Digested peptides were desalted using Phenomenex Strata C18 96-well plates (8E-S001-BGB) following manufacturers’ instructions before being dried in a speed-vacuum concentrator (Thermo Scientific).
Online reverse-phase columns were prepared in-house using a high-pressure packing apparatus previously described.37 In brief, 1.5 μm bridged ethylene hybrid C18 particles were packed at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi into a New Objective PicoTipTM emitter (Stock# PF360-75-10-N-5) with an inner diameter of 75 μm and an outer diameter of 360 μm. During separations, the column was heated to a temperature of 50 °C inside an in-house heater and interfaced with the mass spectrometer via an embedded emitter.
000 psi into a New Objective PicoTipTM emitter (Stock# PF360-75-10-N-5) with an inner diameter of 75 μm and an outer diameter of 360 μm. During separations, the column was heated to a temperature of 50 °C inside an in-house heater and interfaced with the mass spectrometer via an embedded emitter.
Eluted peptides were ionized by electrospray ionization and analyzed on a Thermo Orbitrap Fusion Eclipse. Survey scans of precursors were taken from m/z 300 to 1400 at 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolution while using Advanced Precursor Determination38 with an AGC target of 1E6 and a maximum injection time of 50 ms. Tandem MS was performed using an isolation window of 0.5 Da with 20 ppm mass tolerance and a dynamic exclusion time of 10 s. Selected precursors were fragmented using HCD with a normalized collision energy of 27%. The MS2 AGC target was set at 3E4 with a maximum injection time of 20 ms. Scans were taken in the ion trap using the turbo setting, and only peptides with a charge state of +2 or greater were selected for fragmentation. Samples were analyzed in duplicate.
000 resolution while using Advanced Precursor Determination38 with an AGC target of 1E6 and a maximum injection time of 50 ms. Tandem MS was performed using an isolation window of 0.5 Da with 20 ppm mass tolerance and a dynamic exclusion time of 10 s. Selected precursors were fragmented using HCD with a normalized collision energy of 27%. The MS2 AGC target was set at 3E4 with a maximum injection time of 20 ms. Scans were taken in the ion trap using the turbo setting, and only peptides with a charge state of +2 or greater were selected for fragmentation. Samples were analyzed in duplicate.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 transformation. Paired significance testing and Pearson correlations were performed using base R. Partial least squares discriminant analysis (PLS-DA) was performed using the plsda function from the mixOmics39 package (version 6.10.9) in R. Linear discriminant analyses were performed using the lda function of the MASS package40 (7.3.51.5), with area under the curve (AUC) of the receiver operator characteristic calculated using the auc or the multiclass.auc function of the pROC package41 (1.16.2). Mixed-effect models were generated using the lmer function of the lme4 package42 (1.1.23). Fixed-effect models were generated using lm function in base R. Circular dendrogram was generated using the circlize (0.4.11) and dendextend (1.14.0) packages in R. Other plots generated using base R and ggplot2 (3.3.2).
2 transformation. Paired significance testing and Pearson correlations were performed using base R. Partial least squares discriminant analysis (PLS-DA) was performed using the plsda function from the mixOmics39 package (version 6.10.9) in R. Linear discriminant analyses were performed using the lda function of the MASS package40 (7.3.51.5), with area under the curve (AUC) of the receiver operator characteristic calculated using the auc or the multiclass.auc function of the pROC package41 (1.16.2). Mixed-effect models were generated using the lmer function of the lme4 package42 (1.1.23). Fixed-effect models were generated using lm function in base R. Circular dendrogram was generated using the circlize (0.4.11) and dendextend (1.14.0) packages in R. Other plots generated using base R and ggplot2 (3.3.2).
Results and discussion
The big picture: distinguishing chronic combat-linked stress and enervation
Overall, 2087 proteins were identified based on 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 511 unique peptides. On average, 1332 proteins and 13
511 unique peptides. On average, 1332 proteins and 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 680 peptides were quantified in each sample (ESI,† Fig. S1). Although high variability was observed in peptide identifications across samples, outliers do not appear systematically related to external stimuli, and the protein identifications remain relatively consistent. The analysis quantified 545 protein groups across all 160 samples, with 720 proteins consistently quantified in >50% of all four sample pairs. When examining the gene ontology enrichment using a full human proteome as background, the highest enrichment was observed for the biological processes proteolysis, cell–cell adhesion and complement activation, (ESI,† Fig. S2), which would be expected given the role of saliva as a host-microbe interface and its proximity to the epithelial cells of the mouth. Additionally, enrichment of proteins involved in protein turnover, such as translation initiation and endopeptidase activity, were seen. The 720 proteins consistently quantified here represents ≈20% of the known human salivary proteome.43
680 peptides were quantified in each sample (ESI,† Fig. S1). Although high variability was observed in peptide identifications across samples, outliers do not appear systematically related to external stimuli, and the protein identifications remain relatively consistent. The analysis quantified 545 protein groups across all 160 samples, with 720 proteins consistently quantified in >50% of all four sample pairs. When examining the gene ontology enrichment using a full human proteome as background, the highest enrichment was observed for the biological processes proteolysis, cell–cell adhesion and complement activation, (ESI,† Fig. S2), which would be expected given the role of saliva as a host-microbe interface and its proximity to the epithelial cells of the mouth. Additionally, enrichment of proteins involved in protein turnover, such as translation initiation and endopeptidase activity, were seen. The 720 proteins consistently quantified here represents ≈20% of the known human salivary proteome.43
As part of the physical nature of the field exercise, the hydration level of the participants most likely varies from time point to time point. The effects of dehydration plays a role in physical performance,45 metabolic stress, and cognitive performance.46
As the objective of this work is to determine performance biomarkers during military exercises, any changes that occur as part of changing hydration status are important to monitor.
Time of day vs. mission initiation
In reference to Fig. 1, the following linear analysis was done to compare AM vs. PM collection points and battle ready (Green) vs. samples collected post-mission initiation (Mission-Red & Recovery-Yellow) to determine which variable contributed most to the baseline shift of the proteomic and metabolomic data.Several factors are known contribute to differences in molecular abundances within the saliva, including time of day, level of physical and mental stress, and individual variability. Understanding and controlling for each of these variables is important when attempting to analyze the effects of stress and exercise-induced malaise. When applying a partial least squares discriminant analysis (PLS-DA), proteins exhibit a slight separation between the battle ready time points and the mission/recovery time points (ESI,† Fig. S3A), while more substantial separation occurs between AM and PM collection events (ESI,† Fig. S3C). Metabolomic analyses indicate some separation based on both variables (ESI,† Fig. S3B and D). A principal component analysis (PCA) is included for comparison with an unsupervised technique (ESI,† Fig. S4). Circadian rhythm is known to play a major role in the content of human biofluids including the salivary proteome.47,48 This includes the pulsatile releases of cortisol,49 one of the primary hormonal signalers in stress.50 The discriminatory power of this data based on time of day (morning vs. evening) should increase the capacity to capture hormonal homeostatic systems, such as sleep-wake cycle and stress, both of which interact with the HPA axis in the human brain.4,51
In order to investigate these effects further, a set of linear models was created for both the proteomic and metabolomic datasets. Significance for each protein across time of day (AM vs. PM) and in relation to the mission initiation (Battle Ready vs. Mission/Recovery) was established using a mixed effect model in which time and mission category were treated as fixed effects and individual soldiers were treated as a random effect. Linear models were then constructed for each protein, and comparisons were made using analysis of variance (ANOVA) with and without mission category or time as explanatory variables. As suggested by the discriminant analysis, substantially more proteins were associated with time of day than mission category (Fig. 2A). Small molecule significance was established using ANOVA on fixed-effect linear models with a focus on time of day and mission category. Compound features for metabolites exhibited a more even distribution in significance between these two variables as compared to proteins (Fig. 2B). Normalized metabolite and protein abundances were then correlated using only overlapping samples for which both proteomic and metabolomic data were collected. We identified a single large cluster of metabolites that exhibit strong correlations to two protein clusters: one positively correlated (red box – Fig. 2C) and one negatively correlated (blue box – Fig. 2C). This small molecule cluster includes a high density of compounds found to be significant to both mission initiation and time of day (red box – Fig. 2C), while the anti-correlated protein cluster appears to include a large number of proteins significant to time of day (blue box – Fig. 2C).
When examining the gene ontology terms associated with these two extracted clusters, we observed overrepresentation of proteins related to protein processing/degradation, the endoplasmic reticulum (ER), and amino acid metabolism (Table 1, row 1 and Table 2, row 8). In addition, the metabolite cluster contains more than 40 dipeptides, potentially resulting from protein degradation associated with the proteasome and the ER. The metabolite cluster also includes several isolated canonical amino acids and their precursors.
| GO term | Count | P-Value | |
|---|---|---|---|
| Six most significant biological processes included in protein cluster negatively correlated with small molecule cluster. | |||
| 1 | Negative regulation of cysteine-type endopeptidase activity | 5 | 8.03 × 10−6 |
| 2 | Platelet degranulation | 9 | 1.27 × 10−4 |
| 3 | O-Glycan processing | 4 | 7.48 × 10−3 |
| 4 | Retina homeostasis | 4 | 1.23 × 10−2 |
| 5 | Cell activation | 2 | 1.66 × 10−2 |
| 6 | IRE1-mediated unfolded protein response | 3 | 1.73 × 10−2 |
| GO Term | Count | P-Value | |
|---|---|---|---|
| Eight most significant biological processes included in protein cluster positively correlated with small molecule cluster. | |||
| 1 | Proteolysis involved in cellular protein catabolic processes | 10 | 2.15 × 10−7 |
| 2 | Protein polyubiquitination | 11 | 5.00 × 10−7 |
| 3 | MAPK cascade | 12 | 1.40 × 10−6 |
| 4 | Regulation of mRNA stability | 11 | 1.45 × 10−6 |
| 5 | Stimulatory C-type lectin receptor signaling pathway | 10 | 2.19 × 10−6 |
| 6 | Proteasome | 9 | 3.67 × 10−6 |
| 7 | T cell receptor signaling pathway | 10 | 5.20 × 10−6 |
| 8 | Regulation of cellular amino acid metabolic processes | 9 | 5.43 × 10−6 |
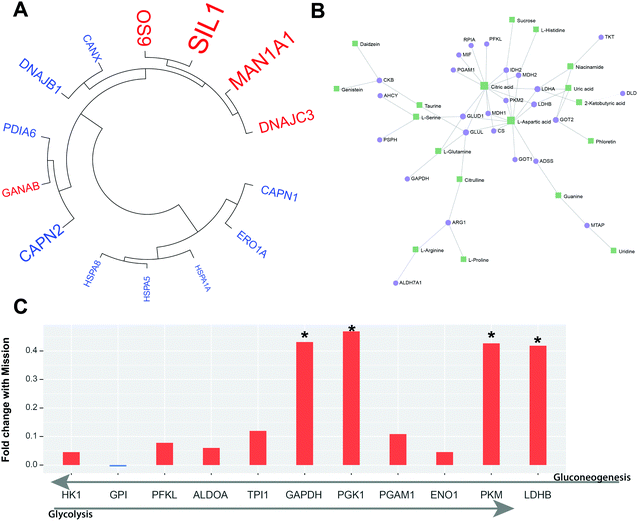
The analysis identified 21 proteins via gene ontology related to protein processing in the ER, with 14 of them found to be significantly associated with mission initiation. Of these 14 proteins, five increase with the initiation of the mission, while the other nine decrease following mission initiation (Fig. 3A). Up regulated proteins exhibit a larger magnitude shift with protein OS9, nucleotide exchange factor SIL1 (SIL1) and mannosyl-oligosaccharide 1,2-alpha-mannosidase IA (MAN1A1) most effected. MAN1A1 and OS9 both function in shuttling glycoproteins out of the endomembrane system, by maturation of their glycan chain or degradation of misfolded proteins, respectively.52,53 Neutral alpha-glucosidase AB (GANAB1) also functions in the maturation process of glycoproteins and increases following mission initiation.54 SIL1 functions as a co-chaperone to ER chaperone BiP (HSPA5), allowing HSPA5 to exchange ADP for ATP. SIL1 has been shown to play a role in cellular sensing of ER stress, with aberrant SIL1 associated with accumulation of ubiquitinated proteins.55 There is a decrease in abundance of proteins, such as PDIA6 and HSPA5, which can repress the unfolded protein response (UPR), along with increases in DNAJC3, which is activated by ER stress and the UPR.56–58 Many of the proteins decreasing following mission initiation participate in the refolding of misfolded proteins including DNAJB1, CANX1, HSPA1B, HSPA8,59–61 although these changes are of a smaller magnitude.
The analysis identified 46 proteins and 120 metabolites related to amino acid biosynthesis or metabolism with 14 and 25 mission-significant proteins and metabolites, respectively (Fig. 3B). When examining this significant protein subset, it was observed that many of the proteins had their primary function in cellular metabolism and ATP generation, rather than acting directly on amino acids or amino acid precursors. Three key enzymes in glycolysis exhibited significantly increased abundance upon initiation of the mission, pyruvate kinase (PKM), phosphoglycerate kinase (PGK1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Additionally, lactate dehydrogenase (LDHB), the enzymatic driver of anaerobic respiration, was also significantly increased. Although not reaching significance, we also detected elevated abundance with mission initiation for seven other enzymes involved in glycolysis: hexokinase (HK1), phosphofructokinase (PFKL), fructose bisphosphate aldolase (ALDOA), triosephosphate isomerase (TPI), enolase (ENO1), and phosphoglycerate mutase (PGAM) (Fig. 3C). These proteins also play a key role in gluconeogenesis which has been shown to be upregulated in situations of acute psychological stress.62,63 Blood glucose elevation has been detected in animals as an early reaction to stressors.2 Increased expression of glycolysis proteins and those associated with anaerobic respiration reflects cellular expression changes in response to continued energetic stress.
Determining a mission-specific protein biomarker panel
In reference to Fig. 1, the following paired analysis was done to determine protein changes that occur when comparing pre/post mission initiation (i.e. Battle Ready vs. Mission/Recovery), controlling for the time of day.Given both the paired structure and observed impact that time of day has on protein expression, the proteomic effects of the mission were also assessed using a paired t-test, across each Battle Ready and Mission/Recovery pair. Overall, proteins trended toward higher abundances in the Mission and Recovery sample groups (i.e. M_6_PM, R_7_AM1/AM3/AM4). Paired t-tests were performed to account for unequal variance and multiple hypotheses (Benjamini Hochberg, 5% FDR) for all proteins identified in at least half of all pairs (720 proteins). From those tests, 302 significant proteins were identified (Fig. 4A). Similar to previous analysis, biological processes such as protein processing and MAPK signaling were observed, but the complement and coagulation cascade was also identified as highly upregulated in the Mission/Recovery samples. Interestingly, the data showed an increase in angiotensinogen, a component of the stress-activated MAPK cascade, which assists in regulation of osmotic balance in the vascular system.64 Stress-related changes to the vascular system have been observed in the literature.2,65 Proteins associated with the complement and coagulation cascade demonstrate a consistent increase in abundance upon mission initiation (Fig. 4B).
When performing a pairwise correlation of expression profiles for all 302 proteins significantly associated with mission initiation, four well-defined clusters were identified (Fig. 4B). Two of these clusters include proteins that modify structures via the cytoskeleton and extracellular matrix. The other two clusters continue the theme of innate immunity and protein processing. Interestingly, the same cluster containing protein chaperones involved in protein folding also contains several proteins for processing reactive oxygen species, compounds that are typically detrimental to the structure of proteins and known to be generated from consistent physical stress66 and ER stress.67 When examining the complement and coagulation cluster more closely (Fig. 4B, bottom right), the data revealed more than 10 complement proteins, including multiple subcomponents of the C1 initiator and the alpha, beta and gamma chain of complement component C8, which forms the membrane attack complex. A comparison of normalized expression across all 160 samples of complement C2, complement C1r, complement 4b, complement C6, complement factor B and complement factor I, reveals highly similar profiles of these proteins. This similarity reflects the tight regulation of the innate immune system at this host–pathogen interface (Fig. 4C).
A paired t-test was also performed against the post mission initiation samples (i.e. M_6_PM, R_7_AM1/AM3/AM4) using only Battle Ready AM or PM samples in order to identify mission-associated proteins specific to a single time of day. These samples yielded an additional 54 significant proteins specific to the AM samples (Fig. 5A) and one additional protein specific to the PM samples (Fig. 5B). The additional protein identified when examining only PM samples was also a complement component (C9). Among the AM specific post mission initiation time points, proteins were detected relating to an increased abundance of hypoxia-up regulated protein 1, which inhibits apoptosis due to oxygen deprivation,68 and 14-3-3 protein eta, which has been shown to be a transcriptional activator of glucocorticoid receptors.69 Alpha-amylase was also observed to be significantly increased with mission initiation in these AM samples. Salivary alpha-amylase has been previously identified as a marker of stress-induced stimulation of the sympathetic-adrenal-medullary system.70–72
Proteomic and metabolomic stress/enervation discrimination power
We sought to identify the molecules most discriminant for mission initiation as indicators of the associated stress and exercise-induced malaise. These compounds could serve as biomarkers of performance and provide clues as to the cellular state. Considering this objective, linear discriminant analysis (LDA) was performed focusing on proteins or small molecules found to be significant in the linear models. We calculated the area-under-the-curve (AUC) for the receiver operator characteristic (ROC), utilizing the leave one out strategy. When performing a linear discriminant analysis using a single protein, periplakin (PPL), galactosidase beta 1 (GLB1), complement component C9 (C9), ubiquitin fold modifier 1 (UFM1), and prelamin A/C (LMNA) provided the best performance, with AUCs of 0.701, 0.697, 0.683, 0.683, 0.674, respectively (Fig. 6A). Among the metabolites, C16H13ClN2O, three pipecolic acid-like compounds (PALC) (ESI,† Table S1), and acetylspermidine exhibited the greatest discriminatory performance for mission initiation with areas under the curve of 0.835, 0.832, 0.819, 0.802 and 0.808, respectively. It should be noted that the discriminatory power of small molecules far outweighs that of even the best performing proteins, with substantially higher AUCs (Fig. 6B).Because human performance deficits manifest as a ‘whole body’ syndrome, a panel of biomarkers will likely offer a better predictive value than a single biomarker. As such, linear discriminant analysis was pursued, relying on five compounds for both proteomic and small molecule analysis. A substantial increase in AUCROC was observed when utilizing a combination of the top five components listed above for each dataset. However, the best performing five-component model was identified by iteratively testing combinations and optimizing area-under-the-curve, starting with periplakin or C16H13ClN2O. The optimal protein combination included periplakin (PPL), heat shock protein alpha family class B member 1 (HSPAB1), myeloperoxidase (MPO), heat shock protein family A member 9 (HSPA9), and transketolase (TKT) yielding a predictive value of 83% (Fig. 6A). The optimal small molecule combination consisted of C16H13ClN2O, gamma-butyrobetaine, L-proline, and proline–glycine, and pipecolic acid yielded a predictive value of 96%.
Increasing the complexity of a model by adding explanatory variables will most likely improve performance. However, it was interesting that, of the nine potentially discriminant proteins identified, two are components of chaperones that participate in the unfolded protein response (UPR) [HSPAB1 and HSPA9] and one mediates the ER stress response (UFM1). In the metabolomic model, gamma-butyrobetaine, L-proline, pipecolic acid, and proline–glycine are all associated with amino acid metabolism. Gamma-butyrobetaine is involved in lysine degradation and a precursor to L-carnitine.73L-Proline is an amino acid itself, with proline-rich proteins playing an important signaling role in human saliva.74 Proline–glycine is involved in protein catabolism but also plays roles in metabolism and coagulation.75–77 The proteins identified here are of interest in understanding the biological effects of stress, but more targeted methods would be required for validating protein signatures as baseline response to stress varies greatly among individuals, thus even the most discriminatory proteins exhibit somewhat minor shifts in abundance overall (ESI,† Fig. S5).
Detection of acute stress by metabolomics
In reference to Fig. 1, we are assessing whether or not there are metabolites that discriminate the two ASE events from the rest of the study (Battle Ready, Mission, and Recovery).Because metabolomics analysis proved to have the most discrimination power for exercised-induced malaise and stress, we sought to investigate if only small molecules could discriminate a short-lived acute stress event (Fig. 1, ASE 1 & 2) during a period of chronic stress. For this study, we included 30 individuals that were sampled at five different time points (Baseline, T0, T+15min, T+30min, and T+45min) during both acute stress events (ASE 1 & ASE 2). The ASE simulates both an acute physical and cognitive stress event, in which subjects must learn a series of signs and symbols before the activity that relate to how they must execute encountered tasks (i.e. recognizing friend versus foe, aim points, accuracy, position: standing, kneeling, or prone). These tasks were then performed while the subjects were timed.
Normalized peak areas for all compound features were averaged for Battle Ready, Mission, Recovery, ASE 1 and ASE 2 then these groups and associated compounds were clustered based on Euclidean distance. Fig. 7A shows the distinct grouping of metabolites that occurs among compounds when comparing these five groups. We specifically observe increased abundance in several compound clusters in the two ASE samples, while the other three time point groups cluster together, with the ASE 1 baseline event located most distally (Fig. 7A). Based on this preliminary result, 200 features were determined to experience significant changes in abundance between the five time point groups, with p-values ≤0.05. These compounds included small molecules involved in a variety of biological pathways largely related to metabolism of amino acids. Metaboanalyst78 pathway enrichment analysis (Fig. 7B) reveals enrichment occurring in 30 different pathways, including arginine biosynthesis, histidine metabolism, beta-alanine and alanine, aspartate and glutamate metabolism.
In order to distinguish potential discriminant biomarkers of different stress states, linear discriminant analysis (LDA) was performed with the objective of separating the ASE samples from all others. Several amino acids were identified as discriminant for these acute stress events including arginine, histidine and the dipeptide alanine–proline (Fig. 7C). Dipeptide alanine–proline is known to interact with angiotensin converting enzyme (ACE).79 This enzyme is a central component of the renin–angiotensin system that controls blood pressure regulation and electrolyte balance, particularly when the “flight or fight” response is observed as a result of exposure to acute stress.80 Chitobiose, an acylaminosugar, has been observed in protein folding assistance, furthering the previous hypothesis that stressor events cause an alteration in protein refolding, degradation, and synthesis.81 Considering the identification of histidine and arginine and the significance of these two pathways in the enrichment analysis (Fig. 7B), arginine biosynthesis and histidine metabolism were extracted and analyzed via a similar clustering analysis. Compounds related to arginine biosynthesis cluster into two high level groups: one with large variability in mean abundance across the different time point events and one with minimal variability. The high variance compounds contribute to the separation of the two ASE samples from the other three time point groups (ESI,† Fig. S6). Upon initial review, the biological variation occurring during the ASE time points appear to stem from increased amino acid metabolism.
Discussion
Presented is a multiomics analysis of raw saliva focused on identifying biomolecular perturbations following two unique stress models: (1) chronic/long-term stress and (2) acute/short-lived stress. Regarding chronic stress, like that experienced during multi-day military exercises, significant abundance changes were observed in molecules related to three biological functions: (1) innate immunity, (2) protein cycling and processing, and (3) metabolism of sugars and amino acids. Within the proteins, increased expression was identified in the complement system upon initiation of the mission, indicating immune activation and inflammation. These observations demonstrated altered abundance of endoplasmic reticulum processing proteins, such as chaperones and those involved in the unfolded protein response (UPR). These changes are accompanied by increased abundance of metabolites associated with amino acid metabolism and translation. Changes are also observed in proteins functioning in sugar metabolism.The proteomic analysis identifies more than 300 proteins significantly affected by mission. Relatively uniform upregulation of complement proteins was observed upon mission initiation, with tightly correlated shifts from sample to sample. The complement system has been shown to be stimulated in situations of both short and long duration exercise.82–84 Moderate physical training provides a beneficial immunomodulatory effect,85 while extreme physical exertion can increase susceptibility to infection86 by depressing immune cell populations and signaling molecules. Similar changes have been detected in cases of psychological stress with increased expression of proinflammatory cytokines in acute psychological stress87,88 and immune dysfunction linked to long-term psychological health impacts, such as post-traumatic stress disorder.89,90 In contrast to the complement system, two other pathways of interest, (1) protein processing in the ER and (2) the stress-activated MAPK cascade, demonstrate varying directions of abundance changes. In the stress activated MAPK cascade, angiotensinogen exhibits increased abundance with mission, likely in response to the associated physical stress. When attempting to construct a discriminant proteomic model, several of the proteins identified were related to innate immunity or protein processing, including complement component C9 and multiple heat shock cognate chaperones.
The metabolomic analysis identified more than 100 compound features associated with mission initiation. Metabolites identified as significantly associated with mission initiation, like proline, arginine, and g-butyrobetaine, are related to amino acid biosynthesis and protein translation. When a discriminatory model for mission initiation was built, those compounds were highly discriminant within the system. As these metabolites are involved in protein translation, effects were further witnessed in the aminoacyl-tRNA biosynthesis pathway. Perturbations of translation via this pathway have been observed as a response to various stressors, including temperature, oxidative environment, and nutrient deprivation.91,92
The strongest correlation between proteomic and metabolomic datasets occurred across two protein clusters and a single small molecule cluster. The cluster of metabolites was comprised of isolated amino acids and short polypeptides. These products are related to protein degradation. Positive correlation was observed between these small molecules and a protein cluster enriched for components of the proteasome, as well as the polyubiquitination process, protein catabolism and regulation of amino acid metabolism. These same small molecules also exhibit an anticorrelation with a second protein cluster containing the ER-associated processes of glycosylation and the unfolded protein response.
Upon initiation of the mission, altered abundances were observed for many key chaperones associated with protein quality control, managing ER stress, and the unfolded protein response (UPR), such as SIL1, PDIA, DNAJC1, DNAJB1, OS9, HSPA5, HSPA1B, and HSPA8.52,53,59–61,93 ER stress and the UPR can be induced by both physical and mental stress. Accelerated metabolic activity in skeletal muscle in response to physical exertion generates increased reactive oxygen species and stimulates the UPR.94,95 Stress-induced stimulation of the hypothalamic–pituitary–adrenal axis increases free radicals in the brain and has been associated with altered chaperone expression.96–98 Similar to immunity, moderate triggering of these quality control system can be beneficially adaptive,99 but constant or extreme pressure on the ER can lead to autophagy or apoptosis.67 These detrimental pressures can also lead to a pathogenic reduction in response in the ER, which can play a role in pathogenesis of diseases such as diabetes.100
When the effected metabolic pathways were examined more closely, the data demonstrated mission-associated increases in glycolysis enzymes: glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate kinase, and pyruvate kinase, as well as lactate dehydrogenase, a key enzyme in anaerobic respiration. Less significant increases were detected in seven other glycolysis-related enzymes: hexokinase, phosphofructokinase, fructose bisphosphate aldolase, triosephosphate isomerase, enolase, and phosphoglycerate mutase. These shifts suggest increased glycolytic activity and anaerobic energy generation. Metabolic changes of this type have been observed in muscle in both endurance training,101 as well as short interval, high intensity training.102–104 Psychological stress can also lead to stimulation of lactate dehydrogenase,105,106 as well as induction of genes that stimulate glycolysis.107–109 This chronic stress often leads to a variety of metabolic diseases, including obesity and diabetes.110,111 These proteins also participate in the reverse reaction of gluconeogenesis, generating glucose. Glucose serves as the only useable energy reservoir for central nervous system function, and elevated blood glucose and gluconeogenesis activity has been observed in animals undergoing acute stress.2
As a result of the metabolite changes seen due to the chronic stress of a multi-day battlefield exercise, a further study was performed in an effort to distinguish long-term stress events from acute stress events. During the field study, subjects underwent two acute stress scenarios, which entailed learning a series of signs and symbols before the activity that were related to how they must execute the task. The signs and symbols relayed to the subject how they should position their bodies at each station (i.e. standing, kneeling, or prone) and where on the target they should aim their weapon (i.e. head vs. center of mass).
When the acute stress time points are compared to all other events within the study (i.e. ASE 1 or 2 vs. Battle Ready, ASE 1 or 2 vs. Mission, ASE 1 or 2 vs. Recovery), more than 200 features are significantly changing within the comparison groups. Metabolites identified as significantly changing within specific time point comparisons were related to amino acid metabolism, with arginine, histidine, aspartate, and glutamate especially impacted. Histidine and arginine metabolites exhibited increased abundance specifically in samples from the two ASE, indicating a possible physiological effect of acute stress in these systems. Previous work has indicated a role of L-arginine as a component molecule of the arginine nitric oxide pathway, which can be stimulated by psychological stress.112 Some evidence also exists for L-arginine as a stress reducing molecule when ingested as a dietary supplement.113 Metabolomic discrimination analysis showed compounds associated with a “flight or fight” response and protein folding can be used to differentiate points of acute stress. Further studies will focus on validating identification of the compounds contributing most to the differentiation of the time point groups and absolute quantitation of selected metabolites to identify state of stress expression thresholds.
Conclusions
Stress responses play a complex role in human physiology, effecting a variety of physical and psychological systems and requiring a multifaceted approach for their study. This effort presents potential biomarkers that could be utilized in diagnostic tools to differentiate types of stress – either (1) chronic stress by monitoring proteins associated with quality control and unfolded protein response and metabolites associated with the glycolytic process or (2) acute stress by monitoring dipeptides associated with “fight or flight” response and small molecules that assist in protein folding. Differentiating these types of stress can lead to better management of their effects in both the short and long-term, leading to better overall health outcomes for the warfighter. The impact of this stress-type differentiation extends beyond the warfighter to other high stress jobs – such as medical personnel, aircraft controllers, EMS, and law enforcement. Research has pointed to higher levels of acute fatigue in nurses and trauma surgeons leading to decreased performance and decision making, even after the period during which the acute stress occurred.114,115 Understanding the overlap of these biomarkers, particularly amino acids, with other non-invasive mediums such as exhaled breath would allow for a breath-based diagnostic that could prevent fatigue-linked accidents and deaths.116 Further research needs to be conducted to validate these markers and assess their availability in human breath.Author contributions
JM and CCJ contributed equally to this work. TGG, JC, and ESD are corresponding authors, with ESD being the primary corresponding author. The field study through which the samples were collected was designed, in part, and coordinated by EKH. ESD, PMM, TGG, and JC designed the analysis plan detailed within this manuscript, and revised the manuscript. ESD and CM performed sample preparation and data collection. JM, CCJ and PMM performed data analysis, and JM and CCJ drafted the manuscript. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE database with the dataset identifier PXD026680. The mass spectrometry metabolomics data have been deposited to the Metabolomics Workbench (http://www.metabolomicsworkbench.org).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank DEVCOM Soldier Center for funding this project. The authors would like to thank members of the 82nd Airborne's 2nd Battalion, 505th Parachute Infantry Regiment of the United States Army for their participation in this study. Conclusions and opinions presented here are those of the authors and are not the official policy of the U.S. Army, DEVCOM CBC, or the U.S. Government. Information in this report is cleared for public release and distribution is unlimited.References
- S. F. De Boer, S. J. Koopmans, J. L. Slangen and J. Van Der Gugten, Plasma catecholamine, corticosterone and glucose responses to repeated stress in rats: Effect of interstressor interval length, Physiol. Behav., 1990, 47(6), 1117–1124 CrossRef CAS PubMed.
- V. A. Viblanc, Q. Schull, T. Cornioley, A. Stier, J. J. Ménard and R. Groscolas, et al., An integrative appraisal of the hormonal and metabolic changes induced by acute stress using king penguins as a model, Gen. Comp. Endocrinol., 2018, 269, 1–10 CrossRef CAS PubMed.
- R. M. Sapolsky, L. M. Romero and A. U. Munck, How Do Glucocorticoids Influence Stress Responses? Integrating Permissive, Suppressive, Stimulatory, and Preparative Actions*, Endocr. Rev., 2000, 21(1), 55–89 CAS , available from: https://academic.oup.com/edrv/article/21/1/55/2423840.
- G. Russell and S. Lightman, The human stress response, Nat. Rev. Endocrinol., 2019, 525–534, available from: http://www.nature.com/nrendo Search PubMed.
- H. Yaribeygi, Y. Panahi, H. Sahraei, T. P. Johnston and A. Sahebkar, The impact of stress on body function: A review, EXCLI J., 2017, 16, 1057–1072 Search PubMed , Leibniz Research Centre for Working Environment and Human Factors, available from: http://file:///pmc/articles/PMC5579396/?report=abstract.
- C. Santone, V. Dinallod, M. Paci, S. D’Ottavio, G. Barbato and S. Bernardini, Saliva metabolomics by NMR for the evaluation of sport performance, J. Pharm. Biomed. Anal., 2014, 88, 441–446 CrossRef CAS PubMed.
- M. L. Cummings, C. Mastracchio, K. M. Thornburg and A. Mkrtchyan, Boredom and distraction in multiple unmanned vehicle supervisory control, Interact. Comput., 2013, 25(1), 34–47 CrossRef.
- J. Langan-Fox, M. J. Sankey and J. M. Canty, Human factors measurement for future air traffic control systems, Hum. Factors, 2009, 51(5), 595–637 CrossRef PubMed , available from: http://journals.sagepub.com/doi/10.1177/0018720809355278.
- C. A. Morgan, S. Wang, S. M. Southwick, A. Rasmusson, G. Hazlett and R. L. Hauger, et al., Plasma neuropeptide-Y concentrations in humans exposed to military survival training, Biol. Psychiatry, 2000, 47(10), 902–909 CrossRef CAS PubMed.
- J. F. O’Hanlon, Boredom: Practical consequences and a theory, Acta Psychol., 1981, 49(1), 53–82 CrossRef.
- M. B. Weinger, Vigilance, boredeom, and sleepiness, J. Clin. Monit. Comput., 1999, 15, 549–552 CrossRef CAS PubMed , available from: https://link-springer-com.ezproxy.library.wisc.edu/article/10.1023/A:1009993614060.
- M. L. Pacella, B. Hruska and D. L. Delahanty, The physical health consequences of PTSD and PTSD symptoms: A meta-analytic review, J. Anxiety Disord., 2013, 27, 33–46 CrossRef PubMed.
- R. Yang, A. Gautam, D. Getnet, B. J. Daigle, S. Miller and B. Misganaw, et al., Epigenetic biotypes of post-traumatic stress disorder in war-zone exposed veteran and active duty males, Mol. Psychiatry, 2020, 1–15 Search PubMed , available from: DOI: http://10.1038/s41380-020-00966-2.
- A. Gautam, P. D’Arpa, D. E. Donohue, S. Muhie, N. Chakraborty and B. T. Luke, et al., Acute and Chronic Plasma Metabolomic and Liver Transcriptomic Stress Effects in a Mouse Model with Features of Post-Traumatic Stress Disorder, PLoS One, 2015, 10(1), e0117092 CrossRef PubMed , available from: https://dx.plos.org/10.1371/journal.pone.0117092.
- E. M. Blessing, V. Reus, S. H. Mellon, O. M. Wolkowitz, J. D. Flory and L. Bierer, et al., Biological predictors of insulin resistance associated with posttraumatic stress disorder in young military veterans, Psychoneuroendocrinology, 2017, 82, 91–97 CrossRef CAS PubMed , available from: http://file:///Users/justinmcketney/Library/Application.
- S. Maguen, B. Cohen, L. Ren, J. Bosch, R. Kimerling and K. Seal, Gender differences in military sexual trauma and mental health diagnoses among iraq and afghanistan veterans with posttraumatic stress disorder, Women's Health, 2012, 22(1), e61–e66 Search PubMed.
- A. O’Donovan, B. E. Cohen, K. H. Seal, D. Bertenthal, M. Margaretten and K. Nishimi, et al., Elevated risk for autoimmune disorders in iraq and afghanistan veterans with posttraumatic stress disorder, Biol. Psychiatry, 2015, 77(4), 365–374 CrossRef PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/25104173/.
- R. Glaser and J. K. Kiecolt-Glaser, Stress-induced immune dysfunction: Implications for health, Nat. Rev. Immunol., 2005, 5, 243–251 CrossRef CAS PubMed , available from: http://www.nature.com/reviews/immunol.
- J. E. Dimsdale, Psychological Stress and Cardiovascular Disease, J. Am. Coll. Cardiol., 2008, 51, 1237–1246 CrossRef PubMed.
- W. Kuo, L. C. Bratzke, L. D. Oakley, F. Kuo, H. Wang and R. L. Brown, The association between psychological stress and metabolic syndrome: A systematic review and meta-analysis, Obes. Rev., 2019, 20(11), 1651–1664 CrossRef PubMed , Available from: https://onlinelibrary.wiley.com/doi/abs/10.1111/obr.12915.
- D. J. Michael, S. Daugherty, A. Santos, B. C. Ruby and J. E. Kalns, Fatigue biomarker index: An objective salivary measure of fatigue level, Accid. Anal. Prev., 2012, 45, 68–73 CrossRef PubMed.
- D. J. Michael, B. Valle, J. Cox, J. E. Kalns and D. L. Fogt, Salivary Biomarkers of Physical Fatigue as Markers of Sleep Deprivation, J. Clin. Sleep Med., 2013, 09(12), 1325–1331 CrossRef PubMed , available from: http://jcsm.aasm.org/doi/10.5664/jcsm.3280.
- X.-H. Su, F.-L. Liu, J.-Z. Wang, Z.-J. Liu and J.-Z. Zhang, Discovery and identification of fatigue-related biomarkers in human saliva, Eur. Rev. Med. Pharmacol. Sci., 2018, 22, 8519–8536 Search PubMed.
- S. G. Ra, S. Maeda, R. Higashino, T. Imai and S. Miyakawa, Metabolomics of salivary fatigue markers in soccer players after consecutive games, Appl. Physiol. Nutr. Metab., 2014, 39(10), 1120–1126 CrossRef CAS PubMed.
- K. Ngamchuea, K. Chaisiwamongkhol, C. Batchelor-Mcauley and R. G. Compton, Chemical analysis in saliva and the search for salivary biomarkers-a tutorial review, Analyst, 2018, 143, 81–99 RSC , available from: https://pubs.rsc.org/en/content/articlehtml/2018/an/c7an01571b.
- E. M. McBride, R. J. Lawrence, K. McGee, P. M. Mach, P. S. Demond and M. W. Busch, et al., Rapid liquid chromatography tandem mass spectrometry method for targeted quantitation of human performance metabolites in saliva, J. Chromatogr. A, 2019, 1601, 205–213 CrossRef CAS PubMed.
- B. L. Schulz, J. Cooper-White and C. K. Punyadeera, Saliva proteome research: Current status and future outlook, Crit. Rev. Biotechnol., 2013, 33, 246–259 CrossRef CAS PubMed , available from: https://www.tandfonline.com/doi/abs/10.3109/07388551.2012.687361.
- I. Messana, T. Cabras, F. Iavarone, B. Manconi, L. Huang and C. Martelli, et al., Chrono-proteomics of human saliva: Variations of the salivary proteome during human development, J. Proteome Res., 2015, 14(4), 1666–1677 CrossRef CAS PubMed , available from: https://pubs.acs.org/sharingguidelines.
- L. M. R. B. Arantes, A. C. De Carvalho, M. E. Melendez and A. L. Carvalho, Serum, plasma and saliva biomarkers for head and neck cancer, Expert Rev. Mol. Diagn., 2017, 18, 85–112 CrossRef PubMed , available from: https://www.tandfonline.com/doi/abs/10.1080/14737159.2017.1404906.
- B. Manconi, B. Liori, T. Cabras, F. Vincenzoni, F. Iavarone and L. Lorefice, et al., Top-down proteomic profiling of human saliva in multiple sclerosis patients, J. Proteomics, 2018, 187, 212–222 CrossRef CAS PubMed.
- M. Bayani, M. Pourali and M. Keivan, Possible interaction between visfatin, periodontal infection, and other systemic diseases: A brief review of literature, Eur. J. Dent., 2017, 11(3), 407–410 CrossRef PubMed , available from: http://www.thieme-connect.de/DOI/DOI?10.4103/ejd.ejd_284_16.
- A. K. Dey, B. Kumar, A. K. Singh, P. Ranjan, R. Thiruvengadam and B. K. Desiraju, et al., Salivary proteome signatures in the early and middle stages of human pregnancy with term birth outcome, Sci. Rep., 2020, 10(1), 1–15 CrossRef PubMed , available from: http://10.1038/s41598-020-64483-6.
- S. Jain, Y. Ahmad and K. Bhargava, Salivary proteome patterns of individuals exposed to High Altitude, Arch. Oral Biol., 2018, 96, 104–112 CrossRef CAS PubMed.
- S. T. O’Donnell, R. P. Ross and C. Stanton, The progress of multi-omics technologies: determining function in lactic acid bacteria using a systems level approach, Front. Microbiol., 2020, 10, 3084 CrossRef PubMed.
- E. Shishkova, A. S. Hebert, M. S. Westphall and J. J. Coon, Ultra-High Pressure (>30,000 psi) Packing of Capillary Columns Enhancing Depth of Shortgun Proteomic Analyses, Anal Chem., 2018, 90(19), 11503–11508 CrossRef CAS PubMed.
- A. J. Steckl and P. Ray, Stress Biomarkers in Biological Fluids and Their Point-of-Use Detection, ACS Sensors, 2018, 3(10), 2025–2044 CrossRef CAS PubMed , available from: https://pubs.acs.org/sharingguidelines.
- E. Shishkova, A. S. Hebert, M. S. Westphall and J. J. Coon, Ultra-High Pressure (>30
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 psi) Packing of Capillary Columns Enhancing Depth of Shotgun Proteomic Analyses, Anal. Chem., 2018, 90(19), 11503–11508 CrossRef CAS PubMed.
000 psi) Packing of Capillary Columns Enhancing Depth of Shotgun Proteomic Analyses, Anal. Chem., 2018, 90(19), 11503–11508 CrossRef CAS PubMed. - A. S. Hebert, C. Thöing, N. M. Riley, N. W. Kwiecien, E. Shiskova and R. Huguet, et al., Improved Precursor Characterization for Data-Dependent Mass Spectrometry, Anal. Chem., 2018, 90(3), 2333–2340 CrossRef CAS PubMed , available from: https://pubs.acs.org/doi/abs/10.1021/acs.analchem.7b04808.
- F. Rohart, B. Gautier, A. Singh and K.-A. Lê Cao, mixOmics: An R package for ‘omics feature selection and multiple data integration, PLoS Comput. Biol., 2017, 13(11), e1005752 CrossRef PubMed , available from: https://dx.plos.org/10.1371/journal.pcbi.1005752.
- F. Kemp, Modern Applied Statistics with S, J. R. Stat. Soc. Ser. D, 2003, 52(4), 704–705 CrossRef , available from: DOI: http://10.1046/j.1467-9884.2003.t01-19-00383_22.x.
- X. Robin, N. Turck, A. Hainard, N. Tiberti, F. Lisacek and J. C. Sanchez, et al., pROC: An open-source package for R and S+ to analyze and compare ROC curves, BMC Bioinf., 2011, 12(1), 77 CrossRef PubMed , available from: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-12-77.
- D. Bates, M. Mächler, B. M. Bolker and S. C. Walker, Fitting linear mixed-effects models using lme4, J. Stat. Softw., 2014, 67, 1–48 Search PubMed.
- P. Sivadasan, M. K. Gupta, G. J. Sathe, L. Balakrishnan, P. Palit and H. Gowda, et al., Human salivary proteome - a resource of potential biomarkers for oral cancer, J. Proteomics, 2015, 127, 89–95 CrossRef CAS PubMed.
- L. W. Sumner, A. Amberg, D. Barrett, M. H. Beale, R. Beger, C. A. Daykin, T. W. Fan, O. Fiehn, R. Goodacre, J. L. Griffin and T. Hankemeier, Proposed minimum reporting standards for chemical analysis, Metabolomics, 2007, 3(3), 211–221 CrossRef CAS PubMed.
- S. M. Shirreffs, Markers of hydration status., Eur. J. Clin. Nutr., 2003, 57(2), S6–S9 CrossRef PubMed.
- N. A. Masento, M. Golightly, D. T. Field, L. T. Butler and C. M. van Reekum, Effects of hydration status on cognitive performance and mood, Br. J. Nutr., 2014, 111(10), 1841–1852 CrossRef CAS PubMed.
- S. K. Al-Tarawneh, M. B. Border, C. F. Dibble and S. Bencharit, Defining salivary biomarkers using mass spectrometry-based proteomics: A systematic review, OMICS: J. Integr. Biol., 2011, 15, 353–361 CrossRef CAS PubMed , available from: https://www.liebertpub.com/doi/abs/10.1089/omi.2010.0134.
- S. Schenk, S. C. Bannister, F. J. Sedlazeck, D. Anrather, B. Q. Minh and A. Bileck, et al., Combined transcriptome and proteome profiling reveals specific molecular brain signatures for sex, maturation and circalunar clock phase, eLife, 2019, 8 DOI:10.7554/eLife.41556.
- B. Gibbison, F. Spiga, J. J. Walker, G. M. Russell, K. Stevenson and Y. Kershaw, et al., Dynamic Pituitary-Adrenal Interactions in Response to Cardiac Surgery, Crit. Care Med., 2015, 43(4), 791–800 CrossRef CAS PubMed.
- S. L. Lightman, The Neuroendocrinology of Stress: A Never Ending Story, J Neuroendocrinol., 2008, 20(6), 880–884 CrossRef CAS PubMed , available from: http://doi.wiley.com/10.1111/j.1365-2826.2008.01711.x.
- T. Roenneberg and M. Merrow, The circadian clock and human health, Curr. Biol., 2016, 26, R432–R443 CrossRef CAS PubMed , available from: http://file:///Users/justinmcketney/Library/Application.
- Y. Wang, X. Fu, S. Gaiser, M. Köttgen, A. Kramer-Zucker and G. Walz, et al., OS-9 regulates the transit and polyubiquitination of TRPV4 in the endoplasmic reticulum, J. Biol. Chem., 2007, 282(50), 36561–36570 CrossRef CAS PubMed , available from: http://www.jbc.org.
- K. Legler, R. Rosprim, T. Karius, K. Eylmann, M. Rossberg and R. M. Wirtz, et al., Reduced mannosidase MAN1A1 expression leads to aberrant N-glycosylation and impaired survival in breast cancer, Br. J. Cancer, 2018, 118(6), 847–856 CrossRef CAS PubMed , available from: http://www.bjcancer.com.
- B. Porath, V. G. Gainullin, E. Cornec-Le Gall, E. K. Dillinger, C. M. Heyer and K. Hopp, et al., Mutations in GANAB, Encoding the Glucosidase II$α$ Subunit, Cause Autosomal-Dominant Polycystic Kidney and Liver Disease, Am. J. Hum. Genet., 2016, 98(6), 1193–1207 CrossRef CAS PubMed , available from: DOI: http://10.1016/j.ajhg.2016.05.004.
- L. Zhao, C. Longo-Guess, B. S. Harris, J. W. Lee and S. L. Ackerman, Protein accumulation and neurodegeneration in the woozy mutant mouse is caused by disruption of SIL1, a cochaperone of BiP, Nat. Genet., 2005, 37(9), 974–979 CrossRef CAS PubMed , available from: http://www.nature.com/naturegenetics.
- D. Eletto, D. Eletto, D. Dersh, T. Gidalevitz and Y. Argon, Protein Disulfide Isomerase A6 Controls the Decay of IRE1$α$ Signaling via Disulfide-Dependent Association, Mol. Cell, 2014, 53(4), 562–576 CrossRef CAS PubMed , available from: DOI: http://10.1016/j.molcel.2014.01.004.
- R. Van Huizen, J. L. Martindale, M. Gorospe and N. J. Holbrook, P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2$α$ signaling, J. Biol. Chem., 2003, 278(18), 15558–15564 CrossRef CAS PubMed , available from: http://www.jbc.org.
- N. Amin-Wetzel, R. A. Saunders, M. J. Kamphuis, C. Rato, S. Preissler and H. P. Harding, et al., A J-Protein Co-chaperone Recruits BiP to Monomerize IRE1 and Repress the Unfolded Protein Response, Cell, 2017, 171(7), 1625–1637 CrossRef CAS PubMed , available from: DOI: http://10.1016/j.cell.2017.10.040.
- J. N. Rauch and J. E. Gestwicki, Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro, J. Biol. Chem., 2014, 289(3), 1402–1414 CrossRef CAS PubMed , available from: http://www.jbc.org/article/S0021925820336206/fulltext.
- V. S. Stronge, Y. Saito, Y. Ihara and D. B. Williams, Relationship between Calnexin and BiP in Suppressing Aggregation and Promoting Refolding of Protein and Glycoprotein Substrates, J. Biol. Chem., 2001, 276(43), 39779–39787 CrossRef CAS PubMed , available from: http://file:///Users/justinmcketney/Library/Application.
- F. Stricher, C. Macri, M. Ruff and S. Muller, HSPA8/HSC70 chaperone protein: Structure, function, and chemical targeting, Autophagy, 2013, 9, 1937–1954 CrossRef CAS PubMed , available from: https://www.tandfonline.com/action/journalInformation?journalCode=kaup20www.landesbioscience.com.
- J. H. Exton and C. R. Park, Control of Gluconeogenesis in Liver, J. Biol. Chem., 1967, 242, 2622–2636 CrossRef CAS.
- S. J. Pilkis and D. K. Granner, Molecular Physiology of the Regulation of Hepatic Gluconeogenesis and Glycolysis, Annu. Rev. Physiol., 1992, 54(1), 885–909 CrossRef CAS PubMed , available from: http://www.annualreviews.org/doi/10.1146/annurev.ph.54.030192.004321.
- C. Delles, E. Carrick, D. Graham and S. A. Nicklin, Utilizing proteomics to understand and define hypertension: where are we and where do we go?, Expert Rev. Proteomics, 2018, 15(7), 581–592 CrossRef CAS PubMed , available from: https://www.tandfonline.com/doi/full/10.1080/14789450.2018.1493927.
- Y. J. Chen, F. Huang, M. Zhang and H. Y. Shang, Psychological stress alters ultrastructure and energy metabolism of masticatory muscle in rats, J. Biomed. Biotechnol., 2010 DOI:10.115/2010/302693.
- K. Sahlin, I. G. Shabalina, C. M. Mattsson, L. Bakkman, M. Fernström and Z. Rozhdestvenskaya, et al., Ultraendurance exercise increases the production of reactive oxygen species in isolated mitochondria from human skeletal muscle, J. Appl. Physiol., 2010, 108(4), 780–787 CrossRef CAS PubMed , available from: https://www.physiology.org/doi/10.1152/japplphysiol.00966.2009.
- J. D. Malhotra, H. Miao, K. Zhang, A. Wolfson, S. Pennathur and S. W. Pipe, et al., Antioxidants reduce endoplasmic reticulum stress and improve protein secretion, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(47), 18525–18530 CrossRef CAS PubMed , available from: http://www.pnas.org/cgi/content/full/.
- K. Ozawa, K. Kuwabara, M. Tamatani, K. Takatsuji, Y. Tsukamoto and S. Kaneda, et al., 150 kDa oxygen-regulated protein (ORP150) suppresses hypoxia-induced apoptotic cell death, J. Biol. Chem., 1999, 274(10), 6397–6404 CrossRef CAS PubMed , available from: http://www.jbc.org.
- Y. S. Kim, S.-W. Jang, H. J. Sung, H. J. Lee, I. S. Kim and D. S. Na, et al., Role of 14-3-3$η$ as a Positive Regulator of the Glucocorticoid Receptor Transcriptional Activation, Endocrinology, 2005, 146(7), 3133–3140 CrossRef CAS PubMed , available from: https://academic.oup.com/endo/article-lookup/doi/10.1210/en.2004-1455.
- E. B. Gordis, D. A. Granger, E. J. Susman and P. K. Trickett, Asymmetry between salivary cortisol and α-amylase reactivity to stress: Relation to aggressive behavior in adolescents, Psychoneuroendocrinology, 2006, 31(8), 976–987 CrossRef CAS PubMed.
- S. Het, D. Schoofs, N. Rohleder and O. T. Wolf, Stress-Induced Cortisol Level Elevations Are Associated With Reduced Negative Affect After Stress, Psychosom. Med., 2012, 74(1), 23–32 CrossRef CAS PubMed , available from: http://journals.lww.com/00006842-201201000-00006.
- S. Het, N. Rohleder, D. Schoofs, C. Kirschbaum and O. T. Wolf, Neuroendocrine and psychometric evaluation of a placebo version of the “Trier Social Stress Test, Psychoneuroendocrinology, 2009, 34(7), 1075–1086 CrossRef CAS PubMed.
- F. M. Vaz and R. J. A. Wanders, Carnitine biosynthesis in mammals, Biochem. J., 2002, 361(3), 417–429 CrossRef CAS , available from: http://file:///biochemj/article/361/3/417/39798/Carnitine-biosynthesis-in-mammals.
- B. Manconi, M. Castagnola, T. Cabras, A. Olianas, A. Vitali and C. Desiderio, et al., The intriguing heterogeneity of human salivary proline-rich proteins: Short title: Salivary proline-rich protein species, J. Proteomics, 2016, 134, 47–56 CrossRef CAS PubMed.
- T. Vahdatpour, H. Valizadeh, N. Mirzakhani and M. Mesgari-Abbasi, Renoprotective Effects of Di- and Tri-peptides Containing Proline, Glycine and Leucine in Diabetes Model of Adult Mice: Enzymology and Histopathology, Int. J. Pept. Res. Ther., 2020, 26(4), 2345–2354 CrossRef CAS , available from: DOI: http://10.1007/s10989-020-10024-8.
- M. E. Grigorieva, T. Y. Obergan, E. S. Maystrenko and M. D. Kalugina, Anticoagulant Effects of Heparin Complexes with Prolyl-Glycine Peptide and Glycine and Proline Amino Acids, Bull. Exp. Biol. Med., 2016, 161(1), 54–57 CrossRef CAS PubMed , available from: https://link.springer.com/article/10.1007/s10517-016-3343-0.
- M. Zhang, J. Xu, T. Wang, X. Wan, F. Zhang and L. Wang, et al., The Dipeptide Pro-Gly Promotes IGF-1 Expression and Secretion in HepG2 and Female Mice via PepT1-JAK2/STAT5 Pathway, Front. Endocrinol., 2018, 9, 424 CrossRef PubMed , available from: https://www.frontiersin.org/article/10.3389/fendo.2018.00424/full.
- J. Chong, O. Soufan, C. Li, I. Caraus, S. Li and G. Bourque, et al., MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis, Nucleic Acids Res., 2018, 46(W1), W486–W494 CrossRef CAS PubMed , available from: http://old.metaboanalyst.ca.
- NCBI. PubChem Gene Summary for NCBI Gene 1636, ACE – angiotensin I converting enzyme (human). National Center for Biotechnology Information, 2021 [cited 2021 Jun 1], available from: https://pubchem.ncbi.nlm.nih.gov/gene/ACE/human.
- C. Ayada, Ü. Toru and Y. Korkut, The relationship of stress and blood pressure effectors, Hippokratia, 2015, 19(2), 99–108 CAS , available from: https://pubmed.ncbi.nlm.nih.gov/27418756.
- M. D. Witte, D. Horst, E. J. H. J. Wiertz, G. A. van der Marel and H. S. Overkleeft, Synthesis and Biological Evaluation of a Chitobiose-Based Peptide N-Glycanase Inhibitor Library, J. Org. Chem., 2009, 74(2), 605–616 CrossRef CAS PubMed , available from: DOI: http://10.1021/jo801906s.
- B. Dufaux and U. Order, Complement activation after prolonged exercise, Clin. Chim. Acta, 1989, 179(1), 45–49 CrossRef CAS.
- J. K. Smith, D. S. Chi, G. Krish, S. Reynolds and G. Cambron, Effect of exercise on complement activity, Ann. Allergy, 1990, 65(4), 304–310 CAS , available from: https://europepmc.org/article/med/2221489.
- B. Dufaux, U. Order and H. Liesen, Effect of a Short Maximal Physical Exercise on Coagulation, Fibrinolysis, and Complement System, Int. J. Sports Med., 1991, 12(1), S38–S42 CrossRef PubMed , available from: http://www.thieme-connect.de/DOI/DOI?10.1055/s-2007-1024748.
- E. Ortega, Neuroendocrine mediators in the modulation of phagocytosis by exercise: Physiological implications, Exerc. Immunol. Rev., 2003, 9, 70–93 Search PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/14686096/.
- D. C. Nieman, Marathon training and immune function, Sports Med., 2007, 412–415 CrossRef PubMed , available from: https://link.springer.com/article/10.2165/00007256-200737040-00036.
- V. E. Burns, K. M. Edwards, C. Ring, M. Drayson and D. Carroll, Complement Cascade Activation After an Acute Psychological Stress Task, Psychosom. Med., 2008, 70(4), 387–396 CrossRef PubMed , available from: http://journals.lww.com/00006842-200805000-00001.
- M. S. Breen, N. Beliakova-Bethell, L. R. Mujica-Parodi, J. M. Carlson, W. Y. Ensign and C. H. Woelk, et al., Acute psychological stress induces short-term variable immune response, Brain, Behav., Immun., 2016, 53, 172–182 CrossRef CAS PubMed.
- M. S. Breen, A. X. Maihofer, S. J. Glatt, D. S. Tylee, S. D. Chandler and M. T. Tsuang, et al., Gene networks specific for innate immunity define post-traumatic stress disorder, Mol. Psychiatry, 2015, 20(12), 1538–1545 CrossRef CAS PubMed , available from: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=.
- L. P. Hovhannisyan, G. M. Mkrtchyan, S. H. Sukiasian and A. S. Boyajyan, Alterations in the complement cascade in post-traumatic stress disorder, Allergy, Asthma, Clin. Immunol., 2010, 6(1), 3 CrossRef PubMed , available from: http://aacijournal.biomedcentral.com/articles/10.1186/1710-1492-6-3.
- T. Pan, Adaptive Translation as a Mechanism of Stress Response and Adaptation, Annu. Rev. Genet., 2013, 47(1), 121–137 CrossRef CAS PubMed , available from: http://www.annualreviews.org/doi/10.1146/annurev-genet-111212-133522.
- N. C. Han, P. Kelly and M. Ibba, Translational quality control and reprogramming during stress adaptation, Exp. Cell Res., 2020, 394, 112161 CrossRef CAS PubMed.
- T. Liu, C. K. Daniels and S. Cao, Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential, Pharmacol. Ther., 2012, 354–374 CrossRef CAS PubMed.
- J. Wu, J. L. Ruas, J. L. Estall, K. A. Rasbach, J. H. Choi and L. Ye, et al., The unfolded protein response mediates adaptation to exercise in skeletal muscle through a PGC-1$α$/ATF6$α$ complex, Cell Metab., 2011, 13(2), 160–169 CrossRef CAS PubMed , available from: http://www.cell.com/article/S1550413111000040/fulltext.
- D. I. Ogborn, B. R. McKay, J. D. Crane, G. Parise and M. A. Tarnopolsky, The unfolded protein response is triggered following a single, unaccustomed resistance-exercise bout, Am. J. Physiol., 2014, 307(6), R664–R669 CAS , available from: https://www.physiology.org/doi/10.1152/ajpregu.00511.2013.
- S. Currie, S. LeBlanc, M. A. Watters and K. M. Gilmour, Agonistic encounters and cellular angst: social interactions induce heat shock proteins in juvenile salmonid fish, Proc. R. Soc. B, 2010, 277(1683), 905–913 CrossRef PubMed , available from: https://royalsocietypublishing.org/doi/10.1098/rspb.2009.1562.
- D. Vassilopoulos and D. Mantzoukis, Dialogue between the Brain and the Immune System in Inflammatory Arthritis, Ann. N. Y. Acad. Sci., 2006, 1088(1), 132–138 CrossRef CAS PubMed , available from: http://doi.wiley.com/10.1196/annals.1366.031.
- T. Hayashi, Conversion of psychological stress into cellular stress response: Roles of the Sigma-1 receptor in the process, Psychiatry Clin. Neurosci., 2015, 69(4), 179–191 CrossRef CAS PubMed , available from: http://doi.wiley.com/10.1111/pcn.12262.
- M. Ost, V. Coleman, J. Kasch and S. Klaus, Regulation of myokine expression: Role of exercise and cellular stress, Free Radical Biol. Med., 2016, 98, 78–89 CrossRef CAS PubMed.
- M. Cnop, F. Foufelle and L. A. Velloso, Endoplasmic reticulum stress, obesity and diabetes, Trends Mol. Med., 2012, 18, 59–68 CrossRef CAS PubMed.
- A. S. Deshmukh, D. E. Steenberg, M. Hostrup, J. B. Birk, J. K. Larsen and A. Santos, et al., Deep muscle-proteomic analysis of freeze-dried human muscle biopsies reveals fiber type-specific adaptations to exercise training, Nat. Commun., 2021, 12(1), 1–15 CrossRef PubMed , available from: DOI: http://10.1038/s41467-020-20556-8.
- T. Abe, Y. Kitaoka, D. M. Kikuchi, K. Takeda, O. Numata and T. Takemasa, High-intensity interval training-induced metabolic adaptation coupled with an increase in Hif-1α and glycolytic protein expression, J. Appl. Physiol., 2015, 119(11), 1297–1302 CrossRef CAS PubMed , available from: https://www.physiology.org/doi/10.1152/japplphysiol.00499.2015.
- I. Jacobs, M. Esbjörnsson, C. Sylvén, I. Holm and E. Jansson, Sprint training effects on muscle myoglobin, enzymes, fiber types, and blood lactate, Med. Sci. Sports Exercise, 1987, 19(4), 368–374 CrossRef CAS , available from: https://europepmc.org/article/med/2958671.
- M.-T. Linossier, D. Dormois, C. Perier, J. Frey, A. Geyssant and C. Denis, Enzyme adaptations of human skeletal muscle during bicycle short-sprint training and detraining, Acta Physiol. Scand., 1997, 161(4), 439–445 CrossRef CAS PubMed , available from: https://onlinelibrary.wiley.com/doi/abs/10.1046/j.1365-<?pdb_no 201X?><?pdb_no 201X?>201X<?pdb END?><?pdb<?db_id PDB?> END?>.1997.00244.x.
- H. Arakawa, H. Kodama, N. Matsuoka and I. Yamaguchi, Stress Increases Plasma Enzyme Activity in Rats: Differential Effects of Adrenergic and Cholinergic Blockades, J. Pharmacol. Exp. Ther., 1997, 280(3), 1296–1303 CAS.
- B. Cui, Y. Luo, P. Tian, F. Peng, J. Lu and Y. Yang, et al., Stress-induced epinephrine enhances lactate dehydrogenase A and promotes breast cancer stem-like cells, J. Clin. Invest., 2019, 129(3), 1030–1046 CrossRef PubMed , available from: DOI: http://10.1172/JCI121685DS1.
- G. Ermak, S. Sojitra, F. Yin, E. Cadenas, A. M. Cuervo and K. J. A. Davies, Chronic expression of RCAN1-1L protein induces mitochondrial autophagy and metabolic shift from oxidative phosphorylation to glycolysis in neuronal cells, J. Biol. Chem., 2012, 287(17), 14088–14098 CrossRef CAS PubMed , available from: http://www.jbc.org/article/S0021925820529803/fulltext.
- G. Ermak, M. A. Pritchard, S. Dronjak, B. Niu and K. J. A. Davies, Do RCAN1 proteins link chronic stress with neurodegeneration?, FASEB J., 2011, 25(10), 3306–3311 CrossRef CAS PubMed , available from: https://onlinelibrary.wiley.com/doi/abs/10.1096/fj.11-185728.
- Y. Hirakawa, L. J. Nary and R. D. Medh, Glucocorticoid evoked upregulation of RCAN1-1 in human leukemic CEM cells susceptible to apoptosis, J. Mol. Signal., 2009, 4(1), 6 CrossRef PubMed , available from: https://link.springer.com/articles/10.1186/1750-2187-4-6.
- M. Picard and B. S. McEwen, Psychological Stress and Mitochondria: A Systematic Review, Psychosom. Med., 2018, 80, 141–153 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/29389736/.
- C. Rabasa and S. L. Dickson, Impact of stress on metabolism and energy balance, Curr. Opin. Behav. Sci., 2016, 9, 71–77 CrossRef , available from: http://file:///Users/justinmcketney/Library/Application.
- M. Reimann, M. Hamer, N. T. Malan, M. P. Schlaich, G. W. Lambert and T. Ziemssen, et al., Effects of acute and chronic stress on the L-arginine nitric oxide pathway in black and white South Africans: The sympathetic activity and ambulatory blood pressure in Africans study, Psychosom. Med., 2013, 75(8), 751–758 CrossRef CAS PubMed , available from: https://pubmed.ncbi.nlm.nih.gov/23960161/.
- M. Smriga, T. Ando, M. Akutsu, Y. Furukawa, K. Miwa and Y. Morinaga, Oral treatment with L-lysine and L-arginine reduces anxiety and basal cortisol levels in healthy humans, Biomed. Res., 2007, 28(2), 85–90 CrossRef CAS PubMed , available from: http://joi.jlc.jst.go.jp/JST.JSTAGE/biomedres/28.85?from=CrossRef.
- L. M. Barker and M. A. Nussbaum, Fatigue, performance and the work environment: a survey of registered nurses, J. Adv. Nurs., 2011, 67(6), 1370–1382 CrossRef PubMed.
- K. Bajunaid, M. A. S. Mullah, A. Winkler-Schwartz, F. E. Alotaibi, J. Fares and M. Baggiani, et al., Impact of acute stress on psychomotor bimanual performance during a simulated tumor resection task, J. Neurosurg., 2017, 126(1), 71–80 Search PubMed.
- T. Bruderer, M. T. Gaugg, L. Cappellin, F. Lopez-Hilfiker, M. Hutterli and N. Perkins, et al., Detection of Volatile Organic Compounds with Secondary Electrospray Ionization and Proton Transfer Reaction High-Resolution Mass Spectrometry: A Feature Comparison, J. Am. Soc. Mass Spectrom., 2020, 31(8), 1632–1640 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1mo00271f |
| ‡ JM and CCM are co-first authors. |
| This journal is © The Royal Society of Chemistry 2022 |