Open Access Article
Open Access ArticleSelenium nanoparticles: a review on synthesis and biomedical applications
Neha
Bisht
,
Priyanka
Phalswal
 and
Pawan K.
Khanna
and
Pawan K.
Khanna
 *
*
Nano Chemistry and Quantum Dots R & D Laboratory, Department of Applied Chemistry, Defence Institute of Advanced Technology (DIAT), Ministry of Defence, Govt. of India, Girinagar, Pune-411025, India. E-mail: pawankhanna2002@yahoo.co.in
First published on 17th December 2021
Abstract
Selenium is a trace and essential micronutrient for the health of humans, animals, and microorganisms. Recently, selenium nanoparticles (SeNPs) attracted the interest of many researchers due to their biocompatibility, bioavailability, and low toxicity. Therefore, due to their higher bioactivity selenium nanoparticles are largely being used in various biomedical applications. Generally, selenium nanoparticles can be synthesized by physical, chemical, and biological methods. However, the biologically synthesized SeNPs demonstrate greater compatibility with human organs and tissues. The effect of size, shape, and the method employed for their synthesis on their applications in biological systems has been explored by many researchers. This review discusses various synthesis methods employed for their preparation and highlights their applications in the biomedical field such as in the treatment of fungal, bacterial, and parasitic infections, cancer, and diabetes. They can also act as chemopreventive agents, anti-inflammatory agents, and antioxidants.
1. Introduction
Elemental selenium (Se) has great importance in the fields of physics, chemistry, and biology. Naturally, selenium exists in two forms: inorganic (selenite and selenate) and organic (selenomethionine and selenocysteine). Selenium is found in the form of both crystalline and amorphous polymorphic structures in nature. Monoclinic and trigonal selenium are the crystalline forms. Monoclinic selenium (m-Se) is red in color and contains rings of Se8. Based on different packings, it exists in three allotropic forms (α, β and γ). Trigonal selenium (t-Se) is black in colour and the most stable crystalline form at room temperature. Red amorphous (a-Se), black amorphous and vitreous selenium are the non-crystalline forms of selenium.1 The crystal structures of t-Se and m-Se are presented in Fig. 1(a and b), respectively.2 Selenium, a part of selenoproteins and selenocompounds within the human body, plays a critical role in reproduction, DNA synthesis, thyroid hormone, metabolism, and protection from infections and oxidative damage. It has many industrial and commercial applications. Due to its high photoconductivity and low melting point, it possesses great catalytic activity towards organic hydration and oxidation reactions. Selenium is an essential trace element in the human body but the margin between its usefulness and toxicity is very slender. The United Kingdom group of vitamins and minerals recommended the daily intake of selenium by women and men should be 60 μg and 70 μg, respectively.3 A daily intake of more than 400 μg could be toxic which leads to a disorder known as selenosis.3–5 Selenium plays a key role as a biochemical component of glutathione peroxidase, an enzyme responsible for the protection of essential SH-groups and for the decomposition of peroxides, thereby acting as an antioxidant. The bactericidal activity of selenium is because of its capacity to catalyse the oxidation of intracellular thiols, causing death of microscopic organisms.6–8 Selenium, as one of the fundamental minor components, is affirmed to improve the action or restore the activity of the seleno-catalyst and glutathione peroxidase in prevention of free radical harm to cells and tissues in vivo.9–12 It is widely utilized in nutritional supplements as well as a potential nutrient in fertilizers.13 A selenium free diet leads to malfunctioning of the liver and hemolytic processes. Se deficiency leads to many diseases such as Kashin–Beck disease,14 neurological disorders,15 and chronic degenerative diseases.16 Selenium supplements can prevent diseases, such as viral infections,17 immune system dysfunction,18 and neural function loss.19 The recent boom in nanotechnology furnished an indefinite number of applications of metal nanoparticles in biomedicine. Metal NPs of Au and Ag, have immense medicinal benefits but are costlier to synthesize, whereas the synthesis of Se nanoparticles (SeNPs) is economical and they can be integrated with other biological agents to enhance their biological properties. Vahdati et al.20 demonstrated the synergistic antimicrobial effect of SeNPs and lysozymes. Due to their higher surface-to-volume ratio at the nano-level, the surface of the particles is more exposed which leads to an enhanced activity of selenium more profoundly in the nano-regime. SeNPs show promising potential as antioxidants, cancer therapeutic agents, and drug carriers in biological applications.21 Several studies have supported their anticancer,22 antioxidant,21 antimicrobial,23–25 and anti-biofilm properties.26 Use of nano-Se medication in the therapy of Huntington's disease has given promising results.27 SeNPs possess unique semiconducting, photoelectric and X-ray-sensing properties, and are used in photocells, photocopying, photometers, and xerography.28 Their importance in renewable energy devices has also been greatly mentioned.29 Additionally, selenium nanoparticles (SeNPs) are environmentally important because of their mercury capturing properties.30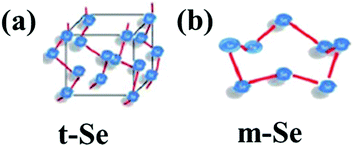 | ||
| Fig. 1 General representation of the crystal structure of (a) t-Se and (b) m-Se. Reproduced from ref. 2 with permission from The Royal Society of Chemistry, copyright 2015. | ||
The absorption profile of selenium indicates that nano-selenium can lead to a blue shift in the absorption spectrum and the range of this shift can vary from preparation to preparation. Thus, it is deduced that the bandgap of Se increases from 1.7 eV in bulk to 3.3 eV in the nano-range.31 The least toxic form of selenium is elemental Se, and hence its nano-form has attracted significant attention. Interestingly, functionalized SeNPs exhibit less cytotoxicity than their other forms such as selenate, selenite, selenoproteins, and inorganic selenium.32,33 In clinical trials, instead of conventional selenium sources, elemental nano-sized selenium can be a better alternative. Fig. 2 demonstrates the decrease of cytotoxicity as the selenium nanoparticle size increases.34 SeNPs are highly biologically active,35 anti-hydroxyl radicals,36 and chemo-preventive,32,37 have a detoxifying effect on heavy metal exposure,38 and prevent DNA oxidation.39
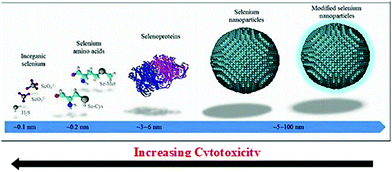 | ||
| Fig. 2 Extent of cytotoxicity from various sources of selenium with respect to their size. Reproduced from ref. 34 with permission from Science Elsevier, copyright 2017. | ||
It has been suggested by various researchers that impetus to conduct research on SeNPs is greatly desired owing to their tremendous applications in the biomedical field due to their excellent antibacterial activity, sometimes even more effective than Ag NPs.40–42 SeNPs have been synthesized in various forms such as nanowires, nanorods, and nanotubes through various methods such as sonochemical,43 refluxing, microwave,44 hydrothermal,45 gamma irradiation,46 pulsed laser ablation,13,47,48 and physical evaporation approaches.49–51 The precursors mostly used in these methods are sodium selenite, sodium selenate, sodium selenosulphate, selenous acid and selenium oxide. Use of these precursors is often preferred because selenium in selenite and selenate forms has a hazardous effect even at extremely low concentrations and gets accumulated in the bio-system. In recent times Khanna et al. have demonstrated exclusive use of 1,2,3-selenadiazole as a source of selenium for a variety of semiconductor NP ink/quantum dots including selenium nanoparticles.2,52–55 Converting them to biologically and technologically important SeNPs can help in revitalizing the environment and protect from contamination of Se compounds. Various reviews on the biological applications of selenium nanoparticles and their compounds56 have been published in recent years while focusing on their biological synthesis methods.57 This review emphasizes the different synthesis methods of SeNPs and their advantages and disadvantages over each other. The authors also present a detailed discussion of various biological applications of SeNPs such as anti-cancerous, antioxidative, antidiabetic, antiparasitic, antimicrobial, antibacterial, antifungal and chemopreventive agents.
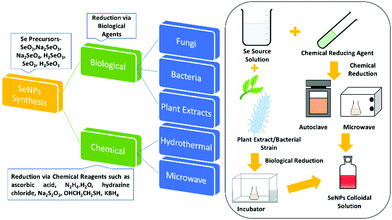
2. Synthesis methods
The synthesis of SeNPs by various methods has been reported. These methods can be broadly divided into two categories, namely biological and chemical reduction. Biological reduction methods include reduction of various organic/inorganic compounds of selenium via biological agents such as bacteria or plant extracts and their conversion to non-toxic and beneficial SeNPs. Chemical reduction uses chemical reducing agents. This method can further be classified depending upon the energy source used or the apparatus employed for the reaction. Researchers have reported mainly hydrothermal, microwave, and sonochemical methods in this category. This broad classification is presented in Fig. 3 and then these methods are further discussed in sub-sections.2.1. Chemical reduction method
Among the vast varieties of synthesis methods reported, reduction of Se salts is the most common and simplest method to synthesize SeNPs.58 The sources can be natural compounds from plants or microorganisms59,60 or reagents/chemicals having the ability to cause reduction in the oxidation state such as ascorbic acid.61 However, the nanoparticles (NPs) synthesized from natural compounds are found to be less toxic than those prepared by employing chemicals.62 In this section, synthesis via the chemical method is discussed and synthesis from natural compounds, i.e., biogenic reduction, will be discussed in a later section. Chemical reduction uses chemical compounds that reduce the element, its salt or compounds and the size is controlled by use of surfactants or growth terminating reagents such as polyvinylchloride (PVP), folic acid, etc.63 to obtain a stable colloidal solution of SeNPs desired for various applications. Taking SeO2 as the precursor, El-ghazaly et al.64 synthesized spherical SeNPs about 13 nm in size where PVP served as the stabilising agent and KBH4 as the reducing agent in ice-cold solution. The appearance of orange colour indicated the formation of α-Se. The particle shape and size was <10 nm in diameter based on transmission electron microscopy (TEM) analysis. Gao et al.36 reported the use of protein molecules to stabilise hollow SeNPs formed via reduction of sodium selenite with mercaptoethanol. The reaction was carried out at a low temperature (<10 °C) and continued for almost 3 days. Particles with an average diameter of nearly 30 nm and a shell thickness of 4 nm were identified and the characteristic binding energy of 55.3 eV corresponding to Se(3d) from the X-ray photoelectron spectroscopy (XPS) spectrum confirmed Se in the zero-valent state.In another study, Tran et al.65 reduced sodium selenite by ascorbic acid using polyvinyl alcohol (PVA) as the stabilising agent, resulting in an average particle size of 70 nm as observed from dynamic light scattering (DLS) measurements. The absorption was