 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The importance of structure property relationship for the designing of biomaterials using liquid crystal elastomers
Grace A. R.
Rohaley
 ab and
Elda
Hegmann
ab and
Elda
Hegmann
 *abcd
*abcd
aAdvanced Materials and Liquid Crystal Institute (AMLCI), Kent State University (KSU), Kent, OH 44242-0001, USA. E-mail: ehegmann@kent.edu
bMaterials Science Graduate Program, KSU, Kent, OH 44242-0001, USA
cDepartment of Biological Sciences, KSU, Kent, OH 44242-0001, USA
dBrain Health Research Institute (BHRI), KSU, Kent, OH 44242-0001, USA
First published on 21st June 2022
Abstract
Regenerative medicine (RM) and tissue engineering (TE) have been at the forefront of the pursuit for repairing, regenerating, restoring diseased or damaged tissues or organs, and restoring their functions. The combined efforts within RM and TE had made it possible to increase the choice of biomaterials created while improving their final properties, study their processes/technologies to engineer scaffolds, and study scaffold-tissue behaviour. The advancement of the RM and TE interdisciplinary areas are fundamentally connected with the success of innovative biomaterials. In short, the success of RM and TE is contingent to biomaterials. To this end, the search for interactive biomaterial-scaffold-host systems led us to use liquid crystal elastomers (LCEs) as biomaterials due to their exceptional intrinsic anisotropy, mechanical properties, and shape actuation. The development of LCEs as biomaterials has been motivated by the chemistries for biocompatibility, easy processing, mechanical studies, and recently 3D printing. In our continuing pursuit to use LCEs as biomaterials we have carefully considered the chemistry and processability overall, while also refining our LCEs to fit precise needs of cells specifically to their size, mechanical needs, and time for growth and regeneration.
1 Introduction
As part of our natural processes, all living organisms have some capacity to self-heal to be able to replace or restore damaged tissues. However, this regeneration process differs from different species. Complex animals, especially mammals, limit their regeneration capacity to for example, repair bone, regrowing hair and skin and forming scars as part of wound healing processes. In humans, the liver can grow back to its full size when a portion of it is removed or damaged.1 Whereas plants and some sea creatures, can fully replace missing parts. Two-dimensional (2D) cell culture has played an important role within biomedical and biological research to help understand biological events from cell metabolic pathways to the formation of tissue as part of tissue engineering (TE). TE is the interdisciplinary field that combines the core values of biology and engineering with the purpose of developing close to ideal alternatives to replace or regenerate damaged or diseased tissue,2,3 and has made strides into moving towards three-dimensional (3D) systems away from static monolayer cell growth observed in 2D traditional cell culture. Despite the overall success creating 3D systems using natural and synthetic polymeric materials, ceramics, and composites,4 most 3D materials still present some restrictions, mostly when providing effective support for cell populations while at the same time conferring sufficient mechanical properties to tissues. In TE, the therapeutic reconstruction of tissue is often made by an external stimulation of selected target cells over a systematic combination of molecular and mechanical signals,5 thus it is important to ensure that 3D systems provide appropriate support, promote extracellular matrix (ECM) formation,6,7 as well as sustained 3D cell growth and proliferation.8Soon after the discovery of liquid crystals (LCs) in 1888, Otto Lehmann in 19099 suggested that LCs can be an artificial muscular driving motor which was later supported by de Gennes in 1997.10 LC is a state of matter with properties between conventional liquids and solid crystals. Liquid crystalline molecules organize themselves in semi-crystalline fashion while retaining liquid flow characteristics. Liquid Crystals can be divided in two main classes, thermotropic (temperature dependent), and lyotropic (can be both temperature and concentration dependent). Depending on the positional ordering of the LC moieties, several liquid crystal phases (or mesophases) are observed, the most common are nematic (N), smectic (Sm), cholesteric, chiral, discotic, etc.11
LCEs combine the orientational order of LC moieties with the rubbery-elastic properties of elastomers. Elastomers are synthetic polymers that mimic rubber properties and can suffer several degrees of deformation under stress and will recover its original state once the stress is eliminated.4 LCEs are classified according to the position of the LC moieties within its bulk structure. If LC molecules are part of the polymer backbone they are recognized as main-chain LCEs. If they are linked as a pendant group to the main chain, then they are recognized as side-chain LCEs. Comparable to any LC moiety, LCEs also present difference LC Phases, the most common are nematic (N) and smectic-A (Sm-A). The applications of liquid crystal elastomers (LCE) have been recognized, among other properties, as materials capable of reversible actuation12 since they combine orientational anisotropy of LCs with elasticity of elastomers. Within the last decade, most of the new LCE applications have geared towards biological and biomedical applications and TE.4,13,14 In this perspective, we summarize our group's journey in the design of biomaterials using LCEs with, as always, being cognizant of the work of others.15 For the focus of this discussion, we will introduce the specific properties sought in a scaffold that address specific needs of cells, the approach used to fulfill those needs while keeping in mind the intrinsic properties of LCEs and their influence on cell behavior. We will also present our recent attempts to use LCEs as inks for 3D printing.
2 Liquid crystal elastomers as biomaterials
LCEs have shown contractile and expansion properties due to their ability to undergo significant shape change in response to external stimuli,16,17 making them exciting bioinspired materials offering optical, mechanical, sensing,18 and actuating behaviors.19–21 LCEs can also mimic vital features of endogenous tissue thus satisfying the prerequisites biocompatibility and biodegradability, mechanical properties, cell spatial growth, cell alignment, and long-term studies that are considered as tissue engineering systems.22–24 LCs have long been used and/or incorporated into materials to create composites, ensure better processing,25–28 introduce LC properties, create chemical sensors,29–31 and biosensors,32,33 soft actuators,34–38 light driven motors,39,40 as responsive building blocks for guiding 2D cell growth,41,42 promoters of cell orientational order,43–45 or to control the dynamics of bacteria.46,472.1 Biocompatibility and biodegradability
Biocompatibility of a material is contingent on its capability to exist with living bodily tissue without causing considerable harm such as through the production of toxic by-products or inducing immunologic responses. As tissues interact continuously with the scaffold during regeneration, it is essential that biomaterial scaffolds promote cellular adhesion and proliferation. Materials for the use of tissue engineering are also expected to be considered biocompatible, and biodegradable. For most polymeric biomaterials, degradation mostly occurs through physical or mechanical processes (wear and tear) and chemical (photo-, thermal-) stimulations.48 Biodegradable polymers allude to materials where degradation is facilitated, to a certain extent, by a biological system. Enzyme-catalyzed hydrolysis is facilitated by hydrolases (i.e., proteases, phosphatases and esterases) that act as biochemical catalysts and are responsible for the catalysis of several reactions in the body. Thus, degradation rate should be controlled and manipulated during material design since its desired degradation rate depends on the targeted tissue of interest.15 Thus, is important to keep in mind features of specific tissue, for example brain is known to be composed of mostly water (80–85%) suggesting that most biomaterials will suffer a fast hydrolytic degradation when in the brain.49 Thus, materials biodegradability rate should be tissue-specific to ensure the time needed for new tissue formation.Our approach started with the synthesis of non-liquid crystalline elastomer that was first established as a potential biomaterial by Younes et al.,50 this particular elastomer was prepared from biodegradable components and its degradation in water was been found to happen without harmful or toxic by-products.51 