Open Access Article
Open Access ArticleHigh precision, high throughput generation of droplets containing single cells†
Jiande
Zhou
 *,
Amaury
Wei
,
Arnaud
Bertsch
and
Philippe
Renaud
*
*,
Amaury
Wei
,
Arnaud
Bertsch
and
Philippe
Renaud
*
Laboratory of Microsystems 4, Swiss Federal Institute of Technology Lausanne (EPFL), Lausanne, Switzerland. E-mail: jiande.zhou@epfl.ch; philippe.renaud@epfl.ch
First published on 21st November 2022
Abstract
The Poisson limit is a major problem for the isolation of single cells in different single-cell technologies and applications. In droplet-based single-cell assays, a scheme that is increasingly popular, the intrinsic randomness during single-cell encapsulation in droplets requires most of the created droplets to be empty, which has a profound impact on the efficiency and throughput of such techniques, and on the predictability of the combinatory droplet assays. Here we present a simple passive microfluidic system overcoming this limitation with unprecedented efficacy, allowing the generation of single-cell droplets for a wide range of operating conditions, with extremely high throughput (more than 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 single-cell loaded droplets per minute) and with an extremely low fault ratio (doublets or empty droplets), applicable to any cells and deformable particles. This versatile technique will shift the paradigm of single-cell encapsulation and will impact single-cell sequencing, rare cell isolation, multicellular/bead studies in immunology or cancer biology, etc.
000 single-cell loaded droplets per minute) and with an extremely low fault ratio (doublets or empty droplets), applicable to any cells and deformable particles. This versatile technique will shift the paradigm of single-cell encapsulation and will impact single-cell sequencing, rare cell isolation, multicellular/bead studies in immunology or cancer biology, etc.
Introduction
Single-cell analysis is of great scientific importance1–5 and has a good potential for clinical practices6–9 and industrial applications.10,11 Recently, droplet-based microfluidics became a popular tool for constructing single-cell assays. By compartmentalizing each cell into a pico-liter size droplet, up-concentrated and well-confined reactions can be performed, allowing single-cell analysis at all levels of the central dogma of molecular biology (including genomic, transcriptomic, proteomic, and metabolomic) in a massively parallel manner at a low cost. Various operations such as high throughput screening, transportation, storage, mixing, and fusion of droplets can be configured with droplet-based microfluidics. To perform single-cell droplet-based assays, the first step required is encapsulating each cell into one droplet. However, the efficacy of single-cell encapsulation is intrinsically limited by the random cell arrival at the droplet generation nozzle, resulting in a random number of cells loaded in each generated droplet, known as the Poisson limit. According to the Poisson statistic,12 a significant number of empty droplets needs to be produced to limit the number of droplets containing multiple cells (cell ≧2), which has a strong impact on the throughput and the cost of the method. Moreover, many arising applications require the co-encapsulation of more than one cell per particle into a droplet. For example, for single-cell mRNA sequencing12–14 and single-cell secretory analysis,15–17 the co-encapsulation of exactly one cell and one functional bead is required; for cell–cell interaction and functional screening, two distinct cell types need to be paired one-to-one in every droplet;18–21 and in terms of cell–cell communication profiling, two types of cells plus one or two distinct types of beads are needed to construct an assay in each droplet.22–24 In these cases, the limitation brought by the Poisson statistics is even more pronounced. For particle concentrations all equal to λ = 0.2 (particle per droplet),12 the ratio of droplets with the right combination is 2.7% for pairing two particles (cells) and 0.44% for pairing three particles (cells), with significant amounts of cell being lost in droplets with incorrect combinations. On the other hand, by constructing a pure population of single-cell (single-bead) droplets, the accurate assembly of different content single-cell assays becomes possible. Therefore, there is a strong need to overcome the Poisson limit to realize deterministic single cell (and bead) encapsulation.To this end, there exist both passive and active methods. A passive method allows automatic cell encapsulations at a low cost. However, current passive solutions for deterministic single-cell encapsulation are not robust enough. An important reason is most of these methods seek to achieve precise synchronization between the flows and the cells for yielding single-cell droplets (i.e., synchronous operation), which is intrinsically difficult to achieve and maintain deterministically. For example, a known passive technique is inertial ordering,25,26 where a distance between cells is generated by the Dean effect and the droplet generation frequency is fine-tuned to match the cell spacing. In practice, fine-tuning causes difficulty to use under the needed high flow speed and high cell concentration, with a demonstrated cell loading efficiency of up to 77%.26 Another passive method is only applicable to deformable beads, where they are densely packed and released at a rate that, again, needs to match the droplet generation rate.27 On the other hand, active methods are based on the sorting of randomly generated single-cell droplets. These methods require complex and expensive systems for real-time detection and fast and synchronized actuation. While high throughput sorting can be achieved if the cells are labelled and can be detected by fluorescence,28 in many applications, pre-labelling is undesirable or not possible. The label-free active sorting currently requires more sophisticated detection methods (e.g., imaging-based, spectrometry-based, etc.), and produces lower throughputs (max 50 Hz sorting rate).29–33 Until now, it is considered difficult, if not impossible, to overcome the Poisson limit of droplet microfluidics with an easy and versatile solution.