observed in the wavelength range 250 to 450 nm, and the non-appearance of a sharp peak was attributed to the non-metallic nature of Se. The formation of elemental selenium was also confirmed by a characteristic XPS peak at 54.2 eV for Se (3d). Khalid et al.66 repeated the same experiment for PVA capped SeNPs to study their application in cellular imaging. They observed an XPS peak at almost the same value as reported by Tran et al.65 A similar UV-Vis absorption band was observed, and the PL showed an emission at 580 nm. By using the same chemical sources and reagents along with chitosan as an additional stabilising agent, Boroumand et al.67 obtained spherical NPs of sizes 136 and 195 nm, respectively, where the UV-Vis spectra showed absorption peaks at 264 and 310 nm. Similarly, by changing the stabilising agent to polysorbate 20, Vahdati et al.20 obtained a characteristic UV absorption peak at 265 nm consistent with a previous report but with a much smaller particle size in the range of 35–45 nm in comparison to that obtained by using PVA or chitosan. Likewise, Shah et al.68 also used PVA as the stabilizer to obtain SeNPs of 50–100 nm size. The authors for the first time reported that sodium selenosulphate responded to the acrylonitrile monomer, forming SeNPs. By replacing acrylonitrile with acetonitrile, the authors could not isolate Se nanoparticles. This suggests that the nitrile group was not directly involved in the reaction. Using hyperbranched polysaccharide as the stabiliser in the reaction of selenious acid with ascorbic acid in water led to formation of highly stable (>1 month) SeNPs with an average size of about 24 nm which was calculated through DSC.69 Lin et al.70 used a very mild reducing agent, i.e., dilute SO2 solution, and selenious acid as the selenium source. Selenous acid being a strong oxidising agent gets easily reduced by SO2. Sodium dodecyl sulfate (SDS) was used as the stabilizer and the reaction temperature was kept at 80 °C. The variation in particle size with time was studied by TEM. The particle size varied from 30 nm to 200 nm in a time span of 30 s to 4 minutes. The article by Yu et al.71 highlighted the use of sodium thiosulfate as the reducing agent. Reaction of selenium dioxide with sodium thiosulfate in the presence of SDS for about 4 h in ambient conditions yielded monodisperse SeNPs of about 70 nm size as estimated from the TEM images. Huang et al.72 studied the effect of the size of SeNPs on their inhibiting action towards methicillin-sensitive and methicillin-resistant Staphylococcus aureus (MSSA and MRSA). For this, they synthesized spherical SeNPs of size ranging from 43 to 205 nm by varying the ratio of SeO2, the reducing agent sodium thiosulphate and the capping agent PVA. From the antibacterial and cytotoxicity studies, they observed the highest efficacy for 81 nm sized particles. The synthesized particles were found to be spherical and mono-disperse from the scanning electron microscopy (SEM) images.72 The SeNPs synthesized using the chemical reduction method by the use of polysaccharides as stabilizing/reducing agents was critically reviewed by Shi et al.73
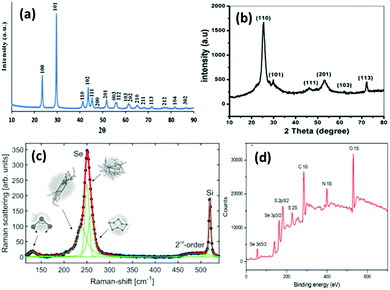 | ||
| Fig. 4 (a) XRD pattern of SeNPs synthesized via the hydrothermal method. Reproduced from ref. 78 with permission from Science Elsevier, copyright 2013, (b) XRD pattern of SeNPs synthesized using drumstick extract. Reproduced from ref. 105 with permission from John Wiley and Sons, copyright 2019, (c) Raman spectra of Se representing the presence of various phases at different energies. Reproduced from ref. 48 with permission from Springer Nature, Copyright 2019. (d) XPS spectrum of L-cysteine capped SeNPs. Reproduced from ref. 109 with permission from RSC Publications, copyright 2017. | ||
2.2. Biogenic reduction
Green synthesis of metal NPs have gained the attention of researchers in the last two decades. Despite the various advantages of SeNPs over organic and inorganic selenium compounds, the main problem is their poor cellular intake. Therefore, it is desirable to synthesize SeNPs via a biological route to offer improved biocompatibility and stability. The previous methods mentioned above require chemical reducing and stabilising agents which may be toxic and can hinder their utilization in biological systems. Generally, it is believed that biological agents from plant extracts and microorganisms represent a superior option in contrast to chemical methods to satisfy the increasing demand for low-cost and non-hazardous preparation methods. It has been reported that biological extracts act as bio-reducing agents, as well as stabilizers for nanoparticles. There is a tremendous amount of literature available on the biosynthesis of SeNPs.80–87 Biological agent mediated synthesis is easy as this does not require special apparatus and conditions. These biological reagents include bacteria, fungi, algae, protein molecules and plant extracts. For instance, several researchers have reported the synthesis of SeNPs using fenugreek extract,88Cassia auriculata extract,89E. officinalis fruit extract,90 microbial polymers,91Bacillus sp. B2,92Saccharomyces cerevisiae var. boulardii yeast cells,93Penicillium chrysogenum,94Penicillium expansum,95Vitis vinifera aq.,96Withania somnifera extract,97Azadirachta indica,98Aloe vera leaf extract,99Burkholderia fungorum,100Diospyros montana leaf extract,101 lemon leaf extract,102 or bacteria grown from Mueller–Hinton broth103 as reducing agents with incubation of the precursor mixture for a day or two. However, the use of plant extracts is even more cost effective than using bacteria or fungi as bacterial culture and isolation requires techniques, experience, and special equipment. With this method, high-quality SeNPs can be produced without requiring specific equipment, with less or no product wastage, and without the need of a large amount of solvent. Hence, such methods are considered as single-step, inexpensive and eco-friendly. Plant extracts contain terpenes, flavonoids, tannins, coumarins, cinnamic acid, phenolic acid, vitamins, sterols, polysaccharides, enzymes, proteins, etc. which can act both as reducing and as stabilizing agents. Recently, a few review articles brought out the importance of plant-based SeNPs, the simplicity in the synthesis and the feasibility for biological applications.57,104 Moreover, preparation of SeNPs using medicinal plants may enhance their beneficial properties. These biological catalysts are vital environmentally as they reduce the toxic oxyanions present in the environment into non-toxic elemental selenium. Fig. 4b shows the XRD pattern of SeNPs synthesized using drumstick extract showing peaks at 2θ values of 25.38°, 29.98°, 46.1°, 53.00°, 62.38°, and 72.00° due to [100], [101], [111], [201], [103], and [113] planes, respectively, showing the crystalline nature of Se.105 However, the XRD diffraction peaks for red Se prepared from Muller–Hinton broth were obtained at 23.78°, 29.79°, and 43.88° indexed to the face centred cubic (fcc) planes [100], [101], and [102], respectively, and UV absorption was observed at 263 nm.103 The most intense peak for the hexagonal phase of SeNPs appears at 29.73°.106 The characteristic Raman peak for elemental Se is observed at 254 cm−1. However, a slight shift may be observed depending upon the polymorphic forms of Se. Fig. 4c shows the Raman spectra of SeNPs synthesized via the laser ablation method which shows a mixture of various phases.48 The authors interpreted that the peak at the lowest frequency of 131 cm−1 corresponds to bending vibrations. The Se–Se stretching in crystalline Se chains appears at 237 cm−1 and 250 cm−1. The peak at 259 cm−1 was reported to be due to the vibration modes of in the Se8 crown structure. In general, depending upon the intensity of a particular peak, one can estimate the type of Se formed. Piacenza et al.107 suggested the use of biologically synthesized SeNPs in bio-imaging. They synthesized SeNPs from yeast extract which exhibited a strong photoluminescence emission in the blue-red region of the visible spectrum and a luminescence lifetime comparable to those of mostly used fluorophores. Afzal et al.108 reported the use of various cyanobacterial strains for the biogenic synthesis of SeNPs and studied their antioxidant properties. Out of the many strains used, Arthrospira indica SOSA-4 was suggested to be the best strain as it yielded SeNPs of 11.8 nm size and improved the antioxidant activity. When heated for a few minutes, reaction of L-cystine with selenious acid yielded cystine capped SeNPs whose size was determined by the DLS technique and valency was confirmed by the XPS spectra. The appearance of peaks at 55.5 eV and 161.5 eV for Se 3d5/2 and Se 3P3/2, respectively, affirms Se in the zero-valent state. The XPS spectrum is shown in Fig. 4d.109In another report, Ramamurthy et al.88 prepared 100–150 nm SeNPs by both chemical and biogenic methods. They observed that the size obtained from the biogenic method was slightly bigger than that achieved by the chemical method, yet the biogenic method proved to be more efficient mainly because biological strains are a combination of various reducing and stabilising agents. For the synthesis of SeNPs, researchers have mostly reported the use of biogenic/biotic reduction. The various biological agents used along with the precursor and reaction conditions are summarized in Table 1.
| S. no. | Se precursor | Biological agent/extract origin | Reaction conditions | Morphology and size of particles | Applications | Ref. |
|---|---|---|---|---|---|---|
| 1 | SeO2 | Bacillus licheniformis | Incubated at 37 °C for 48 h | Spherical with a diameter of 10–50 nm, λabs = 263 nm, colourless to orange-red (red Se) | Antimicrobial against foodborne pathogens | 103 |
| 2 | SeO2 | Bacillus licheniformis | Incubated at 37 °C for 24 h | Spherical with a diameter of 50–80 nm, λabs = 560 nm | Chemopreventive against lung carcinoma | 124 |
| 3 | Na2SeO3 | Azadirachta indica | Incubated for 5 min and 10 min | λ abs = 560 nm, size ∼153 nm | Antibacterial against Gram positive and Gram negative bacteria | 98 |
| λ abs = 580, 670 nm, size ∼287 nm | ||||||
| 4 | SeO2 | Trigonella foenum-graecum leaf extract | Stirring at room temperature for 2–8 h | D = 20 nm, nanospheres with a 5–12 nm diameter, λabs = 355 nm | Anticancer | 125 |
| Degradation of sunset yellow FCF | ||||||
| 5 | Na2SeO3 | Terminalia | Incubated at 30 °C for 72 h | Size = 10-80 nm, λabs = 390 nm, dark red, λem = 595 nm | Effect against As(III) toxicity | 126 |
| Arjuna leaf extract | ||||||
| 6 | H2SeO4 | L-Cystine | Heating at 105 °C for 1 h | Nanospheres ∼60 nm, red Se | Interaction with human serum albumin | 109 |
| 7 | H2SeO3 | Trigonella foenum-graecum L. leaf extract | Incubated at RT for 24 h | Absorption band = 200–400 nm, red Se, 50–150 nm oval particles | Breast cancer | 88 |
| 8 | Na2SeO3 | Glutathione (GSH) | Instant formation of SeNPs | Nanospheres of 40–100 nm size | Prevention of PVC related medical infections | 127 and 128 |
| 9 | Na2SeO3 | Glutathione (GSH) | Instant formation of SeNPs in basic media | Hemispherical, size = 80–200 nm | Cytotoxicity of SeNP coated PVC in rat dermal fibroblasts | 6 |
| 10 | Na2SeO3·5H2O, capped with PVP | Pseudomonas alcaliphila (bacteria) | Incubated for 48 h | λ abs = 280 nm, Raman = 254 cm−1 (m-Se), nanospheres of 40–100 nm size | The role of PVP explained in text | 129 |
| 11 | H2SeO3 | Vitis vinifera extract | Refluxed together for 15 min in water | Nanospheres of 3–18 nm size | — | 130 |
