 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Engineered liver tissue in vitro to mimic liver functions and its biomedical applications
Wenguang
Yang
 *,
Xiaowen
Wang
and
Zhen
Wang
*,
Xiaowen
Wang
and
Zhen
Wang
School of Electromechanical and Automotive Engineering, Yantai University, Yantai 264005, China. E-mail: yangwenguang@ytu.edu.cn
First published on 31st March 2022
Abstract
The liver is an important organ with metabolic functions, and plays the important role of deoxidizing, storing liver sugar, synthesizing secretory proteins and other functions inside the body. The prevalence of liver disease has been increasing in recent years. For acute liver failure, liver cancer and other serious liver diseases, the best treatment is liver transplantation. However, still there is shortage of organs in clinical practice. Under such a severe background, it is very important to cultivate liver tissue in vitro to make up for the shortage of organs and to test the hepatotoxicity of drugs. In this review, we will introduce the structure of liver and the function of liver cells, summarize several methods of liver tissue culture in vitro, and introduce the application of liver tissue culture in vitro. Finally, we forecast the development prospects of the liver tissue.
1. Introduction
The liver plays an important role in deoxidizing, storing liver sugar, and synthesizing secretory proteins.1–3 The surface of the liver is covered by a thin capsule of dense connective tissue. The capsule penetrates the liver to form a reticular scaffold that separates the liver parenchyma into many hepatic lobules, which have similar morphology and the same function. The liver is made up of 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 1 million hexagonal lobules. There is a central vein in the center of the lobule, which is surrounded by a roughly radially arranged plate of liver cells (hepatic lamina). The liver is also composed of different cell types, including hepatocytes, sinusoidal endothelial cells, hepatic stellate cells, Kupffer cells, biliary epithelial cells, and dendritic cells (Fig. 1).4–8 Furthermore, it is important to maintain the polarity of hepatocytes under in vitro culture conditions.9,10 Due to the structural arrangement and cellular heterogeneity of the liver, liver cells are exposed to a gradient of nutrients, hormones, and growth factors through the combined portal vein and hepatic artery blood supply. According to estimation, there are about 90 million people who are infected with HBV (hepatitis B virus) in China11 and there are 7.6 million HCV patients (4.56 million chronic patients).12 Every year, 330
000 to 1 million hexagonal lobules. There is a central vein in the center of the lobule, which is surrounded by a roughly radially arranged plate of liver cells (hepatic lamina). The liver is also composed of different cell types, including hepatocytes, sinusoidal endothelial cells, hepatic stellate cells, Kupffer cells, biliary epithelial cells, and dendritic cells (Fig. 1).4–8 Furthermore, it is important to maintain the polarity of hepatocytes under in vitro culture conditions.9,10 Due to the structural arrangement and cellular heterogeneity of the liver, liver cells are exposed to a gradient of nutrients, hormones, and growth factors through the combined portal vein and hepatic artery blood supply. According to estimation, there are about 90 million people who are infected with HBV (hepatitis B virus) in China11 and there are 7.6 million HCV patients (4.56 million chronic patients).12 Every year, 330![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people die of cirrhosis and primary liver cancer. In this severe situation, it is very urgent to understand the physiological function of liver cells and the related disease mechanism, and to provide effective treatment. Unfortunately, traditional research models have many limitations. Primary human hepatocytes cannot be cultured in vitro for a long time to maintain cell morphology and function.13,14 The genetic metabolic mechanisms of the animal models are different from those of humans, which prevents the results from being translated into clinical practice. Therefore, more closely resembling human hepatocytes are needed for experimental and therapeutic studies.
000 people die of cirrhosis and primary liver cancer. In this severe situation, it is very urgent to understand the physiological function of liver cells and the related disease mechanism, and to provide effective treatment. Unfortunately, traditional research models have many limitations. Primary human hepatocytes cannot be cultured in vitro for a long time to maintain cell morphology and function.13,14 The genetic metabolic mechanisms of the animal models are different from those of humans, which prevents the results from being translated into clinical practice. Therefore, more closely resembling human hepatocytes are needed for experimental and therapeutic studies.
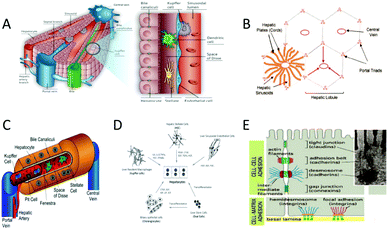 | ||
| Fig. 1 The structure of the liver. (A) The structure of the liver and the distribution of major liver cells. (Reproduced with permission from ref. 4.) (B) The structure of the hepatic lobule. (Reproduced with permission from ref. 26.) (C) Structural diagram of hepatic sinuses and distribution of major cells. (Reproduced with permission from ref. 27.) (D) The types of cells in the liver and their relationships. (Reproduced with permission from ref. 26.) (E) The interaction between cell and cell, and the interaction between cell and the extracellular matrix (ECM). (Reproduced with permission from ref. 26.) | ||
For drug screening, some drugs will still induce human toxicity and side effects after appearing on the market, and about 50% of them are due to the liver toxicity of the drug. It is particularly important to test drugs for hepatotoxicity.15 Initially, researchers used live animal models to investigate treatments or drugs for liver diseases. However, the metabolic rules of the livers of mice, pigs and other animals used in the research are greatly different from those of human livers in many aspects, and the results may not be consistent with the actual effect. In addition, using live animals as models is very cruel to animals, which does not meet the humanitarian spirit. The liver models for drug screening are two-dimensional monolayer cultured cells. However, the two-dimensional monolayer culture model is limited by the distribution of receptors on the surface and cannot simulate the clustering state of the cells, which greatly limits the wide application of this technology. Therefore, the newly emerging liver tissue engineering has become a research hotspot in recent years because of its cell composition and similar three-dimensional structure, which is closed to human physiology. Compared with live animal models, a liver tissue constructed in vitro has almost the same mechanism as the human liver, possessing the genetic characteristics of the individual of origin, and avoids harm to animals. Compared with the two-dimensional monolayer tissue, liver tissue constructed in vitro can maintain cell polarity for a long time, and ensure cell surface protein transmission, avoiding the loss of liver tissue function. In recent years, with the continuous development of life science, material science and engineering science, researchers have made one by one breakthroughs in tissue construction and organ reconstruction in vitro.16–18 Many tissue engineering products have been applied in clinical medicine, such as bone tissue and skin tissue. Liver tissue engineering is an important research field in tissue engineering, whose goal is to construct a transplantable liver tissue or organoid with the function of the human original liver,19 so that patients who have liver injury can be treated and the deficiency of the clinical organ shortage can be made up.20–25
In this review, we will roughly divide the content into five parts. First, we will introduce the structure of the liver, and the types and functions of liver cells. Then, we will outline the cells and materials that are used in liver tissue engineering. With the above content as the basis, we will present eight methods to culture liver tissue in vitro, and list their applications in tissue reconstruction and drug preparation. Finally, the prospects of liver tissue culture will be prospected.
2. Cells and materials for liver tissue engineering
2.1 Cells for liver tissue engineering
The selection of cells is very important in the process of liver tissue culture in vitro. There are several kinds of cells that can be used to grow liver tissue: bone marrow-derived stem cells, liver oval progenitor cells, liver cell lines and mature liver cells, embryonic stem cell (ESC), induced pluripotent stem cells (iPSC), bone marrow-derived very small embryonic-like stem cells (BM VSEL), and hepatic liver stem cells (HLSCs).4,282.2 Materials for liver tissue engineering
2.2.1.1 Alginate. Alginate is a natural polymer found in brown algae and often made into gels, which are similar to the structure of the extracellular matrix. Alginate is already widely used in wound healing, tissue engineering and drug delivery. High purity alginate has good biocompatibility and can be used in humans. Kinasiewicz et al. coated C3A (human immortalized liver cells) with alginate and cultured them statically for 7 days.37 The cells then proliferated to form multicellular aggregates. The research demonstrated that alginate-coated C3A liver cancer cells could produce albumin, which is the main serum protein synthesized by the liver. The result proved that alginate microencapsulation could promote three-dimensional cell interaction and significantly improve cell metabolism. Fluidization allowed for better nutrient and oxygen supply to the intracellular cells, which increased the synthesis function. Lan et al. encapsulated HepG2 cells with alginate.38 They demonstrated the possibility of HepG2 cell lines that could be encapsulated in a 3D alginate for drug screening. Liu et al. used calcium alginate in the study of simulating liver lobules in vitro.39 They proposed a method of preparing calcium alginate cell sheets by electro-deposition, which were used to form liver lobules. This method provided a new “bottom-up” paradigm for constructing three-dimensional macroscopic liver tissue similar to that of a living organism. The assembled liver lobules were expected to serve as extracorporeal models of liver organs, and would promote new applications of electro-deposition methods in tissue engineering. Masumi Yamada et al. proposed an efficient method for the extracorporeal culture of a hepatic cord structure by using anisotropic alginate saline gel microfibers to generate complex hepatic microorgans,40,41 which were composed of primary hepatocytes and parenchymal cells in vivo by microfluidic technique. The method provided an ideal microenvironment for hepatocytes, including cell and cell matrix interactions. Jeon et al. also used alginate to generate multi-layered three-dimensional structures of HepG2 cells by bioprinting.41 After printing, the 3D alginate hydrogel was immersed in 1% calcium chloride solution for solid crosslinking. It turned out that 3D bioprinting could be used to create the liver with 3D structures, which contributed to the stable regeneration of HepG2 cells.
2.2.1.2 Collagen. Collagen is the most abundant protein in mammals, which is widely found in all tissues from the body surface of lower vertebrates to the body of mammals. Collagen mainly exists in the form of collagen fiber in the extracellular matrix such as tissue and bone, which has high tensile strength. It can maintain and stabilize the tissue structure of the body. Brown et al. used collagen to draw colloidal islands on polystyrene plates for tissue culture.42 The liver cells selectively attached to colloidal islands, and parenchymal cells grew on the surrounding areas. Ukairo et al. also tested the bioactivity and toxicity of acetaminophen in rats by preparing the co-culture system of liver cell micrograph.43 Okudaira et al. prepared a fibrous liver tissue by using the bottom-up method of the multilayer spheres.44 Collagen was attached to the spherical aggregates of hepatocytes, and then fibroblasts and human umbilical vein cells were attached to the co-culture with hepatocytes. Then, 3D liver tissue was constructed by high-density stacking cells. The results showed that cells could receive adequate oxygen supply, and could maintain cell viability and liver function for a long period of time. Moreover, this method does not require specialized or expensive materials and equipment. It can also build large-scale liver tissue.
2.2.1.3 Chitosan. Chitosan is the product of the deacetylation of chitin, which mainly exists in the shells of shrimps, crabs, insects and other crustaceans. It has good biocompatibility, and has been widely used in biomedical and pharmaceutical fields. Feng et al. investigated the effects of nanofiber galactosylated chitosan (GC) scaffolds on the formation of primary hepatocyte aggregation and the maintenance of liver function in rats.45 By using chitosan as the raw material, the galactose ligand was successfully combined into chitosan to improve the adhesion and synergistic function of liver cells. A nanofiber galactose chitosan scaffold was prepared by electrostatic spinning technology, and the primary liver cells were attached and cultured on the scaffold. The results of this research showed that the GC nanofiber scaffolds showed good cell activity and higher liver function, and their mechanical stability was better than that of three-dimensional spherical aggregates. Therefore, GC nanofiber scaffolds could be used as biodegradable natural scaffolds for primary liver cell culture in vivo or in vitro. Li et al. prepared microcapsules that encapsulated liver cells with alginate and chitosan as raw materials.46 Microencapsulation of hepatocytes can provide a new technology for the large-scale preparation of hepatocytes, which could also keep the high activity in vitro and long-term cryopreservation of hepatocytes.
2.2.1.4 Gelatin. Gelatin is a kind of macromolecule hydrophilic colloid, which is the product of the partial hydrolysis of collagen, and has been widely used in the medical field. In 3D bioprinting technology, the use of gelatin as a substrate can control the ink thickness and high printing fitness. Mazza et al. added gelatin to the alginate, squeezed a bio-ink mixed with pluripotent stem cell differentiated liver cells at low temperatures, and solidified it with calcium chloride to form liver tissue.47 Nagamoto et al. used gelatin-coated scaffolds to harvest cell sheets, which were prepared by co-culture method.48 The liver cells and fibroblasts were co-cultured for 14 days, placing the gelatin-coated scaffold on the cell sheet, and then the culture temperature was lowered to 20 °C for 60 min. Due to changes in the hydrophobicity of the temperature-sensitive surface, the cell sheet adhered to the gelatin-coated layer and was harvested.
2.2.1.5 Decellularized tissue matrix. Decellularized tissue matrix is a new kind of biological material that can be used to repair damaged tissues, and regenerates by decellularizing allogeneic or heterogeneic tissues. Decellularized tissue has good biocompatibility, good mechanical strength, and can induce and promote cell adhesion, proliferation, differentiation and tissue formation. Mazza et al. used high shear stress vibration-decellularization technology to rapidly produce functional human liver scaffolds, and then cultured liver cells on scaffolds by perfusion method.49 Alejandro et al. removed all cells from isolated rat livers by combining the use of enzymes, detergents, and mechanical methods.50 The decellularized liver is examined by morphological, biochemical, and immunolabeling techniques to preserve the primary matrix structure and components. To ensure maximum cell survival, they also evaluated three different methods of hepatocyte seeding: direct parenchymal infusion, multi-step infusion, and continuous infusion, and found that multi-step infusion produced the best results.
2.2.1.6 Hyaluronic acid. Hyaluronic acid is the main component of connective tissue such as intercellular stroma, ocular vitreous and synovial fluid of joints. Hyaluronic acid can be used as a viscoelastic agent for ophthalmic intraocular lens implantation, as a filler for osteoarthritis and rheumatoid arthritis, and is widely used as a medium in eye drops. Turner et al. used hyaluronic acid to maintain the human hepatoblast phenotype.51 The liver cells inoculated in the hyaluronic acid hydrogel maintained the stability of the early hepatocytes throughout the culture process. Although other culture conditions, such as plastic or embryonic collagen or embryonic feed, were also available, hyaluronic acid was the only culture condition that reliably promoted the survival, proliferation and maintenance of hepatoblasts.
2.2.1.7 Matrigel. The main components of matrix glue are laminin, type IV collagen, nestin, heparin sulfate glycoprotein, growth factors and matrix metalloproteinases. At room temperature, matrix glue polymerization can form a three-dimensional matrix with biological activity, which simulates the structure, composition, physical properties and function of the cell basement membrane in vivo. It is beneficial to cell culture and differentiation in vitro, and can also be used for the study of the cell morphology, biochemical function, migration, infection and gene expression. Khetani et al. prepared the MPCC platform combined with the matrigel covering layer, which was useful to further mature iHeps toward a more adult-like PHH phenotype.52 It also maintained the function of liver cells after several weeks of culture, so it could be used for chronic drug delivery research.