In this work, we are going to present such a solution. The proposed method is based on a recent observation of a new droplet splitting instability in microfluidic T junctions.34 Due to this splitting instability that occurs naturally in microfluidic junctions of specific geometries, a concept of cell-triggered splitting (CTS) is developed to achieve the passive and deterministic single-cell encapsulation in droplets without synchronization. A high throughput, label-free, cost-effective, and robust single-cell encapsulation method will be demonstrated in this paper.
Results
Concept of cell-triggered splitting system (CTS) for single cell encapsulation
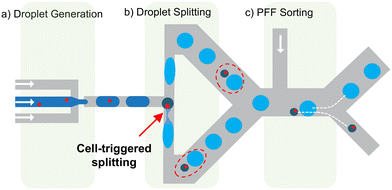
The cell-triggered splitting (CTS) concept for single-cell encapsulation is shown in Fig. 1. It includes three steps: (a) mother droplet generation, (b) droplet splitting, and (c) passive droplet sorting, all performed in one chip. At the start, a random cell encapsulation is performed using a standard flow focusing geometry with low cell density (λ = 0.1–0.2), producing mother droplets that are mostly empty or contain a single cell.12 These droplets then flow through a T junction designed to implement droplet splitting instability conditions.34 In this T junction, empty mother droplets are split in two, but mother droplets containing a cell induce a lateral splitting instability,34 which creates one additional small satellite droplet containing the cell. As a result, two types of droplets are obtained after splitting: large empty droplets and small cell-loaded satellite droplets. The satellite droplets containing cells can be collected by any passive size sorting method. In this work, we perform the sorting step with pinched flow fractionation (PFF)35 (Fig. 1c). The overall chip design for these three units and parameters can be found in ESI† Fig. S1.CTS is based on capillary instabilities in narrow channels
In a recent study, we demonstrated that a droplet splitting T junction with narrow side channels, having an aspect ratio greater than 1, generates a capillary instability which, in certain circumstances, leads to the breakup of the droplet in the lateral side channels. We call it here “lateral breakup” (LB), which creates three new droplets. The lateral instability-caused lateral breakup occurs at specific flow conditions, shown in Fig. 2a with the points in red. On the same geometry at different flow conditions – i.e., with higher flowrates and longer initial droplets (points in blue), this instability cannot develop because the droplets do not stay in the lateral channel long enough for this phenomenon to occur, and due to the suppressed instability by the pressure drop along the droplet. This results in the conventional T junction breakup where the initial droplet is split into two from the center,36 we call it “central breakup” (CB). When the system operates at a CB regime close to the transition to LB, a cell whose diameter is slightly larger than the width of the narrow section (wo) in the droplet can delay the CB and trigger the LB. As the cell is retained temporally outside of the narrow section due to its size, it occupies the droplet rear cap and delays the rear cap pinching responsible for CB, while slowing down the droplet at the junction for the instability to develop (Fig. S2†). In the end, the system is switched to LB breakup and a new satellite droplet containing the cell is formed, together with two empty large droplets (Fig. 2b). As the lateral droplet splitting is only triggered by the presence of a deformable particle such as a cell, all and only satellite droplets will contain single cells. Consequently, satellite droplets are a pure population of cell-loaded droplets. This is achieved automatically and passively by just flowing the mother droplets through the special T junction at a constant flow condition (see Movie S1†).CTS with high efficacy and specificity
In this study, we use an outlet channel width (wo) of 11 μm, and HT-29 cells (average diameter of 13.7 μm) to demonstrate the cell-triggered splitting. As all CB regimes that are close to the transition boundary are suitable for operation, there exist many working flow combinations with different throughputs on the same geometry. We first conducted an experiment at a mother droplet sorting rate of 47 Hz (Qwater = 0.5 μL min−1). The single cell triggering efficiency is defined as the success rate of satellite droplet generation upon the presence of a single cell in the mother droplet. For a total of 473 single cells, we obtained an overall triggering efficiency of 91.3%. The detailed analysis for different cell size categories shows that cells that are sufficiently larger than (wo) have achieved triggering efficiencies close to 100% (Fig. 3a). This high triggering efficiency for large cells remains true when a different flow combination with higher throughput – i.e., at a mother droplet sorting rate of 241 Hz (Qwater = 2.0 μL min−1) is performed and was observed for a total of 808 cells (Fig. S3†). At both throughput conditions, the obtained droplet size difference associated with cell occupancy is significant – over 12 times the difference in droplet volume between small satellite droplets and large empty droplets is observed. The satellite droplets have a good monodispersity, and their diameter is 23 ± 1.5 μm@47 Hz and 22 ± 1 μm@241 Hz (Fig. S4†).The specificity of the method, defined as the number of single cells loaded satellites among the total population of satellite droplets, has been compared to the Poisson distribution under the same cell concentration (λ = 0.12) in Fig. 3b. The percentage of empty droplets is reduced from 89% to around 1%, while the percentage of droplets containing single cells rises from 11% to 94%. Empty satellite droplets rarely exist – satellites loaded with cells comprise >98% of the population. In our case, some doublets and multiplets result from adherent cells obtained during the cell preparation. These cells are partitioned into one satellite droplet as a single (large) particle. Note that for a given T junction geometry, if the fluid properties do not change, the working regimes (i.e., the operational flow conditions) are fixed and reproducible. Therefore, real-time flow tuning is avoided, which allows the system to start operating from the first cell entering the system until the whole sample is processed.