| 12 | Na2SeO3 | Ethanol extract of Bee propolis from Apis mellifera colonies along with ascorbic acid | Stirred at RT for 24 h | Brown to orange colour change | Antioxidant, antimicrobial, antibacterial, antifungal | 131 |
| λ abs = 265 nm | ||||||
| 52–118 nm diameter | ||||||
| 13 | Na2SeO3 | Allium sativum extract | Incubated for 72 h until color change | 7–45 nm, stable up to 2 months | Antioxidant | 132 |
| Dark pink colour of solution, λabs = 400 nm | ||||||
| 14 | SeO2 | Lactobacillus brevis | Incubated at 37 °C for 72 h | — | Anticancer (studies on mice with breast cancer) | 133 and 134 |
| 15 | Na2SeO3 | Various cyanobacterial strains | Incubated at 32 °C for 2–5 days | λ abs = 259–274 nm, red spherical NPs with sizes in the range of 11–60 nm depending upon the strain | Antioxidant | 108 |
| 16 | Na2SeO3 | Moringa oleifera (drumstick) leaf extracts | Incubated at RT for 24 h | λ abs = 299, 400 nm, λem = 599 nm, bandgap = 2.3 eV | Degradation of sunset yellow dye | 105 |
| Mixture of nanospheres and nanorods | Anticancer studies on human colon, breast and hepatic cancers | |||||
| Average size = 18.85 nm (calculated by Scherrer's equation) and 23–35 nm from TEM | ||||||
| 17 | NaHSeO3 | Crispum (parsley) leaf extracts | Kept overnight at room temperature | Orange-red Se | — | 135 |
| λ abs = 270 nm | ||||||
| Spherical with a maximum number of particles with a size around 400 nm (DLS) | ||||||
| Zeta potential = −14.2 mV | ||||||
| 18 | Na2SeO3 | Rhizobacterium Azospirillum brasilense Sp7 | Incubated at 32 °C for 24 h | Nanospheres of 50–100 nm diameter coated with a thin layer of protein | — | 136 |
| 19 | Na2SeO3 | L-Cystine and peptone from soybean | Stirring for 10 h at 25 °C | 36.2 nm (from XRD) | Protection against irradiation used nephropathy | 106 |
| 20–50 nm (TEM) | ||||||
| 20 | H2SeO3 | Diospyros montana leaf extract | Incubated at RT for 24 h | 4–16 nm from TEM and DLS | Antibacterial activity against Gram (+), Gram (−) bacteria and fungi, anticancer activity against human breast cancer | 101 |
| 21 | Na2SeO3 | Mushroom polysaccharide protein complex – stabilising agent, ascorbic acid – reducing agent | Kept at RT for 12 h with stirring | 100 to 600 nm in size depending upon concentration of the stabilising agent | Anticancer activity against human breast cancer cells | 137 |
3. Biomedical applications
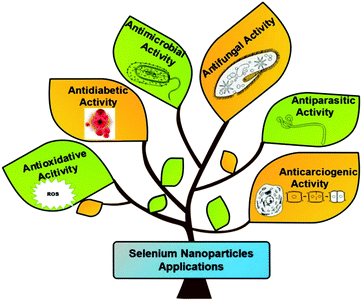
The biomedical applications of SeNPs have attracted global interest of many researchers due to their importance at cellular and tissue levels. It is well known that excessive production of toxic reactive oxygen species (ROS) can be triggered by various abiotic stresses, thus causing several diseases, due to damaged essential nutrients such as carbohydrates, proteins, and lipids.110 The continuous rise in diseases like cancer, diabetes, and bacterial infections is threatening the worldwide healthcare system. Currently, treatment of cancer needs various kinds of therapies such as chemotherapy, radiotherapy, or a combination of both to reduce the global death rate.111,112 The adverse effects of such therapies generally are of great concern.113 It has been documented that Se supplements alongside regular anticancer treatments upgraded the productivity of chemotherapeutic medications, declining results and enhancing the generic state of the patients.114–116 The new clinical preliminaries have assessed the well-being and efficiency of selenium, both in viability and toxicity of normal anticancer treatments.117 Numerous in vivo and in vitro studies at the supranutritional level have exhibited an anticancerous impact by selenium.118,119 Davis et al.120 reported that the frequency of liver malignancy can be decreased by 35% on consuming supplements of selenite salt. A few studies revealed that, by taking 200 μg of selenium each day, as Se yeasts, the risk of colorectal, lung, and prostate cancers has decreased.121 Similarly, Se supplements also reduce the stomach malignancy risk.122 It has been reported that a lower dosage of selenoprotein (SelP) can increase the risk of cancer in prostate, throat, kidney, colon, and lungs.123 Selenium rich Brassica and Allium plants lessen the risk of colon malignant growth.121 Despite having such advantages these organic and inorganic sources of selenium exert higher toxicity and possess lower biocompatibility. To overcome these issues, SeNPs came to the limelight. Nano-selenium in the form of nano-medicine offers excellent properties such as antimicrobial, anticancer, antidiabetic, antiparasitic, and antioxidant as shown in Fig. 5. The antioxidative properties of nano-selenium minimize the impact of ROS and free radicals. The higher efficiency of SeNPs displays augmented medicinal effects as antibiotic, antidiabetic, cytotoxic, and chemoprotective medicines in comparison to the traditional therapeutic medicines.138 They can also be used as therapeutic and theranostic agents.139 It is noteworthy that various studies on SeNPs evidenced reduced toxicity and characteristics of interfacing biomaterials with tissues and cells. SeNPs are biologically active and naturally accessible and take part in numerous oxidoreductive cycles. They also show many regulative outcomes to help the appropriate functioning of plants and living bodies and numerous medical advantages.34,140,141 Researchers have shown that SeNPs can be utilized as a cancer preventive agent by controlling the cell damage caused by free radicals, in anticancer therapies, and as antimicrobial agents. These can also be used as an ingredient in nano-biosensor fabrication. The action mechanism of SeNPs can be defined in two ways: (a) disrupting the integrity of the cell membrane through ROS generation and (b) alteration of the DNA sequence of microorganisms by damaging the cell wall and binding to the cell membrane to inhibit the growth of microorganisms. The cycles associated with SeNP preparation can be utilized in wastewater treatment and bioremediation. They can play a significant role in the treatment of heavy metals by combining with them to suppress their poisonous impact.56 Based on the abovementioned advantages the next section discusses the various applications of SeNPs in detail.3.1. Anti-carcinogenic activity
Nowadays, selenium nanoparticles are widely being investigated for their anti-cancer activity against breast, lung, kidney, and osteosarcoma cancers based on in vivo and in vitro experiments.88,124,134,142,143 However, due to selenium toxicity and bioavailability elemental selenium and naturally sourced selenium (selenocysteine and selenomethionine) cannot be used in cancer therapies.144 Nano-selenium has advantages over other natural sources of selenium because of its size, porosity, and bio-dispersion. Thus, Polyporus umbellatus polysaccharide (PUP) capped SeNPs have been utilized to detect the in vitro anti-proliferation effect with the MTT assay. Particularly, PUP-SeNPs inhibit the cell growth of four types of human cancer: (a) MDA-MB-23 cells causing breast cancer in humans, (b) HepG2 cells responsible for human liver cancer, (c) HeLa cells known for human cervical cancer, and (d) HT 29 for colon cancer. It is shown by researchers that, by using nano-selenium of the above nature, no toxicity to normal human cells such as liver cells (LO2), embryonic kidney cells (293T), and mouse embryonic fibroblast cells (NIH3T3) was observed.145 A schematic of the effect of SeNPs on cancerous and normal cells is shown in Fig. 6.146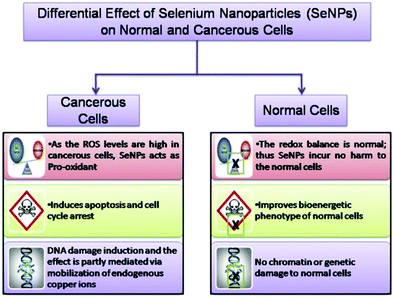 | ||
| Fig. 6 A schematic of the effect of SeNPs on cancerous and normal cells. Reproduced from ref. 146 (A. Khurana, S. Tekula, M. A. Saifi, P. Venkatesh and C. Godugu, Biomed. Pharmacother., 2019, 111, 802–812), copyright authors 2019, open access Science Elsevier. | ||
It has been demonstrated by numerous analyses that incorporation of low Se leads to various hazardous diseases. Se acts as a chemopreventive agent when utilized at optimal doses.147,148 Selenium has therefore been proposed as a disease therapy agent in combination with chemotherapy and radiation.117 The most extreme anticarcinogenic effect of Se has been reported when it is administered at the beginning phase of the disease.149,150 The risk of cancer in humans can increase due to the presence of arsenic (As) in drinking water. However, it has been reported that the generation of As(III)-activated ROS can be minimized by use of Terminalia arjuna (TA) leaf extract encapsulated SeNPs, which fortifies As(III)-induced cell death and DNA catastrophe.102,151 Metastatic breast cancer in a mouse model has been reported to be treated by oral administration of Lactobacillus brevis with SeNPs, which increases interferon generation and delayed hypersensitivity for triggering the immune response.134 Therefore, it has been reported by many researchers that if SeNPs are conjugated with organic moieties and drugs, they can inhibit accumulation of Se nanoparticles, thereby enhancing the anticancer activity and minimizing the antibiotics’ toxic effect.88,143,152–155 The nano-conjugates of SeNPs and doxorubicin show enhanced cytotoxic effect against cancer cells by assisting antibiotic cellular uptake. In a study of lactate dehydrogenase activity and cell viability, Yang et al.143 found that doxorubicin alone was responsible for destroying 20% of cancer cells, whereas more than 50% of cancer cells were destroyed by conjugation of SeNPs and doxorubicin. As an anticancer drug responsible for oxidative stress and DNA cross-linking, cisplatin leads to nephrotoxicity and spermatotoxicity. Interestingly, nephrotoxicity can be reduced using 11-mercapto-1-undecanol functionalized SeNPs by preventing ROS-mediated apoptosis. The antioxidative properties of SeNPs in combination with cisplatin enhance the sperm quality and spermatogenesis.153,154 During breast cancer treatment, anastrozole is used to inhibit the growth of aromatase which leads to side effects such as bone fracture and osteoporosis. These can be prevented by conjugating anastrozole with SeNPs.155,156 Similarly, SeNPs prepared using highly branched β-(1–3)-D-glucan obtained from fruiting plants of Auricularia auricula-judae (AF1-Se) showed strong suppression against human breast adenocarcinoma cancer cell lines such as MCF-7, MDA-MB-468, and MDAMB-231.157 SeNPs decorated using various polysaccharides extracted from Pleurotus tuberregium (PTR), Ganoderma lucidum (GL), Coriolus versicolor (CV), and Polyporus rhinoceros (PR) showed the highest activity for the gastric cancer cell line AGS.158 Induction of cell apoptosis is the key mechanism that plays an important role in the antitumor activities of SeNPs and polysaccharide conjugates. Apoptosis, which is also known as programed cell death, can be described by certain governing properties such as nuclear chromatin condensation, cytoplasm shrinkage, development of apoptotic parts, and membrane dysfunction.159 The cellular growth of human lung cancer cell line A549 can be inhibited by utilizing the complex of Arabinogalactans (LAG) and SeNPs.160 The human breast cancer can be treated by PUP coated SeNPs through stimulating mitochondria mediated and death receptor mediated apoptotic pathways. The anti-cancer activity of these complexes was regulated by the Bax/Bcl-2 ratio, endorsing cytochrome-c release, enhanced caspase-9, -8, and -3 activities and poly(ADP-ribose) polymerase cleavage.161 Therefore, the higher toxicity of SeNPs for cancer cells can be synergistically utilized with anticancer drugs for cancer treatment.