2.2.2.1 Polylactic acid–glycolic acid copolymer (PLGA). Polylactic acid–glycolic acid copolymer is a kind of biodegradable polymer organic compound, which is randomly polymerized by two monomers known as lactic acid and glycolic acid. It has good biocompatibility, non-toxicity and good performance of forming capsules and films. Now, PLGA has been widely used in the pharmaceutical, medical engineering materials and modern industry fields. Jessica H. Brown et al. developed a 3D nanofiber scaffold that was made of PLGA polymer by using the newly optimized wet electrostatic spinning technology.53 It had a high pore structure and could accommodate primary human liver cells, simulating the ECM structure in vivo. Subsequently, it was proved that PLGA wet electrostatic spinning 3D nanofiber scaffolds modified with type I collagen enhanced the synthetic activity of primary human hepatocytes, which might be used as the starting material for the biosynthesis of primary hepatocytes in long-term culture.
2.2.2.2 Polycaprolactone (PCL). Polycaprolactone is a thermoplastic crystalline polyester obtained by ring-opening polymerization of caprolactone with dialcohol. Because PCL has good biodegradability, biocompatibility and nontoxicity, it has been widely used as medical biodegradable materials and drug control release system. Gao et al. prepared electrospun PCL mats by using an electrospinning system.54 A HepG2 cell suspension was inoculated onto the PCL scaffold. Results have shown that the morphology of the fiber can significantly affect the behavior and growth of cells, and the fiber arrangement can affect cell elongation and orientation. Slivac et al. also manufactured electrospun PCL mats, and tested the effects of PCL mats and liver extracellular matrix scaffolds on the bioactivity of HepG2 cells.55 Kim et al. promoted the regenerative treatment of the liver injury by stacking patient-specific progenitor cell sheets onto multiscale electrospun fibers.56 The results showed that the liver patch was a technique for the generation of an artificial liver, which might have potential clinical application value in the future.
2.2.2.3 Polydimethylsiloxane (PDMS). Polydimethylsiloxane is a kind of hydrophobic silicone material. Yu Du et al. introduced the process of manufacturing microfluidic chips by using PDMs as raw materials when introducing the microfluidic chips that simulate the structure and function of liver sine.39,57
Liu et al. used electrodeposition technology to prepare alginate gel cell sheets to assemble multilayer hepatic lobule models.39 After the cell sheets were collected, they were transferred to PDMS molds for assembly. Khetani et al. used PDMS templates made by photolithography to draw an extracellular matrix or collagen on tissue-cultured polystyrene.52 Hepatocytes only attached to the ECM domain and the parenchymal cells were seeded, and attached to areas not occupied by hepatocytes. Pang et al. used PDMS-based cellular microarray culture plates to construct an aggregate of endothelialized rat liver cells.58 Endothelialized rat hepatocytes were aggregated in the microporous apparatus.
2.2.2.4 Polyvinylidene fluoride (PVDF). Polyvinylidene fluoride mainly refers to the partial fluorine ethylene homopolymer or partial fluorine ethylene, and other small amounts of fluoride copolymer of vinyl monomers. PVDF membranes bind proteins and can separate small fragments of proteins. Its stability and corrosion resistance make it ideal for protein sequencing, and it is still in use today. Chu et al. developed a three-dimensional liver model to test sodium nitrite and acrylamide-induced hepatotoxicity,59 which forms cell cylinders in PVDF hollow fibers to mimic the microenvironment of liver tissue.
2.2.2.5 L-Polylactic acid (PLLA). PLLA is an important biodegradable polymer material, which is characterized by nontoxicity, nonirritating property, biodegradable absorption, high strength, good plasticity and easy processing. Its degradation cycle is 2–12 months. The degradation cycle can also be changed according to the addition of modifiers. PLLA is decomposed by enzymes in organisms. It eventually forms carbon dioxide and water, which has good biological compatibility. PLLA can be used to prepare scaffolds in the biological field. Jiang et al. cultured infant liver cells with a 3D PLLA scaffold.60 The results showed that the PLLA scaffold is a good supporting material for infant liver cells, which can be used for in vitro culture of infant liver cells. Bierwolf et al. used PLLA as a raw material to prepare 3D nanofibers for toxicology and medical research.61 The data showed that PALL nanofiber scaffolds provided a good microenvironment for the survival and new tissue formation of primary rat hepatocytes, and retained specific enzyme functions and morphology.
3. Methods of liver tissue culture
3.1 Bottom-up tissue engineering method
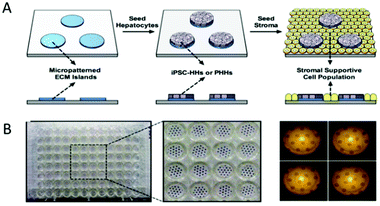 | ||
| Fig. 2 Coculture of hepatocytes and nonparenchymal cells. (A) Polydimethylsiloxane (PDMS) templates are made by photolithography. Round collagen islands are planted on the template. Hepatocytes can only attach to collagen islands, and the hepatocytes on the outside of the collagen islands are washed away. The next day, the parenchymal cells are seeded and attach to areas not occupied by hepatocytes. (B) The polydimethylsiloxane (PDMS) template is made using the photolithography method. (Reproduced with permission from ref. 68.) | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, and loaded into a bioreactor. In these processes, biodegradable PLLA fibers were used as scaffolds that aggregated with aggregates to form a loosely packed mode (Fig. 3B-c). These aggregates exhibited higher levels of cell proliferation and liver-specific function compared with hepatocyte aggregates alone. This study verified the important role of fibroblasts in liver cell co-culture.
1 ratio, and loaded into a bioreactor. In these processes, biodegradable PLLA fibers were used as scaffolds that aggregated with aggregates to form a loosely packed mode (Fig. 3B-c). These aggregates exhibited higher levels of cell proliferation and liver-specific function compared with hepatocyte aggregates alone. This study verified the important role of fibroblasts in liver cell co-culture.
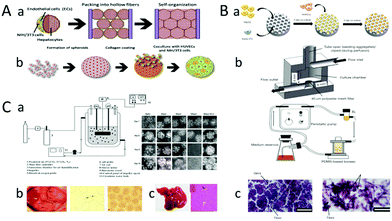 | ||
| Fig. 3 (A) Heap hepatocytes covered by the HUVECs-3T3 spherical aggregate to form liver tissue. (a) The spherical aggregates form liver tissue through the bottom-up accumulation. (b) The fabrication process of the hepatocytes spherical aggregates coated with HUVECs-3T3. (Reproduced with permission from ref. 44.) (B) The spherical aggregates are assembled into liver tissue using bioreactor perfusion. (a) Endothelialized hepatocyte aggregates are prepared by PDMS. (b) Schematic diagram of the perfusion function of a bioreactor. (c) Culture of simple HepG2 aggregates (left). Culture of aggregates of endothelialized hepatocytes (right). (Reproduced with permission from ref. 61.) (C) Liver tissue is cultured by microcapsules. (a) The installation diagram of a bioreactor. Fabrication of chitosan-coated microcapsules of HepG2 cells. The microscopy images of microencapsulated HepG2 cells at an initial cell density of 2 × 106 cells per ml for each microcapsule during 10 days cultivation in the stirred bioreactor (right). (b) Treat acute liver failure by transplant alginate microencapsulated hepatocytes. Distribution of hepatocytes in alginate microspheres. (c) 7 days after transplantation, microbeads spread throughout the abdominal cavity (black arrow). The liver of the surviving mice shows normal lobular structures after 7 days transplantation. (Reproduced with permission from ref. 83.) | ||
Microcapsule technology is also one of the methods used to obtain spherical aggregates. It has been widely used in the fields of immobilization of cells and enzymes, large-scale culture of microbial and animal and plant cells,79 controlling the release of drugs, screening of anticancer drugs,80 and separation of proteins. Cell microcapsule technology is a promising technology that provides a three-dimensional microenvironment for cells to maintain cell vitality and function through the exchange of nutrients and wastes.61,81,82 Khodabakhshaghdam et al. designed a small stirred bioreactor to study the effects of different stirring rates on the proliferation and liver specific functions of HepG2 cells cultured with alginate chitosan core–shell microcapsules (Fig. 3C-a). In this study, precooled type I collagen solution was mixed with alginate solution to make alginate collagen solution, and the microcapsules were cleaned twice. Then, the microcapsules were suspended in 0.3% (wt.) chitosan solution (pH = 6.5–6.6) for 5 min to obtain chitosan-coated microcapsules. The spherical aggregates were assembled into the liver tissue by agitation of flat blades. When the stirring rate increased, a large number of smaller and denser aggregates were formed. Finally, rat liver cell microspheres were transplanted into the abdominal cavity of rats with acute liver injury. Then, they found that the microspheres spread throughout the abdomen 7 days after transplantation (black arrow shows the cell high density culture is realized). These results showed that microencapsulated hydrogels loaded with liver cells were cultured in a small stirred bioreactor at a low mixing rate and had great potential for mass production of liver cells, while maintaining liver specific function. At present, although spherical aggregates culture technology has been used for many years, it is mainly used in bioreactors or artificial liver devices.
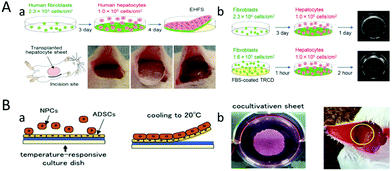 | ||
| Fig. 4 The liver tissue was cultured with thin cell sheets. (A) (a) Liver tissue is cultured from human liver parenchyma cells and human fibroblasts. The cell sheets are transplanted into mice. (b) Rapid tissue culture can be achieved by using high density fibroblast cells. (Reproduced with permission from ref. 44.) (B) (a) Liver tissue is cultured from hepatic non-parenchymal cells (NPCs) and adipose-derived stem cells (ADSCs). (b) The cell sheets are transplanted into mice. (Reproduced with permission from ref. 87.) | ||
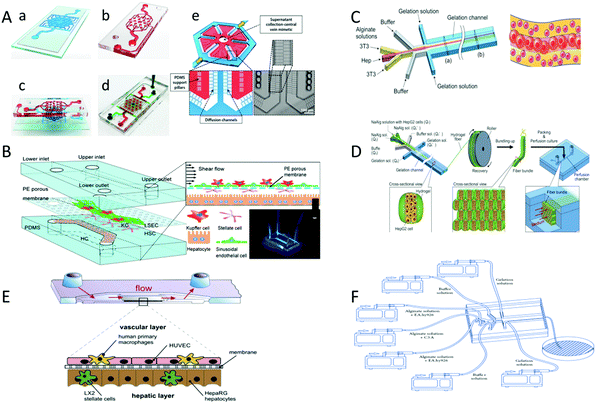 | ||
| Fig. 5 The liver tissue is prepared through microfluidic method. (A) Hepatic lobules are fabricated by microfluidic technique. (a) The lower layer is the culture chamber, and (b) the upper layer provides nutrients to the cells. (c) is the side view of the microfluidic device. (d) is the top view of the device. Red and green food dyes (d) are injected into the tissue lumen and surrounding feed channels. A single lobule binds to the bottom layer of the cell compartment (Red), surrounding flow channels (Blue) and PDMS walls 2 μm (White) (e) diffusion channels. (Reproduced with permission from ref. 88.) (B) Hepatic sinusoids are fabricated through the microfluidic method. The sinusoidal chip consists of two PDMS cavities, which are combined by plasma treatment with the upper and lower channels and membranes. (Reproduced with permission from ref. 57.) (C) The liver plate was fabricated through microfluidic technology. Liver cell fluid, endothelial cell fluid, buffer fluid and gelatin fluid flow through the chip from the inside to the outside. (Reproduced with permission from ref. 99.) (D) The hepatic lobules were fabricated through microfluidic method and perfusion. Fabricated microfibers embedded with HepG2 cells by using microfluidic technology. The fibers are then bundled into bundles by using rollers. (Reproduced with permission from ref. 100.) (E) The liver-like organ were fabricated through the microfluidic method. (F) The hepatic cords were fabricated through microfluidic technology. The sodium alginate solution containing hepatocytes and 3T3 cells is continuously introduced into the microfluidic channel to generate cell-loaded sodium alginate hydrogel microfibers. (Reproduced with permission from ref. 96.) | ||
3.2 Top-down tissue engineering method
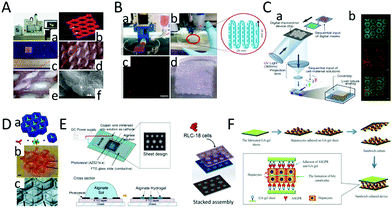 | ||
| Fig. 6 Creation of liver tissue using 3D bioprinting technology. (A) Creation of mini livers using 3D printing technology. (a) The 3D printing equipment. (b) The 3D-printed structure containing pores. (c) A view of the structure obtained through 3D bioprinting technology. (d and e) The picture of the pores. (f) The single triangle refers to the cells, and the double triangles refer to the hydrogel. (Reproduced from ref. 102.) (B) (a) The 3D printing equipment. (b) An alginate containing mCherry-HepG2 cells is extruded utilizing nozzle pressure. (c) The fluorescence micrograph of mCherry-HepG2 cells. (d) Multilayer mCherry-HepG2 cells solidified successively to form tissue. (Reproduced with permission from ref. 104.) (C) (a) Liver lobules are prepared through 3D bioprinting technology. (b) Sertoli cells and HiPSC-derived hepatocytes are fluorescently labeled with red and green dye. (Reproduced with permission from ref. 101.) (D) Multiple layers are printed to form liver tissue. (Reproduced with permission from ref. 103.) (E) Multiple layers stack to form hepatic lobules. (Reproduced with permission from ref. 39.) (F) Liver cell coagulation tablets were prepared using 3D bioprinting technology. Hepatocytes were seeded and adhered to a gel sheet. (Reproduced with permission from ref. 105.) | ||
Hepatic lobule structures could also be obtained by sequential 3D bioprinting of various materials (Fig. 6D).103 Liu et al. used a multilayer photopatterning platform to embed cells in complex hydrogels to make liver tissue. First, a closed chamber was filled with a cell-loaded prepolymer solution. This was exposed to light through a mask, and then photocrosslinked to form patterns with microscale characteristics. The height of the chambers was increased to repeat this process to form a multi-layer cell-rich liver tissue structure with microscale characteristics.