Remarkably, being a passive method without any detection/actuation limit, our method has shown efficient cell-triggered splitting at a mother droplet sorting rate up to 3100 Hz (Qwater = 8 μL min−1), giving 22.3k cell loaded droplets per minute (with λ = 0.12 cells per droplet). An overall single cell triggering efficiency of over 70% is demonstrated (including cell size population smaller than wo), with nearly 100% cell loading efficiency. Such throughput has not been demonstrated by any other passive or active methods so far.
Passive sorting of small cell-loaded droplets
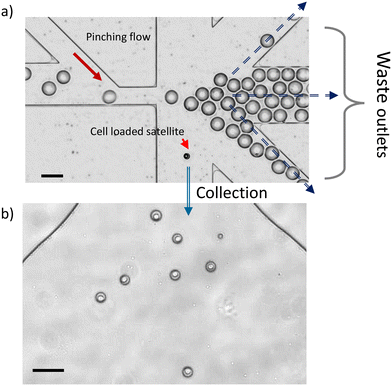
After the T junction splitting, the cell occupancy of the droplets is no more random but linked to the droplet size. The next step is to collect the smaller satellite droplets which contain the cells. Given the significant size difference, many existing sized-based sorting methods can be used. Here we use PFF (pinch flow fractionation), where an oil flow pushes all droplets against a channel wall, such that the smaller satellites align with the streamlines closer to the wall, whereas the larger droplets align with streamlines that are further away from the channel wall. A subsequent expansion geometry separates the two populations. The small ones flow into the bottom channel for collection, and the larger ones towards the middle channels for waste (Fig. 4a). In our demonstration, 89% of the satellites generated upstream went into the collection channel; the rest were misplaced into the waste channels due to occasional droplet congestions. We expect this to be avoided/improved with better geometry design. Remarkably, during 20 min of operation at 241 Hz, over 5 × 105 large droplets were generated, but there was not even one misplaced into the collection channel (Fig. 4b), achieving a 100% purity of satellite droplets population for the final collection. Therefore, after the sorting step, we preserved a droplet population with over 98% cell loading efficiency, among which 92–94% contain single cells.To confirm that the confinement of a cell in a channel slightly smaller than its dimension does not adversely affect the cell's viability, we conducted a cell membrane integrity test after operation at the throughput of 241 Hz. After the cells have passed through the splitting and sorting geometries on the chip, they are recollected from the created droplets and are stained with propidium iodide (PI). The cell damage rate is only 3% higher than the one of the control experiments. Note that the preservation of the plasma membrane after microfluidics squeezing was also confirmed in other studies where cells were pushed through a constriction of half of our channel size (wo) at double of our cell speed.37,38 The collected satellite droplets are stable for transfer and storage. We observe no satellite droplet merging in the droplet collection reservoir during the 5 hours following their production.
Single bead encapsulation
To demonstrate that this method is applicable to any deformable particle, we applied it to deterministically encapsulate deformable gel beads from 10X Genomics, which are currently widely used for single-cell RNA sequencing. Based on the simple working principle and design rule of the device presented in this study (ESI† Section S1), we easily scaled up the geometry to accommodate gel beads with an average diameter of 70 μm. The corresponding cell-triggered splitting is shown in Fig. 5 with a device with a lateral channel width wo = 55 μm. Thanks to the monodisperse size distribution and homogeneous viscoelastic properties of beads, we reached 100% triggering efficacy (10 beads), with 100% single bead droplet specificity (beads do not stick to each other). The created satellite droplets have a diameter of around 90 μm, with a volume of Vdroplet = 0.8 nL.Experimental
Chip fabrication
To create the microchannels, we produce polydimethylsiloxane (PDMS) replicas from silicon moulds, then sealed them with glass slides. For the silicon mould fabrication, AZ ECI 3007 photoresist (1.5) is used during standard photolithography. Then, the wafer with a patterned photoresist is etched with dry reactive ion etching (DRIE) using the Bosch process (Alcatel AMS 200). The etching time determines the height of the channels, the latter is measured with a surface profilometer (Tencor Alpha-Step 500). The obtained Si mould was silanized with trichloro-(1H,1H,2H,2H-perfluorooctyl)silane in a desiccator for five hours, which makes it ready for PDMS moulding (using a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 mixing ratio). After the moulding, we bond the PDMS replica to a glass slide with oxygen plasma (1 min, 29 W). The hydrophobicity of the channels was obtained by placing the freshly bonded device (with the activated surface) in the above-mentioned PFOTS-filled desiccator for five hours.
10 mixing ratio). After the moulding, we bond the PDMS replica to a glass slide with oxygen plasma (1 min, 29 W). The hydrophobicity of the channels was obtained by placing the freshly bonded device (with the activated surface) in the above-mentioned PFOTS-filled desiccator for five hours.
Cell culture and preparation
We used the GFP stable cell line-HT-29 (Cat. No. CSC-RR0119), which was cultured in DMEM/F-12 supplemented with 10% FBS and 1% penicillin–streptomycin at 37 °C in a 5% CO2 atmosphere. For use of single-cell encapsulation, the cells are detached with trypsin (reaction for 5 min) and resuspended into PBS at a concentration of 1.2 M mL−1.Single cell/bead encapsulation
BioRad droplet generation oil (for ddPCR) is used in all experiments. For single cell encapsulation, the aqueous phase is the single cell suspension prepared using above method; for single bead encapsulation, next GEM Single Cell 5′ Gel Beads from 10X Genomics was diluted 20 times in DI water, then used as the aqueous phase.Cell viability test
After preparation of a single cell suspension, half of the suspension is kept at room temperature (control group), the rest is processed in the microfluidics chip at a throughput of 241 Hz (for droplet generation) for 20 min. At the end of the operation, the droplets floating on top of the oil in the collection Eppendorf were recovered and poured onto a super-hydrophobic film (Millipore Membrane filter, 1 pore size), which absorbs the surfactant and the excessive oil for breaking the droplets. After 1 min, the aqueous phase was collected with a pipette (experiment group). The cells from both the experiment and control groups were stained with propidium iodide (PI) and observed under a microscope for evaluating the membrane integrity. The HT-29 cells were modified to express the green fluorescent protein (GFP) gene thus showing green fluorescence. Cells expressing “only red” or “red plus green” fluorescence are considered with compromised membrane.Discussion
In this paper, we presented an extremely simple method for deterministic single-cell encapsulation in droplets. Based on a newly discovered droplet instability,34 we developed an automatic workflow using a cell triggered splitting phenomenon (CTS) to create smaller satellite droplets containing single cells, for downstream separation of the positive droplets. Also for the post-encapsulation droplet size-based sorting, Chabert et al.40 reported a different mechanism for inducing droplet size difference. By creating empty droplets that were smaller than the cells, they ensured that any droplet that included a cell during random encapsulation immediately became larger than the rest. However, the major drawbacks are that the size difference between the positive and the negative droplets is not significant and not robust. Particularly, the volume difference is only about a cell's volume; and the jetting regime that can produce droplets smaller than a cell is a difficult regime that is sensitive to ambient perturbations, which can easily and drastically change the default droplet size. In addition, the arrival of a cell disturbs the thin jetting thread and results in increased droplet size for adjacent droplets. These factors rendered the droplet size difference less robust to use. In comparison, with CTS, the working regimes are robust, and a significant size difference is generated by default, which allows efficient and automatic recovery of single-cell droplets downstream. Because of the cell triggering satellite formation mechanism, the cell occupancy of each satellite droplet is guaranteed – this is the intrinsic high specificity of this method. We show that nearly 100% of the cells that are sufficiently larger than the lateral channel restriction (i.e., Ddroplet > 12 μm) have triggered the formation of satellite droplets, among which more than 98% contain at least one cell, and 92–94% contain exactly one single cell.The versatility of this technique is demonstrated. The CTS working principle applies not only to the cells but also to other deformable particles (e.g., functional beads) and particles of different sizes. The design principle for accommodating the different sizes of particles is simple and purely geometry-dependent34 (ESI† Section S1). Such a mechanism works for a wide range of flow conditions, allowing the throughput of this method to be adapted to different volumes of the samples, and thus to the different initial numbers of cells or particles in the samples. This was demonstrated experimentally with various mother droplet sorting rates ranging from 47 Hz up to 3100 Hz.