3.2. Antioxidant activity
ROS and reactive nitrogen species (RNS) free radicals are generally generated in the human body during several physicochemical and biochemical reactions. The higher amounts of intermediate complexes such as superoxide and hydrogen peroxide produced during these reactions are responsible for cellular damage, leading to generation of lethal diseases. Therefore, antioxidative compounds are utilized to suppress the creation as well as scavenging of these free radicals.162,163 The surface of Se nanoparticles decorated with various organic molecules and plant extracts act as both antioxidants and pro-oxidants on different cells of the human body and their action mechanism is shown in Fig. 7.164 Battin et al.165 reported that selenium nanoparticles can play an important role in minimizing the free radical concentration to prevent the oxidative damage of DNA in both in vivo and in vitro experimental conditions. It has also been documented that selenoprotein is a key source of selenium and produces vital antioxidants such as deiodinase, thioredoxin reductase, and glutathione peroxidase.166 In an investigation, Zhang et al.167 showed that sodium selenium inhibited the growth of Candida utilis by improving the excretion and biosynthesis of glutathione. Similarly, chitosan functionalized SeNPs showed enhanced glutathione peroxidase and anticipated lipofuscin development in mice.168 Additionally, ABTS°+ and superoxide anion radical can be scavenged by utilizing SeNPs in combination with Cordyceps sinensis exopolysaccharide.145 The plant-based synthesis of selenium nanoparticles demonstrated antioxidative activity through ABTS and DPPH assays.169 Further, the Kong group170 presented the enhanced antioxidative activity of gum-arabic functionalized SeNPs (GA-SeNPs) for scavenging hydroxyl radicals and the DPPH assay test. In an investigation, selenium nanoparticles acted as a hepatoprotective component against acetaminophen accelerated hepatotoxicity by refining liver capacity and restraining oxidative stress in rats. In another study Kokila et al.101 demonstrated the conceivable antioxidative properties of SeNPs. It is known from the literature that the shape and size of nanoparticles can play a key role in such studies. For example, hollow spherical selenium nanoparticles have shown antioxidative properties.32,36,37 Thus, plant based SeNPs show better prevention against ROS mediated oxidative stress induced diseases.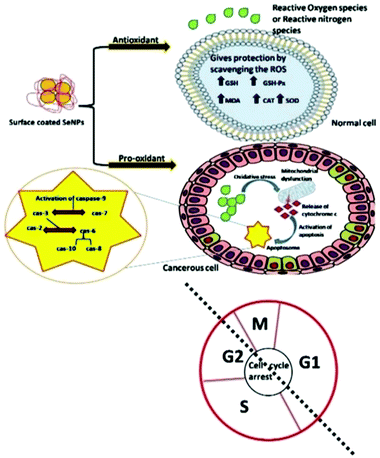 | ||
| Fig. 7 A systematic representation of the action mechanism by surface decorated Se NPs on various cells as an antioxidant and a pro-oxidant. Reproduced from ref. 164 with permission from Science Elsevier, copyright 2020. | ||
3.3. Antidiabetic activity
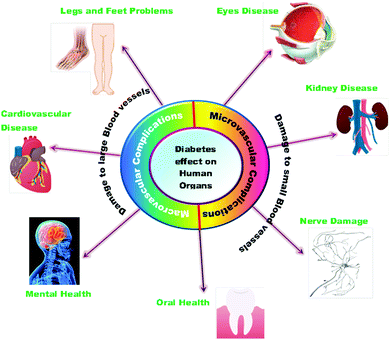
The high amount of glucose in our body due to the absence of enough insulin creation or inadequate usage of insulin leads to chronic diseases like diabetes. The insulin chemical is responsible for maintaining the blood sugar level. Diabetes negatively impacts people of all age groups. Several researchers have reported the use of selenium nanoparticles in the treatment of diabetes because of its effective controlling ability to regulate the blood sugar level.171 The long-term effects of diabetes can damage large and small blood vessels, causing a problem in the functioning of various human organs. Diabetes leads to the generation of various diseases in the heart, legs, brain, eyes, kidneys, skin, digestive system, oral health, and immune system as shown in Fig. 8. Deng et al.172 reported that Se nanoparticles loaded with insulin (INS-SeNPs) can be delivered orally to treat diabetes in mice. In their study the researchers stated that the insulin was released in the blood in a controllable manner and showed excellent stability in digestive foods. The Liu group173 examined the enhanced antidiabetic activity of Catathelesma ventricosum polysaccharides (CVPs) coated Se nanoparticles in streptozocin (STZ) induced diabetic mice. Similarly, the effect of SeNPs in combination with insulin was investigated by Quraishy et al.174 to enhance the activity of hyperglycemia and hyperlipidemia in STZ induced diabetes in mice. At a concentration of 2.0 mg kg−1 body weight, chitosan stabilized SeNPs resulted in improved antidiabetic activity.175 Furthermore, phytomedicines are proven candidates to combat diabetes in an effective way.176 Therefore, Deng et al.177 demonstrated that extracts of mulberry leaf and Pueraria lobata (MPE) (phytomedicines) coated SeNPs exerting a hypoglycemic effect can cure diabetes mellitus. The plasma associated selenium demonstrated a positive effect on prevalent and incident diabetes by redox regulation and signaling of insulin pathways.178 Therefore, selenium nanoparticles functionalized with various moieties are expected to play a crucial role in treating different types of diabetes. A schematic of the effect of SeNPs on various diabetic cells is shown in Fig. 9.146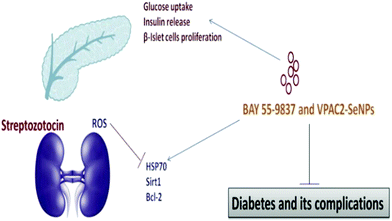 | ||
| Fig. 9 The effect of SeNPs on diabetic cells. Reproduced from ref. 146 (A. Khurana, S. Tekula, M. A. Saifi, P. Venkatesh and C. Godugu, Biomed. Pharmacother., 2019, 111, 802–812), copyright authors 2019, open access Science Elsevier. | ||
3.4. Antimicrobial activity
Living beings are susceptible to microbial infections leading to numerous diseases causing great health concerns. Nowadays, most of the pathogenic organisms have become drug-resistant because of the constant utilization of a wide range of antibiotics. Particularly, multidrug-resistant microorganisms and fungi are profoundly becoming irresistible as they have procured protection from practically all the accessible antimicrobial drugs.179–181 Therefore, anti-infection agents are developed for the treatment of these irresistible diseases. There are a variety of metal nanoparticles as well as metal oxides and their combinations which have been documented as potential antimicrobial agents. SeNPs have also found tremendous scope in biomedical applications as an antimicrobial agent to protect implanted clinical gadgets from bacteria. Tran et al.41 incorporated SeNPs by an in situ method into regular polymers such as polyvinyl chloride, polyurethane, and silicon, which are generally used in biomedical devices. They observed that the antibacterial performance was directly linked with the concentration of selenium as the coating material in combination with different polymers. Their findings stated that nano-selenium coated polymers essentially can hinder Staphylococcus aureus (S. aureus) growth, in contrast to uncoated polymers. Additionally, they discovered that SeNP-covered PVC shows more prominent antibacterial effects compared to commercially available Ag-covered PVC.41 Generally, biofilm development on the surfaces of clinical gadgets by microbes is difficult to treat. Thus, SeNPs act as a suppressing agent for biofilm formation.34,182–184 Wang et al.42 reported biogenic SeNPs utilizing Bacillus licheniformis JS2 to suppress bacterial (S. aureus) adherence, biofilm development, and microcolony formation to various polymer surfaces. These outcomes have possible incentive for application as antibacterial covering prostheses against S. aureus and other clinical gadgets against nosocomial infections.183 Shakibaie et al.26 studied various biofilm forming microbes such as S. aureus, Proteus mirabilis, (P. mirabilis), and Pseudomonas aeruginosa (P. aeruginosa) to check the viability of biogenic SeNPs-Bacillus sp. MSh-1, having spherical morphology with the dimension range of 80–220 nm. For comparison purposes they incubated the biofilm formation assay with both SeNPs and SeO2 with microbes. In both the cases, they observed no significant decrease in the biofilm formation; however, due to the lower toxicity of SeNPs they were preferred over SeO2. It was believed that SeNPs show antibacterial activity due to oxidative stress.185 Huang et al.186 reported a new inorganic antibacterial agent based on quercetin (Qu) and acetylcholine (Ach) functionalized SeNPs (Qu-Ach@SeNPs), which demonstrated improved antimicrobial activity against multidrug-resistant superbugs (MDRs) in contrast to individual Qu@SeNPs and Ach@SeNPs. They observed that when Qu-Ach@SeNPs adhered to the bacterial cell surface, causing disruption of the cytoplasmic membrane, the growth of bacterial cell methicillin resistant S. aureus (MRSA) was inhibited and the DNA structure was damaged by antibacterial action as shown in Fig. 10.186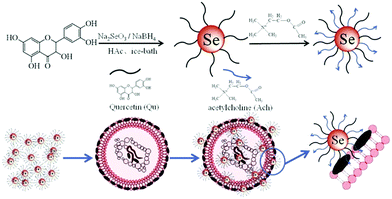 | ||
| Fig. 10 A schematic representation of the synthesis of Qu-Ach SeNPs and their antibacterial action. Reproduced from ref. 186 with permission from Science Elsevier, copyright 2016. | ||
In a separate study researchers40 also found that an in vitro study involving SeNPs emphatically suppressed the growth of S. aureus (60 times higher) at different doses such as 7.8, 15.5, and 31 μg mL−1 for time periods of 3, 4, and 5 h, respectively. However, this study was found unsuitable in microbiology in terms of an effective antimicrobial agent. Therefore, to overcome these issues Mittal et al.187 combined selenium and silver nanoparticles to enhance the antimicrobial activity. Here, Ag-SeNPs having a size range of 30–35 nm were synthesized utilizing bioactive gallic acid and quercetin (QC). The antimicrobial activity of the composite of Qu-gallic acid@Ag-SeNPs was found to be superior to both silver nitrate and sodium selenite against Escherichia coli (E. coli) and Bacillus subtilis (B. subtilis) on agar plates. They also found comparable results of the nanocomposite with an antimicrobial agent, chloramphenicol, for an equivalent dose of 50 μg mL−1. Here, the authors described the antimicrobial mechanism in two ways: in the first assumption, the nanoparticles adhere to the bacterial cell wall, where silver NPs are released and destroy the bacterial cells; in the second assumption, the interconnectivity of AgNPs with DNA and bacterial protein and more activity for sulfur and phosphorus compounds leads to destruction of these compounds. Menon et al.188 reported biosynthesized SeNPs from Z. officinale with spherical morphology and a 100–150 nm size range. They examined the antimicrobial activity of the SeNPs against five microbial species such as E. coli, Klebsiella sp., Pseudomonas sp., S. aureus, and Proteus sp. The Proteus sp. bacterial growth was effectively inhibited by SeNPs at a concentration of 250 μg mL−1. Generally, SeNPs have size and concentration dependent effects against various microorganisms.189 Further, selenium and tellurium nanoparticles exhibit antimicrobial activity and destruction of biofilm formation. These nanoparticles were effective on inhibiting the growth of E. coli JM109, Pseudomonas aeruginosa PAO1, and S. aureus ATCC 25923.190 Consequently, SeNPs can be utilized as an effective antimicrobial agent in pharmaceutical applications.