Another way to construct liver lobules using 3D bioprinting is with micro-patterned electrode devices.103,104 Liu et al. developed a micropatterned electrode device using lithography technology. Two kinds of hepatic lobule patterns with different diameters (1.5 mm and 2.0 mm) were designed. A DC voltage was applied to the device to trigger the electrodeposition process. An alginate solution formed the alginate gel structure according to the principle of electrodeposition. Finally, the prepared cell-loaded alginate sheets were further separated and stacked to form multilayer liver lobule tissue (Fig. 6E).39
In addition to mixing cells with bio-ink to form a cell-printing solution, Lee et al. printed gel sheets using a 3D bio-printer. Liver cells can then be seeded and attached to the gel sheets to obtain liver tissue. The researchers synthesized a polymer of galactosylated alginate (GA) to produce the gel sheet because it contained a sufficient number of galactosylated chains that bind easily to the salivation glycoprotein receptor (ASGPR) of liver cells. For a comparative study, liver cells were individually inoculated on alginate gel sheets and GA-gel sheets. The results showed that the hepatocytes inoculated on the GA-gel sheets showed good adhesion characteristics, but the adhesion of the liver cells inoculated on the alginate gel sheet was poor (Fig. 6F).105
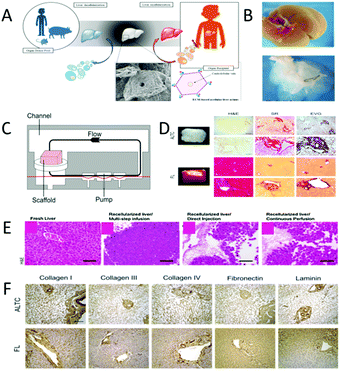 | ||
| Fig. 7 Decellularization–cellularization technology is used to produce liver tissue. (A) Livers from humans, mice and pigs are decellularized, and then inoculated with human liver cells and transplanted into the body. (Reproduced with permission from ref. 108.) (B) Fresh harvested liver (top) and the decellularized liver (bottom). (C) A reperfusion device for regeneration. (D) Rapid production of liver tissue by high shear stress decellularization-recellularization technique. (E) Reaggregation of the acellular liver matrix with adult hepatocytes. Three different cell seeding techniques are used for recellularized liver transplantation. (Reproduced with permission from ref. 50.) (F) Comparison of the expression and distribution of type I collagen, type III collagen, type IV collagen, fibronectin and laminin. The results show that the acellular sample (top) is consistent with the fresh sample (bottom). (Reproduced with permission from ref. 107.) | ||
Gao et al. investigated the response of liver cells to the surface characteristics of electrospinning fibers. PCL scaffolds with large surface depression (2 μm), small surface depression (0.37 μm) and no surface depression (NSD) were prepared. HepG2 was inoculated onto the PCL scaffold for a maximum of 14 days. The comparison and analysis of the cell activity and DNA content in each group showed that the scaffolds could promote the gene expression of albumin, and it was found that the HepG2 cells may prefer small depressions to smooth and large depressions (Fig. 8A).54 In addition, Slivac et al. compared a synthetic polyester PCL with a natural ECM isolated from pig liver55 (Fig. 8B-a). Cells are transfected with a plasmid carrying the red fluorescent protein (RFP) gene. Green cells are living cells, red and orange cells are successfully transfected cells. There is no significant difference in the number of transfections between the two scaffolds. It was found that the cell diffusion rates and the percentage of transfected cells on the two scaffolds were quite similar (Fig. 8B-b). Mechanical and structural analyses showed that the tensile stress resistance of PCL increased by about 12 times compared with the natural ECM. These results indicated that PCL electrospinning pads can be used to simulate natural scaffolds.
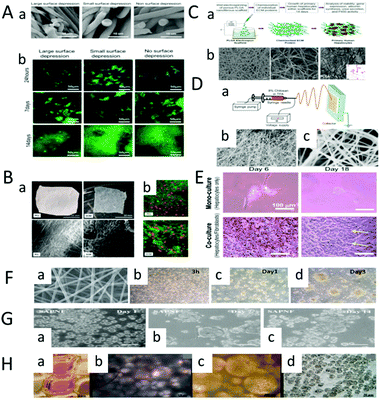 | ||
| Fig. 8 Preparation of liver tissue based on nanofiber scaffolds. (A) Fabrication of the liver tissue based on PCL scaffolds. (a) Scanning electron microscopy (SEM) images of large, small and non-pitted fibers. (b) Seed liver cells on fibrous scaffolds with large, small and no surface depressions. The distribution of cells is observed under a microscope at 24 h, 7 d and 14 d. The nucleus is blue. (Reproduced with a permission from ref. 54.) (B) Comparison of the synthetic polyester PCL scaffold with natural protein-based ECM. (a) Physical and electron microscopic views of the electrospun PCL felt and pig liver ECM. (b) Cells are transfected with a plasmid carrying the red fluorescent protein (RFP) gene. Green cells are living cells, red and orange cells are successfully transfected cells. (Reproduced with permission from ref. 60.) (C) Fabrication of liver tissue based on the PLGA scaffolds. (a) Porous PLGA nanofiber scaffolds are prepared by wet electrospinning. Seed human primary hepatocytes on the scaffolds and culture for 14 days. (b) Polylactic acid scaffolds with different thicknesses (top). Human primary hepatocytes implanted on scaffolds (bottom). (Reproduced with permission from ref. 53.) (D) (a) Diagram of the electrospinning device. (b and c) SEM images of the chitosan nanofiber scaffolds at low and high magnification. (E) Morphology of the single and co-cultured hepatocytes on the surface of the chitosan nanofiber scaffolds. (Reproduced with permission from ref. 115.) (F) Fabrication of liver tissue based on galactosylated chitosan (GC) nanofiber scaffolds. (a) Fabrication galactosylated chitosan (GC) nanofibers by electrostatic spinning. (b) Distribution of human liver cells at different times after inoculation into scaffolds. (Reproduced with permission from ref. 45.) (G) Fabrication of liver tissue based on SAPNF self-assembled peptide nanofiber scaffolds. (a–c) At 24 h after inoculation, they aggregated into twins and triplets. (Reproduced with permission from ref. 116.) (H) Fabrication of liver tissue based on three-dimensional nanofiber PLLA scaffolds. (a and b) Schematic diagram of the scaffold after the implantation of primary rat hepatocytes by centrifugal seeding technique. (c) After 7 days of 3D culture, the nanofiber scaffold is observed by transmission light microscope. (d) Transmission light images of 2D cells culture in a 96-well Petri dish for 7 days. (Reproduced with permission from ref. 60.) | ||
Brown et al. developed a three-dimensional nanofiber scaffold based on the PLGA polymer and used the wet electrostatic spinning technology to create a porous structure. ECM proteins (type I collagen or fibronectin) were chemically linked to electrospun PLGA via amine-coupling to maintain hepatocyte function in vitro (Fig. 8C).53 Rajendran et al. used chitosan nanofiber scaffolds to construct 3D liver models115 (Fig. 8D). The chitosan nanofiber scaffolds can be obtained by applying a DC voltage of 20–25 kV between the syringe needle and the square metal collector. By comparing the single and co-cultured hepatocytes, it was found that the co-cultured hepatocytes formed colonies, and maintained their morphology and function for a long time (Fig. 8E). The 3D liver tissue model established in this study provides a useful tool for drug screening and tissue engineering development of the engineered liver tissue. Feng et al. developed a novel natural nanofiber scaffold with a surface galactose ligand to enhance the biological activity and mechanical stability of the cultured primary hepatocytes. The nanofiber scaffolds were prepared using natural material GC by electrostatic spinning.45
Based on the disintegration assessment and Young's modulus test, GC nanofiber scaffolds were found to degrade slowly and have suitable mechanical properties as ECM. Morphological characterization, dual staining fluorescence analysis and functional analysis showed that the liver cells cultured on the GC nanofiber scaffolds formed stable three-dimensional aggregates, which were similar to human liver in terms of the albumin secretion. The result indicated that this GC-based nanofiber scaffold can be used in a variety of applications, such as the construction of bioartificial liver (Fig. 8F). Alvarez et al. compared the effects of SAPNF on hepatocyte metabolism and secretory activity after hepatocyte isolation (Fig. 8G).116 The isolated porcine liver cells were cultured in SAPNF and type I collagen, respectively, and a morphological evaluation was conducted by scanning electron microscopy at different time points. It was found that the liver cells cultured with SAPNF showed a three-dimensional spherical structure and maintained cell differentiation during 2 weeks of culture, which indicates that SAPNF plays an important role in maintaining the differentiation function of porcine liver cells. Bierwolf et al. cultured primary rat liver cells on a 3D nanofiber PLLA scaffold for 7 days.60 After 7 days of 3D culture, the nanofiber scaffold was observed by transmission light microscope, and the pores were randomly full of hepatocytes and no morphological changes were noticed. T cell viability and hepatocellular specific functions (albumin secretion, glycogen storage capacity) showed that the primary rat hepatocytes cultured on nanofiber scaffolds had high viability and well-preserved glycogen storage. Albumin was secreted during the whole culture process, and hepatocytes were highly differentiated and preserved. These phenomena indicate the applicability of three-dimensional nanofiber PLLA scaffolds in liver tissue construction (Fig. 8H).
Furthermore, the various methods for liver tissue construction in vitro are summarized as shown in Table 1.
| Methods | Advantages | Disadvantages | Ref. |
|---|---|---|---|
| Micromode coculture method | – The cells had good growth status and differentiation characteristics; | – Cell separation technology is high; | 48, 61–64, 66 and 68 |
| – High cell density. | – The cultivation conditions are high. | ||
| Spherical aggregate culture method | – The cells had good growth status and differentiation characteristics; | – High purity cells are needed; | 44, 61, 70, 75–83 and 117–122 |
| – Simple equipment and low cost | – High level of operation. | ||
| – High cell density; | |||
| – No specific carrier is required. | |||
| Cell sheets culture method | – Simple operation; | – Cells cannot be grown on a large scale; | 39, 44, 48, 86, 87 and 105 |
| – The cells had good growth status and differentiation characteristics in a good microenvironment. | – Liver cells can easily lose differentiation phenotype. | ||
| Microfluidic culture method | – The number of cells required is small and the test dose is small; | – It requires special technology and is expensive; | 57, 88, 96, 99, 100 and 123–126 |
| – The cells had good growth status and differentiation characteristics; | – Cell growth is slow and susceptibility is low; | ||
| – Cells can maintain a specific phenotype. | – Cells cannot be grown on a large scale. | ||
| 3D bioprinting culture method | – Provides cells with an in vivo 3D environment; | – The material is required to be photosensitive. | 16, 39, 79, 83, 101–105 and 127–134 |
| – Cells adhere easily. | |||
| Decellularized–recellularized culture method | – Cells adhere easily and grow well. | – Problems such as shear forces and nutrient supply may affect cell growth. | 47, 50, 95, 106–109, 135 and 136 |
| Nanofiber scaffold culture method | – Large and homogeneous space for cultivation; | – Cells easily lose normal morphology and differentiation function; | 45, 53–56, 58, 60, 71, 96, 110–116 and 137–142 |
| – Cells adhere easily and grow well. | – Problems such as shear forces and nutrient supply may affect cell growth. |
4. Applications of liver tissue
4.1 Tissue reconstruction
Although liver transplantation has become a standard treatment, the mortality rate from liver disease remains high. In the research phase of liver cell transplantation, Sakai et al. treated acute liver failure in mice with xenogeneic hepatocyte transplantation (Fig. 9A). First, porcine liver cells embedded in SAPNF or type I collagen were injected directly into 90% of hepatectomy mice with acute liver failure. After transplantation, the researchers followed up with these mice to assess their survival. H&E was used to stain the nucleus and cytoplasm. On the seventh day after transplantation, a considerable number of porcine liver cells were detected in the spleens of mice injected with self-assembled peptide nanofibers, which had trabecular arrangement, well-developed bile ducts and tight intercellular spaces. Moreover, the glycogen storage capacity was detected after staining with PAS (Fig. 9B-a, d). Small clusters of porcine liver cells were observed in the spleen of collagen-injected mice. However, the result of PAS staining was weak, which indicated poor glycogen storage capacity (Fig. 10B-b, e). Few hepatocytes were detected when transplanted alone (Fig. 10B-c, f).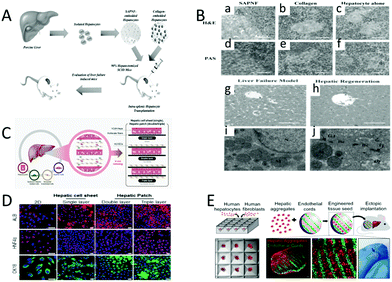 | ||
| Fig. 9 Liver reconstruction. (A) Treatment of acute liver failure in mice by xenogeneic hepatocyte transplantation. (B) Histological findings of mice after hepatocyte transplantation. H&E stains the nucleus and cytoplasm. PAS staining shows glycogen storage capacity. (a and d) Pig liver cells are seeded on the SAPNF scaffold, and then transplanted into the spleen of mice. (b and e) Pig liver cells are seeded on the collagen, and then transplanted into the spleen of mice. (c and f) The liver cells are individually injected into the spleens of the mice. (g and i) Liver injury model. (h and j) Liver repair. Intercellular interstitial junction (Gj) and tight junction (Tj), well-developed bile duct (Bc), and glycogen wreath (Gly). (Reproduced with permission from ref. 143.) (C) Promoting regenerative treatment of the liver injury by stacking the patient-specific progenitor cells on electrospun nanofibers. (D) HCdhs-heps differentiate on single, double, or three-layer electrostatic spun fiber stacked sheets express mature hepatocyte specific markers. (Reproduced with permission from ref. 56.) (E) A treatment that uses autologous (or allogeneic) adult cells (or stem cells) from patients to repair tissues or organs. (Reproduced with permission from ref. 144.) | ||
 | ||
| Fig. 10 Drug efficacy testing. (A) Building a liver model. Micromode coculture (MPCCs) allows the regulation of homotypic interactions between the hepatocytes and heterotypic interfaces between the hepatocytes and fibroblasts, while maintaining a constant cell count/ratio in various mode configurations. (B) (b) Tissue cultured by MPCCs can maintain a high level of CYP450 activity for several weeks, which can greatly reduce the influence of culture time on CYP450 activity. (Reproduced with permission from ref. 132.) | ||
Kim et al. tried to explore the effect of the number of nanofiber layers on liver tissue repair (Fig. 9C).143 The regenerative treatments of liver injury are promoted by stacking patient-specific progenitor cells on electrospun nanofibers. HCdH-Heps cells are seeded onto fiber sheets to form a hepatic cell sheet (single layer). Human umbilical vein cells and liver cells are seeded on the thin sheet of electrospun fibers to form a hepatic cell patch (double layer and triple layer). ALB, CK18, and HNF4 are differentiation products, so the effects of hepatic sheets and hepatic patches on tissue reconstruction can be determined by the detection of ALB (red, top), CK18 (green, bottom), and HNF4 (red, middle). Functional analysis revealed that the hepatic patch synthesized using hCdHs recapitulated a liver tissue-like structure. Therefore, patient-specific hCdHs may be a novel source to develop hepatic cell sheets or hepatic cell patches for application in personalized tissue engineering and cell sheet-based regenerative medicine. It is also obvious that the hepatic cell patch is a better choice to promote tissue regeneration than the hepatic cell sheet (Fig. 9D). Kim et al. investigated the importance of the relationship between liver cells, endothelial cells, and stromal cells in tissue regeneration. After transplantation of liver seeds into mice with liver injury, large amounts of Ki67 protein and cytokeratin-18 were observed (left, white arrow), and mitosis of cells was also observed (right, white arrow), which demonstrated that ectopic liver seed transplantation can repair damaged liver tissue (Fig. 9E).144 Furthermore, the relationship between hepatocytes, endothelial cells and stromal cells proved to be important in tissue regeneration.
