Open Access Article
Open Access ArticleFabrication of substrates for multiple cell patterning using a copolymer with a UV-degradable oligoethylene glycol side chain†
Hiroaki
Ogawa
a,
Yuka
Yamazawa
a,
Tadashi
Nakaji-Hirabayashi
 *abc,
Hiromi
Kitano
ad,
Yoshiyuki
Saruwatari
e and
Kazuyoshi
Matsuoka
e
*abc,
Hiromi
Kitano
ad,
Yoshiyuki
Saruwatari
e and
Kazuyoshi
Matsuoka
e
aGraduate School of Science and Engineering, University of Toyama, Toyama 930-0555, Japan. E-mail: nakaji@eng.u-toyama.ac.jp
bGraduate School of Innovative Life Science, University of Toyama, Toyama 930-0194, Japan
cFaculty of Engineering, Academic Assembly, University of Toyama, Toyama 930-0555, Japan
dInstitute for Polymer-Water Interfaces, Toyama 939-2376, Japan
eR & D Laboratory, Osaka Organic Chemical Industry, Inc., Kashiwara, Osaka, Japan
First published on 10th May 2022
Abstract
A random copolymer of ω-methoxy-(ethylene glycol)8 acrylamide (MEGAm) and 3-methacryloxypropyltrimethoxysilane (MPTMS) was prepared by free radical polymerization, and glass substrates were modified with the copolymer by silane coupling. MEGAm is a bio-inactive material because of its oligoethylene glycol chain, which can be cleaved by UV irradiation. Therefore, the amount of protein adsorption and cell adhesion to the copolymer-modified glass substrate increased after UV irradiation. Furthermore, when a part of the copolymer-modified glass substrate was subjected to UV irradiation and cells were seeded on the substrate, the cells adhered selectively to the UV-irradiated part. The subsequent UV-irradiation of the other part containing the area where cells adhered caused another kind of cell to adhere to the latter part of the substrate. Thus, we could achieve multiple cell patterning.
Introduction
Biological phenomena are dynamic and complicated; therefore, a complete understanding of their detailed mechanisms is challenging. In multicellular organisms, various factors expressed by cell–cell interactions often cause various biological phenomena; for example, the transformation from normal to cancer cells,1 building up of organs or tissues,2 coordination of organ functions,3 and formation of cell–cell interactions. Considering these aspects, understanding cell–cell interactions is essential to control cells and organs, and develop biomaterials that contact the internal organs. Therefore, various studies on cell–cell interactions have been conducted.2–7Theoretically, cells influence their neighboring cells through cell–cell interactions and secretory factors in their boundaries, while individual cells are controlled only by secretion factors in the non-boundary regions. Therefore, it is possible to simultaneously evaluate and understand the relationship and importance of two kinds of actions (cell–cell interactions and secretion factors) for cell control in a single culture system. However, constructing an in vitro experimental system to evaluate cell–cell interaction individually in the same system has been considerably challenging; consequently, it has not been well understood.
We have been studying the construction of surfaces that do not respond to biological components such as cells and proteins. In particular, we have reported that zwitterionic polymers show non-responsiveness to biological phenomena such as cell adhesion and protein adsorption.8–11 Additionally, a substrate could be created that could pattern cell positions, specifically when a UV or ion beam is irradiated on a surface modified with zwitterionic polymers.12,13 Thus, we conceived a multi-cell co-culture system that helps evaluate cell–cell interactions in vitro by applying this patterning technology.
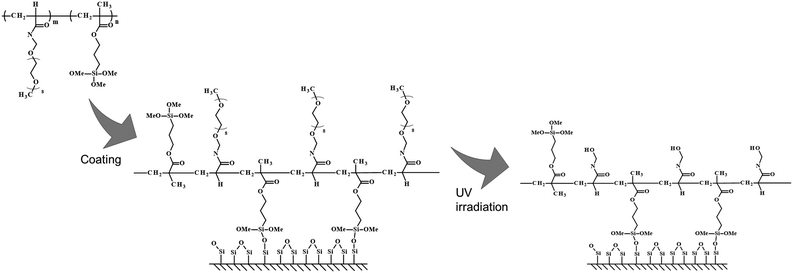
Experimental procedures examined here are shown below (Scheme 1). First, a copolymer of ω-methoxy-(ethylene glycol)8 acrylamide (MEGAm) (Fig. 1a) was covalently fixed to a glass substrate by silane coupling. The oilogoethylene glycol (oligoEG) chain of MEGAm could be cleaved by UV irradiation.14–18 The resulting cleaved oligoEG chain is harmless for cells. Thereafter, a part of the polymer-modified substrate was UV-irradiated, and cells were seeded on the substrate. After cells were cultured to confluence, the substrate was again UV-irradiated. Finally, a different cell type was seeded on the surface (Scheme 1). Using this procedure, it was expected that multi cells could adhere in a position-specific manner. The two types of cells adhered to the substrate were regulated by cell–cell interactions and secretion factors expressed by the cell types. Cells adhered to areas away from boundaries were regulated only by the secretion factor. Conversely, cells adhered on the border of each cell were regulated by both cell–cell interactions and secretion factors.
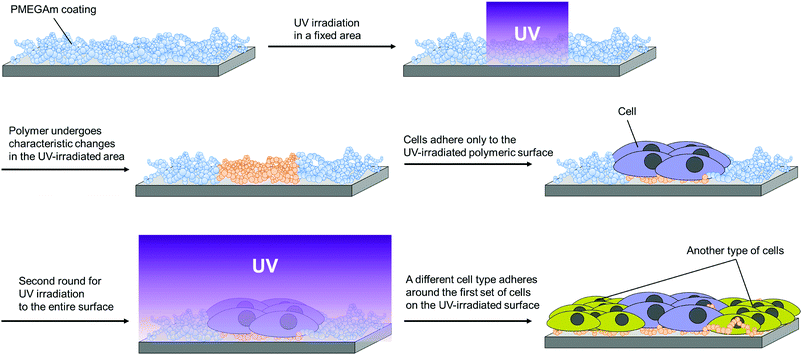 | ||
| Scheme 1 Schematic image of patterning of the MEGAm-MPTMS copolymer surface by UV irradiation and creation of a heterologous cell-patterning surface. | ||
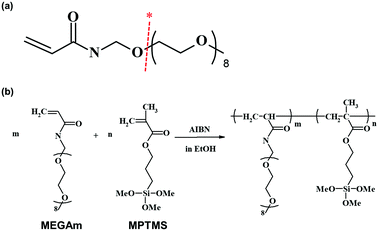 | ||
| Fig. 1 (a) Chemical structure of MEGAm. Note: * cleavage by irradiation at 360–370 nm. (b) Preparation of the random copolymer of MEGAm and MPTMS. | ||
Therefore, it was strongly expected that the regulation of cells by cell–cell interactions and secretion factors could be simultaneously and independently investigated by observing cells both at the junctions and areas away from the boundaries. We aimed to prepare a substrate that could easily pattern multiple cells using a polymer easily degradable by UV irradiation.
Results and discussion
Wettability of polymer-modified substrates evaluated by the sessile drop method
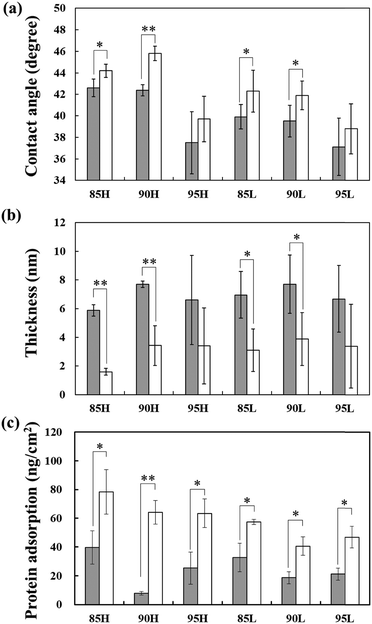
Surfaces modified with the copolymer of ω-methoxy-(ethylene glycol)8 acrylamide (MEGAm) and 3-methacryloxypropyltrimethoxysilane (MPTMS) (P(MEGAm-r-MPTMS)) had contact angles of 37–46° (Fig. 2a). The contact angle of the polyethylene glycol (PEG)-modified surface was reported to be 40–45°,19 which was similar to that of the surfaces examined here, modified with P(MEGAm-r-MPTMS). This suggests that the polymer with oligoEG side chains was coated on the substrates. Both the high and low molecular weight polymer showed the same tendency: their contact angles increased with an increase in the ratio of MPTMS. This occurred because unreacted MPTMS side chains pointed toward the glass surface. Cross-linking between the unreacted silane coupling groups could also be inferred.20 Moreover, the average contact angles of the surfaces modified with 95H and 95L (P(MEGAm95-r-MPTMS5) of high and low molecular weight) were relatively low, and the standard deviations of these data were significantly large compared with those with 85H and 90H. This point was considered based on the results of X-ray photoelectron spectroscopy (XPS) and ellipsometry measurements as described in the section “Measuring the thickness of the substrate surface by ellipsometry”.Additionally, the contact angle of the polymer-modified surfaces irradiated with UV increased compared to those without UV irradiation. This occurred because oligoEG side chains were removed by UV irradiation on the former surface. The contact angles in this study correlated with the contact angle of a surface coated with poly(2-hydroxyethyl methacrylate).20 Therefore, we speculated that irradiation reformed the PEG side chain to a hydroxyl group.
Characterization of the copolymer-modified substrate surface by X-ray photoelectron spectroscopy
The atomic composition of the copolymer-modified surface was analyzed by X-ray photoelectron spectroscopy (XPS) (Table S1 in ESI†). The ratios of carbon on all kinds of surfaces decreased through UV irradiation because the oligoEG side chains were removed by cleavage. The shape of the carbon peaks also changed before and after UV irradiation. The peak intensity of a substrate UV-irradiated at nearly 285 eV decreased compared to substrates without UV irradiation (Fig. S1 in ESI†). Notably, the standard deviation of the data for surfaces modified with several copolymers (95H and low-molecular weight polymers) was relatively large, which is likely related to the surface condition of the copolymers. This information is discussed in detail in the next section.Measuring the thickness of the substrate surface by ellipsometry
The thickness of the UV-irradiated copolymer surface decreased compared to that of the surface not subjected to UV irradiation (Fig. 2b). It is inferred that oligoEG side chains were cleaved by UV irradiation. As the oligoEG chains directed at a liquid phase resembled a “polymer brush” accumulated by the “grafting-to” method,10 it was expected that the coverage ratio of the substrates with the low-molecular weight copolymer would be higher than that on substrates with the high-molecular weight polymer. However, based on the contact angle, XPS, and ellipsometry results, the surfaces modified with 85H and 90H copolymers were uniformly covered, whereas the glass surface was partially exposed on the surface modified with other copolymers. These results can be explained as follows: (1) the surfaces modified with 95H, 85L, 90L, and 95L copolymers showed relatively high wettability, which decreased the contact angle compared with that on surfaces modified with 85H and 90H copolymers, (2) the standard deviations of the contact angle, XPS, and ellipsometry results on these copolymer surfaces were significantly larger than those on the 85H and 90H copolymer surfaces. The surface was uniformly covered by 85H and 90H copolymers likely because many silane coupling side chains were not only used as a surface modification but also acted as cross-linkers with copolymers because of the relatively high content of MPTMS and high-molecular weight polymer.Protein adsorption onto polymer-modified substrates
Protein adsorption onto a substrate surface was investigated by micro bicinchoninic acid (BCA) assay (Fig. 2c). The amount of protein adsorption on the UV-irradiated surface was higher than that on surfaces without UV irradiation. This occurred because the oligoEG side chain was cleaved by UV irradiation. As in the case of PEG, oligoEG could suppress protein adsorption owing to their similar structure yet different degrees of polymerization. PEG has an antifouling property21–25 because it is hydrophilic26 (in other words, it is characterized by low polymer–water interfacial energy27) and has a steric-repulsive effect on proteins.26,28The amount of protein adsorption on the surface modified with 90H (MEGAm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MPTMS = 90
MPTMS = 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, high molecular weight polymer) was the lowest; conversely, the standard deviation of the thickness on the surface was not as high as compared to other copolymer-modified substrates. This occurred because the amount of protein adsorption depends on a high surface coverage and an exclusive volume effect associated with the molecular weight of the polymer. The amount of protein adsorption on the substrates modified with 85H (MEGAm
10, high molecular weight polymer) was the lowest; conversely, the standard deviation of the thickness on the surface was not as high as compared to other copolymer-modified substrates. This occurred because the amount of protein adsorption depends on a high surface coverage and an exclusive volume effect associated with the molecular weight of the polymer. The amount of protein adsorption on the substrates modified with 85H (MEGAm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MPTMS = 85
MPTMS = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, high molecular weight copolymer) was higher than that on substrates with 90H, probably because the ratio of surface coverage was insufficient owing to the coupling reaction between the residual silane coupling groups. The amount of protein adsorption on the substrates modified with 95H (MEGAm
15, high molecular weight copolymer) was higher than that on substrates with 90H, probably because the ratio of surface coverage was insufficient owing to the coupling reaction between the residual silane coupling groups. The amount of protein adsorption on the substrates modified with 95H (MEGAm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MPTMS = 95
MPTMS = 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, high molecular weight copolymer) was also higher than that on substrates with 90H because the coverage ratio of the copolymer on the surface was smaller than that on substrates with 90H due to the low ratio of MPTMS residue. Consequently, the glass substrates were exposed to a larger extent; hence, proteins were adsorbed on the surface.
5, high molecular weight copolymer) was also higher than that on substrates with 90H because the coverage ratio of the copolymer on the surface was smaller than that on substrates with 90H due to the low ratio of MPTMS residue. Consequently, the glass substrates were exposed to a larger extent; hence, proteins were adsorbed on the surface.
Cell adhesion on the surface modified with copolymers
Cells hardly adhered and extended on surfaces modified with the high molecular weight polymer without UV irradiation (Fig. 3 and Fig. S2, ESI†). Cell adhesion on surfaces modified with the low molecular weight polymer was also suppressed. Conversely, cell adhesion and cell extension were observed on all kinds of surfaces after UV irradiation due to the cleavage of the oligoEG side chains by UV irradiation. The surface with modified 90H clearly showed cells not adhering to the substrates without UV irradiation; however, cells adhered to the substrates after UV irradiation. This result was concordant with the amount of protein adsorbed on these surfaces. Therefore, we infer that substrates modified with 90H were the most suitable for multiple cell patterning.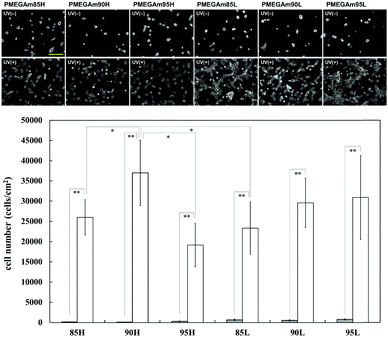 | ||
| Fig. 3 Phase contrast images (scale bar: 100 μm) of adhered cells cultured for 6 h, and the number of adhered cells cultured for a day on substrates modified with the copolymers before (gray bar) and after (white bar) UV irradiation. Phase contrast images of adhered cells cultured for a day on the substrates are shown in Fig. S2 in the ESI.† Sample abbreviation: 85, 90, and 95 represent the molar ratio of MEGAm, and H and L represent the high- and low-molecular weight polymers, respectively. | ||
Multiple cell patterning on copolymer-modified substrate surfaces
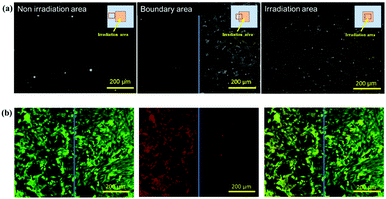
First, a part of the surface modified with 90H copolymer was covered with a UV shutter, which has an inner square window of 2.5 × 2.5 mm, and the entire area of the substrate was UV-irradiated. After rinsing, NIH3T3 was seeded on the substrates and cultured for a day. It was found that cells adhered to the UV-irradiated part of the substrates (Fig. 4a). Then, the substrate surface containing the NIH3T3 cells-adhered area was again subjected to UV irradiation (Scheme 1). Thereafter, HEK293 cells stained by a cell tracker were seeded, and they were cultured for a day and stained by Calcein AM (Fig. 4b). Both NIH3T3 cells and HEK293 cells were stained with green by Calcein AM. Conversely, red cells in Fig. 4b show cells dyed with a cell tracker. Only HEK293 cells were dyed by the cell tracker. Therefore, the left half of the picture in Fig. 4b shows only HEK293 cells, while NIH3T3 cells were mostly present in the right half of the figure. However, a few HEK293 cells were also seen in the right half of the picture. It is considered that HEK293 cells adhered between or on NIH3T3 cells when they were seeded after NIH3T3 cells.For cell patterning, the first seeding cells, which were NIH3T3 cells in this model experiment, were exposed to UV light because of the surface modification. Therefore, the first seeding cells may have been damaged, such as damage to the cell cycle and protein expression and secretion. Hence, we investigated the survival of cells exposed to UV light. As shown in Fig. S3 in the ESI,† almost all cells survived after irradiation with UV light for 15 min under a fluorescence microscope. Additionally, cells cultured for a day after UV irradiation proliferated and reached a confluent state. Although further studies are needed to determine whether this method can be applied to various cells such as stem cells and generally weak and sensitive cells (neural cells etc.), our results show that cell patterning can be performed using our method. We will next evaluate the relationships between regulation by neuron-astrocyte interactions and regulation of neurons by paracrine factors from astrocytes using this cell patterning substrate.
Conclusions
The surface of a glass substrate could be modified with the MEGAm and MPTMS copolymer. OligoEG chains of MEGAm residues were cleaved by UV irradiation; thus, the on/off behavior of cell adhesion could be regulated using this property. In particular, a 90H copolymer-modified surface exhibited this behavior most efficiently. Furthermore, multiple cell patterning was conducted using surfaces modified with the 90H polymer. The patterning was almost achieved using NIH3T3 and HEK293 cells as model cells. The current multiple cell patterning technique requires further optimization to prevent the adherence of one cell over the other during the seeding stage.Perfect patterning could help overcome the abovementioned technical problems. Therefore, a substrate with the ability to pattern multiple cells and to help evaluate cell–cell interactions could be developed. In particular, we strongly expect that the substrate we developed could be used in the detailed investigation of the correlation between stem cells and feeder cells and for co-culturing neutrophils with bronchial epithelia to study inflammation.
Experimental
Materials
ω-Methoxy-(ethylene glycol)8 acrylamide (MEGAm) was donated by Osaka Organic Chemical Industry Co. Ltd (Osaka, Japan). 3-Methacryloxypropyltrimethoxysilane (MPTMS) and 2,2′-azobisisobutyronitrile (AIBN) were obtained from Wako Pure Chemical Industry Co., Ltd (Osaka, Japan). Other reagents and solvents were commercially available. Ultrapure water (18 MΩ cm, Millipore System) was used for preparing sample solutions.Preparation of P(MEGAm-r-MPTMS) by free radical polymerization
Copolymers of MEGAm and MPTMS with a high and low molecular weight were synthesized. MEGAm and MPTMS were dissolved in dehydrated ethanol (10 mL) at the ratio shown in Table 1, while AIBN was added under the conditions shown in Table 1. O2 gas was removed by purging N2 gas. The reaction mixture was incubated at 70 °C for 24 h to prepare the high molecular weight polymer.| Sample | MEGAm![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MPTMS MPTMS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AIBNb AIBNb |
M n (Mw/Mn) |
|---|---|---|
| a Total amount of monomer, 0.011 mol. b Molar ratio. | ||
High Mn Polymer 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (85H) 15 (85H) |
510![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
5.3 × 104 (4.31) |
High Mn Polymer 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (90H) 10 (90H) |
540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6.1 × 104 (5.18) |
High Mn Polymer 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (95H) 5 (95H) |
570![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
5.9 × 104 (4.84) |
Low Mn Polymer 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (85L) 15 (85L) |
14.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.5 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
6.8 × 103 (2.31) |
Low Mn Polymer 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (90L) 10 (90L) |
15.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.7 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
9.1 × 103 (3.21) |
Low Mn Polymer 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 (95L) 5 (95L) |
15.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
8.4 × 103 (2.83) |
Furthermore, during the low molecular weight polymer preparation, AIBN was added at 5 (w/w) % of the total amount of the monomer substance, and the solution was allowed to react at 70 °C for 4 h. AIBN was further added at 1 (w/w) % of the total amount of the monomers and was continuously allowed to polymerize for a further 4 h (Fig. 1b). The prepared polymer solution was shielded from light and stored in a refrigerator.
Surface modification with P(MEGAm-r-MPTMS) through silane coupling reaction
Glass substrates were immersed in piranha solution (H2O2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2SO4 = 3
H2SO4 = 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7) for 1 h. Then, the substrates were continuously washed by water, acetone, and methanol, and the substrates were immersed overnight in dehydrated ethanol containing 1 (w/v)% P(MEGAm-r-MPTMS) at 50 °C. Finally, the substrates were washed with ethanol and water (Scheme 2).
7) for 1 h. Then, the substrates were continuously washed by water, acetone, and methanol, and the substrates were immersed overnight in dehydrated ethanol containing 1 (w/v)% P(MEGAm-r-MPTMS) at 50 °C. Finally, the substrates were washed with ethanol and water (Scheme 2).
 | ||
| Scheme 2 Schematic illustration of the surface modification of a glass substrate with P(MEGAm-r-MPTMS) through silane-coupling. | ||
Moreover, a glass-based dish (AGC Inc., Tokyo, Japan) was used for the experiment on multiple cell patterning. The surface of the glass-based dishes was treated with an oxygen plasma device (PM100, YAMATO Scientific, Co., Ltd, Tokyo, Japan) for 3 min. After washing by water and ethanol, dehydrated ethanol containing 1 (w/v)% P(MEGAm-r-MPTMS) was added to the glass-based dish and incubated overnight at 50 °C. Thereafter, the dish was washed with ethanol and water. The glass-based dish coated with the copolymer was sterilized with 70% ethanol solution before use in cell culture.
Reformation of the copolymer anchored surface by UV irradiation
The surface of the substrate modified with the copolymer was reformed by a UV light system (SP9-250UB, Usio Inc.; center wavelength, 365 nm; equipped SFH lens; strength, 250 W; 5–10 min) and a halogen light system of the fluorescence microscope (U-RFL-T equipped to IX71, Olympus Corporation; band-pass filter, 370 nm; strength. 150 W; 15–20 min). UV light was irradiated on the entire surface of the substrates at an exposure dose of 450 J cm−2 (Scheme 2).Evaluation of surface wettability
Contact angle measurements were performed to evaluate surface wettability (DMs-401, Kyowa Interface Science Co., Ltd). During the sessile drop method, 1 μL of water was put on the substrate surface, and 30 s later, the contact angle was measured. The contact angle measurement was repeated at 10 different positions on the same sample, and the average contact angles were evaluated based on all data obtained from eight independent samples.Surface characterization of the copolymer-modified surface by XPS
Atomic components of a surface modified with the polymer were determined by XPS (ESCALAB 250Xi, Thermo Fisher Scientific, Inc., Waltham, Massachusetts, USA). The measuring conditions were as follows: detection angle: 90 degrees; X-ray source type: monochromated; micro-focused Al K-alpha; and X-ray spot size: 650 μm. The Avantage (Ver. Sl. 05) software (Thermo Fisher Scientific, Waltham, MA, USA) was used for peak analyses. To evaluate the ratio of the atomic composition, five positions on a polymer-coated surface were measured, and eight independent samples were re-measured to ensure reproducibility. These data were averaged and expressed as the mean ± standard deviation.Thickness measurements of copolymer-modified surfaces by ellipsometry
The thickness of the copolymers coated on the substrates was measured in a dry state by a polarization analysis-type film thickness-measuring device (α-SE-T, J. A. Woollam Co. Ltd, Lincoln, NE, USA). A 25 mm × 25 mm silicon wafer was used as a substrate modified with the copolymers. The average thickness of each surface was obtained by measuring 11 points on a sample and re-measurement using eight independent specimens.Evaluation of protein adsorption on the surface modified with polymers
The amount of protein (bovine serum albumin (BSA), pI: 4.7–4.929,30) adsorbed on the surface of the substrates was evaluated by the microBCA method as described in the ESI.† The data obtained from ten independent specimens were averaged and evaluated to determine the standard deviation.Evaluation of cell adhesion on the polymer-modified substrate surface
The substrates modified with P(MEGAm-r-MPTMS) were sterilized with 70% ethanol solution on a clean bench. The substrates immersed in PBS for 10 min for priming were put into a culture medium (Minimum essential medium (MEM) was used as the primary medium) containing 10% fetal bovine serum (FBS) until cell seeding. NIH3T3 cells (passages 131–140) cultured for 3 days on a TCPS dish were harvested. Thereafter, the cells were dislodged from the dish by incubation for 2 min with 0.25% trypsin–EDTA solution. Next, a culture medium including FBS was added to inactivate the trypsin, and the cells were dispersed into single cells with a pipette. After centrifugation for 5 min (500 × g), the cells were dispersed in 2 mL fresh culture medium, and the number of cells was counted by a hemocytometer. The cells were seeded on substrates modified with the copolymer at a cell density of 3 × 104 cell cm−2. Finally, the cells on the substrates were cultured at 37 °C and 5% CO2 in an incubator. Adherent cells were observed at 6 h after seeding (initial adhesion on the surface) and after culture for a day.Surface modification with the polymer for multiple cell-patterning
In this experiment, a glass-based dish (AGC, Inc.) was used as the base material because it is optimal for performing irradiation from the bottom of the dish using an inverted fluorescence microscope. The transmission of UV light (300–400 nm) through various types of glasses and the TCPS dish was measured with a UV spectrophotometer (UV-2600, Shimadzu, Kyoto, Japan). The results showed that the transmission of the glass-based dish was the highest among all substrates (Scheme S4c in ESI†).
Statistical analysis
Data on the contact angle, polymer thickness, protein adsorption, and adhesive cell density were expressed as the mean ± standard deviation. One-way analysis of variance (ANOVA) was used to assess statistical significance, and Tukey's honestly significant difference (HSD) test was used for multiple comparisons. The difference was considered significant at *p < 0.05 and **p < 0.01. All statistical analyses were performed using the JMP Pro 15.2 software (SAS Institute Inc., Cary, NC, USA).Conflicts of interest
The authors declare no conflicts of interest associated with this manuscript.Acknowledgements
We wish to thank Grants-in-Aid from the Japan Society for Promotion of Science (15H05353, 17KK0130, 18K19907 and 22H03951). We are grateful to Osaka Organic Chemical Industry for the donation of MEGAm and their continuous support to pursue this work. We are indebted to Mr Tomohiro Kamada, who works in our department, for his technical assistance.Notes and references
- S. Ohsawa, Y. Sato, M. Enomoto, M. Nakamura, A. Betsumiya and T. Igaki, Nature, 2012, 490, 547–551 CrossRef CAS PubMed.
- T. Takebe, K. Sekine, M. Enomura, H. Koike, M. Kimura, T. Ogaeri, R. R. Zhang, Y. Ueno, Y. W. Zheng, N. Koike, S. Aoyama, Y. Adachi and H. Taniguchi, Nature, 2013, 499, 481–484 CrossRef CAS PubMed.
- S. N. Bhatia, U. J. Balis, M. L. Yarmush and M. Toner, FASEB J., 1999, 13, 1883–1900 CrossRef CAS PubMed.
- T. Iriki, K. Ohnishi, Y. Fujiwara, H. Horlad, Y. Saito, C. Pan, K. Ikeda, T. Mori, M. Suzuki, H. Ichiyasu, H. Kohrogi, M. Takeya and Y. Komohara, Lung Cancer, 2017, 106, 22–32 CrossRef PubMed.
- A. C.-A. Wan, Trends Biotechnol., 2016, 34, 711–721 CrossRef CAS PubMed.
- T. H. Qazi, D. J. Mooney, G. N. Duda and S. Geissler, Biomaterials, 2017, 140, 103–114 CrossRef CAS PubMed.
- P. Shi, E. Ju, Z. Yan, N. Gao, J. Wang, J. Hou, Y. Zhang, J. Ren and X. Qu, Nat. Commun., 2016, 7, 13088 CrossRef CAS PubMed.
- H. Kitano, H. Suzuki, K. Matsuura and K. Ohno, Langmuir, 2010, 26, 6767–6774 CrossRef CAS PubMed.
- H. Suzuki, M. Murou, H. Kitano, K. Ohno and Y. Saruwatari, Colloids Surf., B, 2011, 84, 111–116 CrossRef CAS PubMed.
- M. Nishida, T. Nakaji-Hirabayashi, H. Kitano, K. Matsuoka and Y. Saruwatari, J. Biomed. Mater. Res., Part A, 2016, 104, 2029–2036 CrossRef CAS PubMed.
- M. Nishida, T. Nakaji-Hirabayashi and H. Kitano, Colloids Surf., B, 2017, 152, 302–310 CrossRef CAS PubMed.
- H. Suzuki, L. Li, T. Nakaji-Hirabayashi, H. Kitano, K. Ohno, K. Matsuoka and Y. Saruwatari, Colloids Surf., B, 2012, 94, 107–113 CrossRef CAS PubMed.
- L. Li, T. Nakaji-Hirabayashi, H. Kitano, K. Ohno, T. Kishioka and Y. Usui, Colloids Surf., B, 2016, 144, 180–187 CrossRef CAS PubMed.
- D. Ednah Simangoye Ngobissi, J. Soufi, L. Vanoye and D. Richard, Catal. Lett., 2029, 147, 1608–1614 CrossRef.
- T. Nishiyama, K. Matsuura, E. Sato, N. Kometani and H. Horibe, J. Photopolym. Sci. Technol., 2017, 30, 285–289 CrossRef CAS.
- R. Haseneder, B. Fdez-Navamuel and G. Härtel, Water Sci. Technol., 2007, 55, 83–87 CrossRef CAS PubMed.
- J. A. Giroto, A. C.-S. C. Teixeira, C. A.-O. Nascimento and R. Guardani, Ind. Eng. Chem. Res., 2010, 49, 3200–3206 CrossRef CAS.
- D. F. Dwyer and J. M. Tiedje, Appl. Environ. Microbiol., 1983, 46, 185–190 CrossRef CAS PubMed.
- S. R. Benhabbour, H. Sheardown and A. Adronov, Macromolecules, 2008, 41, 4817–4823 CrossRef CAS.
- Y. Yamazawa, H. Kato, T. Nakaji-Hirabayashi, C. Yoshikawa, H. Kitano, K. Ohno, Y. Saruwatari and K. Matsuoka, J. Mater. Chem. B, 2019, 7, 4280–4291 RSC.
- W. Ma, S. Rajabzadeh, A. R. Shaikh, Y. Kakihana, Y. Sun and H. Matsuyama, J. Membr. Sci., 2016, 514, 429–439 CrossRef CAS.
- Y. Dang, M. Quan, C. M. Xing, Y. B. Wang and Y. K. Gong, J. Mater. Chem. B, 2015, 3, 2350–2361 RSC.
- J. A. Prince, S. Bhuvana, K. V.-K. Boodhoo, V. Anbharasi and G. Singh, J. Membr. Sci., 2014, 454, 538–548 CrossRef CAS.
- A. R. Statz, R. J. Meagher, A. E. Barron and P. B. Messersmith, J. Am. Chem. Soc., 2005, 127, 7972–7973 CrossRef CAS PubMed.
- C. S. Gudipati, C. M. Creenlief, J. A. Johnson, P. Prayoncpan and K. L. Wooley, J. Polym. Sci., Part A: Polym. Chem., 2004, 42, 6193–6208 CrossRef CAS.
- H. Zhang and M. Chiao, J. Med. Biol. Eng., 2015, 35, 143–155 CrossRef PubMed.
- S. Krishnan, C. J. Weinman and C. K. Ober, J. Mater. Chem., 2008, 18, 3405–3413 RSC.
- G. dong Kang and Y. ming Cao, Water Res., 2012, 46, 584–600 CrossRef PubMed.
- D. Voet and J. G. Voet, Biochemistry, Wiley, New York, 4th edition, 2013 Search PubMed.
- T. OOshima, K. Suzuki and Y. Seyama, Biochemistry Data Book, Tokyo Kagaku Dojin, Tokyo, 4th edition, 2007 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ma00198e |
| This journal is © The Royal Society of Chemistry 2022 |