To maintain the biocompatible and biodegradable properties of an LCE, the search for a biocompatible LC led to the selection of the very first LC discovered: cholesterol. With the incorporation of biocompatible liquid crystalline pendants, we were able to introduce new functionalization and tunability to the elastomer's properties, while maintaining its biocompatibility and biodegradability. We then reported52 (Scheme 1) the first synthesis, characterization, and use of a biocompatible, biodegradable, and porous Liquid Crystal Elastomer (LCE) with the use of crosslinked star block-co-polymers (SBC) and interdigitated liquid crystalline cholesterol pendants, using bis-caprolactone (CL) as crosslinker. We were able to confirm that the LCE was fully biocompatible and LC functionalization properties were introduced at low levels of LCs. This functionalization is seen in the ordering of the interdigitated LC moieties into a side-chain Sm-A that remains present over a wide temperature range including room- and physiological temperatures. Later on, we demonstrated that varying the number of arms on the central node of the star-block co-polymer does not alter either the biocompatibility of the LCE or the phase Sm-A observed through central nodes with more arms do see a higher degree of crosslinkage,53 and we reported, that the higher number of arms on the central node promoted a higher degree of cell alignment.53
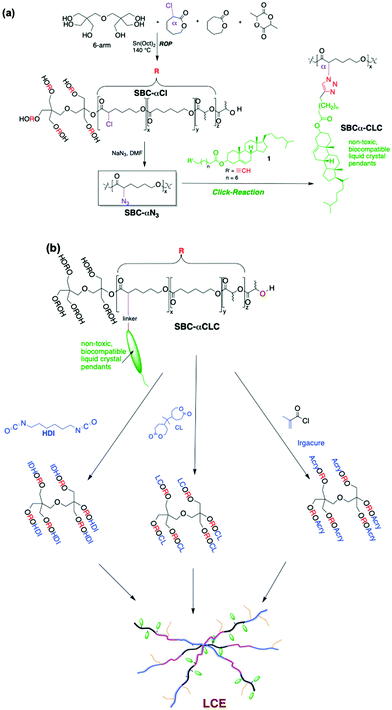 | ||
| Scheme 1 (a) Chemical structure of 6-arm initiators (central nodes) adapted from (b) two pathways for thermal-crosslinking using biscaprolactone (first synthesis path reported) or HDI, and photo-crosslinking by replacing –OH group by an acrylate group and Irgacure to obtain an LCEs. Images adapted from ref. 53, 60, and 80 this is an open access MDPI article distributed under the Creative Commons Attribution License. © 2016 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim and RSC, permission have been requested. | ||
While the LCE has been shown to be biocompatible, it is imperative that they also degrade at a time scale similar to that of target growth tissues. We aim for our scaffolds to degrade progressively, matching the rate of formation for new tissue while keeping the material structure as intact as possible for as long as the forming tissue needs. Caprolactone-lactide co-polymers degradation is well-known to occur by primarily degradation via bulk acid-catalyzed hydrolysis.50,54 This degradation is continuously auto-catalyzed due the hydrolysis reaction that produces oligocarboxylic acids and is expected to begin at the cross-linking sites.50 We presented that biodegradability of the materials was observed by the diffusion of water into caprolactone-lactide elastomers at above Tg temperatures which decreased as the cross-linking density of the elastomer increased. On Scheme 1, we also presented a new crosslinking method by replacing bis-caprolactone with hexamethylene diisocyanate (HDI) as crosslinker (Scheme 1) to significantly reduce crosslinking times.
Comparing our first approach with non-LC elastomers against LCEs, we observed that LCEs degraded faster than its counterpart non-LC elastomer. The rate was contingent to the position of the attached cholesteric pendant on the polymer backbone.52 α-LCE survived for at least 10 weeks before major degradation was observed, affording a suitable time to allow for the generation of tissue within a scaffold of this material. We also validated that tuneable surface characteristics and morphology within our LCEs lead to enhanced cellular proliferation and that elastomers with higher crosslinking density degraded more rapidly.
2.2 Suitable structure and 3D printing
Morphological features of the scaffolds must be tunable to control and affect cell adhesion features such as heterogeneity, (an)isotropy, as well as geometrical features of the porous architecture including pore size and inter-connectivity.3,55,56 When developing new biomaterials, or as in this case, engineering LCEs, as scaffolds for TE and/or biomedical applications, they must have a well-defined porosity and surface properties. These aspects provide support for cell adherence, growth, and mass transport between of the scaffold pores, under physiological conditions, to promote healthy cellular growth. Mass transport is usually related to proper flow and access to oxygen, nutrients, and waste management (mainly to eliminate toxins that can potentially raise pH to toxic levels).57 The porosity of the scaffold supports cell–material interactions, space for extracellular matrix (ECM) formation, and increases the likelihood of connecting molecular entities enhance growth and adhesion. Traditional 2D systems usually allow cells to populate Petri dishes indiscriminately forming exclusive monolayers that do not accurately represent a natural 3D system. This led researchers to find ways to overcome the lack of cell ability to grow in favored and more appropriate 3D orientations that allow for cell motility and metabolic functions. In this way, the systems are made to fully mimic endogenous tissue. Scaffolds should be morphologically and architecturally similar to their host environment. 3D scaffold biomaterials have been made following approaches that vary from particle leaching, gas foaming methods, emulsion freeze drying, to solution casting, melt molding, electrospinning,58etc. Porous foams have a history of being fabricated through many different methods including micellar and microfiber temples, though these methods form only simplified models of vascular conduits. Unfortunately, some limitations are still detected, principally, as geometric constraints show deficiencies in controlling of pore size and the rate of interconnectivity within the pores. Pore size should be adjusted to the different cell sizes and shapes in general as tissue properties of host vary greatly from one another.We first prepared LCE films, comparing film morphologies containing LC moieties in alpha- (α-), or gamma- (γ-) position to the epsilon (ε-)-carbonyl from the ε-caprolactone monomer in the main chain. Fig. 1 presents all the morphologies produced in our quest to find appropriate porosities to fulfill cell needs. Elastomers without the LC presence presented a smoother morphology, the difference can be attributed to the steric demand of the pendant cholesterol groups. We also observed that different internal morphologies of the LCE foams fabricated can be controlled by the position of the cholesterol (LC) moieties along the polymer backbone. An α-position LCE appeared more rough and less elastic compared to the γ-position with a lower pore density.52 These differences are thought to be due to either the higher degree of molecular flexibility or functionalization of the γ-position cholesterol groups leading to LCEs with higher pore density and smoother morphologies. Since films did not provided a truly 3-D porosity, then our first attempts (Fig. 1) to create internal morphologies that will allow for cell attachment was by creating nematic LCEs with globular morphologies.59–61
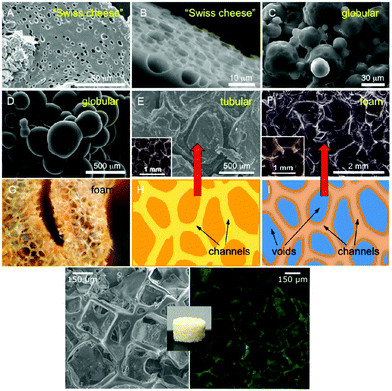 | ||
| Fig. 1 Top: SEM images of the internal porosity of: smectic LCE-γ (A) before, (B) and after 16 weeks of biodegradation in PBS, (C and D) of globular, porous microstructure of nematic LCE (E) LCEPP, (F) LCEPP, (G) optical image of LCEFP + SP. Scheme of type de foams used to fabricate: (H) LCEPP, and (I) LCEFP + SP. Reprinted (adapted) with permission from ACS(C) Symposium series ref. 4. Copyright 2017 American Chemical Society. Bottom: Adapted from ref. 65 RSC Soft Matter. | ||
Nematic LCEs were sought as artificial muscles so we designed globular nematic LCEs with intrinsic porosity to support muscle growth. We demonstrated that nematic LCEs allowed for attachment, growth, and proliferation of skeletal muscle cells (C2C12 myoblasts) making it, at the time, the first study on the successful use of nematic LCEs as scaffolds for muscle cells. We then proceeded to apply a known templating method, previously used to create graphene foams,62 for the creating 3D LCE scaffolds using nickel templates56,63 with interconnected microchannel networks, suitable for enhanced cell proliferation. Foam Sm-A LCEs showed overall high cell proliferation compared to the globular nematic LCEs, we believe that while globular LCEs have high porosity, it was a very dense morphology that did not show fully interconnected pores that would allow for efficient mass transport. The total porosity of foam Sm-A LCEs was determined using Micro CT to be at 77 ± 5%, so close porosity was negligible. Globular LCEs are prepared using microemulsion techniques so pore size/space would be harder to control limiting to the use of small cell lines, whereas nickel templates can be made to create specific pore sizes.
A previously reported method using salt leaching64 was used to distribute salt crystals that were re-engineered with tunable size into the pre-polymer mix before crosslinking. Using this fabrication method, LCE foams had a high percentage of porosity, the percentage of closed pores within the samples was negligible evaluated by micro-CT (μCT) scanning, showcasing that the foam's pores are all interconnected with access from deep inside to the outside of the foam surface.65 This particular salt leaching foam showed mechanical properties similar to neural environments, making it ideal to host brain or spinal cord cells.
Overall porosity degree of the foam can be adjusted by manipulating the ratio of polymer mixture to salt prior crosslinking allowing allow cells to constant access to nutrients, gases, and waste management.
With the desire for fine tunning of scaffold pore size and connectivity in mind, the modern usage of bio-applicative additive manufacturing, or 3D printing, opens a new avenue for novel TE and biomedical applications. Very recently, the combination of medical imaging has enabled researchers to reproduce customized tissues of interest.66,67 To be successful with 3D printing of scaffolds it is required to match of biomaterials and/or composites that provide with specific requirements of the products of interest. Many applications in 3D modeling68 have been engaged for several biology/medical research applications such as, anticancer drug screening, drug delivery and kinetics, and tissue engineering among others.69–74 The main application for 3D printing is to transition from 2D to 3D cell research environments that allows reproducibility of endogenous environment. This is turn, promotes cell growth, proliferation and cell maturation, while ensuring the study in-depth of metabolic processes.75 Several synthetic based bio-inks have been used for 3D printing, and have shown positive cellular responses, from viability and biocompatibility to ECM formation.76 Most of these inks bases are polymers such as polycaprolactone (PCL) and polyethylene glycol (PEG) based. Early studies reported a nematic-based thermally-responsive LCE ink77 which allows for 4D printing of shape-morphing LCE architectures. The LCE was direct-write printed into 3D structures with a controlled molecular order. In this case, the molecular control is locally programmed while controlling the printing path. Kotikian et al.78 later reported the additive manufacturing of nematic ordered LCE actuators (LCEAs) that exhibited large, reversible, and repeatable contraction with high specific work capacity using high operating temperature direct ink writing (HOT-DIW). Most recently, Mistry et al.79 reported how processing can increase the feasibility of external stimulation of LCE shape actuation, as well as with the help of additive manufacturing technologies LCE actuators are not only thin film devices but now can be made in more complex 3D structures. The work presented makes the case of how careful processing of materials is crucial for the preparation of actuators. However, while there are many examples of printed LCEs and their applications, most of the 3D printed LCEs reported have possessed nematic phases. We have, however, reported Sm-A 3D-printed LCE (Fig. 2) where we presented and studied the local orientation of the LCE using time-resolved synchrotron microbeam Small-Angle X-ray Diffraction (μ-SAXD).80
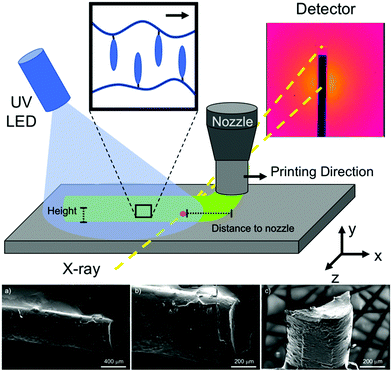 | ||
| Fig. 2 Top: Schematic description of the 3D printing process coupled with in situ m-SAXD, showing the position of the sample compared with the synchrotron X-ray beam. Bottom: SEM images of 3D printed LCE filaments, (a and b) topview, and (c) the cross-section view. Reproduced under the terms of the CC-BY license Copyright 2021,80 this is an open access MDPI article distributed under the Creative Commons Attribution License. | ||
To achieve 3D printing, our thermal-crosslinked LCE formulation52,65 (Scheme 1) was adjusted by replacing the thermal-crosslinking method with the addition of a methacrylate prior to UV curing which then allows for a photo-crosslinked LCE formulation.80 Our in situ μ-SAXD and 3D printing experiments were accomplished on a customized 3D printing platform mounted in-line with the X-ray beam inside the sample chamber of the Soft Matter Interfaces (SMI) beamline at NSLS-II (Fig. 2). Using shear stress during extrusion, and deformation at deposition pursued by relaxation (as external stimuli source) the LCE layering alignment followed the print direction which locked into the final structure. The data obtained on the 3D printed filaments provided insights into the internal structure of the LCE that will guide the future fabrication of LCEs as responsive and anisotropic 3D cell scaffolds and to continue to promote cell proliferation and alignment.
2.3 Mechanical properties
The mechanical properties of the extracellular matrix (ECM) differ between types of tissue, living tissue stiffness vary greatly from tissue to tissue.81 Thus the stiffness and strength of scaffold materials be thoroughly considered and prepared in a way to ensure a better match to the host cell or tissue of interest.82 All mechanical properties of cells are usually founded by cytoskeletal elements that consist of mainly of actin and intermediate filaments, and microtubules.83 Each of these components responds differently to mechanical deformations and varies from tissue.84 Thus efforts are concentrated on studying how cells respond to mechanical stimuli so materials can be made to limit potential or irreversible cell damage.85–87 When preparing materials, Younes et al. confirmed the effects of crosslinking density of biocompatible polycaprolactone based star-copolymer elastomer and the stress of the material.50 We reported that with the incorporation of the LC pendants, the liquid crystalline character of the LCE made it more resistant to strain, however it was still able to mimic the properties of tissue elasticity.52 From uniaxial tensile measurements and calculation of Young's modulus, we demonstrated53 that the number of arms present in the central node the star block-co-polymer did affect not only the Young's modulus (E) of the materials, but also the cellular of cultures grown on films and foams of the materials.52,53,65 Data obtained show that the number of arms on 4-arms-LCE-γ correlated with previous theoretical and experimental studies on tetra-arm polymer hydrogel based systems, that were reported to show a high degree of homogeneous packing resulting in higher stiffness.88 Our LCEs exhibited elastic moduli closer to and slightly lower than those of tissues formed by the cells that were investigated, allowing for cells to attach and thrive in a more “familiar” environment. Indentation measurements of the fabricated foams have also been studied, as materials that interact with regenerating tissue are under constant and dynamic stresses of the ECM. The LCE foams generated by nickel templating and salt leaching were able to recover their initial shape and size immediately after compression deformation.65 Mechanical tuning of materials can be done by increasing the degree of crosslinking, increasing initial monomer concentration, chemically introducing strong molecular interactions, or by the addition of additives, among others.89,90As discussed, direct modulation of mechanical properties is indispensable to match those of tissue on interest. It has been shown that CNCs have successfully being used to adjust the mechanical properties (tensile strength and E values) of several different polymer matrices including polylactic acid (PLA),91–95 inducing shape memory effects of polymers,96 and capable of tuning biodegradable properties of materials.97 CNCs have since been known to be biocompatible and biodegradable and on its own has been used for a myriad of medical applications.98–107 CNCs offers low cytotoxicity for different cell-types,108–113 supporting cell attachment and proliferation.114–117 CNCs are also recognized to form lyotropic liquid crystalline (LLC) phases when in aqueous dispersion.118,119 Knowing all the excellent properties of CNC, we proposed a binary composite of CNC constituent that is incorporated at several weight% (wt%) ratios to our biopolymeric scaffold material quickly enabling the tunability of the mechanical properties to match those of tissues of interest.80,82
To study the mechanical properties of our LCEs, we started by preparing elastomer/cellulose nanocrystals (CNC) composites. The addition of CNCs to the elastomer mix increased bulk stiffness of the composite relative to the pure 6-arm e-caprolactone-based star copolymer (6-arm-PCL) based elastomer, its dependency is seen to be exponential. We presented 6A-PCL/CNC composite where the wt% of CNC was increased from 0 (no CNC present) to 50%, as we increased wt% of CNC content E increased. The tensile values of the composite with 5 wt% and 40 wt% of CNC corresponded to that of gray matter and skin respectively. In addition, during degradation studies it was observed that E increased for both 5 wt% and 40 wt% CNC content after 16 weeks due to the amount of CNC slightly increasing during degradation of the elastomer bulk. This new approach also contributes to providing cell-friendly 3D scaffolds designed for the particular needs of different types of cells and fine-tuned according to the cell response observed. We selected two types of cell lines with different requirements of mechanical properties (human dermal fibroblast -hDFs and neuro-blastomas -SH-SY5Ys) cell lines to compare cell growth and proliferation within two mechanically different 3D foam scaffolds (Fig. 3). We found, in this proof of concept, that the composite with lower E was more suitable for SH-SY5Ys, where as the composite higher E was more suitable for hDFs which naturally possess lower and higher elasticities respectively.
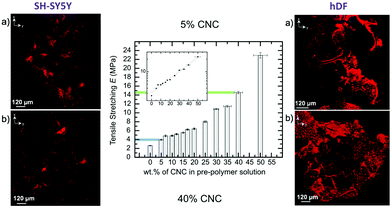 | ||
Fig. 3 Middle: Evolution of the tensile Young's modulus of 6A-PCL/CNC composite films with the addition of CNCs (data measured by tensile stretching). The values in blue and green match reported tensile deformation values of brain and skin tissues, respectively. The inset shows the E in log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 scale, to emphasize the exponential dependency found with the addition of CNCs. Right and left confocal images of SH-SY%Y and hDF (respectively) cells seeded into (a) 5 wt% and (b) 40 wt% 6A-PCL/CNC composite foams after two weeks of proliferation. Adapted from ref. 82, Royal Society of Chemistry. 10 scale, to emphasize the exponential dependency found with the addition of CNCs. Right and left confocal images of SH-SY%Y and hDF (respectively) cells seeded into (a) 5 wt% and (b) 40 wt% 6A-PCL/CNC composite foams after two weeks of proliferation. Adapted from ref. 82, Royal Society of Chemistry. | ||
We demonstrated that our elastomer/CNC composites were stable for 15 weeks before major degradation set in 6arm-PCL based elastomer making these composites more resistant to degradation.82 Further studies utilizing the approach of preparing LCE/CNC composites to better understand the how the LCE's mechanical properties and anisotropy affect cell behavior are underway.
2.4 Cell studies and anisotropy
To determine the viability of cells within our designed materials we first studied mouse skeletal myoblasts (C2C12s) and neuroblastomas (SH-SY5Ys) on films. We found that C2C12s preferred a slightly hydrophobic surface, which was provided by the α-LCE attached inherent property, and cells grew quite well. On the other hand, SH-SY5Ys attached and grew on the γ-LCE well, as a more hydrophilic surface. It was also observed that SH-SY5Ys seeded on non-LC elastomers showed significantly lower cell densities than when seeded on γ-LCE. It was found that non-LC elastomers presented a very smooth morphology that, we believe, made difficult for the cells to properly anchor on the surface of the material. This coupled with lower stress-strain curves values than their LCE counterparts, might have hindered proliferation rates and bio-viability of cells.We proceed to study tunable hydrophilicity of the materials and enhancing water contact angle using O2 plasma etching. O2 plasma etching was found to be a very useful and relevant approach when cell seeding to avoid the additional use of an extra Matrigel™ layer (frequently applied by dipping, drop-casting or spin-coating of materials), required previously in experiments described by others.120 Unfortunately, the addition of an extra layer can, in turn mask the surface topology of any tissue culture material or, in our case, the anisotropic ordering of the LCEs.
As shown, foam scaffolding offers a more “native” environmental replication for tissue growth. C2C12s were then grown on LCE foam scaffolds made using the nickel templating approach. The C2C12s grew within the struts of the foam channel sections and spontaneously aligned with the channel, perhaps aided by the Sm-A phase present of the LCE. They showed a slightly random orientation at the junctions.56 The nickel templated foam showed 4x times higher cell proliferation than previously made scaffolds with spherical templates.55 C2C12s were tested on α- and γ-series of LCE and compared to comparison to non-LC elastomers with the same number of core arms. α-LCEs series provides a better platform for the attachment and proliferation of C2C12s as well as human dermal fibroblasts (hDFs) cells rather than unmodified elastomers.53 This was further confirmed by cell viability assay, showing α- and γ-LCE series outperform the non-LC elastomer counterparts with same central nodes.
Among all α-LCEs, 3-arm showed higher proliferation of both C2C12s and hDFs. among all γ-LCE series, 4-arm showed best proliferation for C2C12s, and γ-LCE 6-arm showed best proliferation for hDFs. This further confirms that different cell types prefer different elasticity properties of the supporting scaffold. Therefore, modified LCEs for cellular growth must be carefully selected based on intended cell types to be grown to provide the most optimal proliferation rates.
We also showed that our LCE foam materials indicated a good scaffold for long term culture of neuronal cells (30 days) and SH-SY5Ys survived on LCE foams for over 60 days. This indicates that utilizing a LCE foam with an adequate system of microchannels similar to that of a microvascular system, and appropriate mechanical properties the in vitro lifespan of neuronal tissue growth is greatly extended.49 Due to the inherent directionality of the liquid crystal moieties attached in a side-chain orientation to the polymer backbone of the LCE, any anisotropy of cells must be recognized Cell anisotropy was studied and observed by identifying nuclei elongation,65 which has also featured by others as an important characteristic for cell growth (Fig. 4).
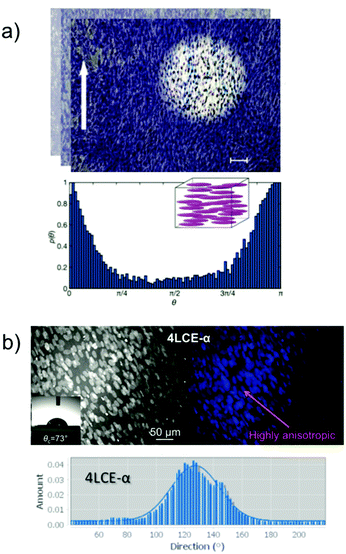 | ||
| Fig. 4 (a) Optical image of myoblast (C2C12) alignment on planar homogenous liquid crystalline networks (LCN) (scale bar: 100 μm) and directionality histogram. The histogram reports the probability density function to find a cell tilted by a certain angle θ with respect to a reference 0° angle (white arrow). Image and figure caption have been obtained and adapted with permission from copyright 2019 Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Advanced HealthCare Materials;41 (b) primary human dermal fibroblast (hDF) cultures grown for 5 d on an 4LCE-α film and directionality histograms of human dermal fibroblast (hDF) Copyright WILEY-VCH Verlag GmbH & Co. KGaA [ref. 53]. | ||
To further understand how anisotropy of scaffolds affect cell behaviour we reported a Sm-A 3D-printed LCE in situ and studied the local orientation of the LCE using time-resolved synchrotron microbeam Small-Angle X-ray Diffraction (μ-SAXD).80 This allowed us to elucidate the local orientational structure of a LCE-based printed scaffold. We observed that the orientation of the Sm-A layering was particularly pronounced at the substrate interface. With the help of μ-SAXD, we obtained a 2D mapping of the Sm-A orientation with layer normal nearly perpendicular to the incident X-ray beam. We have continuously demonstrated that cells sense the anisotropy of the Sm-A LC phase nature of our LCEs, promoting orientation and alignment without external stimuli,4,15,29,49,52,53,65 and our next steps to study cell growth and behaviour on the in situ printed filaments are ongoing.
For that purpose, we generated a vascular tissue model that was generated from a fluorescently post-mortem stained mouse tissue.121 This post-mortem stained tissue was imaged using confocal microscopy and consequently processed to create a digital 3D model suitable for printing. Using our LCE-based bio-ink we achieved a 3D duplication of a highly complex brain vascular tissue structure, using a Digital Light Processing (DLP) stereolithography (SLA) desktop 3D printer. We then proceeded to study the LCE alignment within the printed struts using SAXS. Our approach shown the capability to reproduce model tissues built within hours that can promote cellular alignment credited to the anisotropy of the LCE-based bio-ink. These findings are presently under peer-review.
3 Summary and outlook
Collectively over the last few decades, LCs and LCE research has made a lot of strides within the realms of biological and biomedical applications. While the first LC discovered was cholesterol, countless efforts have been made in the LC display manufacturing area, a new LC renaissance sees the uses of novel and functional materials in wider spheres of technology and biology. Today, there is plenty of research being conducted surrounding LCEs for 3D printing using not only extrusion 3D printing but also digital light processing (DLP). Our hope is that we are closer to fully recognize the specific requirements of most cells, so we can address them while at the same time being able transfer anisotropy of our materials with the ultimate goal of promoting superior tissue performance.Our quest lies in the continuation of developing appropriate LC-based materials for the purpose of aiding the biological and biomedical fields. Finding better models to provide for research applications does not stop at tissue engineering but reaches into improving therapeutics in the future.
This future will see further ex situ and in situ studies that couple X-ray characterization, exploring the Sm-A layering of our 3D printed materials. Gaining a better understanding of how a responsive cell support structure can promote better control over cell directionality and anisotropy will help to drive the realization of these types of novel materials.
Author contributions
All authors have given approval to the final version of the manuscript. GARR, and EH prepared an early manuscript draft. EH directed the research and wrote the manuscript with contributions from both authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research used funds from the KSU Collaborative Research Grant and the National Science Foundation (CHE-1263087). SAXS/WAXS measurements were funded from beamline time applications at Advanced Light Source, Lawrence Berkeley National Laboratory, Berkeley, Beamline 7.3.3 of the Advanced Light Source is supported by the Director of the Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231, and SMI beamline of the National Synchrotron Light Source II, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Brookhaven National Laboratory under Contract No. DE-SC0012704. The authors also want to thank the Materials Characterization Facility at the Advanced Materials and Liquid Crystal Institute (AMLCI) where the SEM data was acquired. The authors are grateful of the Advanced Light Source (ALS) beamline 7.3.3 and National Synchrotron Light Source II (NSLS-II) SMI beamline for their support and help in collecting and analyzing SAXS data.References
- D. Groeneveld, D. Pereyra, Z. Veldhuis, J. Adelmeijer, P. Ottens, A. K. Kopec, P. Starlinger, T. Lisman and J. P. Luyendyk, Blood, 2019, 133, 1245–1256 CrossRef CAS PubMed.
- R. Langer and J. P. Vacanti, Science, 1993, 260, 920–926 CrossRef CAS PubMed.
- F. Xu, C. Dawson, M. Lamb, E. Mueller, E. Stefanek, M. Akbari and T. Hoare, Front. Bioeng. Biotechnol., 2022, 10, 849831 CrossRef PubMed.
- M. E. Prévôt and E. Hegmann, From Biomaterial, Biomimetic, and Polymer to Biodegradable and Biocompatible Liquid Crystal Elastomer Cell Scaffolds, in Advances in Bioinspired and Biomedical Materials, ed. Y. Ito, X. Chen and I.-K. Kang, ACS Symposium Series, ACS, Washington, DC, 2017, vol. 1253, pp. 3–45 Search PubMed.
- U. Novak and A. H. Kaye, J. Clin. Neurosci., 2000, 7, 280–290 CrossRef CAS PubMed.
- H. Cheng, P. S. Hill, D. J. Siegwart, N. Vacanti, A. K. R. Lytton-Jean, S. W. Cho, A. Ye, R. Langer and D. G. Anderson, Adv. Mater., 2011, 23, H95–H100 CrossRef CAS PubMed.
- N. George and H. M. Geller, J. Neuro. Res., 2018, 96, 573–588 CrossRef CAS PubMed.
- P. Bartolo, M. Domingos, A. Gloria and J. Ciurana, CIRP Ann., 2011, 60(1), 271–274 CrossRef.
- O. Lehmann, J. Phys. Theor. Appl., 1909, 8, 713–735 CrossRef.
- P.-G. deGennes, C. R. Acad. Sci., Ser. IIb: Mec., Phys., Chim., Astron., 1997, 324, 343–348 CAS.
- M. E. Prévôt, J. P. Vanegas, E. Hegmann, T. Hegmann, J. Pérez-Prieto and Y. Molard, in 21st Century Nanoscience, A handbook: Exotic Nanostructures and Quantum systems, ed. K. D. Sattler, Taylor & Francis Books, Inc. Published April 29 2020 CRC Press, 2020, ch. 6, vol. 5, pp. 6-1–6-27, ISBN 9780815356264 Search PubMed.
- M. Warner and E. M. Terentjev, Liquid Crystal Elastomers, Clarendon Press, Oxford, 2013 Search PubMed.
- C. M. Yakacki, M. Saed, D. P. Nair, T. Gong, S. M. Reed and C. N. Bowman, RSC Adv., 2015, 5, 18997–19001 RSC.
- D. Martella, P. Paoli, J. M. Pioner, L. Sacconi, R. Coppini, L. Santini, M. Lulli, E. Cerbai, D. S. Wiersma, C. Poggesi, C. Ferrantini and C. Parmeggianni, Small, 2017, 13, 1702677 CrossRef PubMed.
- S. Ustunel, M. E. Prévôt, R. J. Clements and E. Hegmann, Liq. Cryst. Today, 2020, 29(3), 40–52 CrossRef CAS.
- A. Sanchez-Ferrer, T. Fischl, M. Stubenrauch, A. Albrecht, H. Wurmus, M. Hoffmann and H. Finkelmann, Adv. Mater., 2011, 23, 4526–4530 CrossRef CAS PubMed.
- P. Stein, N. Assfalg, H. Finkelmann and P. Martinoty, Eur. Phys. J. E: Soft Matter Biol. Phys., 2001, 4, 255–262 CrossRef CAS.
- N. Herzer, H. Guneysu, D. J. D. Davies, D. Yildirim, A. R. Vaccaro, D. J. Broer, C. W. M. Bastiaansen and A. P. H. J. Schenning, J. Am. Chem. Soc., 2012, 134, 7608 CrossRef CAS PubMed.
- T. H. Ware, M. E. McConney, J. J. Wie, V. P. Tondiglia and T. J. White, Science, 2015, 347, 982–984 CrossRef CAS PubMed.
- C. P. Ambulo, S. Tasmim, S. Wang, M. K. Abelrahman, P. E. Zimmern and T. H. Ware, J. Appl. Phys., 2020, 128, 140901 CrossRef CAS PubMed.
- M. Barnes, S. M. Sajadi, S. Parekh, M. M. Rahman, P. M. Ajayan and R. Verduzco, ACS Appl. Mater. Interfaces, 2020, 12, 28692–28699 CrossRef CAS PubMed.
- C. Ohm, M. Brehmer and R. Zentel, Adv. Polym. Sci., 2012, 250, 49–94 CrossRef CAS.
- H. R. Brand, H. Finkelmann, D. Demus, J. W. Goodby, G. W. Gray, H. W. Spiess and V. Vill, Physical properties of liquid crystalline elastomers, Handbook of Liquid Crystals, Wiley, New York, 2008, vol. 3, pp. 277–302 Search PubMed.
- W. H. E. de Jeu, Liquid crystal elastomers: Materials and applications, Springer, New York, 2012 Search PubMed.
- G. B. Kharas, J. R. Heiskell, J. Herrman, P. T. Kasudia, P. J. Schreiber, L. B. Passe, E. Bravo-Grimaldo, C. G. Bazuin, P. T. Romanowsky and R. M. Schueller, J. Macromol. Sci., Part A: Pure Appl. Chem., 2006, 43, 213–28820 CrossRef CAS.
- K.-S. Jang, J. C. Johnson, E. Hegmann, T. Hegmann and L. T. J. Korley, Liq. Cryst., 2014, 41, 1473–1482 CrossRef CAS.
- W. Zheng and G. H. Milburn, Liq. Cryst., 2000, 27, 1423–1430 CrossRef CAS.
- K.-S. Jang and L. T. J. Korley, Pol. Eng. Sci., 2016, 56, 388–393 CrossRef CAS.
- M. E. Prévôt, A. Nemati, T. R. Cull, E. Hegmann and T. Hegmann, Adv. Mater. Technol., 2020, 5, 2000058 CrossRef.
- C. Esteves, E. Ramou, A. R. P. Porteira, A. J. M. Barbosa and A. C. A. Roque, Adv. Opt. Mater., 2020, 8, 1902117 CrossRef CAS PubMed.
- R. J. Carlton, J. T. Hunter, D. S. Miller, R. Abbasi, P. C. Mushenheim, L. N. Tan and N. L. Abbott, Liq. Cryst. Rev., 2013, 1, 29–51 CrossRef CAS PubMed.
- Z. Wang, T. Xu, A. Noel, Y. Chen and T. Liu, Soft Matter, 2021, 17, 4675–4702 RSC.
- S. J. Woltman, G. D. Jay and G. P. Crawford, Nat. Mater., 2007, 6, 929–938 CrossRef CAS PubMed.
- C. L. van Oosten, C. W. M. Bastiaansen and D. J. Broer, Nat. Mater., 2009, 8, 677 CrossRef CAS PubMed.
- A. Kotikian, C. McMahan, E. C. Davidson, J. M. Muhammad, R. D. Weeks, C. Daraio and J. A. Lewis, Sci. Robot., 2019, 4, eaax7044 CrossRef PubMed.
- H. Zeng, O. M. Wani, P. Wasylczyk, R. Kaczmarek and A. Priimagi, Adv. Mater., 2017, 29, 1701814 CrossRef PubMed.
- M. Schwartz and J. P. F. Lagerwall, 2021, arXiv:2103.11005.
- M. H. Li, P. Keller, B. Li, X. G. Wang and M. Brunet, Adv. Mater., 2003, 15, 569–572 CrossRef CAS.
- M. Yamada, M. Kondo, J. Mamiya, Y. Yu, M. Kinoshita, C. J. Barrett and T. Ikeda, Angew. Chem., Int. Ed., 2008, 47, 4986 CrossRef CAS PubMed.
- A.-I. Bunea, D. Martella, S. Nocentini, C. Parmeggiani, R. Taboryski and D. S. Wiersma, Adv. Intell. Syst., 2021, 3, 2000256 CrossRef.
- D. Martella, L. Pattelli, C. Matassini, F. Ridi, M. Bonini, P. Paoli, P. Baglioni, D. Wiersma and C. Parmeggiani, Adv. Healthcare Mater., 2019, B, 1801489 CrossRef PubMed.
- G. Babakhanova, J. Krieger, B.-X. Li, T. Turiv, M.-H. Kim and O. D. Lavrentovich, J. Biomed. Mater. Res., 2020, 108, 1223 CrossRef CAS PubMed.
- N. A. Lockwood, J. C. Mohr, L. Ji, C. J. Murphy, S. P. Palecek, J. J. de Pablo and N. L. Abbott, Adv. Funct. Mater., 2006, 16, 618 CrossRef CAS.
- N. L. Abbott, J. J. de Pablo, S. P. Palecek, N. A. Lockwood, J. C. MohrC. J. Murphy and E. Huang, Liquid crystalline substrates for culturing cells, US Pat., 7732152, 2010 Search PubMed.
- J. Fang, W. Ma, J. V. Selinger and R. Shashidhar, Imaging biological cells using liquid crystals, Langmuir, 2003, 19, 2865 CrossRef CAS.
- N. P. Dhakal, J. Jiang, Y. Guo and C. Peng, ACS Appl. Mater. Interfaces, 2020, 12, 13680 CrossRef CAS PubMed.
- T. Turiv, R. Koizumi, K. Thijssen, M. M. Genkin, H. Yu, C. Peng, Q.-H. Wei, J. M. Yeomans, I. S. Aranson, A. Doostmohammadi and O. D. Lavrentovich, Nat. Phys., 2020, 16, 481 Search PubMed.
- A. Göpferich, Biomaterials, 1996, 17, 103–114 CrossRef.
- T. Mori, R. Cukelj, M. E. Prévôt, S. Ustunel, A. Story, Y. Gao, K. Diabre, J. A. McDonough, E. J. Freeman, E. Hegmann and R. J. Clements, Macromol. Rapid Commun., 2020, 41, 1900585 CrossRef CAS PubMed.
- H. M. Younes, E. Bravo-Grimaldo and B. G. Amsden, Biomaterials, 2004, 25, 5261–5269 CrossRef CAS PubMed.
- B. Amsden, A. Hatefi, D. Knight and E. Bravo-Grimaldo, Biomacromolecules, 2004, 5, 637–642 CrossRef CAS PubMed.
- A. Sharma, A. Neshat, C. J. Mahnen, Ad Nielsen, J. Snyder, T. L. Stankovich, B. G. Daum, E. M. LaSpina, G. Beltrano, Y. Gao, S. Li, B.-W. Park, R. J. Clements, E. J. Freeman, C. Malcuit, J. A. McDonough, L. T. J. Korley, T. Hegmann and E. Hegmann, Macromol. Biosci., 2015, 15, 200–214 CrossRef CAS PubMed.
- A. Sharma, T. Mori, C. J. Mahnen, H. R. Everson, M. T. Leslie, Ad Nielsen, L. Lussier, C. Zhu, C. Malcuit, T. Hegmann, J. A. McDonough, E. J. Freeman, L. T. J. Korley, R. J. Clements and E. Hegmann, Macromol. Biosci., 2017, 17, 1600278 CrossRef PubMed.
- H. Cheng, P. S. Hill, D. J. Siegwart, N. Vacanti, A. K. R. Lytton-Jean, S.-W. Cho, A. Ye, R. Langer and D. G. Anderson, Adv. Mater., 2011, 23, H95–H100 CrossRef CAS PubMed.
- T. Bera, E. J. Freeman, J. A. McDonough, R. J. Clements, A. Aladlaan, C. Malcuit, T. Hegmann and E. Hegmann, ACS Appl. Mater. Interfaces, 2015, 7, 14528–14535 CrossRef CAS PubMed.
- Y. Gao, S. Manning, Y. Zhao, Ad Nielsen, T. Mori, A. Neshat, A. Sharma, C. J. Mahnen, H. Everson, S. Crotty, R. J. Clements, C. Malcuit and E. Hegmann, ACS Macro Lett., 2016, 5, 4–9 CrossRef CAS PubMed.
- Y. Martin and P. Vermette, Biomaterials, 2005, 26, 7481 CrossRef CAS PubMed.
- S. Chung, A. K. Moghe, G. A. Montero, S. H. Kim and M. W. King, Biomed. Mater., 2009, 1, 105019 Search PubMed.
- M. E. Prévôt, S. Ustunel and E. Hegmann, Materials, 2018, 11, 377 CrossRef PubMed.
- M. E. Prévôt, L. E. Bergquist, A. Sharma, T. Mori, Y. Gao, T. Bera, C. Zhu, M. T. Leslie, R. Cukelj, L. T. J. Korley, E. J. Freeman, J. A. McDonough, R. J. Clements and E. Hegmann, Proc. SPIE, 2017, 10361(103610T), 1–11 Search PubMed.
- T. Bera, C. Malcuit, R. J. Clements and E. Hegmann, Front. Mater., 2016, 3, 31 CrossRef section Biomaterials.
- C. Martinez-Ramos, A. Valles-Lluch, J. M. G. Verdugo, J. L. G. Ribelles, J. A. Barcia, A. B. Orts, J. M. S. Lopez and M. M. J. Pradas, J. Biomed. Mater. Res., Part A, 2012, 100, 3276–3328 CrossRef PubMed.
- M. E. Prévôt, S. Ustunel, L. E. Bergquist, R. Cukelj, Y. Gao, T. Mori, L. Pauline, R. J. Clements and E. Hegmann, J. Vis. Exp., 2017, 122, e55452 Search PubMed.
- A. J. Hoyt, C. M. Yakacki, S. R. Fertig, R. D. Carpenter and C. P. Frick, J. Mech. Behav. Biomed. Mater., 2015, 41, 136–148 CrossRef CAS PubMed.
- M. E. Prévôt, H. Andro, S. L. M. Alexander, S. Ustunel, C. Zhu, Z. Nikolov, S. T. Rafferty, M. T. Brannum, B. Kinsel, L. T. J. Korley, E. J. Freeman, J. A. McDonough, R. J. Clements and E. Hegmann, Soft Matter, 2018, 14, 354–360 RSC.
- T. Xu, J. I. Rodriguez-Devora, D. Reyna-Soriano, B. Mohammod, L. Zhu, K. Wang and Y. Yuan, Bioprinting for constructing microvascular systems for organs, in Rapid Prototyping of Biomaterials - Principles and Applications, ed. R. Narayan, Woodhead Publishing, 2014, pp. 201–220 Search PubMed.
- V. Tran and X. Wen, Rapid prototyping technologies for tissue regeneration, in Rapid Prototyping of Biomaterials - Principles and Applications, ed. R. Narayan, Woodhead Publishing, 2014, pp. 97–155 Search PubMed.
- R. Langer and J. P. Vacanti, Science, 1993, 260, 920 CrossRef CAS PubMed.
- B. Tian, W. Y. Wong, E. Hegmann, K. Gaspar, P. Kumar and H. Chao, Bioconjugate Chem., 2015, 26, 1144–1155 CrossRef CAS PubMed.
- M. Ravi, V. Paramesh, S. R. Kaviya, E. Anuradha and S. F. D. Paul, J. Cell. Physiol., 2015, 230, 16 CrossRef CAS PubMed.
- N. Chaicharoenaudomrung, P. Kunhorm and P. Noisa, World J. Stem Cells, 2019, 11, 1065 CrossRef PubMed.
- H. W. Kang, S. J. Lee, I. K. Ko, C. Kengla, J. J. Yoo and A. A. Atala, 3D bioprinting system to produce human-scale tissue constructs with structural integrity, Nat. Biotechnol., 2016, 34, 312 CrossRef CAS PubMed.
- D. B. Kolesky, K. A. Homan, M. A. Skylar-Scott and J. A. Lewis, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3179 CrossRef CAS PubMed.
- S. Massa, M. A. Sakr, J. Seo, P. Bandaru, A. Arneri, S. Bersini, E. Zare-Eelanjegh, E. Jalilian, B.-H. Cha, S. Antona, A. Enrico, Y. Gao, S. Hassan, J. P. Acevedo, M. R. Dokmeci, Y. S. Zhang, A. Khademhosseini and S. R. Shin, Biomicrofluidics, 2017, 11, 044109 CrossRef PubMed.
- J. W. Haycock, 3D Cell Culture: A Review of Current Approaches and Techniques, Methods Mol, Biol., 2010, 1–15 Search PubMed.
- P. S. Gungor-Ozkerim, I. Inci, Y. S. Zhang, A. Khademhosseini and M. R. Dokmeci, Biomater. Sci., 2018, 6, 915 RSC.
- C. Ambulo, J. J. Burroughs, J. M. Boothby, H. Kim, M. R. Shankar and T. H. Ware, ACS Appl. Mater. Interfaces, 2017, 9, 37332 CrossRef CAS PubMed.
- A. Kotikian, R. L. Truby, J. W. Boley, T. J. White and J. A. Lewis, Adv. Mater., 2018, 30, 1706164 CrossRef PubMed.
- D. Mistry, N. A. Traugutt, K. Yu and C. M. Yakacki, J. Appl. Phys., 2021, 129, 130901 CrossRef CAS.
- M. E. Prévôt, S. Ustunel, B. M. Yavitt, G. Freychet, C. R. Webb, M. Zhernenkov, E. Hegmann and R. Pindak, Crystals, 2021, 11(5), 523 CrossRef.
- C. T. McKee, J. A. Last, P. Russell and C. J. Murphy, Tissue Eng., Part B, 2011, 17, 155–164 CrossRef PubMed.
- S. Ustunel, M. E. Prévôt, G. A. R. Rohaley, C. R. Webb, B. Yavitt, G. Freychet, M. Zhernenkov, R. Pindak, E. Schaible, C. Zhu, T. Hegmann, R. J. Clements and E. Hegmann, Mater. Adv., 2021, 2, 464–476 RSC.
- D. Fletcher and R. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed.
- S. Kumar, I. Z. Maxwell, A. Heisterkamp, T. R. Polte, T. P. Lele, M. Salanga, E. Mazur and D. E. Ingber, Biophys. J., 2006, 90, 3762–3773 CrossRef CAS PubMed.
- R. Bernal, M. V. Hemelryck, B. Gurchenkov and D. Cuvelier, Int. J. Mol. Sci., 2022, 23, 5095 CrossRef CAS PubMed.
- E. Kassianidou and S. Kumar, Biochim. Biophys. Acta, 2015, 1853, 3065–3074 CrossRef CAS PubMed.
- C. E. Keating and D. K. Cullen, Neurobiol. Dis., 2021, 148, 105210 CrossRef CAS PubMed.
- T. Sakai, Polym. J., 2014, 46, 517–523 CrossRef CAS.
- Y. Takeoka, S. Liu and F. Asai, Sci. Technol. Adv. Mater., 2020, 1, 817–832 CrossRef PubMed.
- D. Vigolo, S. N. Ramakrishna and A. J. Demello, Sci. Rep., 2019, 9, 7125 CrossRef PubMed.
- S. J. Eichhorn, A. Dufresne, M. Aranguren, N. E. Marcovich, J. R. Capadona, S. J. Rowan, C. Weder, W. Thielemans, M. Roman, S. Renneckar, W. Gindl, S. Veigel, J. Keckes, H. Yano, K. Abe, M. Nogi, A. N. Nakagaito, A. Mangalam, J. Simonsen, A. S. Benight, A. Bismarck, L. A. Berglund and T. Peijs, J. Mater. Sci., 2010, 45, 1–33 CrossRef CAS.
- S. Mueller, J. Sapkota, A. Nicharat, T. Zimmermann, P. Tingaut, C. Weder and E. J. Foster, J. Appl. Polym. Sci., 2015, 132, 41740 CrossRef.
- M. Jorfi, M. N. Roberts, E. J. Foster and C. Weder, ACS Appl. Mater. Interfaces, 2013, 5, 1517–1526 CrossRef CAS PubMed.
- N. W. Awang, D. Ramasamy, K. Kadirgama, G. Najafi and N. Azwadi Che Sidik, Int. J. Heat Mass Transfer, 2019, 131, 1196–1204 CrossRef CAS.
- J. J. Valle-Delgado, L. S. Johansson and M. Österberg, Colloids Surf., B, 2016, 138, 86–93 CrossRef CAS PubMed.
- A. Nicharat, A. Shirole, E. J. Foster and C. Weder, J. Appl. Polym. Sci., 2017, 134, 45033 CrossRef.
- E. M. Fernandes, R. A. Pires, J. F. Mano and R. L. Reis, Prog. Pol. Sci., 2013, 38, 1415–1441 CrossRef CAS.
- S. Bottan, F. Robotti, P. Jayathissa, A. Hegglin, N. Bahamonde, J. A. Heredia-Guerrero, I. S. Bayer, A. Scarpellini, H. Merker, N. Lindenblatt, D. Poulikakos and A. Ferrari, ACS Nano, 2015, 9, 206–219 CrossRef CAS PubMed.
- J. M. Dugan, R. F. Collins, J. E. Gough and S. J. Eichhorn, Acta Biomater., 2013, 9, 4707–4715 CrossRef CAS PubMed.
- E. Entcheva, H. Bien, L. Yin, C.-Y. Chung, M. Farrell and Y. Kostov, Biomaterials, 2004, 25, 5753–5762 CrossRef CAS PubMed.
- M. Märtson, J. Viljanto, T. Hurme, P. Laippala and P. Saukko, Biomaterials, 1999, 20, 1989–1995 CrossRef.
- A. D. French, N. R. Bertoniere, R. M. Brown, H. Chanzy, D. G. Gray, K. Hattori and W. Glasser, in Kirk-Othmer Encyclopedia of Chemical Technology, ed., A. Seidel, John Wiley & Sons Inc., New York, 5th edn, 2004, vol. 5, pp. 360–394 Search PubMed.
- J. K. Jackson, K. Letchford, B. Z. Wasserman, L. Ye, W. Y. Hamad and H. M. Burt, Int. J. Nanomed., 2011, 6, 321–330 CAS.
- X.-M. Fan, H.-Y. Yu, D.-C. Wang, J. Yao, H. Lin, C.-X. Tang and K. C. Tam, ACS Appl. Mater. Interfaces, 2019, 11, 48192–48201 CrossRef CAS PubMed.
- J. V. Edwards, N. Prevost, K. Sethumadhavan, A. Ullah and B. Condon, Cellulose, 2013, 20, 1223–1235 CrossRef CAS.
- R. J. Moon, A. Martini, J. Nairn, J. Simonsen and J. Youngblood, Chem. Soc. Rev., 2011, 40, 3941–3994 RSC.
- B. G. Rånby, Acta Chem. Scand., 1949, 3, 649–650 CrossRef.
- T. Kovacs, V. Naish, B. O'Connor, C. Blaise, F. Gagné, L. Hall, V. Trudeau and P. Martel, Nanotoxicology, 2010, 4, 255–270 CrossRef CAS PubMed.
- M. Roman, S. Dong, A. Hirani and Y. W. Lee, Cellulose Nanocrystals for Drug Delivery, in Polysaccharide Materials: Performance by Design, ed. K. J. Edgar, T. Heinze and C. M. Buchanan, ACS, Washington, DC, 2009, pp. 81–91 Search PubMed.
- S. Dong, A. A. Hirani, K. R. Colacino, Y. W. Lee and M. Roman, Nano LIFE, 2012, 02, 1241006 CrossRef.
- K. A. Mahmoud, J. A. Mena, K. B. Male, S. Hrapovic, A. Kamen and J. H. T. Luong, ACS Appl. Mater. Interfaces, 2010, 2, 2924–2932 CrossRef CAS PubMed.
- K. B. Male, A. C. W. Leung, J. Montes, A. Kamen and J. H. T. Luong, Nanoscale, 2012, 4, 1373 RSC.
- J. K. Jackson, K. Letchford, B. Z. Wasserman, L. Ye, W. Y. Hamad and H. M. Burt, Int. J. Nanomed., 2011, 6, 321–330 CAS.
- P. Pooyan, R. Tannenbaum and H. Garmestani, J. Mech. Behav. Biomed. Mater., 2012, 7, 50–59 CrossRef CAS PubMed.
- R. Dash, M. Foston and A. J. Ragauskas, Carbohydr. Polym., 2013, 91, 638–645 CrossRef CAS PubMed.
- N. Lin, C. Bruzzese and A. Dufresne, ACS Appl. Mater. Interfaces, 2012, 4, 4948–4959 CrossRef CAS PubMed.
- Y. Wang, C. Chang and L. Zhang, Macromol. Mater. Eng., 2010, 295, 137–145 CrossRef CAS.
- R. Marchessault, F. Morehead and N. Walter, Nature, 1959, 184, 632–633 CrossRef CAS.
- J.-F. Revol, H. Bradford, J. Giasson, R. H. Marchessault and D. G. Gray, Int. J. Biol. Macromol., 1992, 14, 170–172 CrossRef CAS.
- N. A. Lockwood, J. C. Mohr, L. Ji, C. J. Murphy, S. P. Palecek, J. J. de Pablo and N. L. Abbott, Adv. Funct. Mater., 2006, 16, 618 CrossRef CAS.
- M. Prévôt, S. Ustunel, G. Freychet, C. Webb, M. Zhernenkov, R. Pindak, R. J. Clements and E. Hegmann, ACS Appl. Bio Mater., 2022 Search PubMed , Manuscript ID: mt-2022-00462p, in review.
| This journal is © The Royal Society of Chemistry 2022 |