Liver cell transplantation is one of the most effective ways to treat liver failure caused by alcohol consumption, hepatotoxic drugs or viral infection. However, the use of human liver cells has been limited by the shortage of donors, the limited number of cells suitable for transplantation, and the low efficiency of transplantation. In addition, human liver cells may lose their reproductive potential during in vitro culture, making it difficult to grow and reproduce in vitro. These above problems have been largely resolved with the use of HCdHs with proliferative and differentiated potential and cell sheets. The discovery of HCdHs solved the problem of a shortage of donors and a limited number of cells suitable for transplantation. Tissue engineering based on cell sheets solves the problem of low transplantation efficiency. Previous studies have shown that the function and therapeutic effect of transplanted liver cell sheets are significantly higher than that of direct cell perfusion. The liver sheets made up of HCdHs and HUVECs not only have the function of the original human liver, but also have remarkable therapeutic effect on acute liver injury. It is expected that replacing HUVECs with hepatic sinusoidal endothelial cells and hepatic stellate cells will have a better effect.
However, growing a tissue with a fully functional vascular system in vitro has still been a major challenge. A developed vascular system allows the implanted tissue to overcome diffuse limitations, ensuring the long-term survival of the implanted cells. In liver tissue engineering, due to the high metabolic activity of liver cells requiring adequate oxygen supply, the generation of a vascular system is more important. So far, no one has successfully developed a perfused microvascular system. A preliminary approach is to develop vascularized organoids by the co-culture of liver tissue directly with endothelial cells. Inamori et al. filled hollow fibers for plasma separation with hepatocyte spheres covered by human umbilical vein endothelial cells. The dense spheres then connected to each other to form a large tissue with regular distribution of HUVECs. On the ninth day, HUVECs invaded the hepatocytes spheres and formed a dense vascular network.145 LeCluyse et al. also reconstructed the multicellular structure and hemodynamic characteristics of the liver using this method.74 Another approach that seemed to work was to coat liver cells, endothelial cells and pericytes in a hydrogel. The results showed that these cells remained active for a month, and formed a network of blood vessels with the hydrogel. By using PCL as a skeleton material, Lee et al. successfully induced the formation of vascular networks and hepatocyte growth by injecting collagen bioinks containing three different cell types into the channels of the PCL framework.146 Jeon et al. also experimentally verified the feasibility of this approach.104 In addition to the above two methods, the decellularized matrix shows good effects in the construction of liver tissue vascularization in vitro. In Soto-Gutierrez's experiment, the resulting hepatic matrix preserved the entire vascular bed and bile duct after the decellularization treatment.50 Researchers are currently developing many similar methods to build vascular networks, but there is still room for improvement.
4.2 Drug screening
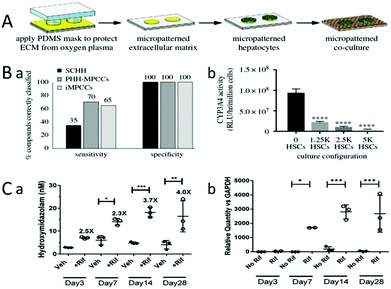
At present, drug-induced liver injury (DILI) remains one of the major causes of preclinical and clinical drug consumption, and the emergence of marketed drugs withdrawal from the market. Since animal drug screening cannot accurately reflect the effects of drugs on the human body, the field of in vitro human tissue engineering is becoming increasingly important.12,42,52,64,127,132,147–152Rifampicin has a strong antibacterial effect on tuberculosis bacilli, and also has a curative effect on Gram-positive or negative bacteria and viruses.42 Rifampicin can activate the CYP enzyme, which is known as cytochrome oxidase P450, and is a group of isoenzymes encoded by superfamily genes related by structure and function. They are monooxygenases mainly existing in the liver and intestine, and mostly located in the endoplasmic reticulum. Furthermore, they are terminal enzymes of mixed functional oxidase system on the endoplasmic reticulum. It is involved in the metabolism of endogenous substances (such as fatty acids, vitamins and cholic acids), detoxification of exogenous substances (such as drugs), activation of pre-carcinogens (such as aromatic substances), and plays an important role in drug metabolism. CYP3A4 and CYP2D6 account for a large proportion of CYP enzymes, and their activities can also be activated by rifampicin. The activity of CYP3A4 can be assessed by metabolizing testosterone into 4-hydroxymidazolam,132 and the activity of CYP2D6 can be assessed by metabolism of dextromethorphan into cathane. Brown et al. constructed a bioengineered liver model by the co-culture of 3T3-J2 fibroblasts with primary hepatocytes. The ECM, such as collagen, was evenly coated on polystyrene (or glass) and protected with a PDMS stamp. The exposed ECM region was ablated under oxygen plasma, leaving tiny ECM islands that matched the geometry of the PDMS stamp. Hepatocytes selectively attached to the ECM island, and parenchymal cells (NPCs) filled the surrounding area. So, a bioengineered liver model can be obtained (Fig. 10A). Tissue cultured by MPCCs can maintain a high level of CYP2D6 (Cytochrome P450 isozymes) activity for several weeks, which can greatly reduce the influence of the culture time on CYP2D6 activity, ensuring that changes in the CYP2D6 activity were caused by rifampicin (Fig. 10B). Then, the activity of CYP2D6 (Cytochrome P450 isozymes) can be assessed by the metabolism of dextromethorphan into coffee alkanes. Finally, the efficacy of rifampicin can be evaluated by testing the content of the metabolites.
Malaria is an insect-borne infection caused by the bite of anopheles mosquitoes or the transmission of plasmodium into the blood of people carrying the parasite. At present, there are many kinds of drugs used to treat malaria, such as artemisinin, aminoquinoline and quinoline alcohol. Primaquine is mainly used to control the recurrence of malaria. Its mechanism of action is mainly by interfering with the reduction of coenzyme II, so that the plasmodium cannot carry out the metabolic oxidation of sugar. The gametophyte of the plasmodium in the infrared phase is then quickly killed, so as to prevent the recurrence and spread of malaria. Primaquine should be screened before it is used for clinical treatment. First, Sandra March et al. made micropatterned collagen islands in the 24-well or 96-well plates.64 Then, MPCCs were formed by seeding primary human hepatocytes continuously onto the collagen island and seeding 3T3-J2 fibroblasts onto the spaces around the island (Fig. 11A). Under different experimental interferences, infected MPCCs by Plasmodium falciparum sporozoa were cryopreserved to detect their sensitivity to primaquine (Fig. 11B-a and b). They reproduced the liver life cycles of the plasmodium pathogens Plasmodium falciparum and Plasmodium vivax in vitro. The results showed that MPCCs had the ability to distinguish live spores from attenuated spores (Fig. 11B-c). The MPCC platform can be used to uncover various aspects of host–pathogen interactions, and has the potential for drug and vaccine development.
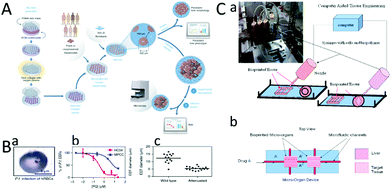 | ||
| Fig. 11 Application of MPCC in the infection of hepatophilic pathogens in vitro. (A) The preparation, sowing and cultivation of MPCC. (B) (a) MPCC is infected with cryopreserved Plasmodium falciparum sporozoan, and its sensitivity to primaquine is tested. (b) Detection of the susceptibility of virus-infected MPCC to primaquine. (c) MPCC has the ability to distinguish between live and attenuated spores. Model of drug metabolism in vitro. (Reproduced from ref. 64.) (C) (a) Cells write directly. (b) Diagram of a device used to detect drug metabolism. (Reproduced with permission from ref. 127.) | ||
In addition to drug efficacy and hepatotoxicity detection, drug metabolism detection is very necessary.127 Chang et al. developed three-dimensional liver micro-organs as an in vitro drug metabolism model. They obtained the liver tissue by 3D bioprinting, and tested its ability to metabolize drugs through an in vitro dynamic drug infusion (Fig. 11C-a).52 Finally, the drug EFC (7-ethoxy-4-trifluoromethyl Coumarin) was injected into the tissue system, and the metabolic degree of the drug was determined by observing the fluorescently-labeled metabolite HFC (7-hydroxy-4-trifluoromethyl Coumarin) (Fig. 11C-b). This research confirmed the feasibility of the model in drug metabolism.
Tetracycline, diclofenac, methotrexate, cyclophosphamide and acetaminophen are known as five hepatotoxic drugs, while propranolol and buspirone are two non-drug liver injury compounds. Spherical aggregates composed of hepatocyte were exposed to the above drugs individually for a long term. Starting from the 7th day after sowing, the shape of the sphere is shown in Fig. 12A. The spheres were treated with a single dose once within 48 h (circle) or repeated every 2 days within 8 d (triangle) to observe the coordinate diagram of the effect of the drug content on the hepatocyte activity153 (Fig. 12B). In both cases, the ATP levels are used as a marker of viability. The IC50 is the half-inhibitory concentration, which indicates the concentration required for a drug to inhibit cell growth, viral replication, or other properties, at 50% of the required concentration. After a single dose of methotrexate and acetaminophen, the cell viability decreased slightly and IC50 values were higher. In contrast, the IC50 value of re-administration was lower and consistent with the previously reported Cmax values in mice. Roproxolol and buspirone produced fairly stable dose–response curves with similar IC50 values for single or repeated dosing. The IC50 values of tetracycline, diclofenac and cyclophosphamide were 10 times lower after repeated treatment than after a single treatment. The corresponding concentrations are correlated with the maximum plasma concentration (Cmax), suggesting that spherical aggregates may be suitable for studying hepatotoxicity in vivo (Fig. 12C). From the substances tested, tetracycline, diclofenac and cyclophosphamide were labeled as hepatotoxic after 48 hours, while methotrexate and acetaminophen showed hepatotoxicity only after repeated exposure. As expected, both propranolol and buspirone were correctly classified as non-pharmacological liver injury compounds in both short-and long-term exposure trials. The research indicated the applicability of liver tissue engineering in the testing of drug hepatotoxicity.
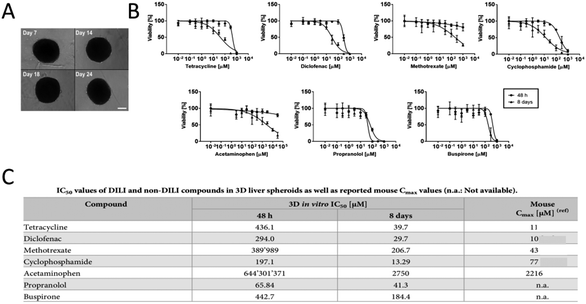 | ||
| Fig. 12 Drug screening for DILI. (A) Spherical aggregates of hepatocytes. (B) Long-term exposure tests were performed on globular cells composed of parenchymal cells using the known hepatotoxic drugs tetracycline, diclofenac, methotrexate, cyclophosphamide, acetaminophen, and the non-pharmaceutical liver injury compounds propranolol and buspirone. From the 7th day after seeding, the balls are treated with a single dose once within 48 h (round) or repeated every 2 days within 8 d (triangle). (C) IC50 values of DILI and non-DILI compounds in 3D liver spheroids, as well as reported mouse Cmax values. (Reproduced with permission from ref. 153.) | ||
Although drug screening based on liver engineering has been widely used, its development process is the joint efforts of many researchers. At an early stage, Elaut et al. found that the cell survival, phenotype and liver specific function were rapidly lost when primary isolated hepatocytes were cultured in a 2D model with simple construction. These phenotypic changes are mainly due to changes in gene expression caused by reduced transcription of relevant liver-specific genes, which also can be called dedifferentiation of isolated hepatocytes.154 Rowe et al. also demonstrated that when liver cells were isolated and cultured by conventional methods, they developed dedifferentiation as matured.155 Then, they quantitatively compared the proteome of human fetal liver, adult hepatocytes, and HepG2 cell lines. They also demonstrated the proteomic changes typical of hepatocyte dedifferentiation in the ECM interlayer culture, a new culture method that can better preserve the phenotype and function of hepatocytes. Nudischer et al. measured cell viability in single and co-culture cells by assessing intracellular ATP levels. It was found that the ATP level of the co-cultured cells was about 40% higher than that of the single cultured cells on the seventh day.153 Nguyen et al. evaluated the toxicity of terovasfloxacin in 2D and 3D hepatocyte cultures. First, both models were administered continuously for 7 days, and the cell activity and albumin production were assessed. It was found that after seven days of administration, the albumin and ATP levels produced by the 3D model were significantly reduced. Meanwhile, the 2D model under the same treatment conditions showed only the highest dose of terovasfloxacin toxicity.132 Chu et al. conducted toxicity tests of sodium nitrite and acrylamide in 3D and monolayer liver models, respectively, to study the sensitivity of the 3D model to toxicity. As expected, HepG2 cells in the 3D model reacted with a lower concentration and shorter exposure time than cells in the monolayer model.59
The results of the above studies indicated that liver tissue cultured by traditional methods lacked the complexity of natural tissue in drug toxicity screening and drug metabolism identification, so the predictive ability was limited and the sensitivity was about 30–50%. However, the liver tissue model cultured by the co-culture method can significantly improve the detection of toxicity and metabolites, and increase the sensitivity to 70–75%. The function of liver tissue cultured in vitro by this method can be maintained for several weeks. There is evidence that the co-culture of different liver cells results in better tissue stability and function than culture alone.127,152,153 Therefore, 3D liver tissues constructed in vitro are more suitable for the accurate screening of drugs than traditional 2D tissues.