3.5. Antifungal activity
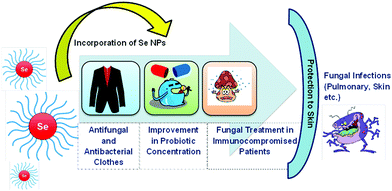
SeNPs acquire antifungal properties for various biological applications. To name a few, SeNPs can be used for the treatment of fungal infections in immunity compromised patients, improvement in probiotic concentration, and fabrication of antifungal and antibacterial cloths for protecting skin from S. aureus and Tinea pedis infections191 as shown in Fig. 11. To prove this, Yip et al.192 demonstrated that nano-Se modified with biogenic polysaccharide–protein (PSP) complexes collected from Pleurotus tuber-regium deposited on a fabric was effective in inhibiting the growth of S. aureus and Trichophyton rubrum bacteria. Similarly, Shakibaie et al.193 examined the antifungal activity of Bacillus sp. Msh-1 functionalized SeNPs against Aspergillus fumigatus (pulmonary infection) and Candida albicans (skin infection). Shahverdi et al.24 reported SeNPs prepared from Klebsiella pneumoniae to treat antifungal infections from Malassezia sympodialis, Malassezia furfur, and Aspergillus terreus in the concentration range from 10 to 260 μg mL−1. Particularly, SeNPs can provide antifungal activity in patients having less immunity against nystatin-immune Candida sp.194 and a more constructive therapy to encounter intrusive aspergillosis in comparison to the amphotericin B drug.193 However, conflicting results have been obtained by the Kazempour group on SeNPs prepared from Klebsiella pneumoniae. They found that SeNPs lack in treating the post-antifungal effect of Aspergillus niger (A. niger) and Candida albicans (C. albicans) and a lower concentration of Se stimulated the growth of A. niger. It was assumed that the reason behind this mechanism is ’the physiological impact of selenium on specific proteins in A. niger. The outcomes may have direct ramifications on dermatitis caused by A. niger or C. albicans. These pathogenic microorganisms can regrow on the skin by exposing to SeNPs’ environment. Generally, the post-antifungal effect (PAE) is a vital pharmacodynamic boundary which helps in setting up measurement regimes by demonstrating inhibition of antimicrobe development after exposing microorganisms to antimicrobial agents for a short period of time.195–1973.6. Antiparasitic activity
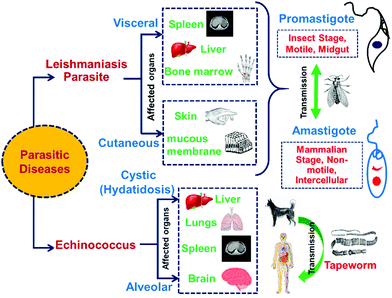
In the past few years, different research groups have made aggressive efforts on exploring the antiparasitic properties of selenium nanoparticles.25,198,199 Various types of parasitic diseases and their effects on human organs are shown in Fig. 12. By in vivo and in vitro studies, it was found that biosynthesized SeNPs from Bacillus sp. Msh-1 can be used in the treatment of Leishmania major parasites such as Promastigote and Amastigote.25 Similarly, in vitro experiments of leishmanicidal activity to combat Leishmania tropica (L. tropica)200 and Leishmania infantum (L. infantum)199 have been reported by use of Bacillus sp. Msh-1 functionalized SeNPs. The gel electrophoresis method has been reported for in vitro investigation of selenium nanoparticles to understand the time-dependent decrease in Promastigote multiplication due to the presence of DNA fragmentation.201 Biosynthesized SeNPs with a size range of 80–220 nm showed a strong scolicidal activity against parasitic infections (Cystic echinococcus) through an in vitro study.202 A comparable time-dependent cytotoxic outcome was also illustrated against intracellular amastigotes. A study on male mice (BALB/c) infected with Leishmania major showed a reduced progression in lesion growth by ejaculation of SeNPs. Soflaei et al.199 reported the antileishmanial properties of SeNPs in both Promastigotes and Amastigotes of L. infantum in contrast to SeO2. The study demonstrated that SeNPs showed improved performance and less cytotoxicity than SeO2 in a time and dose dependent antileishmanial activity. In another study, Mahmoudvand et al.200 reported that SeNPs with a particular size range were effective in inhibiting the resistance of Meglumine antimoniate (MA) and MA-sensitive L. tropica in vitro. Essentially, these outcomes indicated suppression of promastigote depending on the dose, decreased intramacrophage amastigote feasibility in the two strains, and outstanding impact on prophylaxis treatment. Additionally, SeNPs exhibited improved antileishmanicidal activity in conjugation with MA, in contrast to SeNPs or MA alone.138Hence, it is concluded that biosynthesized SeNPs with a particular particle size range shows higher resistant against Leishmania parasitic diseases.
3.7. Other biological applications
Besides the antibacterial properties, SeNPs have also been utilized as wound dressing materials. Biswas et al.203 confirmed the antibacterial properties of permeable chitosan/polyvinyl alcohol (PVA) (CS) in situ mediated Se nanorods compared to Ag nanoparticles (CS–Ag). Both the nanoparticles CS–Se and CS–Ag exhibited antibacterial activity against Gram-negative (E. coli), Gram-positive (S. aureus), and methicillin-resistant S. aureus (MRSA). The slow release of Ag and Se nanoparticles was dependent on the delivery medium utilized during treatment. From the study it was found that CS–Se scaffolds are less cytotoxic toward mammalian cells when compared with the CS–Ag system. Similarly, Ramya et al.204 explored the in vivo impact of biosynthesized Se nanoparticles stabilized with Streptomyces minutiscleroticus M10A62 (spherical size) on Swiss albino mice for wound treatment. The obtained results demonstrated that a high dose of SeNPs (10%) ointment effectively and quickly healed the wound.Selenium has been additionally found to have a chemoprotective effect against chemotherapy-prompted toxicity. Bhattacharjee et al.205 examined Swiss albino mice for treating cyclophosphamide (CP)-prompted hepatotoxicity and genotoxicity by utilizing SeNPs. In the mice, SeNPs switched or potentially enhanced the effects of CP-instigated toxicity: rebuilding of cell reinforcement protein activity, decrease in reactive oxygen species, glutathione levels, and chromosomal variations in bone marrow, and DNA damage.205 Further, in another study they found that SeNPs also possess chemosensitizing and chemoprotective activities. After injecting SeNP adjuvant with cyclophosphamide into Ehrlich's ascites carcinoma (EAC) containing Swiss albino mice, the following changes were observed: a critical decrease in practical tumor cell count, packed cell and tumor volume, while an increase in tumor-bearing hosts’ survivability.206 Gao et al.207 reported SeNPs as an effective chemotherapy protective agent against chemotherapy-prompted toxicity of irinotecan by both in vivo and in vitro studies.
In another study, it was found that the red-allotrope of selenium nanoparticles (rSeNPs) show higher cytotoxicity towards head and neck squamous cell carcinoma (HNSCC) than human dermal fibroblast (HDF) cells, which resulted in cell proliferation.208 Additionally, SeNPs can be used as a potential candidate in chemoradiotherapy and radiation sensitizer. Generally, to treat cancer cells external radiation therapy has been utilized broadly. However, these unpredictable radiations can damage adjacent healthy cells. To overcome this unavoidable impact, chemo-radiotherapy can be utilized with a radioactive seed to deliver enhanced, efficient and precise radiation. Based on this concept, Bhattacharjee et al.205 performed a combined in vivo and in vitro experiment by utilizing folic acid capped SeNPs (FA-SeNPs) in conjugation with radioactive 125I seeds to impart enhanced anticancer activity. In comparison with individual treatment, the combination of (FA-SeNPs) and 125I seeds demonstrated a better in vivo antitumor effect and lower cytotoxicity in MCF-7 infected mice. Yu et al.209 reported that Se nanoparticles in combination with radiation reduces the damage to nearby tissues and enhances the carcinogenic cell affectability to the harmful impacts of illumination on MCF-7 breast cancer cells by in vitro studies. Karami et al.106 reported the radioprotective effects of SeNPs and sodium selenite on gamma radiation (0, 2, and 8 Gy) incited nephropathy in mice for 14 days. They concurrently treated the mice by administrating SeNPs (spherical and 20–50 nm in size) or sodium selenite (0.1 mg kg−1). After 48 hour exposure they found that SeNPs were more viable than sodium selenite on controlling the factors responsible for nephropathy.
4. Conclusion and future prospects
Selenium is an essential micronutrient required for proper functioning of biological and metabolic mechanism within the human body. Deficiency of selenium leads to the generation of several harmful disorders such as cancer, neurological, muscular, immune, etc. Generally, selenium can be depicted within a very narrow concentration range due to its deficiency, physiological effect, and toxic doses. At optimal doses Se acts as an antioxidant, whereas at higher doses it shows pro-oxidant activity. To overcome this issue, a precisely controlled dosage of Se is generally suggested. Selenium has various health advantages because of its bioavailability in the form of selenoproteins as well as lower molecular weight. Various selenoproteins (Se1P, Se1F, Se1S, Se1M), thioredoxin reductases, and glutathione peroxidases show redox activity and control redox reactions in cells. However, concerns related to selenium toxicity lead researchers to focus on nano-selenium. The present review emphasized the various methods used for the synthesis of selenium nanoparticles mainly focused on biogenic methods and their biological applications. Biogenic methods have more advantages over chemical reduction methods due to their higher biocompatibility, bioactivity, and lower cytotoxicity. Biological synthesis is economical, eco-friendly and safe unlike chemical reduction techniques requiring hazardous chemicals, high temperatures and acidic pH. Use of plant extracts, however, is more favourable in comparison to the bacterial path as it eliminates the tedious procedures and cost for maintaining the cell cultures.SeNPs have various biological applications due to their excellent properties. SeNPs play a key role in various biological applications due to their association with various moieties such as selenoproteins, selenocysteine, selenomethionine, etc. Several studies showed that SeNPs are excellent in combating fatal diseases like cancer, diabetes, Alzheimer's, drug-induced toxicity, etc. However, nanoparticle induced toxicity is still a major concern for researchers. The authors concluded from the literature that mostly SeNPs ranging from 50 to 200 nm were effective for their use as a therapeutic agent in cancer treatment and antioxidant and antimicrobial applications.
The near future potential of selenium nanoparticles can be considered in pharmaceutical applications and nutritional supplements. Biogenic SeNPs can be explored for their use as anti-TB and anti-viral agents, drug delivery systems, and as catalysts. More precise clinical trials are required for checking the viability of SeNPs on human health. Extensive research is required to develop less toxic and low cost synthesis methods and to understand the role of nano-selenium in cancer therapy, chemotherapy, and radiation therapy to modulate their efficiency and cytotoxicity. Particularly, the development of new nanoparticle delivery systems can provide substantial dietary and therapeutic potential by offering transport of selenium in organs which can modify the physiochemical properties of the nanoparticles, and higher stability within the gastrointestinal tract by permitting precise release of selenium.
Author contributions
All the authors contributed equally.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the Vice-Chancellor, Defence Institute of Advanced Technology (DIAT), Girinagar, Pune for encouragement and permission. PKK thanks DMSRDE, DRDO India for support.References
- M. Zhu, G. Niu and J. Tang, J. Mater. Chem. C, 2019, 7, 2199–2206 RSC.
- A. A. Jadhav and P. K. Khanna, RSC Adv., 2015, 5, 44756–44763 RSC.
- U. K. EGVM, Revised Review of Selenium, United Kingdom Expert Gr. Vitam. Miner. (EVM/99/17. REVISEDAUG2002).
- Joint FAO/WHO Consultation on Preparation and Use of Food-Based Dietary Guidelines (1995: Nicosia C). and World Health Organization, Geneva, 1998, p. 880.
- N. I. Krinsky, G. R. Beecher, R. F. Burk, A. C. Chan, J. W. Erdman, R. A. Jacob, I. Jialal, L. N. Kolonel, J. R. Marshall and P. R. L. Taylor Mayne, Dietary reference intakes for vitamin C, vitamin E, selenium, and carotenoids, The National Academies Press Washington, DC, USA, 2000 Search PubMed.
- J. F. Ramos and T. J. Webster, Int. J. Nanomed., 2012, 7, 3907–3914 CAS.
- J. E. Spallholz, Free Radic. Biol. Med., 1994, 17, 45–64 CrossRef CAS.
- F. Sieber, J.-P. Daziano, W. H. H. Günther, M. Krieg, K. Miyagi, R. W. Sampson, M. D. Ostrowski, G. S. Anderson, I. Tsujino and R. J. Bula, Phosphorus Sulfur Silicon Relat. Elem., 2005, 180, 647–657 CrossRef CAS PubMed.
- F. Islam, S. Zia, I. Sayeed, K. S. Zafar and A. S. Ahmad, Biol. Trace Elem. Res., 2002, 90, 203–214 CrossRef CAS PubMed.
- K. Venardos, G. Harrison, J. Headrick and A. Perkins, Redox Rep., 2004, 9, 317–320 CrossRef CAS PubMed.
- K. Venardos, G. Harrison, J. Headrick and A. Perkins, J. Trace Elem. Med. Biol., 2004, 18, 81–88 CrossRef CAS PubMed.
- J. Vanderlelie, K. Venardos and A. V. Perkins, Reproduction, 2004, 128, 635–641 CAS.
- S. V. Gudkov, G. A. Shafeev, A. P. Glinushkin, A. V. Shkirin, E. V. Barmina, I. I. Rakov, A. V. Simakin, A. V. Kislov, M. E. Astashev and V. A. Vodeneev, ACS Omega, 2020, 5, 17767–17774 CrossRef CAS PubMed.
- D. Huang, B. Ou and R. L. Prior, J. Agric. Food Chem., 2005, 53, 1841–1856 CrossRef CAS PubMed.
- R. Pillai, J. H. Uyehara-Lock and F. P. Bellinger, IUBMB Life, 2014, 66, 229–239 CrossRef CAS PubMed.
- M. P. Rayman, Lancet, 2000, 356, 233–241 CrossRef CAS.
- M. P. Look, J. K. Rockstroh, G. S. Rao, K. A. Kreuzer, S. Barton, H. Lemoch, T. Sudhop, J. Hoch, K. Stockinger and U. Spengler, Eur. J. Clin. Nutr., 1997, 51, 266–272 CrossRef CAS PubMed.
- M. A. Ansari, A. S. Ahmad, M. Ahmad, S. Salim, S. Yousuf, T. Ishrat and F. Islam, Biol. Trace Elem. Res., 2004, 101, 73–86 CrossRef CAS.
- K. S. Zafar, A. Siddiqui, I. Sayeed, M. Ahmad, S. Salim and F. Islam, J. Neurochem., 2003, 84, 438–446 CrossRef CAS PubMed.
- M. Vahdati and T. Tohidi Moghadam, Sci. Rep., 2020, 10, 510 CrossRef CAS PubMed.
- H. Forootanfar, M. Adeli-Sardou, M. Nikkhoo, M. Mehrabani, B. Amir-Heidari, A. R. Shahverdi and M. Shakibaie, J. Trace Elem. Med. Biol., 2014, 28, 75–79 CrossRef CAS PubMed.
- B. Yu, Y. Zhang, W. Zheng, C. Fan and T. Chen, Inorg. Chem., 2012, 51, 8956–8963 CrossRef CAS PubMed.
- H. Hariharan, N. Al-Harbi, P. Karuppiah and S. Rajaram, Chalcogenide Lett., 2012, 9, 509–515 CAS.
- A. R. Shahverdi, A. Fakhimi, G. Mosavat, P. Jafari-Fesharaki, S. Rezaie and S. M. Rezayat, World Appl. Sci. J., 2010, 10, 918–922 CAS.
- N. Beheshti, S. Soflaei, M. Shakibaie, M. H. Yazdi, F. Ghaffarifar, A. Dalimi and A. R. Shahverdi, J. Trace Elem. Med. Biol., 2013, 27, 203–207 CrossRef CAS PubMed.
- M. Shakibaie, H. Forootanfar, Y. Golkari, T. Mohammadi-Khorsand and M. R. Shakibaie, J. Trace Elem. Med. Biol., 2015, 29, 235–241 CrossRef CAS PubMed.
- W. Cong, R. Bai, Y.-F. Li, L. Wang and C. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 34725–34735 CrossRef CAS PubMed.
- J. A. Johnson, M.-L. Saboungi, P. Thiyagarajan, R. Csencsits and D. Meisel, J. Phys. Chem. B, 1999, 103, 59–63 CrossRef CAS.
- O. Vidal, B. Goffé and N. Arndt, Nat. Geosci., 2013, 6, 894–896 CrossRef CAS.
- R. Jain, G. Gonzalez-Gil, V. Singh, E. D. Van Hullebusch, F. Farges and P. N. L. Lens, Nanobiotechnology, 2014, 10, 365–394 Search PubMed.
- M. Panahi-Kalamuei, M. Salavati-Niasari and S. M. Hosseinpour-Mashkani, J. Alloys Compd., 2014, 617, 627–632 CrossRef CAS.
- J.-S. Zhang, X.-Y. Gao, L.-D. Zhang and Y.-P. Bao, BioFactors, 2001, 15, 27–38 CrossRef CAS PubMed.
- J. Zhang, X. Wang and T. Xu, Toxicol. Sci., 2008, 101, 22–31 CrossRef CAS PubMed.
- S. Skalickova, V. Milosavljevic, K. Cihalova, P. Horky, L. Richtera and V. Adam, Nutrition, 2017, 33, 83–90 CrossRef CAS PubMed.
- J. Zhang, H. Wang, X. Yan and L. Zhang, Life Sci., 2005, 76, 1099–1109 CrossRef CAS PubMed.
- X. Gao, J. Zhang and L. Zhang, Adv. Mater., 2002, 14, 290–293 CrossRef CAS.
- H. Wang, J. Zhang and H. Yu, Free Radic. Biol. Med., 2007, 42, 1524–1533 CrossRef CAS PubMed.
- T. Ikemoto, T. Kunito, H. Tanaka, N. Baba, N. Miyazaki and S. Tanabe, Arch. Environ. Contam. Toxicol., 2004, 47, 402–413 CrossRef CAS PubMed.
- B. Huang, J. Zhang, J. Hou and C. Chen, Free Radic. Biol. Med., 2003, 35, 805–813 CrossRef CAS PubMed.
- P. A. Tran and T. J. Webster, Int. J. Nanomed., 2011, 6, 1553 CAS.
- P. A. Tran and T. J. Webster, Nanotechnology, 2013, 24, 155101 CrossRef PubMed.
- Q. Wang and T. J. Webster, J. Biomed. Mater. Res., Part A, 2012, 100, 3205–3210 CrossRef PubMed.
- B. T. Mayers, K. Liu, D. Sunderland and Y. Xia, Chem. Mater., 2003, 15, 3852–3858 CrossRef CAS.
- B. Yu, P. You, M. Song, Y. Zhou, F. Yu and W. Zheng, New J. Chem., 2016, 40, 1118–1123 RSC.
- A. H. Shar, M. N. Lakhan, J. Wang, M. Ahmed, K. T. Alali, R. Ahmed, I. Ali and A. Q. Dayo, Dig. J. Nanomater. Biostruct., 2019, 14, 867–872 Search PubMed.
- Y. Zhu, Y. Qian, H. Huang and M. Zhang, Mater. Lett., 1996, 28, 119–122 CrossRef CAS.
- M. Quintana, E. Haro-Poniatowski, J. Morales and N. Batina, Appl. Surf. Sci., 2002, 195, 175–186 CrossRef CAS.
- T. Vasileiadis, V. Dracopoulos, M. Kollia, L. Sygellou and S. N. Yannopoulos, J. Nanopart. Res., 2019, 21, 1–8 CrossRef CAS.
- B. Gates, Y. Yin and Y. Xia, J. Am. Chem. Soc., 2000, 122, 12582–12583 CrossRef CAS.
- B. Mayers and Y. Xia, Adv. Mater., 2002, 14, 279–282 CrossRef CAS.
- X.-L. Li, G.-H. Cao, C.-M. Feng and Y.-D. Li, J. Mater. Chem., 2004, 14, 244–247 RSC.
- A. A. Jadhav, P. V. More and P. K. Khanna, RSC Adv., 2015, 5, 76733–76742 RSC.
- P. K. Khanna, A. Jadhav and P. V. More, J. Nanosci. Nanotechnol., 2020, 20, 2946–2954 CrossRef CAS.
- P. K. Khanna, P. More, B. G. Bharate and A. K. Vishwanath, J. Lumin., 2010, 130, 18–23 CrossRef CAS.
- V. Singh, Priyanka, P. V. More, E. Hemmer, Y. K. Mishra and P. K. Khanna, Mater. Adv., 2021, 2, 1204–1228 RSC.
- A. Kumar and K. S. Prasad, J. Biotechnol., 2021, 325, 152–163 CrossRef CAS.
- M. Ikram, B. Javed, N. I. Raja and Z. U. R. Mashwani, Int. J. Nanomed., 2021, 16, 249–268 CrossRef.
- T. Nie, H. Wu, K.-H. Wong and T. Chen, J. Mater. Chem. B, 2016, 4, 2351–2358 RSC.
- H. Fernández-Llamosas, L. Castro, M. L. Blázquez, E. Díaz and M. Carmona, Microb. Cell Fact., 2016, 15, 1–10 CrossRef PubMed.
- S. Shirsat, A. Kadam, M. Naushad and R. S. Mane, RSC Adv., 2015, 5, 92799–92811 RSC.
- C. Kim, J. Hong and J.-W. Park, Polymers, 2019, 11, 1052 CrossRef CAS PubMed.
- Y.-Y. Xia, Mater. Lett., 2007, 61, 4321–4324 CrossRef CAS.
- Y. Zhan, Y. Liu, H. Zu, Y. Guo, S. Wu, H. Yang, Z. Liu, B. Lei, J. Zhuang, X. Zhang, D. Huang and C. Hu, Nanoscale, 2018, 10, 5997–6004 RSC.
- M. A. El-Ghazaly, N. Fadel, E. Rashed, A. El-Batal and S. A. Kenawy, Can. J. Physiol. Pharmacol., 2017, 95, 101–110 CrossRef CAS PubMed.
- P. A. Tran, N. O’Brien-Simpson, E. C. Reynolds, N. Pantarat, D. P. Biswas and A. J. O’Connor, Nanotechnology, 2016, 27, 45101 CrossRef PubMed.
- A. Khalid, P. A. Tran, R. Norello, D. A. Simpson, A. J. O’Connor and S. Tomljenovic-Hanic, Nanoscale, 2016, 8, 3376–3385 RSC.
- S. Boroumand, M. Safari, E. Shaabani, M. Shirzad and R. Faridi-Majidi, Mater. Res. Express, 2019, 6, 0850d8 CrossRef CAS.
- C. P. Shah, M. Kumar, K. K. Pushpa and P. N. Bajaj, Cryst. Growth Des., 2008, 8, 4159–4164 CrossRef CAS.
- Y. Zhang, J. Wang and L. Zhang, Langmuir, 2010, 26, 17617–17623 CrossRef CAS.
- Z. Lin, F. Lin and C. R. C. Wang, J. Chinese Chem. Soc., 2004, 51, 239–242 CrossRef CAS.
- S. Yu, W. Zhang, W. Liu, W. Zhu, R. Guo, Y. Wang, D. Zhang and J. Wang, Nanotechnology, 2015, 26, 145703 CrossRef PubMed.
- T. Huang, J. A. Holden, D. E. Heath, N. M. O’Brien-Simpson and A. J. O’Connor, Nanoscale, 2019, 11, 14937–14951 RSC.
- X. D. Shi, Y. Q. Tian, J. L. Wu and S. Y. Wang, Crit. Rev. Food Sci. Nutr., 2020, 1–12 Search PubMed.
- B. Fardsadegh, H. Vaghari, R. Mohammad-Jafari, Y. Najian and H. Jafarizadeh-Malmiri, Green Process. Synth., 2019, 8, 191–198 CAS.
- C. Mellinas, A. Jiménez and M. C. Garrigós, Molecules, 2019, 24, 4048 CrossRef CAS PubMed.
- Y. Shin, J. M. Blackwood, I.-T. Bae, B. W. Arey and G. J. Exarhos, Mater. Lett., 2007, 61, 4297–4300 CrossRef CAS.
- R. Abbasian and H. Jafarizadeh-Malmiri, Open Agric., 2020, 5, 761–767 CrossRef.
- M. Iranifam, M. Fathinia, T. S. Rad, Y. Hanifehpour, A. R. Khataee and S. W. Joo, Talanta, 2013, 107, 263–269 CrossRef CAS PubMed.
- H. Chen, J. B. Yoo, Y. Liu and G. Zhao, Electron. Mater. Lett., 2011, 7, 333–336 CrossRef CAS.
- C. Xu, L. Qiao, L. Ma, Y. Guo, X. Dou, S. Yan, B. Zhang and A. Roman, Int. J. Nanomed., 2019, 14, 4491–4502 CrossRef CAS.
- X. Wang, X. Pan and G. M. Gadd, Sci. Total Environ., 2019, 668, 303–309 CrossRef CAS PubMed.
- T. Wang, L. Yang, B. Zhang and J. Liu, Colloids Surf., B, 2010, 80, 94–102 CrossRef CAS PubMed.
- A. V. Tugarova and A. A. Kamnev, Talanta, 2017, 174, 539–547 CrossRef CAS PubMed.
- A. V. Tugarova, P. V. Mamchenkova, Y. A. Dyatlova and A. A. Kamnev, Spectrochim. Acta, Part A, 2018, 192, 458–463 CrossRef CAS PubMed.
- A. V. Tugarova, P. V. Mamchenkova, V. A. Khanadeev and A. A. Kamnev, New Biotechnol., 2020, 58, 17–24 CrossRef CAS PubMed.
- S. K. Torres, V. L. Campos, C. G. León, S. M. Rodríguez-Llamazares, S. M. Rojas, M. González, C. Smith and M. A. Mondaca, J. Nanopart. Res., 2012, 14, 1–9 Search PubMed.
- S. Shoeibi and M. Mashreghi, J. Trace Elem. Med. Biol., 2017, 39, 135–139 CrossRef CAS PubMed.
- C. H. Ramamurthy, K. S. Sampath, P. Arunkumar, M. S. Kumar, V. Sujatha, K. Premkumar and C. Thirunavukkarasu, Bioprocess Biosyst. Eng., 2013, 36, 1131–1139 CrossRef CAS PubMed.
- K. Anu, S. Devanesan, R. Prasanth, M. S. AlSalhi, S. Ajithkumar and G. Singaravelu, J. King Saud Univ. - Sci., 2020, 32, 2520–2526 CrossRef.
- L. Gunti, R. S. Dass and N. K. Kalagatur, Front. Microbiol., 2019, 10, 931 CrossRef PubMed.
- M. Abinaya, B. Vaseeharan, R. Rekha, S. Shanthini, M. Govindarajan, N. S. Alharbi, S. Kadaikunnan, J. M. Khaled and M. N. Al-Anbr, J. Photochem. Photobiol. B Biol., 2019, 192, 55–67 CrossRef CAS PubMed.
- S. Bharathi, S. Kumaran, G. Suresh, M. Ramesh, V. Thangamani, S. R. Pugazhvendan and K. Sathiyamurthy, Biocatal. Agric. Biotechnol., 2020, 27, 101655 CrossRef.
- M. Borowska, E. Pawlik and K. Jankowski, Mon. Chem. – Chem. Mon., 2020, 151, 1283–1290 CrossRef CAS.
- G. S. El-Sayyad, H. S. El-Bastawisy, M. Gobara and A. I. El-Batal, Biol. Trace Elem. Res., 2020, 195, 323–342 CrossRef CAS.
- A. H. Hashem, A. M. A. Khalil, A. M. Reyad and S. S. Salem, Biol. Trace Elem. Res., 2021, 199, 3998–4008 CrossRef CAS PubMed.
- M. Kapur, K. Soni and K. Kohli, Adv. Tech. Biol. Med., 2017, 5, 1–7 Search PubMed.
- V. Alagesan and S. Venugopal, Bionanoscience, 2019, 9, 105–116 CrossRef.
- N. A. Mulla, S. V. Otari, R. A. Bohara, H. M. Yadav and S. H. Pawar, Mater. Lett., 2020, 264, 127353 CrossRef CAS.
- B. Fardsadegh and H. Jafarizadeh-Malmiri, Green Process. Synth., 2019, 8, 399–407 CAS.
- N. S. Khoei, S. Lampis, E. Zonaro, K. Yrjälä, P. Bernardi and G. Vallini, New Biotechnol., 2017, 34, 1–11 CrossRef CAS PubMed.
- K. Kokila, N. Elavarasan and V. Sujatha, New J. Chem., 2017, 41, 7481–7490 RSC.
- K. S. Prasad, H. Patel, T. Patel, K. Patel and K. Selvaraj, Colloids Surf., B, 2013, 103, 261–266 CrossRef CAS PubMed.
- G. M. Khiralla and B. A. El-Deeb, LWT – Food Sci. Technol., 2015, 63, 1001–1007 CrossRef CAS.
- P. Korde, S. Ghotekar, T. Pagar, S. Pansambal, R. Oza and D. Mane, J. Chem. Rev., 2020, 2, 157–168 CAS.
- R. Hassanien, A. A. I. Abed-Elmageed and D. Z. Husein, ChemistrySelect, 2019, 4, 9018–9026 CrossRef CAS.
- M. Karami, S. Asri-Rezaei, B. Dormanesh and A. Nazarizadeh, Int. J. Radiat. Biol., 2018, 94, 17–27 CrossRef CAS PubMed.
- E. Piacenza, A. Presentato, B. Heyne and R. J. Turner, Nanophotonics, 2020, 9, 3615–3628 CrossRef CAS.
- B. Afzal, D. Yasin, S. Husain, A. Zaki, P. Srivastava, R. Kumar and T. Fatma, Biocatal. Agric. Biotechnol., 2019, 21, 101307 CrossRef.
- S. Prasanth and C. Sudarsanakumar, New J. Chem., 2017, 41, 9521–9530 RSC.
- H. Kumar, K. Bhardwaj, E. Nepovimova, K. Kuča, D. S. Dhanjal, S. Bhardwaj, S. K. Bhatia, R. Verma and D. Kumar, Nanomaterials, 2020, 10, 1–31 Search PubMed.
- X. Ma and H. Yu, Yale J. Biol. Med., 2006, 79, 85–94 Search PubMed.
- M. J. Thun, J. O. DeLancey, M. M. Center, A. Jemal and E. M. Ward, Carcinogenesis, 2010, 31, 100–110 CrossRef CAS PubMed.
- E. J. Devlin, L. A. Denson and H. S. Whitford, J. Pain Symptom Manage, 2017, 54, 245–258 CrossRef PubMed.
- Q. Tan, J. Li, H. Yin, L. Wang, W. Tang, F. Zhao, X. Liu and H. Zeng, Invest. New Drugs, 2010, 28, 205–215 CrossRef CAS PubMed.
- I. M. Puspitasari, R. Abdulah, C. Yamazaki, S. Kameo, T. Nakano and H. Koyama, Radiat. Oncol., 2014, 9, 125 CrossRef PubMed.
- E. Yakubov, M. Buchfelder, I. Y. Eyüpoglu and N. E. Savaskan, Biol. Trace Elem. Res., 2014, 161, 246–254 CrossRef CAS.
- S. O. Evans, P. F. Khairuddin and M. B. Jameson, Anticancer Res., 2017, 37, 6497 LP–6509 LP Search PubMed.
- H. Song, I. Hur, H. Park, J. Nam, G. Bin Park, K. H. Kong, Y. M. Hwang, Y. S. Kim, D. H. Cho, W. J. Lee and D. Y. Hur, Immune Netw., 2009, 9, 236 CrossRef PubMed.
- Y.-C. Chen, K. S. Prabhu, A. Das and A. M. Mastro, Int. J. Cancer, 2013, 133, 2054–2064 CrossRef CAS PubMed.
- C. D. Davis, P. A. Tsuji and J. A. Milner, Annu. Rev. Nutr., 2012, 32, 73–95 CrossRef CAS.
- R. V. S. Lavu, T. Van De Wiele, V. L. Pratti, F. Tack and G. Du Laing, Food Chem., 2016, 197, 382–387 CrossRef CAS PubMed.
- S. J. Fairweather-Tait, R. Collings and R. Hurst, Am. J. Clin. Nutr., 2010, 91, 1484S–1491S CrossRef CAS PubMed.
- D. N. Ekoue, S. Zaichick, K. Valyi-Nagy, M. Picklo, C. Lacher, K. Hoskins, M. A. Warso, M. G. Bonini and A. M. Diamond, J. Trace Elem. Med. Biol., 2017, 39, 227–233 CrossRef CAS PubMed.
- E. N. Ali, S. M. El-Sonbaty and F. M. Salem, Int. J. Pharmacol. Biol. Sci., 2013, 2, 38–46 Search PubMed.
- A. V. Pansare, D. K. Kulal, A. A. Shedge and V. R. Patil, Dalt. Trans., 2016, 45, 12144–12155 RSC.
- K. S. Prasad and K. Selvaraj, Biol. Trace Elem. Res., 2014, 157, 275–283 CrossRef CAS PubMed.
- J. F. Ramos, P. A. Tran and T. J. Webster, in 2012 38th Annual Northeast Bioengineering Conference (NEBEC), IEEE, 2012, pp. 185–186 Search PubMed.
- T. H. D. Nguyen, B. Vardhanabhuti, M. Lin and A. Mustapha, Food Control, 2017, 77, 17–24 CrossRef CAS.
- W. Zhang, Z. Chen, H. Liu, L. Zhang, P. Gao and D. Li, Colloids Surf., B, 2011, 88, 196–201 CrossRef CAS PubMed.
- G. Sharma, A. Sharma, R. Bhavesh, J. Park, B. Ganbold, J.-S. Nam and S.-S. Lee, Molecules, 2014, 19, 2761–2770 CrossRef PubMed.
- R. Shubharani, M. Mahesh and V. N. Yogananda Murthy, J. Nanomed. Nanotechnol., 2019, 10, 1–7 Search PubMed.
- J. Vyas and S. Rana, Int. J. Phytomed., 2017, 9, 634 CrossRef CAS.
- M. H. Yazdi, M. Mahdavi, E. Faghfuri, M. A. Faramarzi, Z. Sepehrizadeh, Z. M. Hassan, M. Gholami and A. R. Shahverdi, Iran J. Biotechnol., 2015, 13, 1–9 CrossRef PubMed.
- M. H. Yazdi, M. Mahdavi, N. Setayesh, M. Esfandyar and A. R. Shahverdi, DARU J. Pharm. Sci., 2013, 21, 33 CrossRef CAS PubMed.
- L. Fritea, V. Laslo, S. Cavalu, T. Costea and S. I. Vicas, Stud. Univ. Vasile Goldis Arad. Ser. Stiint. Vietii (Life Sci. Ser.), 2017, 27, 203–208 CAS.
- A. A. Kamnev, P. V. Mamchenkova, Y. A. Dyatlova and A. V. Tugarova, J. Mol. Struct., 2017, 1140, 106–112 CrossRef CAS.
- H. Wu, X. Li, W. Liu, T. Chen, Y. Li, W. Zheng, C. W.-Y. Man, M.-K. Wong and K.-H. Wong, J. Mater. Chem., 2012, 22, 9602 RSC.
- V. C. Tan, A. Hinchman, R. Williams, P. A. Tran and K. Fox, Biointerphases, 2018, 13, 06D301 CrossRef PubMed.
- C. Ferro, H. F. Florindo and H. A. Santos, Adv. Healthcare Mater., 2021, 10, 2100598 CrossRef CAS PubMed.
- H. El-Ramady, N. Abdalla, H. S. Taha, T. Alshaal, A. El-Henawy, S. E.-D. A. Faizy, M. S. Shams, S. M. Youssef, T. Shalaby, Y. Bayoumi, N. Elhawat, S. Shehata, A. Sztrik, J. Prokisch, M. Fári, É. Domokos-Szabolcsy, E. A. Pilon-Smits, D. Selmar, S. Haneklaus and E. Schnug, Environ. Chem. Lett., 2016, 14, 123–147 CrossRef CAS.
- G. N. Schrauzer, J. Nutr., 2000, 130, 1653–1656 CrossRef CAS PubMed.
- M. Shakibaie, M. R. Khorramizadeh, M. A. Faramarzi, O. Sabzevari and A. R. Shahverdi, Biotechnol. Appl. Biochem., 2010, 56, 7–15 CrossRef CAS PubMed.
- F. Yang, Q. Tang, X. Zhong, Y. Bai, T. Chen, Y. Zhang, Y. Li and W. Zheng, Int. J. Nanomed., 2012, 7, 835 CrossRef CAS PubMed.
- C. Ip, J. Nutr., 1998, 128, 1845–1854 CrossRef CAS.
- X. D. Shi, Y. Q. Tian, J. L. Wu and S. Y. Wang, Crit. Rev. Food Sci. Nutr., 2020, 0, 1–12 Search PubMed.
- A. Khurana, S. Tekula, M. A. Saifi, P. Venkatesh and C. Godugu, Biomed. Pharmacother., 2019, 111, 802–812 CrossRef CAS PubMed.
- M. P. Rayman, Proc. Nutr. Soc., 2005, 64, 527–542 CrossRef CAS PubMed.
- M. Wallenberg, S. Misra and M. Björnstedt, Basic Clin. Pharmacol. Toxicol., 2014, 114, 377–386 CrossRef CAS PubMed.
- L. V. Papp, J. Lu, A. Holmgren and K. K. Khanna, Antioxid. Redox Signal., 2007, 9, 775–806 CrossRef CAS PubMed.
- J. F. Combs Gerald, Nutrients, 2015, 7, 2209–2236 CrossRef PubMed.
- K. S. Prasad and K. Selvaraj, Biol. Trace Elem. Res., 2014, 157, 275–283 CrossRef CAS PubMed.
- M. S. Ahmad, M. M. Yasser, E. N. Sholkamy, A. M. Ali and M. M. Mehanni, Int. J. Nanomed., 2015, 10, 3389 CAS.
- Y. Li, X. Li, Y.-S. Wong, T. Chen, H. Zhang, C. Liu and W. Zheng, Biomaterials, 2011, 32, 9068–9076 CrossRef CAS PubMed.
- M. A. Rezvanfar, M. A. Rezvanfar, A. R. Shahverdi, A. Ahmadi, M. Baeeri, A. Mohammadirad and M. Abdollahi, Toxicol. Appl. Pharmacol., 2013, 266, 356–365 CrossRef CAS PubMed.
- K. K. Vekariya, J. Kaur and K. Tikoo, Toxicol. Appl. Pharmacol., 2013, 268, 212–220 CrossRef CAS PubMed.
- S. A. Wadhwani, U. U. Shedbalkar, R. Singh and B. A. Chopade, Appl. Microbiol. Biotechnol., 2016, 100, 2555–2566 CrossRef CAS PubMed.
- Z. Ping, T. Liu, H. Xu, Y. Meng, W. Li, X. Xu and L. Zhang, Nano Res., 2017, 10, 3775–3789 CrossRef CAS.
- D. Zeng, J. Zhao, K.-H. Luk, S.-T. Cheung, K.-H. Wong and T. Chen, J. Agric. Food Chem., 2019, 67, 2865–2876 CrossRef CAS PubMed.
- A. Thubagere and B. M. Reinhard, ACS Nano, 2010, 4, 3611–3622 CrossRef CAS PubMed.
- S. Tang, T. Wang, M. Jiang, C. Huang, C. Lai, Y. Fan and Q. Yong, Int. J. Biol. Macromol., 2019, 128, 444–451 CrossRef CAS PubMed.
- X. Gao, X. Li, J. Mu, C.-T. Ho, J. Su, Y. Zhang, X. Lin, Z. Chen, B. Li and Y. Xie, Int. J. Biol. Macromol., 2020, 152, 605–615 CrossRef CAS PubMed.
- T. P. A. Devasagayam, J. C. Tilak, K. K. Boloor, K. S. Sane, S. S. Ghaskadbi and R. D. Lele, J. Assoc. Phys., 2004, 52, 794–804 CAS.
- J. M. C. Gutteridge, Chem. – Biol. Interact., 1994, 91, 133–140 CrossRef CAS.
- S. Menon and V. K. Shanmugam, J. Trace Elem. Med. Biol., 2020, 62, 126549 CrossRef CAS PubMed.
- E. E. Battin, M. T. Zimmerman, R. R. Ramoutar, C. E. Quarles and J. L. Brumaghim, Metallomics, 2011, 3, 503–512 CrossRef CAS PubMed.
- J. T. Rotruck, A. L. Pope, H. E. Ganther, A. B. Swanson, D. G. Hafeman and W. G. Hoekstra, Science, 1973, 179, 588–590 CrossRef CAS PubMed.
- G. Zhang, X. Yao, C. Wang, D. Wang and G. Wei, J. Biotechnol., 2019, 304, 89–96 CrossRef CAS PubMed.
- X. Zhai, C. Zhang, G. Zhao, S. Stoll, F. Ren and X. Leng, J. Nanobiotechnology, 2017, 15, 1–12 CrossRef PubMed.
- L. Gunti, R. S. Dass and N. K. Kalagatur, Front. Microbiol., 2019, 10, 1–17 CrossRef PubMed.
- H. Kong, J. Yang, Y. Zhang, Y. Fang, K. Nishinari and G. O. Phillips, Int. J. Biol. Macromol., 2014, 65, 155–162 CrossRef CAS PubMed.
- A. Kumar and K. S. Prasad, J. Biotechnol., 2021, 325, 152–163 CrossRef CAS PubMed.
- W. Deng, Q. Xie, H. Wang, Z. Ma, B. Wu and X. Zhang, Nanomedicine, 2017, 13, 1965–1974 CrossRef CAS PubMed.
- Y. Liu, S. Zeng, Y. Liu, W. Wu, Y. Shen, L. Zhang, C. Li, H. Chen, A. Liu, L. Shen, B. Hu and C. Wang, Int. J. Biol. Macromol., 2018, 114, 632–639 CrossRef CAS PubMed.
- S. Al-Quraishy, M. Adkhil and A. E. A. Moneim, Int. J. Nanomed., 2015, 10, 6741–6756 CAS.
- S. Zeng, Y. Ke, Y. Liu, Y. Shen, L. Zhang, C. Li, A. Liu, L. Shen, X. Hu, H. Wu, W. Wu and Y. Liu, Colloids Surf., B, 2018, 170, 115–121 CrossRef CAS PubMed.
- Q. Ge, L. Chen, M. Tang, S. Zhang, L. Liu, L. Gao, S. Ma, M. Kong, Q. Yao, F. Feng and K. Chen, Eur. J. Pharmacol., 2018, 833, 50–62 CrossRef CAS PubMed.
- W. Deng, H. Wang, B. Wu and X. Zhang, Acta Pharm. Sin. B, 2019, 9, 74–86 CrossRef PubMed.
- I. Galan-Chilet, M. Grau-Perez, G. De Marco, E. Guallar, J. C. Martin-Escudero, A. Dominguez-Lucas, I. Gonzalez-Manzano, R. Lopez-Izquierdo, L. S. Briongos-Figuero, J. Redon, F. J. Chaves and M. Tellez-Plaza, Redox Biol., 2017, 12, 798–805 CrossRef CAS PubMed.
- H. Grundmann, M. Aires-de-Sousa, J. Boyce and E. Tiemersma, Lancet, 2006, 368, 874–885 CrossRef.
- G. A. Pankey, J. Antimicrob. Chemother., 2005, 56, 470–480 CrossRef CAS PubMed.
- J. N. Steenbergen, J. Alder, G. M. Thorne and F. P. Tally, J. Antimicrob. Chemother., 2005, 55, 283–288 CrossRef CAS PubMed.
- A. A. Ionin, A. K. Ivanova, R. A. Khmel’nitskii, Y. V. Klevkov, S. I. Kudryashov, A. O. Levchenko, A. A. Nastulyavichus, A. A. Rudenko, I. N. Saraeva, N. A. Smirnov, D. A. Zayarny, S. A. Gonchukov and E. R. Tolordava, Laser Phys. Lett., 2018, 15, 15604 CrossRef.
- P. Sonkusre and S. Singh Cameotra, Colloids Surf., B, 2015, 136, 1051–1057 CrossRef CAS PubMed.
- S. Qayyum and A. U. Khan, MedChemComm, 2016, 7, 1479–1498 RSC.
- L. Yan, Z. Gu and Y. Zhao, Chem. – Asian J., 2013, 8, 2342–2353 CrossRef CAS PubMed.
- X. Huang, X. Chen, Q. Chen, Q. Yu, D. Sun and J. Liu, Acta Biomater., 2016, 30, 397–407 CrossRef CAS PubMed.
- A. K. Mittal, S. Kumar and U. C. Banerjee, J. Colloid Interface Sci., 2014, 431, 194–199 CrossRef CAS PubMed.
- S. Menon, K. S. S. Devi, H. Agarwal and V. K. Shanmugam, Colloid Interface Sci. Commun., 2019, 29, 1–8 CrossRef CAS.
- H. Hariharan, N. A. Al-Harbi, P. Karuppiah and S. K. Rajaram, Chalcogenide Lett, 2012, 9, 509–515 CAS.
- E. Zonaro, S. Lampis, R. J. Turner, S. Junaid and G. Vallini, Front. Microbiol., 2015, 6, 1–11 Search PubMed.
- E. Kheradmand, F. Rafii, M. H. Yazdi, A. A. Sepahi, A. R. Shahverdi and M. R. Oveisi, DARU, J. Pharm. Sci., 2014, 22, 1–6 CrossRef PubMed.
- J. Yip, L. Liu, K. H. Wong, P. H. M. Leung, C. W. M. Yuen and M. C. Cheung, J. Appl. Polym. Sci., 2014, 131, 8886–8893 CrossRef.
- M. Shakibaie, N. S. Mohazab and S. A. Ayatollahi Mousavi, Jundishapur J. Microbiol., 2015, 8, 1–4 Search PubMed.
- J. W. Baddley, T. P. Stroud, D. Salzman and P. G. Pappas, Clin. Infect. Dis., 2001, 32, 1319–1324 CrossRef CAS PubMed.
- B. Vogelman, S. Gudmundsson, J. Leggett, J. Turnidge, S. Ebert and W. A. Craig, J. Infect. Dis., 1988, 158, 831–847 CrossRef CAS PubMed.
- W. Craig, Eur. J. Clin. Microbiol. Infect. Dis., 1993, 12, S6–S8 CrossRef CAS PubMed.
- W. A. Craig, Ann. Intern. Med., 1987, 106, 900 CrossRef CAS PubMed.
- P. Jamshidi, S. Soflaei and H. Kooshki, Adv. Environ. Biol., 2014, 1228–1239 Search PubMed.
- S. Soflaei, A. Dalimi, A. Abdoli, M. Kamali, V. Nasiri, M. Shakibaie and M. Tat, Comp. Clin. Path., 2014, 23, 15–20 CrossRef CAS.
- H. Mahmoudvand, M. Shakibaie, R. Tavakoli, S. Jahanbakhsh and I. Sharifi, Iran. J. Parasitol., 2014, 9, 452–460 Search PubMed.
- D. Wlodkowic, W. Telford, J. Skommer and Z. Darzynkiewicz, Methods Cell Biol., 2011, 103, 55–98 CrossRef CAS PubMed.
- H. Mahmoudvand, M. Fasihi Harandi, M. Shakibaie, M. R. Aflatoonian, N. ZiaAli, M. S. Makki and S. Jahanbakhsh, Int. J. Surg., 2014, 12, 399–403 CrossRef PubMed.
- D. P. Biswas, N. M. O’Brien-Simpson, E. C. Reynolds, A. J. O’Connor and P. A. Tran, J. Colloid Interface Sci., 2018, 515, 78–91 CrossRef CAS PubMed.
- S. Ramya, T. Shanmugasundaram and R. Balagurunathan, J. Trace Elem. Med. Biol., 2015, 32, 30–39 CrossRef CAS PubMed.
- A. Bhattacharjee, A. Basu, P. Ghosh, J. Biswas and S. Bhattacharya, J. Biomater. Appl., 2014, 29, 303–317 CrossRef CAS PubMed.
- A. Bhattacharjee, A. Basu, J. Biswas, T. Sen and S. Bhattacharya, Mol. Cell. Biochem., 2017, 424, 13–33 CrossRef CAS PubMed.
- F. Gao, Q. Yuan, L. Gao, P. Cai, H. Zhu, R. Liu, Y. Wang, Y. Wei, G. Huang, J. Liang and X. Gao, Biomaterials, 2014, 35, 8854–8866 CrossRef CAS PubMed.
- C. E. Hassan and T. J. Webster, Int. J. Nanomed., 2016, 11, 3641–3654 CrossRef CAS PubMed.
- B. Yu, T. Liu, Y. Du, Z. Luo, W. Zheng and T. Chen, Colloids Surf., B, 2016, 139, 180–189 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |