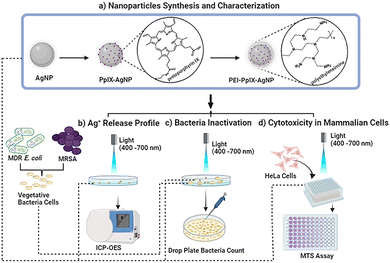
Open Access Article
Open Access ArticleInfluence of silver ion release on the inactivation of antibiotic resistant bacteria using light-activated silver nanoparticles†
Adeola Julian
Sorinolu‡
a,
Varsha
Godakhindi‡
bc,
Paolo
Siano
b,
Juan L.
Vivero-Escoto
 *bc and
Mariya
Munir
*a
*bc and
Mariya
Munir
*a
aDepartment of Civil and Environmental Engineering, University of North Carolina at Charlotte, Charlotte, NC 28223, USA. E-mail: mmunir@uncc.edu; Tel: +1 (704)-687-1623
bDepartment of Chemistry, University of North Carolina at Charlotte, Charlotte, NC 28223, USA. E-mail: jviveroe@uncc.edu; Tel: +1 (704)-687-5239
cNanoscale Science Program, University of North Carolina at Charlotte, Charlotte, NC 28223, USA
First published on 8th November 2022
Abstract
The widespread increase in antibiotic resistance (AR), in an extensive range of microorganisms, demands the development of alternative antimicrobials with novel non-specific low-mutation bacterial targets. Silver nanoparticles (AgNPs) and photosensitizers (PSs) are promising antimicrobial agents with broad-spectrum activity and low tendency for antimicrobial resistance development. Herein, we investigated the light-mediated oxidation of AgNPs for accelerated release of Ag+ in the antibacterial synergy of PS–AgNP conjugates using protoporphyrin IX (PpIX) as a PS. Also, the influence of polyethyleneimine (PEI) coated AgNPs in promoting antibacterial activity was examined. We synthesized, characterized and tested the antimicrobial effect of three nanoparticles: AgNPs, PpIX-AgNPs, and PEI-PpIX-AgNPs against a methicillin-resistant Staphylococcus aureus strain (MRSA) and a wild-type multidrug resistant (MDR) E. coli. PpIX-AgNPs were the most effective material achieving >7 log inactivation of MRSA and MDR E. coli. The order of bacterial log inactivation was PpIX-AgNPs > PEI-PpIX-AgNPs > AgNPs. This order correlates with the trend of Ag+ concentration released by the NPs (PpIX-AgNPs > PEI-PpIX-AgNPs > AgNPs). Our study confirms a synergistic effect between PpIX and AgNPs in the inactivation of AR pathogens with about 10-fold increase in inactivation of ARB relative to AgNPs only. The concentration of Ag+ released from NPs determined the log inactivation of MRSA and MDR E. coli more than either the phototoxic effect or the electrostatic interaction promoted by surface charge of nanoparticles with bacteria cells. All NPs showed negligible cytotoxicity to mammalian cells at the bacterial inhibitory concentration after 24 h exposure. These observations confirm the crucial role of optimized Ag+ release for enhanced performance of AgNP-based antimicrobials against AR pathogens.
Introduction
The emergence and spread of antibiotic resistance (AR) are one of the greatest global public health issues of the 21st century.1–3 The prevalence of AR is linked to the indiscriminate use of antibiotics for human and animal treatment, because antibiotics impose selective pressure by killing susceptible bacteria while resistant bacteria proliferate.4 AR challenges therapeutic potential against pathogens of humans and animals resulting in overwhelming healthcare costs and deaths.5,6 Moreover, advancements in modern medicine become incapacitated with dwindling efficacy of antibacterial drugs.7 The AR problem is compounded by the fact that the evolution of antibiotic-resistant bacterial (ARB) strains has been faster than the development of new classes of antibiotics.6–8 Typically, conventional antibiotics kill or inhibit microorganisms by interfering with a specific cellular function or metabolic pathway resulting in their cell death.6,9 Common antibiotics targets include inhibition of cell wall synthesis, cell membrane functions, protein or nucleic acid synthesis, folic acid or mycolic acid production, and other metabolic processes.10 Bacteria has devised mechanisms of evading these bactericidal effects shrinking the pipeline of effective antibiotics.5,7 Bacteria develop resistance to antibiotics by modifying the target site for antibiotics activity, modifying the pathway of the target synthesis, reducing the antibiotics concentration in the cell or destroying the antibiotics.11 There is evidence of at least one mechanism of bacterial resistance to all the classes of antibiotics available.7,11With the rise in multidrug-resistant (MDR) bacteria, there is an urgent need for new antibacterial agents with targets that have low possibility of developing resistance through mutation, and that can overcome latest bacterial resistance mechanisms.6,7 It is desired that novel antibacterial agents inhibit bacteria via multiple mechanisms with bactericidal actions that do not involve specific biochemical pathways. In recent years, great attention has been directed to the use of antimicrobial nanoparticles (NPs) as an alternative with novel non-specific low-mutation bacterial targets.5,6,12 Antimicrobial NPs inactivate microorganisms via non-specific pathways with multiple targets involving a combination of cell membrane lysis and reactive oxygen species (ROS) generation to degrade cellular components.5,9,13 Thus, antimicrobial NPs have a high barrier against resistance development. Moreover, the use of nanoscale antimicrobials allows for increased bioavailability by promoting transport through the cell membrane to the target site.5,12,14 Silver nanoparticles (AgNPs) have gained wide use in different antimicrobial research areas because they significantly inhibit several Gram-positive and Gram-negative bacteria and have lower tendency for antibacterial resistance development.9,15,16 Consequently, several studies have been conducted to fully understand the antibacterial mechanisms of AgNPs to promote its antimicrobial applications.9,17,18 These studies have elucidated that the antibacterial action of AgNPs is mainly based on the localized release of silver ions (Ag+) from the surface of AgNPs. The released Ag+ afford bacterial inactivation due to interaction with enzymes and proteins, and high ROS production.9,12,16,18–20 Therefore, AgNPs act as a reservoir of Ag+ that are released by oxidative dissolution in aerobic or other oxidative conditions.21 Hence, the design of novel strategies that increase the oxidation of AgNPs to enhance the release of Ag+ ions can have a major impact on the antimicrobial features of AgNPs.
Photosensitizers (PSs) are molecules that upon the absorption of light generate ROS. PSs have also been used to inactivate bacteria by causing oxidative stress or damage to cellular components when irradiated with visible light, this approach is called photodynamic inactivation (PDI).15,16,20 Recent studies have shown that the PS–AgNP combination have synergistic antimicrobial effect. These studies demonstrated that the synergistic antimicrobial effect of PS–AgNP conjugates in bacteria (non-resistant) is concentration dependent and contributed by the oxidative stress due to enhanced ROS production.19,22–25 However, there is a lack of understanding of the impact of the light-mediated release of Ag+ in these systems, which is also a critical parameter that contributes to the antibacterial synergy in PSs and AgNPs conjugates. Herein, we report an in-depth study to understand the role of PS-mediated ROS generation in Ag+ release and its antimicrobial effect against antibiotic resistant bacteria (ARB).
In this work, we synthesized PS-modified AgNPs using protoporphyrin IX (PpIX) as PS. PpIX has been extensively used as an effective antimicrobial agent either as single molecule or attached to different materials.26 The surface of AgNPs were functionalized with a thiol derivative of PpIX to afford conjugated PpIX-AgNPs (Scheme 1). Moreover, to evaluate the effect of positive surface charge on the antimicrobial properties of these nanomaterials, PpIX-AgNPs were further modified with polyethyleneimine (PEI) to afford PEI-PpIX-AgNPs. The physicochemical and photophysical properties of these NPs were characterized. We studied the release of Ag+ from these materials in the presence or absence of light. The synergistic effect of this approach was successfully evaluated against Gram-positive and Gram-negative bacteria, a methicillin resistant Staphylococcus aureus (MRSA) strain and a MDR Escherichia coli strain, respectively. It is noteworthy to mention that over 50% of all clinically reported infections from seven organisms are caused by S. aureus and E. coli.8,27 Finally, the biocompatibility of the three nanoparticles against mammalian cells was examined at the observed bacterial inhibitory concentration. We envision that the results obtained in this work will help to optimize the performance of light activated AgNP platforms. Moreover, it could have a major impact to reduce some of the recent safety concerns associated with colloidal AgNPs such as argyria and other adverse effect leading to human death.16
Experimental protocols
Synthesis and characterization of nanoparticles: AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min to remove excess of tannic acid and further washed with water three times under the same centrifugation conditions. As-synthesized AgNPs are stored in nanopure water.
000 rpm for 15 min to remove excess of tannic acid and further washed with water three times under the same centrifugation conditions. As-synthesized AgNPs are stored in nanopure water.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 mass ratio) (Scheme S2, ESI†). A solution of cysPpIX in DMF (1 mg mL−1) was prepared. AgNPs (1 mg) were centrifuged down at 12
0.5 mass ratio) (Scheme S2, ESI†). A solution of cysPpIX in DMF (1 mg mL−1) was prepared. AgNPs (1 mg) were centrifuged down at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min to remove the water. Following this, 1 mL of DMF was added and sonication was used to disperse the AgNPs. This solution was transferred to a 20 mL scintillation glass vial and 500 μL of previously prepared cysPpIX solution in DMF was added. An additional 3.5 mL of DMF was added and the reaction mixture was vigorously stirred for 48 h at room temperature. After the reaction, the unreacted cysPpIX was collected via centrifugation at 12
000 rpm for 15 min to remove the water. Following this, 1 mL of DMF was added and sonication was used to disperse the AgNPs. This solution was transferred to a 20 mL scintillation glass vial and 500 μL of previously prepared cysPpIX solution in DMF was added. An additional 3.5 mL of DMF was added and the reaction mixture was vigorously stirred for 48 h at room temperature. After the reaction, the unreacted cysPpIX was collected via centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. Nanoparticles were washed three times with DMF and separated under the same centrifugation conditions. The supernatant collected was analyzed using UV-Vis spectrophotometer to determine the amount of PpIX conjugated on AgNPs surface. A typical batch fabricated under the above-mentioned conditions produced 1.2 ± 0.1 mg of PpIX-AgNPs. The synthesized PpIX-AgNPs were stored in DMF under dark conditions.
000 rpm for 15 min. Nanoparticles were washed three times with DMF and separated under the same centrifugation conditions. The supernatant collected was analyzed using UV-Vis spectrophotometer to determine the amount of PpIX conjugated on AgNPs surface. A typical batch fabricated under the above-mentioned conditions produced 1.2 ± 0.1 mg of PpIX-AgNPs. The synthesized PpIX-AgNPs were stored in DMF under dark conditions.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mass ratio. Briefly, 1 mg of PpIX-AgNPs were centrifuged down and dispersed in 1 mL of nanopure water using sonication. Separately, 10 mg of PEI was weighed in a scintillation vial and dispersed in 5 mL of nanopure water to get a concentration of 2 mg mL−1. Finally, 5 mL of the PEI solution was mixed with 1 mg mL−1 of PpIX-AgNPs and the solution was vigorously stirred at room temperature for 24 h. Following this, the material was washed three times with nanopure water and stored at room temperature under dark condition. PEI coating was confirmed via zeta potential measurement.
10 mass ratio. Briefly, 1 mg of PpIX-AgNPs were centrifuged down and dispersed in 1 mL of nanopure water using sonication. Separately, 10 mg of PEI was weighed in a scintillation vial and dispersed in 5 mL of nanopure water to get a concentration of 2 mg mL−1. Finally, 5 mL of the PEI solution was mixed with 1 mg mL−1 of PpIX-AgNPs and the solution was vigorously stirred at room temperature for 24 h. Following this, the material was washed three times with nanopure water and stored at room temperature under dark condition. PEI coating was confirmed via zeta potential measurement.
Light-mediated release profile of Ag+ ions from nanoparticles
The Ag+ quantification was evaluated using inductively coupled plasma – optical emission spectroscopy (ICP-OES) (Optima 3000, Agilent). Samples obtained from digested and release experiments were introduced into plasma by peristaltic pump and discharged as an aerosol suspended in argon gas. The data acquisition was performed in triplicates with the torch assembly in the axial mode. The default acquisition parameters used are RF = 1.2 kW; auxiliary gas flow = 1 L min−1, nebulizer gas flow = 0.7 L min−1; plasma flow = 12 L min−1, pump speed = 12 rpm; stabilization time = 15 s; sample uptake time = 25 s; Rinse time = 30 s and Ag analytical line = 328.068 nm.To determine the total amount of Ag+ in all samples, aliquots of the nanoparticles were digested by performing a “cold digestion” followed by “hot digestion”.28 The digestion protocol was optimized and modified in terms of time and volume. Cold digestion included mixing 50 μg of NPs with 2.5 mL of concentrated HNO3 and incubating at room temperature for 30 min. Later, this mixture was heated under 150 °C using an oil bath for 4–6 h to allow excess HNO3 to evaporate (hot digestion). The remaining volume was measured using a graduated cylinder and diluted using 2% HNO3. The samples were filtered through a 0.22 μm polyethersulfone (PES) membrane before ICP-OES analysis. A calibration curve was obtained using an Ag silver standard diluted in 2% HNO3 to prepare standard concentration of 10, 25, 50, 100 μg L−1 (Fig. S6a, ESI†). A new set of calibration curves were generated for each run. The total amount of Ag+ digested (in μg) was calculated using the calibration curve and expressed in terms of Ag+ amount per μg of NP (%) (Table S1, ESI†). The concentration of AgNPs and PpIX-AgNPs (1.5 μg mL−1) used for release and bacterial experiments was also digested using the same protocol to calculate the total Ag+ digested (Fig. S6b, ESI†).
The release kinetics of Ag+ was tested in Dulbecco's phosphate buffer solution (DPBS, 1×) and nanopure water. Stock solutions of AgNPs (water), PpIX-AgNPs (DMF) and PEI-PpIX-AgNPs (water) equivalent to 150 μg mL−1 of AgNPs were prepared for the analysis. The stock samples were diluted 100 times with the relevant media (PBS or water) to obtain 3 mL of sample. A Petri dish (60 × 15 mm) containing 3 mL of each sample was prepared using the stock solution such that the final concentration is 1.5 μg mL−1 in terms of AgNPs. Each Petri dish containing the samples was irradiated with a white light source (400–700 nm; 56 ± 2 mW cm−2) for 20 min. Before irradiation, 200 μL sample was withdrawn indicating the time point as 0 min. Then, 200 μL was collected at specific time points from each sample at 5, 10, 20, 30 min, 1, 2, 4, 6, 12, 24 h. The collected samples were immediately centrifuged for 10 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and the supernatant was collected for ICP-OES analysis. The same experiments were repeated in the dark as control by covering the Petri dishes with aluminum foil to avoid unwanted light exposure. The Ag+ concentration obtained for each time point was normalized based on the lowest initial amount of Ag+, which is associated with PEI-PpIX-AgNPs. The kinetic release rates were determined by performing linear fit on the Ag+ release kinetic plots using OriginPro 2021 (Academic version). The R2 and slope values corresponding to burst and slow release were recorded (Table S4, ESI†).
000 rpm and the supernatant was collected for ICP-OES analysis. The same experiments were repeated in the dark as control by covering the Petri dishes with aluminum foil to avoid unwanted light exposure. The Ag+ concentration obtained for each time point was normalized based on the lowest initial amount of Ag+, which is associated with PEI-PpIX-AgNPs. The kinetic release rates were determined by performing linear fit on the Ag+ release kinetic plots using OriginPro 2021 (Academic version). The R2 and slope values corresponding to burst and slow release were recorded (Table S4, ESI†).
Bacterial inactivation experiment
The antimicrobial activities of AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs were examined against a MRSA strain (BAA 44) purchased from ATCC and a wild-type MDR E. coli strain (accession number PRJNA806466). The MDR E. coli was isolated from Class B biosolids amended soil.29 Details of the MDR E. coli sequence protocol and data analysis are provided in the ESI.† It was confirmed to be resistant to tetracycline, gentamicin, ampicillin, sulfamethoxazole-trimethoprim and ciprofloxacin using the CLSI M100 Performance Standards.30 Stock solutions of AgNPs and PpIX-AgNPs were prepared in DMF, while PEI-PpIX-AgNPs was prepared in nanopure water (18 MΩ cm). A single bacterial colony of each strain was aseptically picked from the top of a Luria Bertani (LB) agar plate using a sterile loop and inoculated into sterile LB broth media. Bacterial cells were grown overnight (∼18 h) at 37 °C under continuous gentle shaking at 200 rpm. Overnight cells were harvested the following day by centrifugation at 7000 rpm for 5 min, washed twice with PBS and resuspended in 1× DPBS to achieve an absorbance of 0.5 McFarland turbidity standard (∼1.5 × 108 CFU mL−1). Before light irradiation, bacteria cells in 2,970 μL of 1× DPBS were incubated at room temperature in the dark for 30 min with 30 μL of each sample to achieve a working concentration of 1.5 μg mL−1 for AgNPs, and 1 μM cysPpIX-1.5 μg mL−1 AgNPs for PpIX-AgNPs. The resulting DMF concentration in the final reaction volume was ≤1%. Thereafter, cells were irradiated for 20 min without stirring in a Petri dish (60 × 15 mm) with a white light source (400–700 nm; 56 ± 2 mW cm−2). The surviving cells were counted after 0, 4, and 24 h post irradiation in triplicate using the drop plate colony count method.31 At specific time point, 100 μL of the sample was withdrawn, serially diluted and 20 μL of each dilution was spotted on LB agar. The bacterial log inactivation was estimated using eqn (1). In all experimental groups, negative control and dark control samples were counted. A physical mixture of AgNO3 and cysPpIX was used as a positive control. The concentration of AgNO3 used corresponds to the amount of maximum Ag+ (μg L−1) released from the conjugate samples. | (1) |
Cytotoxicity in mammalian cells
AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs were tested in HeLa cells to assess their cytotoxicity in mammalian cells. HeLa cells obtained from ATCC were cultured in RPMI media supplemented with 10% FBS and 1% pen-strep at 37 °C with 5% CO2 exposure. The in vitro cytotoxicity was carried out using the CellTiter 96 Aqueous Assay (MTS assay). HeLa cells were grown in 96-well plate at 1000 cells per well for 24 h before inoculation. Post cell adherence, media was discarded and cell media containing the nanoparticles (1.5 μg mL−1) were inoculated. Post 30 min incubation, cells inoculated with nanoparticles were irradiated with Biotable power source with RGB LED array for 20 min (450–475 nm; 34.5 mW cm−2) and incubated for 24 h at 37 °C with 5% CO2 exposure. After incubation, the media was removed, and the cells were washed with PBS. To each well, 100 μL of media was added followed by the addition of 20 μL of CellTiter 96 Aqueous assay. The plates were incubated for 2.5 h at 37 °C and the absorbance at 490 nm was measured using plate reader Multiskan FC. Cell viability was calculated by subtracting absorbance with that of media and using eqn (2).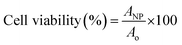 | (2) |
Statistical analysis
Graphs and statistical analyses were performed using OriginPro 2021 (Academic Version). Statistical significance between disinfection trials was assessed by one-way analysis of variance (ANOVA). All the statistical analyses were performed with α = 0.05 and reported as stars assigned to the p values: ***p ≤ 0.0001, **p ≤ 0.001, *p ≤ 0.05, and ns p > 0.05.Results and discussion
Nanoparticles: synthesis and characterization
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching at 1739 cm−1 indicative of ester group and C
O stretching at 1739 cm−1 indicative of ester group and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching at 1810 cm−1 and 1778 cm−1 associated with NHS molecule confirming the synthesis of sePpIX (Fig. S1a, ESI†). After nucleophilic acyl substitution with cysteamine, a C
O stretching at 1810 cm−1 and 1778 cm−1 associated with NHS molecule confirming the synthesis of sePpIX (Fig. S1a, ESI†). After nucleophilic acyl substitution with cysteamine, a C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching at 1643 cm−1 and N–H stretching at 3312 cm−1 were observed, which correspond to formation of amide bond (Fig. S1a, ESI†). UV-Vis spectra indicate minimal difference for Soret-band at 405 nm between PpIX, sePpIX and cysPpIX (Fig. S1b, ESI†). However, the Q-bands observed at 510, 550, 625 and 650 nm in PpIX and sePpIX underwent a slight red-shift for cysPpIX (Fig. S1b, ESI†). 1H NMR analysis also confirmed the successful synthesis of sePpIX and cysPpIX (Fig. S2, ESI†). The synthesis of sePpIX and cysPpIX was further confirmed based on the MALDI-MS peaks at 758.44 (calculated = 756.30) and 678.90 (calculated = 680.93) respectively (Fig. S3, ESI†).
O stretching at 1643 cm−1 and N–H stretching at 3312 cm−1 were observed, which correspond to formation of amide bond (Fig. S1a, ESI†). UV-Vis spectra indicate minimal difference for Soret-band at 405 nm between PpIX, sePpIX and cysPpIX (Fig. S1b, ESI†). However, the Q-bands observed at 510, 550, 625 and 650 nm in PpIX and sePpIX underwent a slight red-shift for cysPpIX (Fig. S1b, ESI†). 1H NMR analysis also confirmed the successful synthesis of sePpIX and cysPpIX (Fig. S2, ESI†). The synthesis of sePpIX and cysPpIX was further confirmed based on the MALDI-MS peaks at 758.44 (calculated = 756.30) and 678.90 (calculated = 680.93) respectively (Fig. S3, ESI†).
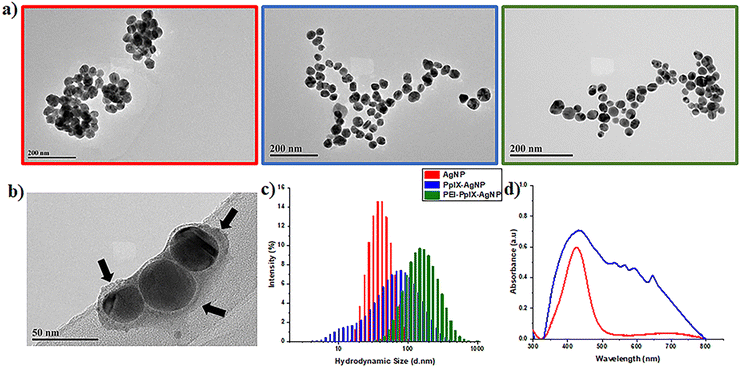
UV-Vis spectroscopy of AgNPs depicts an absorption band with a maximum wavelength at 427 nm (Fig. 1(d)), which is characteristic of the surface plasmon resonance (SPR) associated with metallic nanoparticles.35 UV-Vis spectrum of PpIX-AgNPs clearly indicates the presence of PpIX corresponding to the Soret- and Q-bands. PpIX-AgNPs shows a strong absorption band from 350–500 nm with a maximum at 430 nm, which is the result of the overlap between the SPR and the Soret-band of PpIX. The amount of PpIX loaded on the nanoparticle was determined using a calibration curve of cysPpIX at 500 nm as indicated in the experimental section (Fig. S5, ESI†). The percentage of PpIX loading on AgNPs was found to be around 45 ± 0.57% (n = 6), which corresponds to 240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 620 PpIX molecules per nanoparticle (see ESI† for calculations). This % loading was further confirmed by determining the amount of Ag in the PpIX-AgNPs using ICP-OES (Table S1, ESI†).
620 PpIX molecules per nanoparticle (see ESI† for calculations). This % loading was further confirmed by determining the amount of Ag in the PpIX-AgNPs using ICP-OES (Table S1, ESI†).
The ability of PpIX attached to AgNPs to generate singlet oxygen (1O2) was evaluated by determining the 1O2 quantum yield (ΦΔ). For this experiment, 9,10 dimethylanthracene (DMA) was used as the 1O2 probe and its decay at 380 nm with respect to the irradiation time used to calculate ΦΔ (Table S3, ESI†).36 The lower ΦΔ value for PpIX AgNPs (ΦΔ = 0.01) compared to the parent porphyrin cysPpIX (ΦΔ = 0.38) could be attributed to the fact that the 1O2 released by PpIX is in close proximity to AgNPs; therefore, it is selectively oxidizing the surface of the nanoparticles to release Ag+ ions as demonstrated in the section below. This surface effect will leave a very low amount of 1O2 remaining to react with the DMA probe.
Investigation of the release profile of Ag+ from AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs
Previous reports have described the combination of PSs with AgNPs.19,22–25,37 Most of these studies are limited to demonstrating the increased antimicrobial effect of PS–AgNPs in terms of the enhanced PS photochemical properties. As far as we know, only one study has investigated the oxidative effect of PSs on the release of Ag+ after light irradiation.37 In this work, the increase in Ag+ release from phthalocyanine-AgNPs was analysed only at 1 h post irradiation. A synergistic effect was observed for the elimination of bacteria. A kinetic study detailing the Ag+ release during and post light irradiation is critical to understand their role in antibacterial synergy. Herein, we performed an in-depth study of the impact of light-activated generation of ROS through PpIX on the release kinetics of Ag+. Two different media were investigated; nanopure water and DPBS, and a broad white light (400–700 nm; 56 ± 2 mW cm−2) was used as light source. For these experiments, the Ag+ release kinetics from AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs was evaluated in two distinctive stages; first, the one associated to light irradiation for 20 min, “burst release”; and the second one, due to the long-term dissolution of Ag+ in the two different aqueous media, “steady release.” As seen in Fig. 2(a) and Fig. S7 (ESI†), after light irradiation in water, PpIX-AgNPs showed a burst release within the first 20 minutes (release rate = 8.56 μg L−1 min−1) followed by a constant increase in Ag+ release in the next 24 h (release rate = 0.02 μg L−1 min−1). On the contrary, minimal release of Ag+ was observed in water for PEI-PpIX-AgNPs (Fig. 2(a)). Most likely due to the strong interaction between Ag+ ions and amine groups in the PEI.38 Moreover, a reduction in the oxidative effect of the ROS produced by PpIX on the surface of the AgNPs could be also associated to the presence of PEI.39 In the case of AgNPs, a slow release (release rate = 0.04 μg L−1 min−1) was observed after 720 min. The total Ag+ release achieved for PpIX-AgNPs in water was 22% wt. Control experiment in water without light irradiation show a minimal release of Ag+ from these nanoparticles (Fig. S8a, ESI†). The release of Ag+ from AgNPs in water in the absence of a PS has already been studied.40,41 The presence of oxygen in water affords the oxidation of AgNPs resulting in a slow release of Ag+.41–43 PpIX-AgNPs platform in the presence of light increases the release of Ag+ by 25-fold. The burst release is most likely directly associated with the oxidation of the AgNPs surface in presence of 1O2, which is the main ROS produced by PpIX.44 The lifetime of 1O2 in water is 3–5 μs with diffusion coefficient 2 × 10−5 cm2 s−1, which limits their activity to a distance of 125 nm.45,46 However, in biologicals systems, this distance further reduces to less than half of the one mentioned above.46,47 Therefore, the proximity of the generation of 1O2 to the surface of AgNPs accounts for the fast Ag+ release kinetics in the first 20 min during the time of light irradiation (Scheme S3, ESI†).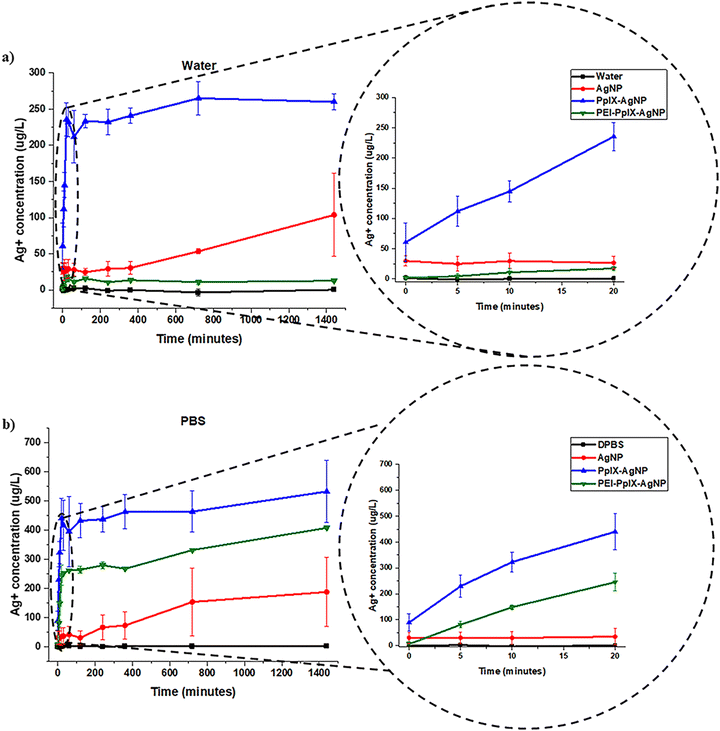 | ||
| Fig. 2 Ag+ release kinetics for AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs measured in (a) water or (b) DPBS. A zoom-in of the first 20 minutes are localized on the right. | ||
To confirm the degradation of AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs associated with the release of Ag+, TEM images of aliquots at two different times post-irradiation, 20 min and 24 h, were taken (Fig. S9, ESI†). Moreover, the sizes of nanoparticles for those samples were measured using image J. A slight increase in PpIX-AgNPs with smaller diameters were measured after 20 min. However, after 24 h almost 90% of the nanoparticles have a diameter below 10 nm (Fig. S9b, ESI†). The original size of the nanoparticles is 42.2 nm; therefore, a size of 10 nm means that they have been degraded by more than 75%. This clearly demonstrate the degradation of PpIX-AgNPs after light irradiation, which is also directly related to the release of Ag+ as shown above. In the case of PEI-PpIX-AgNPs, a slower reduction in the size of the nanoparticles was determined at 20 min, but with less than 10% of the particles measuring 10 nm or below after 24 h (Fig. S9c, ESI†). This confirms our hypothesis that the presence of PEI prevents the oxidation and release of Ag+. AgNPs also showed degradation in water as indicated by the presence of 60% of nanoparticles with diameters at 10 nm or below after 24 h (Fig. S9a, ESI†).
To take in account the effect of the media, the release of Ag+ from AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs were also examined in DPBS. DPBS is a buffer solution composed of potassium chloride (KCl), potassium phosphate monobasic (KH2PO4), sodium phosphate dibasic (Na2HPO4–7H2O) and sodium chloride (NaCl) that is widely used to simulate physiological conditions. A similar trend was observed for the release profile in DPBS associated with PpIX-AgNPs after light irradiation with a burst release in the first 20 min (release rate = 19.18 μg L−1 min−1) followed by a steady release (release rate = 0.08 μg L−1 min−1) during the rest of the experiment (Fig. 2(b)). In the case of PEI-PpIX-AgNPs, a similar two stages release behavior was observed; first the burst release (release rate = 13.53 μg L−1 min−1) followed by a steady phase (release rate = 0.11 μg L−1 min−1) (Fig. 2(b) and Fig. S7, ESI†). Interestingly, in DPBS the interaction between Ag+ and amine groups seems that is not strong enough to completely stop the release of the Ag+ ions. It has been reported that the different ions in DPBS (e.g., KCl, NaCl, KH2PO4, and Na2HPO4) can have a screening effect on the interaction between Ag+ and the amines.42,48,49 Finally, a constant release of Ag+ from AgNPs is observed (release rate = 0.12 μg L−1 min−1) during the whole experiment (Fig. 2(b)). The total amount of Ag+ released was 25, 60, and 50% wt for AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs, respectively (Fig. S10, ESI†). These amounts are at least 2.5 times higher than the one obtained in water, as an indication that different media has a significant impact in the release Ag+.28,50 It has been reported that the presence of NaCl in the aqueous media could have an impact on the rate of dissolution of Ag+.51 Therefore, we hypothesize that the presence of Cl− ions can account for some of the Ag+ release differences between water and DPBS observed in this work (Scheme S3, ESI†).52 Control experiments for PpIX-AgNPs and PEI-PpIX-AgNPs in DPBS without light irradiation only show a steady release of Ag+ ions, similar to the one observed for AgNPs (Fig. S8b, ESI†).
Similar to the Ag+ release experiments in water, we confirmed the degradation of AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs using TEM. Images of aliquots at two different times pot-irradiation, 20 min and 24 h, were taken (Fig. S11, ESI†). The influence of the medium (DPBS) is also corroborated in the degradation of the nanoparticles. PpIX-AgNPs at 20 min post-irradiation already showed 35% of nanoparticles with diameters below 10 nm and more than 70% at 24 h (Fig. S11b, ESI†). PEI-PpIX-NPs are still the material that present less degradation with less than 45% of the nanoparticles having diameters below 10 nm after 24 h; however, at 20 min already 20% of nanoparticles were observed. In the case of AgNPs, there is a fast degradation with almost 50% and 65% of nanoparticles with diameters below 10 nm at 20 min and 24 h, respectively (Fig. S11a, ESI†).
Bacteria inactivation
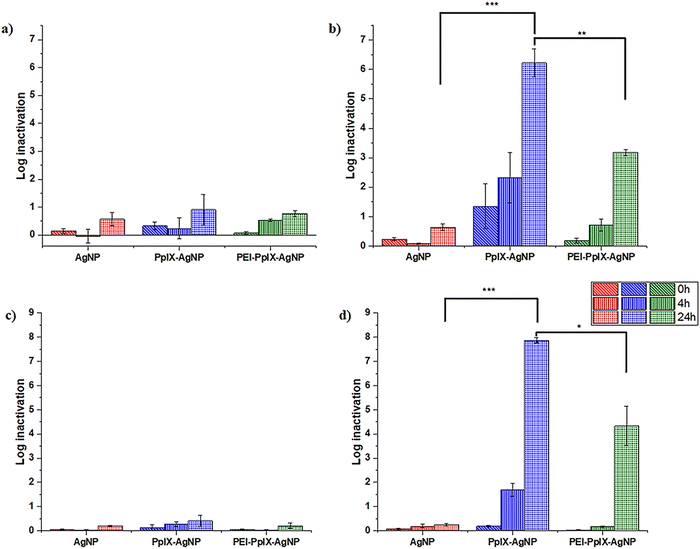
MRSA and MDR E. coli are one of the most common causes of human and animal antibiotic resistant infections.8,20,53 These pathogens are considered as serious threats by the Centers for Disease Control and Prevention (CDC) and are part of the “nine bacteria of international concern”.1,3 Therefore, MDR E. coli and MRSA are ranked in the Priority 1 and 2 of the World Priority List of ARB for R&D of new antibiotics by the World Health Organization (WHO).1 The antimicrobial properties of AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs were tested against MRSA (ATCC BAA 44 strain) and a wild type MDR E. coli. The log bacterial inactivation achieved using these NPs under dark and light conditions are presented in Fig. 3. For all cases, the concentration of AgNPs in all samples was kept constant at 1.5 μg mL−1 while cysPpIX concentration was 1 μM in PpIX-AgNPs and PEI-PpIX-AgNPs. These concentrations were chosen based on the results from preliminary concentration optimization experiments. All microbes were incubated with NPs in the dark for 30 min prior to visible light irradiation (400–700 nm; 56 ± 2 mW cm−2) for 20 min. Subsequently, samples were collected post-irradiation at 0 (i.e., immediately after irradiation), 4 and 24 h post-irradiation. A similar protocol was followed for the dark controls. The results indicate that 1.5 μg mL−1 of AgNPs achieved <1 log inactivation of MRSA both under light and dark conditions even after 24 h of exposure (Fig. 3(a) and (b)). For PpIX-AgNPs, under dark conditions at times 0, 4 and 24 h, the log inactivation was similar to that obtained for AgNPs. This low inactivation by AgNPs at 1.5 μg mL−1 was expected because typical minimum inhibitory concentrations (MIC) reported for MRSA using AgNPs of similar sizes used in this study are about 3–10 folds higher.25,54 However, MRSA inactivation by PpIX-AgNPs increased to ∼1.5 log following immediate irradiation for 20 min (i.e., 0 h) and continuously increased to 2.3 log and 6.2 log after 4 and 24 h contact time, respectively. PpIX-AgNPs achieved the highest inactivation of MRSA amongst the three NPs examined regardless of the contact time (p ≤ 0.0001) (Fig. 3(b)). Control experiments using only the parent porphyrin (cysPpIX) in the presence of light show an inactivation of ∼2.5 log following immediate irradiation (Fig. S10b, ESI†). Nevertheless, this effect remained constant during the rest of the experiment. Most likely due to the short half-life (<10−6 s) of the ROS species generated, which are the antibacterial agents.55The inactivation of the Gram-negative MDR E. coli showed a similar trend as the inactivation of the Gram-positive MRSA. Under dark conditions, the log inactivation was <0.5 log for all three NPs for time 0, 4 and 24 h (Fig. 3(c)). Similarly, PpIX-AgNPs achieved the highest inactivation of ∼8 logs after 24 h under light conditions (p ≤ 0.0001), which is the limit of quantification since the starting bacteria concentration ∼108 CFU per mL (Fig. 3(d)). Interestingly, control experiments using only cysPpIX in the presence of light showed no inactivation of MDR E. coli (Fig. S12d, ESI†) as an indication that for this microbe PDI is not effective. Previous studies have shown that some bacteria show resistance against PDI.31,49,56 The increase in bacterial inactivation of MRSA and MDR E. coli by PpIX-AgNPs following light irradiation is consistent with the patterns recorded in literature.19,22–25,37 Also, other studies have reported complete elimination of MRSA and E. coli using AgNPs only; however, MIC are at least 5 μg mL−1 depending on the physiochemical properties of the examined AgNPs.25,54,57 Whereas, in this study, only 1.5 μg mL−1 of AgNPs in PpIX-AgNPs resulted in complete inactivation of MRSA and MDR E. coli. These results confirm the synergistic effect between PpIX and AgNPs to effectively eliminate MRSA and MDR E. coli.
To deconvolute the impact on the inactivation of MRSA and MDR E. coli by using the nanoparticulate platform PpIX-AgNPs, we evaluated the physical mixture of cysPpIX and Ag+ (AgNO3) at the same concentrations used for the experiments with the NPs (Fig. S12, ESI†). For MRSA, under dark conditions the log inactivation was <0.5 log, <1.0 log and 4.5 log for times 0, 4 and 24 h. In the presence of light irradiation, the inactivation values for the same times are 4.0 log, 4.0 log and 6.0 log (Fig. S12b, ESI†). These data show: first, that Ag+ ions by themselves require time to produce inactivation on MRSA; and second, that PDI has an additive effect at short times (0 and 4 h) of the experiment. In the case of MDR E. coli in the absence of light, the log inactivation was zero, <1.0 log and 6.5 log for times 0, 4 and 24 h, which are the same inactivation values in the presence of light irradiation (Fig. S12d, ESI†). These results confirmed the lack of PDI effect on the elimination of MDR E. coli. PpIX-AgNPs have a similar behaviour against MRSA as the physical mixture (Fig. S9b, ESI†); nevertheless, it performs better in the case of MDR E. coli (Fig. S12d, ESI†).
The incorporation of positive charges in PSs or NPs has been an important strategy to enhance their antimicrobial properties.31,58 The rationale is that an electrostatic interaction between the positively charged agents and the negatively charged surface of bacteria will bring them in close proximity, resulting in an improved antimicrobial effect.31,49 In this work we functionalized PpIX-AgNPs with PEI polymer (MW = 10 kDa) (PEI-PpIX-AgNPs), which contains amine groups that are protonated under physiological conditions. Therefore, it is expected that this strategy would enhance the antimicrobial effect of PpIX-AgNPs.59 For MRSA, under dark conditions, the log inactivation was <1.0 log for PEI-PpIX-AgNPs at times 0, 4 and 24 h (Fig. 3(a)). In the presence of light, only at 24 h a significant increase of inactivation was observed (3.2 log), but not comparable with the value for PpIX-AgNPs (p ≤ 0.001) (Fig. 3(b)). PEI-PpIX-AgNPs depicted a similar performance in the case of MDR E. coli with minimal inactivation effect in the absence of light (<0.5 log) (Fig. 3(c)) and lower increase after light irradiation (4.0 log at 24 h) as compared with PpIX-AgNPs (p ≤ 0.05) (Fig. 3(d)). Control experiments using only PEI polymer depicted ∼0.5 log and ∼1.0 log inactivation of MRSA and MDR E. coli, respectively, under light or dark conditions (Fig. S12, ESI†). The experimental results reveal that by modifying the surface of PpIX-AgNPs with PEI did not enhance their antimicrobial effect; on the contrary, a reduction in the inactivation was observed. This lower antimicrobial activity of PEI-PpIX-AgNPs compared to PpIX-AgNPs can be explained from the lower concentration of Ag+ released by PEI-PpIX-AgNPs as illustrated in the previous section (Fig. 2(b)). This unexpected outcome suggests that different considerations other than functionalizing the surface of nanoparticles with groups that can render positive charge to the surface such as amines needs to be taken in account in the case of AgNP-based platforms that depends on the release of Ag+ to inactivate ARB.
Overall, the log inactivation of MRSA and MDR E. coli was in the order PpIX-AgNPs > PEI-PpIX-AgNPs > AgNPs under light conditions irrespective of the contact time (Fig. 3(b) and (d)). The order of bacterial log inactivation correlates with the trend of Ag+ release profile (PpIX-AgNPs > PEI-PpIX-AgNPs > AgNPs) in DPBS (Fig. 2(a) and (b)) as a clear indication that the antimicrobial activity of this platform is driven by photo-active release of Ag+.