4.3 Disease modeling
In recent years, the incidence of fatty liver, hepatitis, and alcoholic liver has increased sharply.156 Various disease models have been produced in laboratory animals by various methods. However, the main problem is that there are considerable differences in the types of lesions between animals and humans.157 For example, cirrhosis induced in animals is rarely accompanied by the decompensated features of human disease, namely portal hypertension, ascites, encephalopathy and esophageal varicose veins. Therefore, it is necessary to construct the model of human liver disease in vitro, and then study the drug research of the disease.52 In the establishment of the hepatitis C model, a normal liver tissue model was obtained by co-culture of liver cells and fibroblasts. The model was then infected with hepatitis C virus to obtain a hepatitis C model. The model was stably infected in vitro for several weeks. The zigzag pattern was the result of medium change every 2 days, viral replication, and luciferase secretion in the supernatant (Fig. 13A). The MPCCs model can then be treated with interferon or various antiviral protein drugs, and the effect of these drugs on hepatitis C can be observed (Fig. 13B). In addition, the development of therapeutics and vaccines for human liver disease requires the study of models of host–pathogen interactions. However, in vitro models of liver infection usually use physiologically abnormal hepatocellular carcinoma cell lines. However, this model will rapidly lose liver characteristics under in vitro culture conditions. To achieve a stable and robust in vitro model of human primary hepatocytes, March et al. developed MPCCs, which consisted of primary human hepatocytes and fibroblasts. This system can be used to establish hepatitis B and C and malaria models in vitro. The MPCC platform can simulate multiple liver disease models in vitro, which can be used to uncover host–pathogen interactions and has the potential for drug and vaccine development.64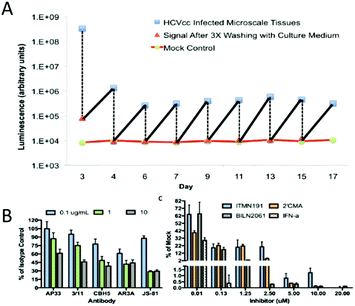 | ||
| Fig. 13 (A) A disease model of infectious disease HCV was obtained. The zigzag pattern is the result of medium change every 2 days, viral replication, and luciferase secretion from the supernatant. MPCCs are treated with antibody or inhibitor to prevent the virus from entering cells. (B) Establishment of a cirrhosis model. Application of DMNA to the liver tissue to induce liver cirrhosis. The surface of the liver capsule is irregular and the reticular bands intersect with the liver parenchyma. Early nodular formation can be seen at the lower left. Hepatocytes vary considerably in size with the presence of several necrotic hepatocytes (marked with arrows). (Reproduced with permission from ref. 52.) | ||
In conclusion, the results of the studies suggest that the mechanisms associated with diseases such as HCV and HBV can be reproduced in a liver tissue model constructed in vitro, which can remain active and functional for several weeks. However, the current disease model also faces an important challenge, which is the low pathogen infection rate (for example, HCV infection efficiency is 5%), which will limit the application of disease models. To overcome this challenge, the researchers studied interactions between the host and virus in greater depth. One interesting example is the use of broad-spectrum Janus kinase (JAK) inhibitors to attenuate the immune response, making HCV and HBV pathogens more susceptible to infection. We believe that with the continuous development of technology, liver tissue models constructed in vitro will be widely used in clinical medicine in the future.
5. Conclusion
The liver is an organ of metabolism in the body, which plays an important role in our body. Construction of liver tissue in vitro provides an effective approach for liver drug screening and transplantation. Nowadays, many techniques involving spherical aggregates of liver cells, nanofiber scaffolds, 3D bioprinting and microfluidic technology, have emerged to construct 3D liver tissue. As a multifunctional model at the intersection of in vivo and in vitro experiments, liver tissue in vitro has a good application prospect in disease model construction, drug screening, and cell therapy. Liver tissue can partially reproduce the process of liver development in vitro, maintaining the genetic characteristics of the source individual for a long time. Liver tissue reconstructed in vitro can perfectly simulate the cell-to-cell and cell-to-matrix contact environment in vivo, which helps to reconstruct the polarity of liver cells and improve the growth activity of liver cells. In vitro cultured liver tissue can survive for several months. After transplantation into the body, there is no immune rejection. It can rapidly develop into mature liver cells.In terms of drug screening, the sensitivity of the 3D liver tissue model constructed in vitro is more than twice that of the 2D liver tissue model cultured by traditional methods. It can effectively detect the hepatotoxicity and metabolic characteristics of drugs, reduce the cost of drug research and development, and reduce the adverse reactions of drugs to the human body. In terms of liver cell transplantation, the use of thin sheets of HCdHs with proliferative and differentiated potential can largely solve the problems of donor shortage, low transplantation efficiency and loss of function. Previous studies have shown that the function and therapeutic effect of transplanted thin sheets are significantly higher than that of direct cell perfusion. The liver sheets composed of HCdHs and HUVECs not only has the function of the original human liver, but also has a significant therapeutic effect on acute liver injury. In terms of tissue models, liver models will provide pharmaceutical companies with more predictability, lower drug development costs, and reduce drug toxicity to humans. These models will also help identify new molecular targets affecting liver disease, which could lead to the development of treatments with fewer side effects.
However, the construction of liver tissue still faces many challenges. First, there is no single chamber for the collection of secreted bile. Bile secreted by liver cells in the body flows slowly through the hepatic tubules, usually in the opposite direction to the blood flow in the hepatic sinuses, and flows out of the bile duct. Bile is a complex liquid that contains bile acids, cholesterol and bilirubin, which is a toxic breakdown product of hemoglobin. Due to the lack of a functional clearance mechanism for bile, these toxic products currently accumulate and mix with the base culture medium in vitro, potentially causing significant damage to cultured liver cells over time. One current solution is to inoculate liver cells in microchannels spaced 50 μm apart, limiting the tissue to two neatly arranged rows of liver cells. The liver cells will then be exposed to the shear stream by layers of collagen and monolayer endothelial cells. This device can clear the secreted bile from the liver cells below, which can also be used to study the reaction of bile acids to various stimuli. In addition, bile ducts can be obtained by incubating hepatic progenitor cells or bile duct cells with hydrogel. Tanimizu et al. explored the influence of matrix composition, matrix composition, and growth factors on bile duct formation.158 Katsuda et al. co-cultured ABECs and FLCs to form functional bile duct structures. Experimental results showed that ABECs could help to establish a bile duct network.159 However, the integration of the bile duct network and its application in 3D implantable structures remain to be explored. Second, liver organs have not yet included all the cell types in the liver. Naturally, it cannot fully realize the functions of the mature liver, as well as the vascularization, immunization and nerve innervation of the organ. To overcome this problem, Inamori et al. filled hollow fibers with hepatocyte spheres covered by human umbilical vein endothelial cells to form a dense vascular network.145 LeCluyse et al. also reconstructed the multicellular structure and hemodynamic characteristics of the liver using this method.74 In addition, Lee et al. successfully induced the formation of vascular networks by injecting collagen bioinks containing three different cell types into the channels of the PCL framework.146 Researchers are developing many similar methods to build vascular networks, but there is still room for improvement. Third, due to the limited source of liver cells, xenogeneic liver cells have to be introduced. However, xenogeneic liver cells will face immune rejection and infection. Fourth, the functional expression of cultured liver tissue requires the co-culture of various functional cells and the interaction of various growth factors. Although some problems still exist, the results of liver tissue engineering research have been remarkable. Although implantable complete livers cannot be constructed at present, it is believed that with the continuous development of science and technology, implantable complete livers will be manufactured in the next few years, which will become a good experimental tool and clinical treatment model.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the National Natural Science Foundation of China (Project No. 61803323), Shan-dong Province Natural Science Foundation (Project No. ZR2019BF049), and Joint fund of Science & Technology Department of Liaoning Province and State Key Laboratory of Robotics (Project No. 2021-KF-22-03).References
- M. R. Murthy, T. J. Reid III, A. Sicignano, N. Tanaka and M. G. J. J. O. M. B. Rossmann, Structure of beef liver catalase, J. Mol. Biol., 1981, 152, 465–499 CrossRef CAS PubMed.
- H. Eklund, B. Nordström, E. Zeppezauer, G. Söderlund, I. Ohlsson, T. Boiwe, B.-O. Söderberg, O. Tapia, C.-I. Brändén and Å. J. J. O. M. B. Åkeson, Three-dimensional structure of horse liver alcohol dehydrogenase at 2.4 Å resolution, J. Mol. Biol., 1976, 102, 27–59 CrossRef CAS PubMed.
- H. Tanaka, K. Kato, E. Yamashita, T. Sumizawa, Y. Zhou, M. Yao, K. Iwasaki, M. Yoshimura and T. J. S. Tsukihara, The structure of rat liver vault at 3.5 angstrom resolution, Science, 2009, 323, 384–388 CrossRef CAS PubMed.
- S. N. Bhatia, G. H. Underhill, K. S. Zaret and I. J. Fox, Cell and tissue engineering for liver disease, Sci. Transl. Med., 2014, 6, 245sr242 Search PubMed.
- S. Bhatia, U. Balis, M. Yarmush and M. J. T. F. J. Toner, Effect of cell–cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells, FASEB J., 1999, 13, 1883–1900 CrossRef CAS PubMed.
- Y.-S. Li, H.-J. Harn, D.-K. Hsieh, T.-C. Wen, Y.-M. Subeq, L.-Y. Sun, S.-Z. Lin and T.-W. Chiou, Cells and Materials for Liver Tissue Engineering, Cell Transplant., 2013, 22, 685–700 Search PubMed.
- S. Shetty, P. F. Lalor and D. H. Adams, Liver sinusoidal endothelial cells — gatekeepers of hepatic immunity, Nat. Rev. Gastroenterol. Hepatol., 2018, 15, 555–567 CrossRef CAS PubMed.
- O. Krenkel and F. Tacke, Liver macrophages in tissue homeostasis and disease, Nat. Rev. Immunol., 2017, 17, 306–321 CrossRef CAS PubMed.
- P. Gissen and I. M. J. J. O. H. Arias, Structural and functional hepatocyte polarity and liver disease, J. Hepatol., 2015, 63, 1023–1037 CrossRef PubMed.
- P. J. Meier, E. S. Sztul, A. Reuben and J. L. J. T. J. O. C. B. Boyer, Structural and functional polarity of canalicular and basolateral plasma membrane vesicles isolated in high yield from rat liver, J. Cell Biol., 1984, 98, 991–1000 CrossRef CAS PubMed.
- M. Tsuge, N. Hiraga, H. Takaishi, C. Noguchi, H. Oga, M. Imamura, S. Takahashi, E. Iwao, Y. Fujimoto, H. Ochi, K. Chayama, C. Tateno and K. Yoshizato, Infection of human hepatocyte chimeric mouse with genetically engineered hepatitis B virus, Hepatology, 2005, 42, 1046–1054 CrossRef CAS PubMed.
- M. Dorner, J. A. Horwitz, B. M. Donovan, R. N. Labitt, W. C. Budell, T. Friling, A. Vogt, M. T. Catanese, T. Satoh, T. Kawai, S. Akira, M. Law, C. M. Rice and A. Ploss, Completion of the entire hepatitis C virus life cycle in genetically humanized mice, Nature, 2013, 501, 237–241 CrossRef CAS PubMed.
- R. Grant, D. Hay and A. Callanan, From scaffold to structure: the synthetic production of cell derived extracellular matrix for liver tissue engineering, Biomed. Phys. Eng. Express, 2018, 4 Search PubMed.
- S. P. Harrison, R. Siller, Y. Tanaka, Y. Xiang, B. Patterson, H. Kempf, E. Melum, K. S. Åsrud, M. E. Chollet, E. Andersen, P. M. Sandset, S. Baumgarten, F. Bonanini, D. Kurek, S. Mathapati, R. Almaas, K. Sharma, S. R. Wilson, F. S. Skottvoll, I. C. Boger, I. L. Bogen, T. A. Nyman, J. J. Wu, A. Bezrouk, D. Cizkova, J. Mokry, R. Zweigerdt, I.-H. Park and G. J. Sullivan, Scalable production of tissue-like vascularised liver organoids from human PSCs, bioRxiv, 2020 DOI:10.1101/2020.12.02.406835.
- P. Gunness, D. Mueller, V. Shevchenko, E. Heinzle, M. Ingelman-Sundberg and F. Noor, 3D organotypic cultures of human HepaRG cells: a tool for in vitro toxicity studies, Toxicol. Sci., 2013, 133, 67–78 CrossRef CAS PubMed.
- J. W. Nichol and A. Khademhosseini, Modular Tissue Engineering: Engineering Biological Tissues from the Bottom Up, Soft Matter, 2009, 5, 1312–1319 RSC.
- R. Sudo, Multiscale tissue engineering for liver reconstruction, Organogenesis, 2014, 10, 216–224 CrossRef PubMed.
- L. Cordero-Espinoza and M. Huch, The balancing act of the liver: tissue regeneration versus fibrosis, J. Clin. Invest., 2018, 128, 85–96 CrossRef PubMed.
- K. Matsumoto and T. J. C. R. I. O. Nakamura, Hepatocyte growth factor: molecular structure, roles in liver regeneration, and other biological functions, Crit. Rev. Oncog., 1992, 3, 27–54 CAS.
- T. Agarwal, B. Subramanian and T. K. Maiti, Liver Tissue Engineering: Challenges and Opportunities, ACS Biomater. Sci. Eng., 2019, 5, 4167–4182 CrossRef CAS PubMed.
- V. Hosseini, N. F. Maroufi, S. Saghati, N. Asadi, M. Darabi, S. N. S. Ahmad, H. Hosseinkhani and R. Rahbarghazi, Current progress in hepatic tissue regeneration by tissue engineering, J. Transl. Med., 2019, 17, 383 CrossRef PubMed.
- A. Boyd, O. Cain, A. Chauhan and G. J. Webb, Medical liver biopsy: background, indications, procedure and histopathology, Frontline Gastroenterol., 2020, 11, 40 CrossRef CAS PubMed.
- Z. Heydari, M. Najimi, H. Mirzaei, A. Shpichka, M. Ruoss, Z. Farzaneh, L. Montazeri, A. Piryaei, P. Timashev, R. Gramignoli, A. Nussler, H. Baharvand and M. Vosough, Tissue Engineering in Liver Regenerative Medicine: Insights into Novel Translational Technologies, Cells, 2020, 9 Search PubMed.
- E. S. Mirdamadi, D. Kalhori, N. Zakeri, N. Azarpira and M. Solati-Hashjin, Liver Tissue Engineering as an Emerging Alternative for Liver Disease Treatment, Tissue Eng., Part B, 2020, 26, 145–163 CrossRef PubMed.