Compared to the current state of the art, we provide a practical passive and asynchronous solution to the single-cell encapsulation problem. Unlike traditional synchronous methods,25,26 the novel asynchronous CTS working principle permits passiveness and robustness at the same time. The simple “plug and play” operation exempts the critical flow rate tuning for precise synchronization. The deterministic encapsulation is thus self-running throughout the experiments, for both long- and short-time applications. For this to happen, only a basic flow generation setup (syringe/pressure pumps) is required. On the other hand, the active methods for sorting single-cell droplets from the random populations, including fluorescence-activated droplet sorting (FADS), Raman-activated droplet sorting (RADS), image-processing-based droplet sorting, UV-vis spectra-activated droplet sorting, etc., require detectors, active sorters, related electronics, and control software for real-time analysis, actuation, integration, and synchronization. Their complexity not only adds to the system cost, limits its range of application, but also limits their throughput. In our case, without any detection/actuation/synchronization limit, a label-free droplet sorting can be performed at a rate higher than 3 kHz, corresponding to a single-cell droplets production rate of more than 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets per minute (with an average cell concentration of λ = 0.12 cells per droplet). This means one million of single-cell droplets can be generated within less than an hour, an unprecedented throughput achieved particularly with an intrinsic high cell loading efficiency. This throughout might not yet be the throughput limit of this device. The passive working principle also allows the parallelization of several splitting geometries to further scale up the throughput.
000 droplets per minute (with an average cell concentration of λ = 0.12 cells per droplet). This means one million of single-cell droplets can be generated within less than an hour, an unprecedented throughput achieved particularly with an intrinsic high cell loading efficiency. This throughout might not yet be the throughput limit of this device. The passive working principle also allows the parallelization of several splitting geometries to further scale up the throughput.
The intrinsic high specificity of single-cell droplets brought by this method has important implications for applications. By carefully designing the critical channel dimension wo with respect to the cell size distribution, the T junction can be used to encapsulate a full cell population with a wo smaller than the smallest cell size in the population, or a subset of the population whose size is larger than the critical threshold set by the wo, for rare sample enrichment and encapsulation. For example, it could be used to isolate circulating tumor cells (CTC) from blood samples.39 In addition, by creating pure populations of single-cell or single-bead droplets, the CTS method enables the deterministic construction of multi-cellular/multi-cell-bead droplet assays. This need is increasingly critical for a wide range of applications but is currently unmet. For example, due to the Poisson limit, current single-cell RNA sequencing is not applicable to rare cell samples that are often of scientific importance and clinical relevance. The Poisson limit also interferes with data analysis and causes more than half of the cell population wasted in droplets containing the wrong combinations.41 This situation is made worse by pairing two distinct types of single cell into one droplet (for immunology, cancer biology studies, etc.). A completely random co-encapsulation generates mostly undesired droplets, comprising 86.5% to more than 99% of the total population.18–21 What's more, the probability of obtaining the desired droplet composition will further decrease with an increased number of objects to be uniquely encapsulated. Clearly, the accurate construction of multi-cellular/multi-cell-particle droplet assays needs to start from a pure population of single-cell (or particle) droplets. Our method is particularly suited for acquiring the latter, which is also compatible with existing droplet merging strategies.
One merging strategy uses wells of different sizes to sequentially trap distinct types of droplets in a close vicinity prior to their electrocoalescence.42–44 This technique is efficient and has the potential to pair and merge more than two types of droplets by carefully designing the wells. Another example is based on the alternating re-injection of two sizes (types) of droplets. After the smaller size droplet (moving faster) catches the larger size droplet, electrocoalescence happens when the droplet pairs pass through an electric-field-ON channel area.45–47 Here, the synchronous effort is less critical: if the larger droplets (e.g., barcode bead-loaded droplet) are injected 10% more frequently than the smaller droplets (e.g., single-cell loaded droplet), 10% of the bead-loaded droplets will be unpaired (wasted), but the cell-loaded droplets remain 100% one-bead–one-cell paired, provided that both droplet populations are purely single-particle-loaded, which can be aided with the CTS single cell encapsulation method. Therefore, a wide range of single cell assays can now be deterministically constructed at a low cost. In short, we have presented CTS as an efficient, versatile, and extremely simple single cell deterministic encapsulation method. We expect this easy solution to the long-standing Poisson limit would impact droplet-based microfluidics and single cell study profoundly.
Conclusions
In this study, we present a novel passive method for high throughput and deterministic encapsulation of single cells or other deformable particles in droplets. Relying on the capillary instability in narrow microfluidic T junctions, which enables the cell-triggered splitting (CTS) of droplets, this method allows an asynchronous and easy-to-use workflow for highly precise and low-cost encapsulation of single cells, meeting different application requirements from low throughput scenarios to unprecedentedly high throughput scenarios (two orders of magnitude faster than existing label-free sorting methods). Such an efficient, versatile, and simple encapsulation tool opens new perspectives for single-cell assays and combinatorial cell/beads encapsulation in the era of single-cell analysis.Author contributions
Conceptualization: JZ, PR. Methodology: JZ, AW. Investigation: JZ, PR, AW. Visualization: JZ, PR, AB. Supervision: PR, AB. Writing – original draft: JZ, PR, AB, AW. Writing – review & editing: JZ, PR, AB.Conflicts of interest
Authors declare that they have no competing interests.Acknowledgements
The authors thank Personalized Health and Related Technologies (PHRT), a strategic focus area of the Swiss ETH Domain for financial support. The authors thank Weida Chen, Antoine Herzog from Parithera® for device and experimental help over technology development. The authors thank Cheyenne Rechsteiner for providing help in HT-29 cells culture and cell viability test. The authors thank Christopher Tse for proof reading.Notes and references
- R. C. Bandler, I. Vitali, R. N. Delgado, M. C. Ho, E. Dvoretskova and J. S. Ibarra Molinas, et al., Single-cell delineation of lineage and genetic identity in the mouse brain, Nature, 2022, 601(7893), 404–409 CrossRef CAS PubMed.
- T. Sapiens, R. C. Jones, J. Karkanias, M. A. Krasnow, A. O. Pisco and S. R. Quake, et al., The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans, Science, 2022, 376(6594), eabl4896 CrossRef PubMed.
- M. M. Rothová, A. V. Nielsen, M. Proks, Y. F. Wong, A. R. Riveiro and M. Linneberg-Agerholm, et al., Identification of the central intermediate in the extra-embryonic to embryonic endoderm transition through single-cell transcriptomics, Nat. Cell Biol., 2022, 1–12 Search PubMed.
- M. Lange, V. Bergen, M. Klein, M. Setty, B. Reuter and M. Bakhti, et al., CellRank for directed single-cell fate mapping, Nat. Methods, 2022, 19(2), 159–170 CrossRef CAS PubMed.
- E. Fiskin, C. A. Lareau, L. S. Ludwig, G. Eraslan, F. Liu and A. M. Ring, et al., Single-cell profiling of proteins and chromatin accessibility using PHAGE-ATAC, Nat. Biotechnol., 2022, 40(3), 374–381 CrossRef CAS PubMed.
- P. Zhang, A. M. Kaushik, K. Hsieh, S. Li, S. Lewis and K. E. Mach, et al., A Cascaded Droplet Microfluidic Platform Enables High-Throughput Single Cell Antibiotic Susceptibility Testing at Scale, Small Methods, 2022, 6(1), 2101254 CrossRef CAS PubMed.
- A. K. Dutta, J. B. Alberge, R. Sklavenitis-Pistofidis, E. D. Lightbody, G. Getz and I. M. Ghobrial, Single-cell profiling of tumour evolution in multiple myeloma—Opportunities for precision medicine, Nat. Rev. Clin. Oncol., 2022, 19(4), 223–236 CrossRef PubMed.
- R. K. Perez, M. G. Gordon, M. Subramaniam, M. C. Kim, G. C. Hartoularos and S. Targ, et al., Single-cell RNA-seq reveals cell type–specific molecular and genetic associations to lupus, Science, 2022, 376(6589), 1970 CrossRef PubMed.
- E. Kobayashi, A. Jin, H. Hamana, K. Shitaoka, K. Tajiri and S. Kusano, et al., Rapid cloning of antigen-specific T-cell receptors by leveraging the cis activation of T cells, Nat. Biomed. Eng., 2022, 1–13 Search PubMed.
- P. Prashar, S. Swain, N. Adhikari, P. Aryan, A. Singh and M. Kwatra, et al., A novel high-throughput single B-cell cloning platform for isolation and characterization of high-affinity and potent SARS-CoV-2 neutralizing antibodies, Antiviral Res., 2022, 105349 CrossRef CAS PubMed.
- W. L. Matochko, C. Nelep, W. C. Chen, S. Grauer, K. McFadden and V. Wilson, et al., CellCelector™ as a platform in isolating primary B cells for antibody discovery, Antibody Ther., 2022, 5(1), 11–17 CrossRef CAS PubMed.
- R. Zilionis, J. Nainys, A. Veres, V. Savova, D. Zemmour and A. M. Klein, et al., Single-cell barcoding and sequencing using droplet microfluidics, Nat. Protoc., 2017, 12(1), 44–73 CrossRef CAS PubMed.
- F. Salmen, J. De Jonghe, T. S. Kaminski, A. Alemany, G. E. Parada and J. Verity-Legg, et al., High-throughput total RNA sequencing in single cells using VASA-seq, Nat. Biotechnol., 2022, 1–14 Search PubMed.
- E. Z. Macosko, A. Basu, R. Satija, J. Nemesh, K. Shekhar and M. Goldman, et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets, Cell, 2015, 161(5), 1202–1214 CrossRef CAS PubMed.
- J. Park, Y. Shin, J. M. Kim, S. Kweon, A. Y. Song and Y. Baek, et al., Multifunctional Microparticles with stimulation and sensing capabilities for facile NK cell activity assay, ACS Sens., 2021, 6(3), 693–697 CrossRef CAS PubMed.
- M. Wang, M. H. Nai, R. Y. J. Huang, H. L. Leo, C. T. Lim and C. H. Chen, High-throughput functional profiling of single adherent cells via hydrogel drop-screen, Lab Chip, 2021, 21(4), 764–774 RSC.
- W. Liu, R. Zhang, S. Huang, X. Li, W. Liu and J. Zhou, et al., Quantification of Intracellular Proteins in Single Cells Based on Engineered Picoliter Droplets, Langmuir, 2022, 38(26), 7929–7937 CrossRef CAS PubMed.
- S. Wang, Y. Liu, Y. Li, M. Lv, K. Gao and Y. He, et al., High-Throughput Functional Screening of Antigen-Specific T Cells Based on Droplet Microfluidics at a Single-Cell Level, Anal. Chem., 2021, 94(2), 918–926 CrossRef PubMed.
- Y. Wang, R. Jin, B. Shen, N. Li, H. Zhou and W. Wang, et al., High-throughput functional screening for next-generation cancer immunotherapy using droplet-based microfluidics, Sci. Adv., 2021, 7(24), eabe3839 CrossRef CAS PubMed.
- S. Sarkar, P. Sabhachandani, D. Ravi, S. Potdar, S. Purvey and A. Beheshti, et al., Dynamic analysis of human natural killer cell response at single-cell resolution in B-Cell Non-Hodgkin Lymphoma, Front. Immunol., 2017, 1736 CrossRef PubMed.
- S. Sarkar, W. Kang, S. Jiang, K. Li, S. Ray and E. Luther, et al., Machine learning-aided quantification of antibody-based cancer immunotherapy by natural killer cells in microfluidic droplets, Lab Chip, 2020, 20(13), 2317–2327 RSC.
- S. Antona, T. Abele, K. Jahnke, Y. Dreher, K. Göpfrich and I. Platzman, et al., Droplet-Based Combinatorial Assay for Cell Cytotoxicity and Cytokine Release Evaluation, Adv. Funct. Mater., 2020, 30(46), 2003479 CrossRef CAS.
- Y. Zhou, N. Shao, R. B. de Castro, P. Zhang, Y. Ma and X. Liu, et al., Evaluation of single-cell cytokine secretion and cell-cell interactions with a hierarchical loading microwell chip, Cell Rep., 2020, 31(4), 107574 CrossRef CAS PubMed.
- J. L. Madrigal, N. G. Schoepp, L. Xu, C. S. Powell, C. L. Delley and C. A. Siltanen, et al., Characterizing cell interactions at scale with made-to-order droplet ensembles (MODEs), Proc. Natl. Acad. Sci. U. S. A., 2022, 119(5), e2110867119 CrossRef CAS PubMed.
- J. F. Edd, D. Di Carlo, K. J. Humphry, S. Köster, D. Irimia and D. A. Weitz, et al., Controlled encapsulation of single-cells into monodisperse picolitre drops, Lab Chip, 2008, 8(8), 1262–1264 RSC.
- E. W. Kemna, R. M. Schoeman, F. Wolbers, I. Vermes, D. A. Weitz and A. Van Den Berg, High-yield cell ordering and deterministic cell-in-droplet encapsulation using Dean flow in a curved microchannel, Lab Chip, 2012, 12(16), 2881–2887 RSC.
- A. R. Abate, C. H. Chen, J. J. Agresti and D. A. Weitz, Beating Poisson encapsulation statistics using close-packed ordering, Lab Chip, 2009, 9(18), 2628–2631 RSC.
- J. C. Baret, O. J. Miller, V. Taly, M. Ryckelynck, A. El-Harrak and L. Frenz, et al., Fluorescence-activated droplet sorting (FADS): efficient microfluidic cell sorting based on enzymatic activity, Lab Chip, 2009, 9(13), 1850–1858 RSC.
- T. A. Duncombe, A. Ponti, F. P. Seebeck and P. S. Dittrich, UV–Vis spectra-activated droplet sorting for label-free chemical identification and collection of droplets, Anal. Chem., 2021, 93(38), 13008–13013 CrossRef CAS PubMed.
- X. Wang, Y. Xin, L. Ren, Z. Sun, P. Zhu and Y. Ji, et al., Positive dielectrophoresis–based Raman-activated droplet sorting for culture-free and label-free screening of enzyme function in vivo, Sci. Adv., 2020, 6(32), eabb3521 CrossRef CAS PubMed.
- S. Hasan, M. E. Blaha, S. K. Piendl, A. Das, D. Geissler and D. Belder, Two-photon fluorescence lifetime for label-free microfluidic droplet sorting, Anal. Bioanal. Chem., 2022, 414(1), 721–730 CrossRef CAS PubMed.
- X. Wang, L. Ren, Y. Su, Y. Ji, Y. Liu and C. Li, et al., Raman-activated droplet sorting (RADS) for label-free high-throughput screening of microalgal single-cells, Anal. Chem., 2017, 89(22), 12569–12577 CrossRef CAS PubMed.
- M. Sesen and G. Whyte, Image-based single cell sorting automation in droplet microfluidics, Sci. Rep., 2020, 10(1), 1–14 CrossRef PubMed.
- J. Zhou, Y. M. Ducimeti'ere, D. Migliozzi, L. Keiser, A. Bertsch and F. Gallaire, et al., Breaking one into three: surface-tension-driven droplet breakup in T-junctions, arXiv, 2022, preprint, arXiv:2207.00077, DOI:10.48550/arXiv.2207.00077.
- M. Yamada, M. Nakashima and M. Seki, Pinched flow fractionation: continuous size separation of particles utilizing a laminar flow profile in a pinched microchannel, Anal. Chem., 2004, 76(18), 5465–5471 CrossRef CAS PubMed.
- C. Haringa, C. De Jong, D. A. Hoang, L. M. Portela, C. R. Kleijn and M. T. Kreutzer, et al., Breakup of elongated droplets in microfluidic T-junctions, Phys. Rev. Fluids, 2019, 4(2), 024203 CrossRef.
- X. Ding, M. P. Stewart, A. Sharei, J. C. Weaver, R. S. Langer and K. F. Jensen, High-throughput nuclear delivery and rapid expression of DNA via mechanical and electrical cell-membrane disruption, Nat. Biomed. Eng., 2017, 1(3), 1–7 Search PubMed.
- A. Kollmannsperger, A. Sharei, A. Raulf, M. Heilemann, R. Langer and K. F. Jensen, et al., Live-cell protein labelling with nanometre precision by cell squeezing, Nat. Commun., 2016, 7(1), 1–7 Search PubMed.
- N. M. Karabacak, P. S. Spuhler, F. Fachin, E. J. Lim, V. Pai and E. Ozkumur, et al., Microfluidic, marker-free isolation of circulating tumor cells from blood samples, Nat. Protoc., 2014, 9(3), 694–710 CrossRef CAS PubMed.
- M. Chabert and J. L. Viovy, Microfluidic high-throughput encapsulation and hydrodynamic self-sorting of single cells, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(9), 3191–3196 CrossRef CAS PubMed.
- J. Bues, M. Biočanin, J. Pezoldt, R. Dainese, A. Chrisnandy and S. Rezakhani, et al., Deterministic scRNA-seq captures variation in intestinal crypt and organoid composition, Nat. Methods, 2022, 19(3), 323–330 CrossRef CAS PubMed.
- M. T. Chung, D. Núñez, D. Cai and K. Kurabayashi, Deterministic droplet-based co-encapsulation and pairing of microparticles via active sorting and downstream merging, Lab Chip, 2017, 17(21), 3664–3671 RSC.
- M. Duchamp, M. Arnaud, S. Bobisse, G. Coukos, A. Harari and P. Renaud, Microfluidic Device for Droplet Pairing by Combining Droplet Railing and Floating Trap Arrays, Micromachines, 2021, 12(9), 1076 CrossRef PubMed.
- M. T. Chung, K. Kurabayashi and D. Cai, Single-cell RT-LAMP mRNA detection by integrated droplet sorting and merging, Lab Chip, 2019, 19(14), 2425–2434 RSC.
- Y. Qiao, J. Fu, F. Yang, M. Duan, M. Huang and J. Tu, et al., An efficient strategy for a controllable droplet merging system for digital analysis, RSC Adv., 2018, 8(60), 34343–34349 RSC.
- E. Brouzes, M. Medkova, N. Savenelli, D. Marran, M. Twardowski and J. B. Hutchi-son, et al., Droplet microfluidic technology for single-cell high-throughput screening, Proc. Natl. Acad. Sci. U. S. A., 2009, 106(34), 14195–14200 CrossRef CAS PubMed.
- J. A. Wippold, H. Wang, J. Tingling, J. L. Leibowitz, P. de Figueiredo and A. Han, PRESCIENT: platform for the rapid evaluation of antibody success using integrated microfluidics enabled technology, Lab Chip, 2020, 20(9), 1628–1638 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2lc00841f |
| This journal is © The Royal Society of Chemistry 2022 |