Cytotoxicity in mammalian cells
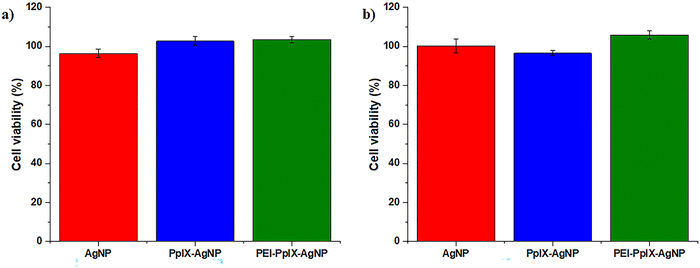
Antimicrobial agents should effectively eliminate bacteria without major toxicity toward mammalian cells. This feature is critical in biomedical applications such as wound healing.23 In this work, an in vitro assay was carried out to evaluate the cytotoxicity of AgNPs, PpIX-AgNPs and PEI-PpIX-AgNPs at bacterial inhibitory concentration in HeLa cells. After 30 min incubation with the NPs, cells were irradiated for 20 min, followed by an additional 24 h of incubation at 37 °C. The viability of HeLa cells after treatment with any of the NPs did not show any phototoxic effect at the concentrations used to eliminate MRSA and MDR E. coli (Fig. 4). The same behaviour was observed under dark conditions. Control experiments using cysPpIX, AgNO3 and the physical mixture of both show a major cytotoxic effect for both cysPpIX and the physical mixture in the presence of light (Fig. S13, ESI†). This phototoxic effect is most likely due to the photodynamic therapy (PDT) properties of cysPpIX.60 Interestingly, neither PpIX-AgNPs nor PEI-PpIX-AgNPs show PDT effect against HeLa cells, most likely because most of the 1O2 generated by the PS reacts with the surface of AgNPs. Similar “quenching” effects have been observed in other NPs carrying PSs.23,24,37,61 Control experiments also demonstrate that the concentration of Ag+ used to eliminate bacteria is not toxic to HeLa cells. It is reported that the IC50 of Ag+ is usually higher for mammalian cells than for bacteria.23–25,37 Therefore, the PpIX-AgNP platform not only offers an effective approach to eliminate MRSA and MDR E. coli, but also shows improved biocompatibility properties against mammalian cell.Conclusions
In this work, we synthesized, characterized, and applied a light-activated PpIX-AgNP platform for the elimination of ARB. PpIX-AgNPs demonstrated broad-spectrum antibacterial action resulting in >7 log inactivation of MRSA and MDR E. coli. The bacterial inactivation achieved was independent of the Gram-stain classification of the examined bacterial strains. The ROS generated, mainly 1O2, due to the irradiation of PpIX increased the release of Ag+ from the surface of the PpIX-AgNPs. We also investigated the influence of the media composition on the release kinetics of Ag+ and degradation of AgNPs. The ionic composition of DPBS increased Ag+ release and degradation with light activated PpIX-AgNPs than in nanopure water. The cationic surface charge associated with PEI-PpIX-AgNPs did not improve bacterial inactivation since it did not allow similar Ag+ release in comparison with PpIX-AgNPs. The amount of released Ag+ drives the inactivation of MDR E. coli and MRSA over electrostatic interaction from PpIX-AgNPs. In addition, PpIX-AgNPs overcame the limitations of PpIX molecules in the PDI of MDR E. coli. All NPs showed negligible cytotoxicity to HeLa cells at the bacterial inhibitory concentration after 24 h exposure. The successful formulation of light-activated PpIX-AgNPs with increased potential for Ag+ release would reduce the MIC of AgNPs for therapeutic applications.Author contributions
Investigation: A. J. S. and V. G., conceptualization: A. J. S., V. G., M. M. and J. V.-E., supervision: M. M. and J. V.-E., funding acquisition: M. M. and J. V.-E., Writing-original draft: A. J. S. and V. G., writing-review & editing: A. J. S., V. G., M. M. and J. V.-E.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was funded by the National Institute of General Medical Sciences (1R16GM145434) and the University of North Carolina Charlotte Faculty Research Grant. We appreciate Dr Navab-Daneshmand from Oregon State University for kindly donating the MDR E. coli isolate. The authors are thankful to Dr Kevin Lambirth, Dr Cynthia Gibas, and Dr Jessica Schlueter in the Bioinformatics Department at UNCC for helping with the sequencing of the MDR E. coli strain. The authors would like to thank Dr Xiuli Lin and Dr Rui He for their assistance with the ICP-OES instrument.References
- WHO. World Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. 2017. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf (accessed 2018 April 3).
- WHO. Antimicrobial resistance global report on surveillance. 2014. https://apps.who.int/iris/bitstream/handle/10665/112642/9789241564748_eng.pdf;sequence=1 (accessed 2021 17 March).
- CDC. Antibiotic Resistance Threats in the United States. U.S. Department of Health and Human Services, 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed 2021 17 March).
- T. U. Berendonk, C. M. Manaia, C. Merlin, D. Fatta-Kassinos, E. Cytryn, F. Walsh, H. Bürgmann, H. Sørum, M. Norström and M. N. Pons, et al., Tackling antibiotic resistance: The environmental framework, Nat. Rev. Microbiol., 2015, 13(5), 310–317, DOI:10.1038/nrmicro3439.
- M. M. Mamun, A. J. Sorinolu, M. Munir and E. P. Vejerano, Nanoantibiotics: Functions and properties at the nanoscale to combat antibiotic resistance, Front. Chem., 2021, 9, 687660, DOI:10.3389/fchem.2021.687660.
- J. A. Jackman, B. K. Yoon, D. Li and N. J. Cho, Nanotechnology formulations for antibacterial free fatty acids and monoglycerides, Molecules, 2016, 21(3), 305, DOI:10.3390/molecules21030305.
- K. M. O’Connell, J. T. Hodgkinson, H. F. Sore, M. Welch, G. P. Salmond and D. R. J. A. C. I. E. Spring, Combating multidrug-resistant bacteria: Current strategies for the discovery of novel antibacterials, Angew. Chem., Int. Ed., 2013, 52(41), 10706–10733 CrossRef PubMed.
- S.-J. Richards, K. Isufi, L. E. Wilkins, J. Lipecki, E. Fullam and M. I. J. B. Gibson, Multivalent antimicrobial polymer nanoparticles target mycobacteria and Gram-negative bacteria by distinct mechanisms, Biomacromolecules, 2018, 19(1), 256–264 CrossRef CAS PubMed.
- D. M. Aruguete, B. Kim, M. F. Hochella Jr., Y. Ma, Y. Cheng, A. Hoegh, J. Liu and A. Pruden, Antimicrobial nanotechnology: Its potential for the effective management of microbial drug resistance and implications for research needs in microbial nanotoxicology, Environ. Sci.: Processes Impacts, 2013, 15(1), 93–102, 10.1039/c2em30692a.
- R. Anderson; P. W. Groundwater; A. Todd and A. Worsley, Antibacterial agents: chemistry, mode of action, mechanisms of resistance and clinical applications, John Wiley & Sons, 2012 Search PubMed.
- S. B. Levy and B. Marshall, Antibacterial resistance worldwide: Causes, challenges and responses, Nat. Med., 2004, 10(12 Suppl), S122–S129, DOI:10.1038/nm1145.
- S. Yougbaré, C. Mutalik, G. Okoro, I. H. Lin, D. I. Krisnawati, A. Jazidie, M. Nuh, C.-C. Chang and T.-R. Kuo, Emerging trends in nanomaterials for antibacterial applications, Int. J. Nanomed., 2021, 16, 5831–5867, DOI:10.2147/ijn.s328767.
- S. Yougbaré, H.-L. Chou, C.-H. Yang, D. I. Krisnawati, A. Jazidie, M. Nuh and T.-R. Kuo, Facet-dependent gold nanocrystals for effective photothermal killing of bacteria, J. Hazard. Mater., 2021, 407, 124617, DOI:10.1016/j.jhazmat.2020.124617.
- N. Abed, F. Saïd-Hassane, F. Zouhiri, J. Mougin, V. Nicolas, D. Desmaële, R. Gref and P. Couvreur, An efficient system for intracellular delivery of beta-lactam antibiotics to overcome bacterial resistance, Sci. Rep., 2015, 5(1), 13500, DOI:10.1038/srep13500; W. Gao, S. Thamphiwatana, P. Angsantikul and L. Zhang, Nanoparticle approaches against bacterial infections, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2014, 6(6), 532–547 Search PubMed.
- A. A. Abdellatif, H. N. Alturki and H. M. J. S. Tawfeek, Different cellulosic polymers for synthesizing silver nanoparticles with antioxidant and antibacterial activities, Sci. Rep., 2021, 11(1), 1–18 CrossRef PubMed.
- F. Cao, E. Ju, Y. Zhang, Z. Wang, C. Liu, W. Li, Y. Huang, K. Dong, J. Ren and X. Qu, An efficient and benign antimicrobial depot based on silver-infused MoS2, ACS Nano, 2017, 11(5), 4651–4659 CrossRef CAS PubMed.
- K. Mijnendonckx, N. Leys, J. Mahillon, S. Silver and R. J. B. Van Houdt, Antimicrobial silver: Uses, toxicity and potential for resistance, Biometals, 2013, 26(4), 609–621 CrossRef CAS PubMed.
- X. Yan, B. He, L. Liu, G. Qu, J. Shi, L. Hu and G. J. M. Jiang, Antibacterial mechanism of silver nanoparticles in Pseudomonas aeruginosa: Proteomics approach, Metallomics, 2018, 10(4), 557–564 CrossRef CAS PubMed.
- R. Elashnikov, M. Radocha, I. Panov, S. Rimpelova, P. Ulbrich, A. Michalcova, V. Svorcik and O. J. M. S. Lyutakov, Porphyrin-silver nanoparticles hybrids: Synthesis, characterization and antibacterial activity, Mater. Sci. Eng., C, 2019, 102, 192–199 CrossRef CAS PubMed.
- R. Salomoni, P. Léo, A. Montemor, B. Rinaldi and M. J. N. Rodrigues, Antibacterial effect of silver nanoparticles in Pseudomonas aeruginosa, Nanotechnol., Sci. Appl., 2017, 10, 115 CrossRef CAS PubMed.
- K. Siriwardana, N. Suwandaratne, G. S. Perera, W. E. Collier, F. Perez and D. J. Zhang, Contradictory dual effects: Organothiols can induce both silver nanoparticle disintegration and formation under ambient conditions, J. Phys. Chem. C, 2015, 119(36), 20975–20984 CrossRef CAS; C. M. Ho, S. K. W. Yau, C. N. Lok, M. H. So and C. M. Che, Oxidative dissolution of silver nanoparticles by biologically relevant oxidants: A kinetic and mechanistic study, Chem. – Asian. J., 2010, 5(2), 285–293 CrossRef PubMed.
- S. M. Shabangu, B. Babu, R. C. Soy, J. Oyim, E. Amuhaya and T. Nyokong, Susceptibility of Staphylococcus aureus to porphyrin-silver nanoparticle mediated photodynamic antimicrobial chemotherapy, J. Lumin., 2020, 222, 117158 CrossRef CAS.
- X. Xie, C. Mao, X. Liu, Y. Zhang, Z. Cui, X. Yang, K. W. Yeung, H. Pan, P. K. Chu and S. J. A. Wu, et al., Synergistic bacteria killing through photodynamic and physical actions of graphene oxide/Ag/collagen coating, ACS Appl. Mater. Interfaces, 2017, 9(31), 26417–26428 CrossRef CAS PubMed.
- M. Ghasemi, K. Khorsandi and Z. Kianmehr, Photodynamic inactivation with curcumin and silver nanoparticles hinders Pseudomonas aeruginosa planktonic and biofilm formation: Evaluation of glutathione peroxidase activity and ROS production, World J. Microbiol. Biotechnol., 2021, 37(9), 1–10 CrossRef PubMed.
- Z. Malá, L. Žárská, R. Bajgar, K. Bogdanová, M. Kolář, A. Panáček, S. Binder and H. J. P. Kolářová, The application of antimicrobial photodynamic inactivation on methicillin-resistant S. aureus and ESBL-producing K. pneumoniae using porphyrin photosensitizer in combination with silver nanoparticles, Photodiagn. Photodyn. Ther., 2021, 33, 102140 CrossRef PubMed.
- A. Almeida, A. Cunha, M. Faustino, A. Tomé and M. J. P. Neves, Porphyrins as antimicrobial photosensitizing agents, in Photodynamic Inactivation of Microbial Pathogens: Medical and Environmental Applications, ed. M. R. Hamlin and G. Jori, 2011, pp. 83–160 CrossRef CAS PubMed; A. N. Vzorov, D. W. Dixon, J. S. Trommel, L. G. Marzilli and R. W. Compans, Inactivation of human immunodeficiency virus type 1 by porphyrins, Antimicrob. Agents Chemother., 2002, 46(12), 3917–3925 CrossRef CAS PubMed.
- B. S. Reisner and G. L. Woods, Times to detection of bacteria and yeasts in BACTEC 9240 blood culture bottles, J. Clin. Microbiol., 1999, 37(6), 2024–2026 CrossRef CAS PubMed.
- S. W. Brittle, J. D. Baker, K. M. Dorney, J. M. Dagher, T. Ebrahimian, S. R. Higgins and I. E. Pavel Sizemore, Measuring the silver composition of nanocolloids by inductively coupled plasma–optical emission spectroscopy: A laboratory experiment for chemistry and engineering students, J. Chem. Educ., 2015, 92(6), 1061–1065, DOI:10.1021/ed500707k.
- C. Mays, G. L. Garza, J. Waite-Cusic, T. S. Radniecki and T. Navab-Daneshmand, Impact of biosolids amendment and wastewater effluent irrigation on enteric antibiotic-resistant bacteria–a greenhouse study, Water Res.: X, 2021, 13, 100119 CAS.
- M. P. Weinstein, B. Limbago, J. Patel, A. Mathers, S. Campeau, T. Mazzulli, G. Eliopoulos, R. Patel, M. Galas and S. J. W. Richter, Performance standards for antimicrobial susceptibility testing, CLSI supplement M100, 2018 Search PubMed.
- A. N. Hurst, B. Scarbrough, R. Saleh, J. Hovey, F. Ari, S. Goyal, R. J. Chi, J. M. Troutman and J. L. Vivero-Escoto, Influence of cationic meso-substituted porphyrins on the antimicrobial photodynamic efficacy and cell membrane interaction in Escherichia coli, Int. J. Mol. Sci., 2019, 20(1), 134, DOI:10.3390/ijms20010134.
- R. Juneja, Z. Lyles, H. Vadarevu, K. A. Afonin and J. L. Vivero-Escoto, Multimodal polysilsesquioxane nanoparticles for combinatorial therapy and gene delivery in triple-negative breast cancer, ACS Appl. Mater. Interfaces, 2019, 11(13), 12308–12320, DOI:10.1021/acsami.9b00704; J. L. Vivero-Escoto and D. L. Vega, Stimuli-responsive protoporphyrin IX silica-based nanoparticles for photodynamic therapy in vitro, RSC Adv., 2014, 4(28), 14400–14407, 10.1039/c4ra01135j.
- N. G. Bastus, F. Merkoci, J. Piella and V. Puntes, Synthesis of highly monodisperse citrate-stabilized silver nanoparticles of up to 200 nm: Kinetic control and catalytic properties, Chem. Mater., 2014, 26(9), 2836–2846, DOI:10.1021/cm500316k.
- M. N. Aktara, S. Nayim, N. K. Sahoo and M. Hossain, The synthesis of thiol-stabilized silver nanoparticles and their application towards the nanomolar-level colorimetric recognition of glutathione, New J. Chem., 2019, 43(34), 13480–13490, 10.1039/c9nj01360a; H. S. Toh, C. Batchelor-Mcauley, K. Tschulik and R. G. Compton, Chemical interactions between silver nanoparticles and thiols: A comparison of mercaptohexanol against cysteine, Sci. China: Chem., 2014, 57(9), 1199–1210, DOI:10.1007/s11426-014-5141-8.
- X.-F. Zhang, Z.-G. Liu, W. Shen and S. Gurunathan, Silver nanoparticles: Synthesis, characterization, properties, applications, and therapeutic approaches, Int. J. Mol. Sci., 2016, 17(9), 1534, DOI:10.3390/ijms17091534.
- A. N. Hurst, B. Scarbrough, R. Saleh, J. Hovey, F. Ari, S. Goyal, R. J. Chi, J. M. Troutman and J. L. Vivero-Escoto, Influence of cationic meso-substituted porphyrins on the antimicrobial photodynamic efficacy and cell membrane interaction in Escherichia coli, Int. J. Mol. Sci., 2019, 20(1), 134, DOI:10.3390/ijms20010134; P. Siano, A. Johnston, P. Loman-Cortes, Z. Zhin and J. L. Vivero-Escoto, Evaluation of polyhedral oligomeric silsesquioxane porphyrin derivatives on photodynamic therapy, Molecules, 2020, 25(21), 4965, DOI:10.3390/molecules25214965.
- J. Chen, L. Yang, J. Chen, W. Liu, D. Zhang, P. Xu, T. Dai, L. Shang, Y. Yang and S. J. C. E. J. Tang, Composite of silver nanoparticles and photosensitizer leads to mutual enhancement of antimicrobial efficacy and promotes wound healing, Chem. Eng. J., 2019, 374, 1373–1381 CrossRef CAS.
- W. Yang, C. Wang and V. Arrighi, Effects of amine types on the properties of silver oxalate ink and the associated film morphology, J. Mater. Sci.: Mater. Electron., 2018, 29(24), 20895–20906, DOI:10.1007/s10854-018-0233-4.
- K. Das and A. Roychoudhury, Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants, Front. Environ. Sci, 2014, 2, 53, DOI:10.3389/fenvs.2014.00053.
- C. S. Caires, L. A. Farias, L. E. Gomes, B. P. Pinto, D. A. Gonçalves, L. F. Zagonel, V. A. Nascimento, D. C. Alves, I. Colbeck and C. J. M. S. Whitby, et al., Effective killing of bacteria under blue-light irradiation promoted by green synthesized silver nanoparticles loaded on reduced graphene oxide sheets, Mater. Sci. Eng., C, 2020, 113, 110984 CrossRef CAS PubMed; B. Le Ouay and F. J. Stellacci, Antibacterial activity of silver nanoparticles: A surface science insight, Nano Today, 2015, 10(3), 339–354 CrossRef.
- S. Kittler, C. Greulich, J. Diendorf, M. Koller and M. J. C. Epple, Toxicity of silver nanoparticles increases during storage because of slow dissolution under release of silver ions, Chem. Mater., 2010, 22(16), 4548–4554, DOI:10.1021/cm100023p.
- K. Loza, J. Diendorf, C. Sengstock, L. Ruiz-Gonzalez, J. Gonzalez-Calbet, M. Vallet-Regi, M. Köller and M. Epple, The dissolution and biological effects of silver nanoparticles in biological media, J. Mater. Chem. B, 2014, 2(12), 1634–1643 RSC.
- W. Zhang, Y. Yao, K. Li, Y. Huang and Y. J. E. P. Chen, Influence of dissolved oxygen on aggregation kinetics of citrate-coated silver nanoparticles, Environ. Pollut., 2011, 159(12), 3757–3762 CrossRef CAS PubMed.
- T. J. Dougherty, C. J. Gomer, B. W. Henderson, G. Jori, D. Kessel, M. Korbelik, J. Moan and Q. Peng, Photodynamic therapy, JNCI, J. Natl. Cancer Inst., 1998, 90(12), 889–905, DOI:10.1093/jnci/90.12.889; P. Agostinis, K. Berg, K. A. Cengel, T. H. Foster, A. W. Girotti, S. O. Gollnick, S. M. Hahn, M. R. Hamblin, A. Juzeniene and D. Kessel, et al., Photodynamic therapy of cancer: An update, Ca-Cancer J. Clin., 2011, 61(4), 250–281, DOI:10.3322/caac.20114.
- F. Wilkinson, W. P. Helman and A. B. Ross, Rate constants for the decay and reactions of the lowest electronically excited singlet state of molecular oxygen in solution. An expanded and revised compilation, J. Phys. Chem. Ref. Data, 1995, 24, 663–677, DOI:10.1063/1.555965.
- S. Hatz, L. Poulsen and P. R. Ogilby, Time-resolved singlet oxygen phosphorescence measurements from photosensitized experiments in single cells: Effects of oxygen diffusion and oxygen concentration, Photochem. Photobiol., 2008, 84(5), 1284–1290, DOI:10.1111/j.1751-1097.2008.00359.x.
- M. Klaper, W. Fudickar and T. Linker, Role of distance in singlet oxygen applications: A model system, J. Am. Chem. Soc., 2016, 138(22), 7024–7029, DOI:10.1021/jacs.6b01555.
- B. Le Ouay and F. Stellacci, Antibacterial activity of silver nanoparticles: A surface science insight, Nano Today, 2015, 10(3), 339–354, DOI:10.1016/j.nantod.2015.04.002.
- R. Yin, T. Agrawal, U. Khan, G. K. Gupta, V. Rai, Y.-Y. Huang and M. R. J. N. Hamblin, Antimicrobial photodynamic inactivation in nanomedicine: Small light strides against bad bugs, Nanomedicine, 2015, 10(15), 2379–2404 CrossRef CAS PubMed.
- F. Laborda, J. Jiménez-Lamana, E. Bolea and J. R. Castillo, Selective identification, characterization and determination of dissolved silver(I) and silver nanoparticles based on single particle detection by inductively coupled plasma mass spectrometry, J. Anal. At. Spectrom., 2011, 26(7), 1362, 10.1039/c0ja00098a.
- K. A. Johnston, L. M. Stabryla, A. M. Smith, X. Y. Gan, L. M. Gilbertson and J. E. Millstone, Impacts of broth chemistry on silver ion release, surface chemistry composition, and bacterial cytotoxicity of silver nanoparticles, Environ. Sci.: Nano, 2018, 5(2), 304–312, 10.1039/c7en00974g.
- I. De Leersnyder, L. De Gelder, I. Van Driessche and P. Vermeir, Influence of growth media components on the antibacterial effect of silver ions on Bacillus subtilis in a liquid growth medium, Sci. Rep., 2018, 8(1), 9325, DOI:10.1038/s41598-018-27540-9.
- J. S. Weese, Methicillin-resistant Staphylococcus aureus in animals, ILAR J., 2010, 51(3), 233–244 CrossRef CAS PubMed.
- S. Agnihotri, S. Mukherji and S. Mukherji, Size-controlled silver nanoparticles synthesized over the range 5–100 nm using the same protocol and their antibacterial efficacy, RSC Adv., 2014, 4(8), 3974–3983, 10.1039/C3RA44507K.
- A. Agarwal; S. Gupta and R. Sharma, Reactive oxygen species (ROS) measurement, in Andrological evaluation of male infertility, Springer, 2016, pp. 155–163 CrossRef CAS PubMed; A. Phaniendra, D. B. Jestadi and L. Periyasamy, Free radicals: Properties, sources, targets, and their implication in various diseases, Indian J. Clin. Biochem., 2015, 30(1), 11–26 CrossRef CAS PubMed.
- M. Merchat, G. Bertolini, P. Giacomini, A. Villaneuva, G. Jori and P. B. Biology, Meso-substituted cationic porphyrins as efficient photosensitizers of Gram-positive and Gram-negative bacteria, J. Photochem. Photobiol., B, 1996, 32(3), 153–157 CrossRef CAS.
- J. L. Martínez, Antibiotics and antibiotic resistance genes in natural environments, Science, 2008, 321(5887), 365–367, DOI:10.1126/science.1159483; V. Pareek, R. Gupta and J. Panwar, Do physico-chemical properties of silver nanoparticles decide their interaction with biological media and bactericidal action? A review, Mater. Sci. Eng., C, 2018, 90, 739–749, DOI:10.1016/j.msec.2018.04.093.
- N. Hasan, J. Cao, J. Lee, S. P. Hlaing, M. A. Oshi, M. Naeem, M.-H. Ki, B. L. Lee, Y. Jung and J.-W. J. P. Yoo, Bacteria-targeted clindamycin loaded polymeric nanoparticles: Effect of surface charge on nanoparticle adhesion to MRSA, antibacterial activity, and wound healing, Pharmaceutics, 2019, 11(5), 236 CrossRef CAS PubMed; F. Nederberg, Y. Zhang, J. P. Tan, K. Xu, H. Wang, C. Yang, S. Gao, X. D. Guo, K. Fukushima and L. J. N. C. Li, Biodegradable nanostructures with selective lysis of microbial membranes, Nat. Chem., 2011, 3(5), 409–414 CrossRef PubMed.
- X.-Z. Zhang, X. Zeng and Y.-X. Sun, et al., Bioactive materials in gene therapy, in Bioactive Materials in Medicine, ed. X. Zhao, J. M. Courtney and H. Qian, Woodhead Publishing, 2011, pp. 179–219 Search PubMed; F. Meng, S. Kwon, J. Wang and Y. Yeo, Immunoactive drug carriers in cancer therapy, in Biomaterials for Cancer Therapeutics, Elsevier, 2020, pp. 53–94 Search PubMed.
- A. T. P. C. Gomes, M. G. P. M. S. Neves and J. A. S. Cavaleiro, Cancer, photodynamic therapy and porphyrin-type derivatives, An. Acad. Bras. Cienc., 2018, 90, 993–1026 CrossRef CAS PubMed; D. J. Granville, B. McManus and D. J. H. Hunt, Photodynamic therapy: Shedding light on the biochemical pathways regulating porphyrin-mediated cell death, Histol. Histopathol., 2001, 16(1), 309–317 Search PubMed.
- V. Alt, T. Bechert, P. Steinrücke, M. Wagener, P. Seidel, E. Dingeldein, E. Domann and R. J. B. Schnettler, An in vitro assessment of the antibacterial properties and cytotoxicity of nanoparticulate silver bone cement, Biomaterials, 2004, 25(18), 4383–4391 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Extended materials and methods sections; MDR E. coli preparation, sequencing and classification protocols; FT-IR, UV-vis, 1H-NMR and MALDI-MS spectra; release kinetics calculations; antibacterial activity control experiments; and HeLa cells control experiments. See DOI: https://doi.org/10.1039/d2ma00711h |
| ‡ Equal contributors. |
| This journal is © The Royal Society of Chemistry 2022 |