- M. A. Pearen, H. K. Lim, F. D. Gratte, M. A. Fernandez-Rojo, S. K. Nawaratna, G. N. Gobert, J. K. Olynyk, J. E. E. Tirnitz-Parker and G. A. Ramm, Murine Precision-Cut Liver Slices as an Ex Vivo Model of Liver Biology, J. Visualized Exp., 2020, 157, e60992 Search PubMed.
- E. L. LeCluyse, R. P. Witek, M. E. Andersen and M. J. J. C. R. I. T. Powers, Organotypic liver culture models: meeting current challenges in toxicity testing, Crit. Rev. Toxicol., 2012, 42, 501–548 CrossRef CAS PubMed.
- Y. Nahmias, F. Berthiaume and M. L. Yarmush, Integration of technologies for hepatic tissue engineering, Adv. Biochem. Eng. Biotechnol., 2007, 103, 309–329 CrossRef PubMed.
- H. Morales-Navarrete, H. Nonaka, F. Segovia-Miranda, M. Zerial and Y. Kalaidzidis, Automatic recognition and characterization of different non-parenchymal cells in liver tissue, in 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI), IEEE, 2016, pp. 536–540 Search PubMed.
- W. Yang, H.-X. Yan, L. Chen, Q. Liu, Y.-Q. He, L.-X. Yu, S.-H. Zhang, D.-D. Huang, L. Tang and X.-N. J. C. R. Kong, Wnt/β-catenin signaling contributes to activation of normal and tumorigenic liver progenitor cells, Cancer Res., 2008, 68, 4287–4295 CrossRef CAS PubMed.
- M. L. Dumble, E. J. Croager, G. C. Yeoh and E. A. J. C. Quail, Generation and characterization of p53 null transformed hepatic progenitor cells: oval cells give rise to hepatocellular carcinoma, Carcinogenesis, 2002, 23, 435–445 CrossRef CAS PubMed.
- N. J. H. Fausto, Liver regeneration and repair: hepatocytes, progenitor cells, and stem cells, Hepatology, 2004, 39, 1477–1487 CrossRef PubMed.
- H. Malhi, A. N. Irani, S. Gagandeep and S. J. J. O. C. S. Gupta, Isolation of human progenitor liver epithelial cells with extensive replication capacity and differentiation into mature hepatocytes, J. Cell Sci., 2002, 115, 2679–2688 CrossRef CAS PubMed.
- G. Nowak, B. Ericzon, S. Nava, M. Jaksch, M. Westgren and S. J. G. Sumitran-Holgersson, Identification of expandable human hepatic progenitors which differentiate into mature hepatic cells in vivo, Gut, 2005, 54, 972–979 CrossRef CAS PubMed.
- G. Darlington, [3] Liver cell lines, 1987.
- R. F. Clayton, A. Rinaldi, E. E. Kandyba, M. Edward, C. Willberg, P. Klenerman and A. H. J. L. I. Patel, Liver cell lines for the study of hepatocyte functions and immunological response, Liver Int., 2005, 25, 389–402 CrossRef PubMed.
- S. Knasmüller, V. Mersch-Sundermann, S. Kevekordes, F. Darroudi, W. Huber, C. Hoelzl, J. Bichler and B. J. T. Majer, Use of human-derived liver cell lines for the detection of environmental and dietary genotoxicants; current state of knowledge, Toxicology, 2004, 198, 315–328 CrossRef PubMed.
- A. Kinasiewicz, A. Gautier, D. Lewinska, J. Bukowski, C. Legallais and A. Weryński, Culture of C3A cells in alginate beads for fluidized bed bioartificial liver, Transplant. Proc., 2007, 39, 2911–2913 CrossRef CAS PubMed.
- S.-F. Lan, B. Safiejko-Mroczka and B. J. T. I. V. Starly, Long-term cultivation of HepG2 liver cells encapsulated in alginate hydrogels: a study of cell viability, morphology and drug metabolism, Toxicol. In Vitro, 2010, 24, 1314–1323 CrossRef CAS PubMed.
- Z. Liu, M. Lu, M. Takeuchi, T. Yue, Y. Hasegawa, Q. Huang and T. Fukuda, 2017.
- M. Yamada, R. Utoh, K. Ohashi, K. Tatsumi, M. Yamato, T. Okano and M. J. B. Seki, Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions, Biomaterials, 2012, 33, 8304–8315 CrossRef CAS PubMed.
- H. Jeon, K. Kang, S. A. Park, W. D. Kim, S. S. Paik, S.-H. Lee, J. Jeong and D. J. G. Choi, Generation of multilayered 3D structures of HepG2 cells using a bio-printing technique, Gut Liver, 2017, 11, 121 CrossRef PubMed.
- G. E. Brown and S. R. Khetani, Microfabrication of liver and heart tissues for drug development, Philos. Trans. R. Soc., B, 2018, 373 Search PubMed.
- O. Ukairo, M. McVay, S. Krzyzewski, S. Aoyama, K. Rose, M. E. Andersen, S. R. Khetani, E. L. J. J. O. B. LeCluyse and M. Toxicology, Bioactivation and toxicity of acetaminophen in a rat hepatocyte micropatterned coculture system, J. Biochem. Mol. Toxicol., 2013, 27, 471–478 CrossRef CAS PubMed.
- T. Okudaira, R. Yabuta, H. Mizumoto and T. Kajiwara, Fabrication of a fiber-type hepatic tissue by bottom-up method using multilayer spheroids, J. Biosci. Bioeng., 2017, 123, 739–747 CrossRef CAS PubMed.
- Z. Q. Feng, X. Chu, N. P. Huang, T. Wang, Y. Wang, X. Shi, Y. Ding and Z. Z. Gu, The effect of nanofibrous galactosylated chitosan scaffolds on the formation of rat primary hepatocyte aggregates and the maintenance of liver function, Biomaterials, 2009, 30, 2753–2763 CrossRef CAS PubMed.
- L. Li, Y. Zhang and X. Pan, Cell Microencapsulation, Springer, 2017, pp. 199–206 Search PubMed.
- G. Mazza, W. Al-Akkad, K. Rombouts and M. J. H. C. Pinzani, Liver tissue engineering: from implantable tissue to whole organ engineering, Hepatol. Commun., 2018, 2, 131–141 CrossRef PubMed.
- Y. Nagamoto, K. Tashiro, K. Takayama, K. Ohashi, K. Kawabata, F. Sakurai, M. Tachibana, T. Hayakawa, M. K. Furue and H. Mizuguchi, The promotion of hepatic maturation of human pluripotent stem cells in 3D co-culture using type I collagen and Swiss 3T3 cell sheets, Biomaterials, 2012, 33, 4526–4534 CrossRef CAS PubMed.
- G. Mazza, W. Al-Akkad, A. Telese, L. Longato, L. Urbani, B. Robinson, A. Hall, K. Kong, L. Frenguelli and G. J. S. R. Marrone, Rapid production of human liver scaffolds for functional tissue engineering by high shear stress oscillation-decellularization, Sci. Rep., 2017, 7, 1–14 CrossRef CAS PubMed.
- A. Soto-Gutierrez, L. Zhang, C. Medberry, K. Fukumitsu, D. Faulk, H. Jiang, J. Reing, R. Gramignoli, J. Komori, M. Ross, M. Nagaya, E. Lagasse, D. Stolz, S. C. Strom, I. J. Fox and S. F. Badylak, A whole-organ regenerative medicine approach for liver replacement, Tissue Eng., Part C, 2011, 17, 677–686 CrossRef CAS PubMed.
- W. S. Turner, E. Schmelzer, R. McClelland, E. Wauthier, W. Chen and L. M. Reid, Human hepatoblast phenotype maintained by hyaluronan hydrogels, J. Biomed. Mater. Res., Part B, 2007, 82, 156–168 CrossRef PubMed.
- S. R. Khetani, D. R. Berger, K. R. Ballinger, M. D. Davidson, C. Lin and B. R. Ware, Microengineered liver tissues for drug testing, J. Lab. Autom., 2015, 20, 216–250 CrossRef CAS PubMed.
- J. H. Brown, P. Das, M. D. DiVito, D. Ivancic, L. P. Tan and J. A. Wertheim, Nanofibrous PLGA electrospun scaffolds modified with type I collagen influence hepatocyte function and support viability in vitro, Acta Biomater., 2018, 73, 217–227 CrossRef CAS PubMed.
- Y. Gao and A. Callanan, Influence of surface topography on PCL electrospun scaffolds for liver tissue engineering, J. Mater. Chem. B, 2021, 9, 8081–8093 RSC.
- I. Slivac, E. Zdraveva, F. Ivančić, B. Žunar, T. Holjevac Grgurić, V. Gaurina Srček, I. K. Svetec, T. Dolenec, E. G. Bajsić, M. Tominac Trcin and B. Mijović, Bioactivity Comparison of Electrospun PCL Mats and Liver Extracellular Matrix as Scaffolds for HepG2 Cells, Polymers, 2021, 13 Search PubMed.
- Y. Kim, Y. W. Kim, S. B. Lee, K. Kang, S. Yoon, D. Choi, S. H. Park and J. Jeong, Hepatic patch by stacking patient-specific liver progenitor cell sheets formed on multiscale electrospun fibers promotes regenerative therapy for liver injury, Biomaterials, 2021, 274, 120899 CrossRef CAS PubMed.
- Y. Du, N. Li, H. Yang, C. Luo, Y. Gong, C. Tong, Y. Gao, S. Lü and M. Long, Mimicking liver sinusoidal structures and functions using a 3D-configured microfluidic chip, Lab Chip, 2017, 17, 782–794 RSC.
- Y. Pang, K. Montagne, M. Shinohara, K. Komori and Y. Sakai, Liver tissue engineering based on aggregate assembly: efficient formation of endothelialized rat hepatocyte aggregates and their immobilization with biodegradable fibres, Biofabrication, 2012, 4, 045004 CrossRef CAS PubMed.
- Q. Chu, Y. Zhao, X. Shi, W. Han, Y. Zhang, X. Zheng and J. Zhu, In vivo-like 3-D model for sodium nitrite- and acrylamide-induced hepatotoxicity tests utilizing HepG2 cells entrapped in micro-hollow fibers, Sci. Rep., 2017, 7, 14837 CrossRef PubMed.
- J. Bierwolf, M. Lutgehetmann, K. Feng, J. Erbes, S. Deichmann, E. Toronyi, C. Stieglitz, B. Nashan, P. X. Ma and J. M. Pollok, Primary rat hepatocyte culture on 3D nanofibrous polymer scaffolds for toxicology and pharmaceutical research, Biotechnol. Bioeng., 2011, 108, 141–150 CrossRef CAS PubMed.
- J. He, Y. Pang, H. Yang, K. Montagne, M. Shinohara, Y. Mao, W. Sun and Y. Sakai, Modular assembly-based approach of loosely packing co-cultured hepatic tissue elements with endothelialization for liver tissue engineering, Ann. Transl. Med., 2020, 8, 1400 CrossRef CAS PubMed.
- S. F. Abu-Absi, L. K. Hansen and W.-S. Hu, Three-dimensional co-culture of hepatocytes and stellate cells, Cytotechnology, 2004, 45, 125–140 CrossRef PubMed.
- A. Xiong, T. W. Austin, E. Lagasse, N. Uchida, S. Tamaki, B. B. Bordier, I. L. Weissman, J. S. Glenn and M. T. Millan, Isolation of human fetal liver progenitors and their enhanced proliferation by three-dimensional coculture with endothelial cells, Tissue Eng., Part A, 2008, 14, 995–1006 CrossRef CAS.
- S. March, V. Ramanan, K. Trehan, S. Ng, A. Galstian, N. Gural, M. A. Scull, A. Shlomai, M. M. Mota, H. E. Fleming, S. R. Khetani, C. M. Rice and S. N. Bhatia, Micropatterned coculture of primary human hepatocytes and supportive cells for the study of hepatotropic pathogens, Nat. Protoc., 2015, 10, 2027–2053 CrossRef CAS PubMed.
- S. Hong, S. J. Oh, D. Choi, Y. Hwang and S. H. Kim, Self-Organized Liver Microtissue on a Bio-Functional Surface: The Role of Human Adipose-Derived Stromal Cells in Hepatic Function, Int. J. Mol. Sci., 2020, 21 Search PubMed.
- S. N. Bhatia, M. L. Yarmush and M. Toner, Controlling cell interactions by micropatterning in co-cultures: hepatocytes and 3T3 fibroblasts, J. Biomed. Mater. Res., 1997, 34, 189–199 CrossRef CAS PubMed.
- Y. Liu, C. Meyer, C. Xu, H. Weng, C. Hellerbrand, P. ten Dijke and S. Dooley, Animal models of chronic liver diseases, Am. J. Physiol.: Gastrointest. Liver Physiol., 2013, 304, G449–468 CrossRef CAS PubMed.
- B. R. Ware, D. R. Berger and S. R. Khetani, Prediction of Drug-Induced Liver Injury in Micropatterned Co-cultures Containing iPSC-Derived Human Hepatocytes, Toxicol. Sci., 2015, 145, 252–262 CrossRef CAS PubMed.
- J. Landry, D. Bernier, C. Ouellet, R. Goyette and N. Marceau, Spheroidal aggregate culture of rat liver cells: histotypic reorganization, biomatrix deposition, and maintenance of functional activities, J. Cell Biol., 1985, 101, 914–923 CrossRef CAS PubMed.
- J. M. Pollok, D. Kluth, R. A. Cusick, H. Lee, H. Utsunomiya, P. X. Ma, R. Langer, C. E. Broelsch and J. P. Vacanti, Formation of spheroidal aggregates of hepatocytes on biodegradable polymers under continuous-flow bioreactor conditions, Eur. J. Pediatr. Surg., 1998, 8, 195–199 CrossRef CAS PubMed.
- D. Rogozhnikov, W. Luo, S. Elahipanah, P. J. O'Brien and M. N. Yousaf, Generation of a Scaffold-Free Three-Dimensional Liver Tissue via a Rapid Cell-to-Cell Click Assembly Process, Bioconjugate Chem., 2016, 27, 1991–1998 CrossRef CAS PubMed.
- J. Landry, D. Bernier, C. Ouellet, R. Goyette and N. Marceau, Spheroidal aggregate culture of rat liver cells: histotypic reorganization, biomatrix deposition, and maintenance of functional activities, J. Cell Biol., 1985, 101, 914–923 CrossRef CAS PubMed.
- P. Godoy, N. J. Hewitt, U. Albrecht, M. E. Andersen, N. Ansari, S. Bhattacharya, J. G. Bode, J. Bolleyn, C. Borner, J. Böttger, A. Braeuning, R. A. Budinsky, B. Burkhardt, N. R. Cameron, G. Camussi, C. S. Cho, Y. J. Choi, J. Craig Rowlands, U. Dahmen, G. Damm, O. Dirsch, M. T. Donato, J. Dong, S. Dooley, D. Drasdo, R. Eakins, K. S. Ferreira, V. Fonsato, J. Fraczek, R. Gebhardt, A. Gibson, M. Glanemann, C. E. Goldring, M. J. Gómez-Lechón, G. M. Groothuis, L. Gustavsson, C. Guyot, D. Hallifax, S. Hammad, A. Hayward, D. Häussinger, C. Hellerbrand, P. Hewitt, S. Hoehme, H. G. Holzhütter, J. B. Houston, J. Hrach, K. Ito, H. Jaeschke, V. Keitel, J. M. Kelm, B. Kevin Park, C. Kordes, G. A. Kullak-Ublick, E. L. LeCluyse, P. Lu, J. Luebke-Wheeler, A. Lutz, D. J. Maltman, M. Matz-Soja, P. McMullen, I. Merfort, S. Messner, C. Meyer, J. Mwinyi, D. J. Naisbitt, A. K. Nussler, P. Olinga, F. Pampaloni, J. Pi, L. Pluta, S. A. Przyborski, A. Ramachandran, V. Rogiers, C. Rowe, C. Schelcher, K. Schmich, M. Schwarz, B. Singh, E. H. Stelzer, B. Stieger, R. Stöber, Y. Sugiyama, C. Tetta, W. E. Thasler, T. Vanhaecke, M. Vinken, T. S. Weiss, A. Widera, C. G. Woods, J. J. Xu, K. M. Yarborough and J. G. Hengstler, Recent advances in 2D and 3D in vitro systems using primary hepatocytes, alternative hepatocyte sources and non-parenchymal liver cells and their use in investigating mechanisms of hepatotoxicity, cell signaling and ADME, Arch. Toxicol., 2013, 87, 1315–1530 CrossRef CAS PubMed.
- E. L. LeCluyse, R. P. Witek, M. E. Andersen and M. J. Powers, Organotypic liver culture models: meeting current challenges in toxicity testing, Crit. Rev. Toxicol., 2012, 42, 501–548 CrossRef CAS PubMed.
- Y. Sakai, K. Furukawa, T. Ushida, T. Tateishi and M. Suzuki, In vitro organization of biohybrid rat liver tissue incorporating growth factor- and hormone-releasing biodegradable polymer microcapsules, Cell Transplant., 2001, 10, 479–483 Search PubMed.
- J. Lee, M. J. Cuddihy, G. M. Cater and N. A. Kotov, Engineering liver tissue spheroids with inverted colloidal crystal scaffolds, Biomaterials, 2009, 30, 4687–4694 CrossRef CAS PubMed.
- R. McClelland, K. Tech and J. M. Macdonald, Construction of a multicoaxial hollow fiber bioreactor, Methods Mol. Biol., 2013, 1001, 215–226 CrossRef CAS PubMed.
- S. Zhang, L. Chen, T. Liu, Z. Wang and Y. Wang, Integration of single-layer skin hollow fibers and scaffolds develops a three-dimensional hybrid bioreactor for bioartificial livers, J. Mater. Sci.: Mater. Med., 2014, 25, 207–216 CrossRef CAS PubMed.
- S. Jitraruch, A. Dhawan, R. D. Hughes, C. Filippi, D. Soong, C. Philippeos, S. C. Lehec, N. D. Heaton, M. S. Longhi and R. R. Mitry, Alginate microencapsulated hepatocytes optimised for transplantation in acute liver failure, PLoS One, 2014, 9, e113609 CrossRef PubMed.
- C. C. Bell, D. F. G. Hendriks, S. M. L. Moro, E. Ellis, J. Walsh, A. Renblom, L. Fredriksson Puigvert, A. C. A. Dankers, F. Jacobs, J. Snoeys, R. L. Sison-Young, R. E. Jenkins, Å. Nordling, S. Mkrtchian, B. K. Park, N. R. Kitteringham, C. E. P. Goldring, V. M. Lauschke and M. Ingelman-Sundberg, Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease, Sci. Rep., 2016, 6, 25187 CrossRef CAS PubMed.
- T. M. Rahman, C. Selden, M. Khalil, I. Diakanov and H. J. Hodgson, Alginate-encapsulated human hepatoblastoma cells in an extracorporeal perfusion system improve some systemic parameters of liver failure in a xenogeneic model, Artif. Organs, 2004, 28, 476–482 CrossRef PubMed.
- G. H. Lee, J. S. Lee, G. H. Lee, W. Y. Joung, S. H. Kim, S. H. Lee, J. Y. Park and D. H. Kim, Networked concave microwell arrays for constructing 3D cell spheroids, Biofabrication, 2017, 10, 015001 CrossRef PubMed.
- S. Khodabakhshaghdam, A. B. Khoshfetrat and R. Rahbarghazi, Alginate-chitosan core-shell microcapsule cultures of hepatic cells in a small scale stirred bioreactor: impact of shear forces and microcapsule core composition, J. Biol. Eng., 2021, 15, 14 CrossRef CAS PubMed.
- L.-Q. Chu, X.-N. Zou, W. Knoll, R. J. S. Förch and C. Technology, Thermosensitive surfaces fabricated by plasma polymerization of N,N-diethylacrylamide, Surf. Coat. Technol., 2008, 202, 2047–2051 CrossRef CAS.
- J. Baek, Y. Cho, H. J. Park, G. Choi, J. S. Lee, M. Lee, S. J. Yu, S. W. Cho, E. Lee and S. G. J. A. M. Im, A Surface-Tailoring Method for Rapid Non-Thermosensitive Cell-Sheet Engineering via Functional Polymer Coatings, Adv. Mater., 2020, 32, 1907225 CrossRef CAS PubMed.
- M. Harimoto, M. Yamato, M. Hirose, C. Takahashi, Y. Isoi, A. Kikuchi and T. Okano, Novel approach for achieving double-layered cell sheets co-culture: overlaying endothelial cell sheets onto monolayer hepatocytes utilizing temperature-responsive culture dishes, J. Biomed. Mater. Res., 2002, 62, 464–470 CrossRef CAS PubMed.
- M. Fujii, K. Yamanouchi, Y. Sakai, Z. Baimakhanov, I. Yamaguchi, A. Soyama, M. Hidaka, M. Takatsuki, T. Kuroki and S. Eguchi, In vivo construction of liver tissue by implantation of a hepatic non-parenchymal/adipose-derived stem cell sheet, J. Tissue Eng. Regener. Med., 2018, 12, e287–e295 CrossRef CAS PubMed.
- A. A. Banaeiyan, J. Theobald, J. Paukštyte, S. Wölfl, C. B. Adiels and M. Goksör, Design and fabrication of a scalable liver-lobule-on-a-chip microphysiological platform, Biofabrication, 2017, 9, 015014 CrossRef PubMed.
- R. Shank, G. Morrison, C. H. Cheng, I. Karl and R. Schwartz, Cell heterogeneity within the hepatic lobule (quantitative histochemistry), J. Histochem. Cytochem., 1959, 7, 237–239 CrossRef CAS PubMed.
- Z. Liu, M. Takeuchi, M. Nakajima, C. Hu, Y. Hasegawa, Q. Huang and T. Fukuda, Three-dimensional hepatic lobule-like tissue constructs using cell-microcapsule technology, Acta Biomater., 2017, 50, 178–187 CrossRef CAS PubMed.
- W. Ekataksin and K. Wake, Liver units in three dimensions: I. organization of argyrophilic connective tissue skeleton in porcine Liver with particular reference to the “compound hepatic lobule”, Am. J. Anat., 1991, 191, 113–153 CrossRef CAS PubMed.
- X. Fu, J. P. Sluka, S. G. Clendenon, K. W. Dunn, Z. Wang, J. E. Klaunig and J. A. J. P. O. Glazier, Modeling of xenobiotic transport and metabolism in virtual hepatic lobule models, PLoS one, 2018, 13, e0198060 CrossRef PubMed.
- J. Poisson, S. Lemoinne, C. Boulanger, F. Durand, R. Moreau, D. Valla and P.-E. Rautou, Liver sinusoidal endothelial cells: Physiology and role in liver diseases, J. Hepatol., 2017, 66, 212–227 CrossRef CAS PubMed.
- L. D. J. T. J. O. C. I. DeLeve, Liver sinusoidal endothelial cells and liver regeneration, J. Clin. Invest., 2013, 123, 1861–1866 CrossRef CAS PubMed.
- K. Rennert, S. Steinborn, M. Gröger, B. Ungerböck, A. M. Jank, J. Ehgartner, S. Nietzsche, J. Dinger, M. Kiehntopf, H. Funke, F. T. Peters, A. Lupp, C. Gärtner, T. Mayr, M. Bauer, O. Huber and A. S. Mosig, A microfluidically perfused three dimensional human liver model, Biomaterials, 2015, 71, 119–131 CrossRef CAS PubMed.
- M. Yamada, R. Utoh, K. Ohashi, K. Tatsumi, M. Yamato, T. Okano and M. Seki, Controlled formation of heterotypic hepatic micro-organoids in anisotropic hydrogel microfibers for long-term preservation of liver-specific functions, Biomaterials, 2012, 33, 8304–8315 CrossRef CAS PubMed.
- S. Kakinuma, Y. Tanaka, R. Chinzei, M. Watanabe, K. Shimizu-saito, Y. Hara, K. Teramoto, S. Arii, C. Sato, K. Takase, T. Yasumizu and H. Teraoka, Human Umbilical Cord Blood as a Source of Transplantable Hepatic Progenitor Cells, Stem Cells, 2003, 21, 217–227 CrossRef PubMed.
- S. Zhang, L. Chen, T. Liu, B. Zhang, D. Xiang, Z. Wang and Y. Wang, Human Umbilical Cord Matrix Stem Cells Efficiently Rescue Acute Liver Failure Through Paracrine Effects Rather than Hepatic Differentiation, Tissue Eng., Part A, 2012, 18, 1352–1364 CrossRef CAS PubMed.
- Z. Jia, Y. Cheng, X. Jiang, C. Zhang, G. Wang, J. Xu, Y. Li, Q. Peng and Y. Gao, 3D Culture System for Liver Tissue Mimicking Hepatic Plates for Improvement of Human Hepatocyte (C3A) Function and Polarity, BioMed Res. Int., 2020, 2020, 6354183 Search PubMed.
- Y. Yajima, C. N. Lee, M. Yamada, R. Utoh and M. Seki, Development of a perfusable 3D liver cell cultivation system via bundling-up assembly of cell-laden microfibers, J. Biosci. Bioeng., 2018, 126, 111–118 CrossRef CAS PubMed.
- S. Caddeo, M. Boffito and S. Sartori, Tissue Engineering Approaches in the Design of Healthy and Pathological In Vitro Tissue Models, Front. Bioeng. Biotechnol., 2017, 5 Search PubMed.
- C. Zhong, H. Y. Xie, L. Zhou, X. Xu and S. S. Zheng, Human hepatocytes loaded in 3D bioprinting generate mini-liver, Hepatobiliary Pancreatic Dis. Int., 2016, 15, 512–518 CrossRef.
- V. Liu Tsang, A. A. Chen, L. M. Cho, K. D. Jadin, R. L. Sah, S. DeLong, J. L. West and S. N. Bhatia, Fabrication of 3D hepatic tissues by additive photopatterning of cellular hydrogels, FASEB J., 2007, 21, 790–801 CrossRef PubMed.
- H. Jeon, K. Kang, S. A. Park, W. D. Kim, S. S. Paik, S.-H. Lee, J. Jeong and D. Choi, Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique, Gut. Liver, 2017, 11, 121–128 CrossRef PubMed.
- K. Arai, T. Yoshida, M. Okabe, M. Goto, T. A. Mir, C. Soko, Y. Tsukamoto, T. Akaike, T. Nikaido, K. Zhou and M. Nakamura, Fabrication of 3D-culture platform with sandwich architecture for preserving liver-specific functions of hepatocytes using 3D bioprinter, J. Biomed. Mater. Res., Part A, 2017, 105, 1583–1592 CrossRef CAS PubMed.
- K. M. Park, K. H. Hussein, S. H. Hong, C. Ahn, S. R. Yang, S. M. Park, O. K. Kweon, B. M. Kim and H. M. Woo, Decellularized Liver Extracellular Matrix as Promising Tools for Transplantable Bioengineered Liver Promotes Hepatic Lineage Commitments of Induced Pluripotent Stem Cells, Tissue Eng., Part A, 2016, 22, 449–460 CrossRef CAS PubMed.
- G. Mazza, W. Al-Akkad, A. Telese, L. Longato, L. Urbani, B. Robinson, A. Hall, K. Kong, L. Frenguelli, G. Marrone, O. Willacy, M. Shaeri, A. Burns, M. Malago, J. Gilbertson, N. Rendell, K. Moore, D. Hughes, I. Notingher, G. Jell, A. Del Rio Hernandez, P. De Coppi, K. Rombouts and M. Pinzani, Rapid production of human liver scaffolds for functional tissue engineering by high shear stress oscillation-decellularization, Sci. Rep., 2017, 7, 5534 CrossRef PubMed.
- S. Croce, A. Peloso, T. Zoro, M. A. Avanzini and L. Cobianchi, A Hepatic Scaffold from Decellularized Liver Tissue: Food for Thought, Biomolecules, 2019, 9 Search PubMed.
- E. A. Rossi, L. F. Quintanilha, C. K. V. Nonaka and B. S. F. Souza, Advances in Hepatic Tissue Bioengineering with Decellularized Liver Bioscaffold, Stem Cells Int., 2019, 2019, 2693189 Search PubMed.
- S. Kazemnejad, Hepatic tissue engineering using scaffolds: state of the art, Avicenna J. Med. Biotechnol., 2009, 1, 135–145 CAS.
- S. Kazemnejad, A. Allameh, M. Soleimani, A. Gharehbaghian, Y. Mohammadi, N. Amirizadeh and M. Jazayery, Biochemical and molecular characterization of hepatocyte-like cells derived from human bone marrow mesenchymal stem cells on a novel three-dimensional biocompatible nanofibrous scaffold, J. Gastroenterol. Hepatol., 2009, 24, 278–287 CrossRef CAS PubMed.
- R. Grant, J. Hallett, S. Forbes, D. Hay and A. Callanan, Blended electrospinning with human liver extracellular matrix for engineering new hepatic microenvironments, Sci. Rep., 2019, 9, 6293 CrossRef PubMed.
- S. Nagam Hanumantharao and S. Rao, Multi-Functional Electrospun Nanofibers from Polymer Blends for Scaffold Tissue Engineering, Fibers, 2019, 7 Search PubMed.
- K. N. Chua, W. S. Lim, P. Zhang, H. Lu, J. Wen, S. Ramakrishna, K. W. Leong and H. Q. Mao, Stable immobilization of rat hepatocyte spheroids on galactosylated nanofiber scaffold, Biomaterials, 2005, 26, 2537–2547 CrossRef CAS PubMed.
- D. Rajendran, A. Hussain, D. Yip, A. Parekh, A. Shrirao and C. H. Cho, Long-term liver-specific functions of hepatocytes in electrospun chitosan nanofiber scaffolds coated with fibronectin, J. Biomed. Mater. Res., Part A, 2017, 105, 2119–2128 CrossRef CAS PubMed.
- N. Navarro-Alvarez, A. Soto-Gutierrez, J. D. Rivas-Carrillo, Y. Chen, T. Yamamoto, T. Yuasa, H. Misawa, J. Takei, N. Tanaka and N. Kobayashi, Self-assembling peptide nanofiber as a novel culture system for isolated porcine hepatocytes, Cell Transplant, 2006, 15, 921–927 Search PubMed.
- S. M. Coward, C. Selden, A. Mantalaris and H. J. Hodgson, Proliferation rates of HepG2 cells encapsulated in alginate are increased in a microgravity environment compared with static cultures, Artif. Organs, 2005, 29, 152–158 CrossRef CAS PubMed.
- A. Kinasiewicz, A. Gautier, D. Lewinska, J. Bukowski, C. Legallais and A. Weryński, Culture of C3A cells in alginate beads for fluidized bed bioartificial liver, Transplant. Proc., 2007, 39, 2911–2913 CrossRef CAS PubMed.
- S.-F. Lan, B. Safiejko-Mroczka and B. J. T. I. V. A. I. J. P. I. A. W. B. Starly, Long-term cultivation of HepG2 liver cells encapsulated in alginate hydrogels: a study of cell viability, morphology and drug metabolism, Toxicol. In Vitro, 2010, 24(4), 1314–1323 CrossRef CAS PubMed.
- L. Li, Y. Zhang and X. Pan, Preparation and Characterization of Alginate-Chitosan Microcapsule for Hepatocyte Culture, Methods Mol. Biol., 2017, 1479, 199–206 CrossRef CAS PubMed.
- Z. Liu, M. Lu, M. Takeuchi, T. Yue, Y. Hasegawa, Q. Huang and T. Fukuda, In vitro mimicking the morphology of hepatic lobule tissue based on Ca-alginate cell sheets, Biomed. Mater., 2018, 13, 035004 CrossRef PubMed.
- J. Meseguer-Ripolles, A. Kasarinaite, B. Lucendo-Villarin and D. C. Hay, Protocol for automated production of human stem cell derived liver spheres, STAR Protocols, 2021, 2, 100502 CrossRef PubMed.
- P. J. Lee, P. J. Hung and L. P. Lee, An artificial liver sinusoid with a microfluidic endothelial-like barrier for primary hepatocyte culture, Biotechnol. Bioeng., 2007, 97, 1340–1346 CrossRef CAS PubMed.
- Y. C. Toh, T. C. Lim, D. Tai, G. Xiao, D. van Noort and H. Yu, A microfluidic 3D hepatocyte chip for drug toxicity testing, Lab Chip, 2009, 9, 2026–2035 RSC.
- X. Ma, X. Qu, W. Zhu, Y.-S. Li, S. Yuan, H. Zhang, J. Liu, P. Wang, C. S. E. Lai, F. Zanella, G.-S. Feng, F. Sheikh, S. Chien and S. Chen, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2206 CrossRef CAS PubMed.
- C. R. Pimentel, S. K. Ko, C. Caviglia, A. Wolff, J. Emnéus, S. S. Keller and M. Dufva, Three-dimensional fabrication of thick and densely populated soft constructs with complex and actively perfused channel network, Acta Biomater., 2018, 65, 174–184 CrossRef PubMed.
- R. Chang, K. Emami, H. Wu and W. Sun, Biofabrication of a three-dimensional liver micro-organ as an in vitro drug metabolism model, Biofabrication, 2010, 2, 045004 CrossRef PubMed.
- A. Faulkner-Jones, C. Fyfe, D.-J. Cornelissen, J. Gardner, J. King, A. Courtney and W. Shu, Bioprinting of human pluripotent stem cells and their directed differentiation for the generation of 3D liver-like micro-tissues, Biofabrication, 2015, 7, 044102 CrossRef PubMed.
- H. Jeon, K. Kang, S. Park, W. Kim, S. Paik, S.-H. Lee, J. Jeong and D. Choi, Generation of Multilayered 3D Structures of HepG2 Cells Using a Bio-printing Technique, Gut. Liver, 2016, 11 Search PubMed.
- S. Knowlton and S. Tasoglu, A Bioprinted Liver-on-a-Chip for Drug Screening Applications, Trends Biotechnol., 2016, 34, 681–682 CrossRef CAS PubMed.
- X. Ma, X. Qu, W. Zhu, Y. S. Li, S. Yuan, H. Zhang, J. Liu, P. Wang, C. Lai, F. Zanella, G.-S. Feng, F. Sheikh, S. Chien and S. Chen, Deterministically patterned biomimetic human iPSC-derived hepatic model via rapid 3D bioprinting, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 2206–2211 CrossRef CAS PubMed.
- D. G. Nguyen, J. Funk, J. B. Robbins, C. Crogan-Grundy, S. C. Presnell, T. Singer and A. B. Roth, Bioprinted 3D Primary Liver Tissues Allow Assessment of Organ-Level Response to Clinical Drug Induced Toxicity In Vitro, PLoS One, 2016, 11, e0158674 CrossRef PubMed.
- I. T. Ozbolat and M. Hospodiuk, Current advances and future perspectives in extrusion-based bioprinting, Biomaterials, 2016, 76, 321–343 CrossRef CAS PubMed.
- N. Sears, P. Dhavalikar, D. Seshadri and E. Cosgriff-Hernandez, A Review of 3D Printing of Tissue Engineering Constructs, Tissue Eng., Part B, 2016, 22 Search PubMed.
- W. Hassanein, M. C. Uluer, J. Langford, J. D. Woodall, A. Cimeno, U. Dhru, A. Werdesheim, J. Harrison, C. Rivera-Pratt, S. Klepfer, A. Khalifeh, B. Buckingham, P. S. Brazio, D. Parsell, C. Klassen, C. Drachenberg, R. N. Barth and J. C. LaMattina, Recellularization via the bile duct supports functional allogenic and xenogenic cell growth on a decellularized rat liver scaffold, Organogenesis, 2017, 13, 16–27 CrossRef CAS PubMed.
- H. Shimoda, H. Yagi, H. Higashi, K. Tajima, K. Kuroda, Y. Abe, M. Kitago, M. Shinoda and Y. Kitagawa, Decellularized liver scaffolds promote liver regeneration after partial hepatectomy, Sci. Rep., 2019, 9, 12543 CrossRef PubMed.
- Y. Kino, M. Sawa, S. Kasai and M. Mito, Multiporous cellulose microcarrier for the development of a hybrid artificial liver using isolated hepatocytes, J. Surg. Res., 1998, 79, 71–76 CrossRef CAS PubMed.
- J. Jiang, N. Kojima, T. Kinoshita, A. Miyajima, W. Yan and Y. Sakai, Cultivation of fetal liver cells in a three-dimensional poly-L-lactic acid scaffold in the presence of oncostatin M, Cell Transplant, 2002, 11, 403–406 Search PubMed.
- C. E. Semino, J. R. Merok, G. G. Crane, G. Panagiotakos and S. Zhang, Functional differentiation of hepatocyte-like spheroid structures from putative liver progenitor cells in three-dimensional peptide scaffolds, Differentiation, 2003, 71, 262–270 CrossRef CAS PubMed.
- B. Zavan, P. Brun, V. Vindigni, A. Amadori, W. Habeler, P. Pontisso, D. Montemurro, G. Abatangelo and R. Cortivo, Extracellular matrix-enriched polymeric scaffolds as a substrate for hepatocyte cultures: in vitro and in vivo studies, Biomaterials, 2005, 26, 7038–7045 CrossRef CAS PubMed.
- A. L. Larkin, R. R. Rodrigues, T. M. Murali and P. Rajagopalan, Designing a multicellular organotypic 3D liver model with a detachable, nanoscale polymeric Space of Disse, Tissue Eng., Part C, 2013, 19, 875–884 CrossRef CAS PubMed.
- S. Nemati, S.-j. Kim, Y. M. Shin and H. Shin, Current progress in application of polymeric nanofibers to tissue engineering, Nano Converge., 2019, 6, 36 CrossRef PubMed.
- T. Yamamoto, N. Navarro-Alvarez, A. Soto-Gutierrez, T. Yuasa, M. Iwamuro, Y. Kubota, M. Seita, H. Kawamoto, S. M. Javed, E. Kondo, H. Noguchi, S. Kobayashi, S. Nakaji and N. Kobayashi, Treatment of acute liver failure in mice by hepatocyte xenotransplantation, Cell Transplant., 2010, 19, 799–806 Search PubMed.
- K. R. Stevens, M. A. Scull, V. Ramanan, C. L. Fortin, R. R. Chaturvedi, K. A. Knouse, J. W. Xiao, C. Fung, T. Mirabella and A. X. J. S. T. M. Chen, In situ expansion of engineered human liver tissue in a mouse model of chronic liver disease, Sci. Transl. Med., 2017, 9 Search PubMed.
- M. Inamori, H. Mizumoto and T. J. T. E. P. A. Kajiwara, An approach for formation of vascularized liver tissue by endothelial cell–covered hepatocyte spheroid integration, Tissue Eng., Part A, 2009, 15, 2029–2037 CrossRef CAS PubMed.
- J. W. Lee, Y.-J. Choi, W.-J. Yong, F. Pati, J.-H. Shim, K. S. Kang, I.-H. Kang, J. Park and D.-W. J. B. Cho, Development of a 3D cell printed construct considering angiogenesis for liver tissue engineering, Biofabrication, 2016, 8, 015007 CrossRef PubMed.
- A. Kern, A. Bader, R. Pichlmayr and K. F. Sewing, Drug metabolism in hepatocyte sandwich cultures of rats and humans, Biochem. Pharmacol., 1997, 54, 761–772 CrossRef CAS PubMed.
- A. Nocito, J. Kononen, O. P. Kallioniemi and G. Sauter, Tissue microarrays (TMAs) for high-throughput molecular pathology research, Int. J. Cancer, 2001, 94, 1–5 CrossRef CAS PubMed.
- S. Zhang, W. Tong, B. Zheng, T. A. Susanto, L. Xia, C. Zhang, A. Ananthanarayanan, X. Tuo, R. B. Sakban, R. Jia, C. Iliescu, K. H. Chai, M. McMillian, S. Shen, H. Leo and H. Yu, A robust high-throughput sandwich cell-based drug screening platform, Biomaterials, 2011, 32, 1229–1241 CrossRef CAS PubMed.
- T. Ohkura, K. Ohta, T. Nagao, K. Kusumoto, A. Koeda, T. Ueda, T. Jomura, T. Ikeya, E. Ozeki, K. Wada, K. Naitoh, Y. Inoue, N. Takahashi, H. Iwai, H. Arakawa and T. Ogihara, Evaluation of human hepatocytes cultured by three-dimensional spheroid systems for drug metabolism, Drug Metab. Pharmacokinet., 2014, 29, 373–378 CrossRef PubMed.
- W. Lin, K. Chang, M. Chiang, S. Fan and C. Liu, Three-dimensional biomimetic liver tissue platform for drug testing, IEEE 16th International Conference on Nanotechnology, 2016, pp. 55–58 Search PubMed.
- Y. Wang, R. K. Kankala, J. Zhang, L. Hao, K. Zhu, S. Wang, Y. S. Zhang and A. Chen, Modeling Endothelialized Hepatic Tumor Microtissues for Drug Screening, Adv. Sci., 2020, 7, 2002002 CrossRef CAS PubMed.
- R. Nudischer, K. Renggli, A. Hierlemann, A. B. Roth and C. Bertinetti-Lapatki, Characterization of a long-term mouse primary liver 3D tissue model recapitulating innate-immune responses and drug-induced liver toxicity, PLoS One, 2020, 15, e0235745 CrossRef CAS PubMed.
- G. Elaut, T. Henkens, P. Papeleu, S. Snykers, M. Vinken, T. Vanhaecke and V. Rogiers, Molecular mechanisms underlying the dedifferentiation process of isolated hepatocytes and their cultures, Curr. Drug Metab., 2006, 7, 629–660 CrossRef CAS PubMed.
- C. Rowe, D. T. Gerrard, R. Jenkins, A. Berry, K. Durkin, L. Sundstrom, C. E. Goldring, B. K. Park, N. R. Kitteringham, K. P. Hanley and N. A. Hanley, Proteome-wide analyses of human hepatocytes during differentiation and dedifferentiation, Hepatology, 2013, 58, 799–809 CrossRef CAS PubMed.
- L. Broutier, G. Mastrogiovanni, M. M. A. Verstegen, H. E. Francies, L. M. Gavarró, C. R. Bradshaw, G. E. Allen, R. Arnes-Benito, O. Sidorova, M. P. Gaspersz, N. Georgakopoulos, B.-K. Koo, S. Dietmann, S. E. Davies, R. K. Praseedom, R. Lieshout, J. N. M. Ijzermans, S. J. Wigmore, K. Saeb-Parsy, M. J. Garnett, L. J. W. van der Laan and M. Huch, Human primary liver cancer–derived organoid cultures for disease modeling and drug screening, Nat. Med., 2017, 23, 1424–1435 CrossRef CAS PubMed.
- S. A. Jenkins, A. Grandison, J. N. Baxter, D. W. Day, I. Taylor and R. Shields, A dimethylnitrosamine-induced model of cirrhosis and portal hypertension in the rat, J. Hepatol., 1985, 1, 489–499 CrossRef CAS PubMed.
- N. Tanimizu, A. Miyajima and K. E. J. M. B. O. T. C. Mostov, Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture, Molecular biology of the cell, 2007, 18, 1472–1479 CrossRef CAS PubMed.
- T. Katsuda, N. Kojima, T. Ochiya and Y. J. T. E. P. A. Sakai, Biliary epithelial cells play an essential role in the reconstruction of hepatic tissue with a functional bile ductular network, Tissue Eng., Part A, 2013, 19, 2402–2411 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |



